Abstract
Aging is marked by a collapse of protein homeostasis and deterioration of adaptive stress responses that often lead to disease. During aging, the induction of stress responses decline along with protein quality control. Here, we have shown that the ability to mount an adaptive response by pretreatment with minor oxidative stress is abrogated in aged Caenorhabditis elegans. We have identified a defect in SKN-1 signaling sensitivity during aging and have also found an aging-related increase in basal proteasome expression and in vitro activity, however, adaptation of the 20S proteasome in response to stress is lost in old animals. Interestingly, increased activation of SKN-1 promotes stress resistance, but is unable to rescue declining adaptation during aging. Our data demonstrate that the aging-dependent decline in SKN-1 signaling negatively impacts adaptation of the 20S proteasome in response to acute oxidative stress.
Keywords: Adaptive homeostasis, Hydrogen peroxide, Proteolysis, Protein oxidation, Nrf2
A hallmark of aging is the decline in the quality control mechanisms of the cellular proteome (1). Loss of protein homeostasis results in an increase in protein oxidation, protein misfolding, and protein aggregation that can lead to aging-related diseases, including cardiovascular disease (2), ischemic stroke (3), and numerous neurodegenerative disorders (4). Several lines of evidence have indicated that oxidative stress is central to aging and the development of many diseases associated with aging (5). A major source for cellular oxidative stress is electron leakage from mitochondrial respiration, resulting in the production of superoxide and hydrogen peroxide (H2O2). Although not highly reactive, superoxide quickly reacts with nitric oxide radical, which leads to the formation of a potent oxidant known as peroxynitrite. In addition, H2O2 can decompose to form hydroxyl radicals in the presence of metal ions (6). Thus, cellular oxidative damage is an inevitable byproduct of aerobic metabolism. Consequently, protein oxidation results in the unfolding of proteins revealing hydrophobic residues that are prone to aggregation, leading to the accumulation of protein aggregates with age. Misfolded proteins are managed by the induction of evolutionary ancient stress response mechanisms, including the heat shock response, oxidative stress response, and proteasome-dependent degradation that triage, refold, or degrade proteins prior to aggregate formation.
Adaptation to mild stress allows cells and organisms to cope with more severe stress by triggering the production of cytoprotective genes that promote cell survival. As oxidative stress increases with age, the ability to mount an adaptive response declines, resulting in the collapse of protein homeostasis and an increase in protein aggregation (7–9). Recent work in our laboratory has shown that induction of the proteasome during oxidative stress is dependent on Nrf2, the master regulator of the oxidative stress response, and that this mechanism is conserved for the Caenorhabditis elegans homolog, SKN-1. H2O2 increases Nrf2 translocation to the nucleus, resulting in increased expression of the 20S proteasome and the subsequent increase in proteolytic capacity. H2O2-induced proteolytic activity is blocked by the inhibition of Nrf2 in C. elegans, Drosophila melanogaster, and mouse embryonic fibroblast cell culture (10,11). To our knowledge, there are currently no studies to address how the adaptive capacity to induce expression of the proteasome may change with age. It is also well established that adaptation to numerous stressors declines during aging, but the nature of this defect in stress response remains unclear. Therefore, we sought to characterize the decline of 20S proteasome-dependent adaptation to oxidative stress in response to SKN-1 signaling during aging in the model organism, C. elegans.
Due to their short life span, C. elegans have been used extensively in aging studies and to model aging-dependent protein aggregation, as demonstrated in neurodegenerative diseases such as Huntington’s and Parkinson’s disease (12). Although SKN-1 has diverged from its mammalian ortholog in regards to some aspects of its regulation, such as DNA binding and negative feedback, it has extremely similar functionality to Nrf2 and activates many of the same gene family targets. In addition to typical Nrf functions, growing evidence suggests that SKN-1 is regulated by metabolic and nutrient signaling and SKN-1 has been shown to play a central role in C. elegans’ life span. The conserved nature of these orthologs suggests that the mechanisms that regulate SKN-1 may also be conserved in vertebrates, making C. elegans a useful organism to understand mammalian aging (13).
In our current study, we have found that the ability to mount an adaptive response by pretreatment with acute oxidative stress is abrogated in aged C. elegans and that SKN-1 signaling and adaptive proteasomal degradation decline with age. Furthermore, we have shown that increased activation of SKN-1 results in increased stress resistance and decreased protein oxidation, but is unable to restore the aging-related decline of proteasome-dependent adaptation. These results demonstrate that the aging-dependent decline in SKN-1 signaling negatively impacts adaptation of the 20S proteasome in response to oxidative stress, suggesting that maintaining tighter control of signaling for adaptive responses may partially alleviate the collapse of protein homeostasis during aging. Essentially all diseases of aging have a protein homeostasis defect, making the investigation of the regulation of proteome quality control mechanisms essential to prolonging health span.
Methods
C. elegans Strains Utilized in This Study
Strains were cultured at 20°C using standard techniques (14). All experiments were performed on age-matched hermaphrodites synchronized by egg lay. Day 3 corresponds to 3 days post egg lay and represents the first day of adulthood. The wild-type control strain was N2 Bristol. The SKN-1::GFP strain was provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. The skn-1 gain-of-function (gf) lax120 strain was a gift from the Sean Curran laboratory. The following chromosomal arrays were also used: P.pbs-5::GFP and P.pbs-5(ARE)::GFP. Due to recent evidence that the DNA synthesis inhibitor fluorodeoxyuridine, commonly used to block egg laying in aging experiments, induces adaptation and promotes cytoprotection in response to stress (15–17), we chose to transfer adults to new plates on a daily basis to avoid confounding the current generation with its progeny. We also chose to avoid the use of sterile mutants as studies have found that blocking germ line development promotes protein homeostasis (18).
Transgenic Animals
The genomic pbs-5 promoter fragment was amplified by polymerase chain reaction (PCR) from mixed-stage genomic DNA. The P.pbs-5(ARE)::GFP mutant was produced using PCR site-directed mutagenesis, thereby interchanging a span of 10 nucleotides including two adjacent putative antioxidant response element (ARE) motifs into a PSTI endonuclease restriction site. For a detailed list of all primers used for molecular cloning, refer to Supplementary Table 1. Chromosomal arrays were generated by injecting N2 animals with the expression construct RR10 or 11 (10ng/µL) and co-injection marker KP708 (Pttx-3-rfp, 40ng/µL). Microinjection was performed using standard techniques (19).
Oxidative Stress Assays
For survival assays, animals were treated with a H2O2 bolus in M9 buffer for 20 minutes and allowed to recover on seeded nematode growth medium (NGM) plates for 2–3 hours. Animals were scored for survival with a platinum pick. For tolerance assays, animals were pretreated with H2O2 as indicated for 20 minutes and allowed to adapt on seeded NGM plates for 16 hours. The next day, animals were treated with an LD50 dose of H2O2 determined by the survival assay. Experiments were performed in 3–7 replicates with N = 30–90 animals. The stability of H2O2 in M9 was validated via spectrophotometry by measuring absorbance at 240nm and extrapolating with an extinction coefficient of 43.6M−1 cM−1 (Supplementary Figure 1).
Microscopy and Analysis
Fluorescence microscopy experiments were performed essentially as previously described (20). Day 3 and Day 10 animals were paralyzed using 30mg/mL 2,3-butanedione monoxime and mounted on 2% agarose pads for imaging. Images were acquired with a Nikon eclipse 90i microscope equipped with a Nikon PlanApo 60× or 100× objective (NA = 1.4) and a Photometrics Coolsnap ES2 camera controlled by Metamorph 7.0 software (Universal Imaging/Molecular Devices). For quantitative analysis, each animal was outlined and the average fluorescence was quantified using Metamorph. At least 30–70 animals were imaged for all experiments. To provide standardization for images acquired on different days, fluorescence values were normalized to the values of 0.5 µM FluoSphere yellow-green beads (Life Technologies, cat. F-8813) acquired during each imaging session.
Quantitative RT-PCR
Approximately 100 animals of the indicated genotypes and age group were collected, washed in M9 buffer, and homogenized in TRIzol reagent (Invitrogen, cat. 15596-026). RNA was extracted according to the manufacturer’s instructions. RNA was reverse transcribed to cDNA using TaqMan Reverse Transcription Reagents (Life Technologies, cat. N8080234) and quantitative PCR was performed using iTaq SYBR Green (Bio-Rad, cat. 172–5121). Samples were normalized to the relative mRNA expression of untreated N2 animals at Day 3 and calculated from five biological replicates. Primers utilized in this study are listed in Supplementary Table 1.
Chromatin Immunoprecipitation
SKN-1::GFP animals were treated at Day 3 and Day 10 with ±2-mM H2O2 for 20 minutes and collected immediately for chromatin immunoprecipitation (ChIP) with a GFP antibody. ChIP was performed as described previously with modifications as specified in the Supplementary Methods (21). Immunoprecipitated DNA was purified and quantitative PCR (qPCR) was performed using primers specific to ARE elements within the pbs-5 and gst-4 promoter, in addition to upstream or downstream nonspecific sites (Supplementary Table 1).
Western Blotting
N2 animals were collected in M9 and sonicated in RIPA buffer (50-mM Tris pH 8, 150-mM NaCl, 1% NP-40, 1% sodium deoxycholate, 1% sodium dodecyl sulfate (SDS), and protease inhibitor cocktail) until fragmented (approximately three 10-second pulses at 30% amplitude). Twenty micrograms of lysate was run on a 10% SDS–PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) gel (Biorad, cat. 456–1034) and blotted to a PVDF membrane (Biorad, cat. 162–0177) prior to immunostaining with PAS-7 (Developmental Studies Hybridoma Bank, cat. CePAS7-c) and ACTIN (Santa Cruz, cat. sc-1616 HRP) antibodies. Western blots were performed in 2–3 replicates and quantified using Image J.
Carbonyl Assay
Oxidatively damaged proteins were measured by the detection of protein carbonyl groups. Animals were collected as described for Western blotting, and the supernatant was then used for protein carbonyl assays using reagents from the Oxyblot kit (Millipore, cat. S7150) per manufacturer’s instructions. Briefly, 10 µg of protein was brought to a 5-µL volume with RIPA buffer, and 5 µL of 12% SDS was added for a total concentration of 6% SDS. Samples were incubated with 10 µL of 2,4-dinitrophenylhydrazine for 20 minutes, after which 7.5 µL of neutralization buffer was added to halt the derivatization reaction. Samples were reduced with 1.5-µL β-mercaptoethanol prior to running on an 8% hand cast SDS–PAGE gel. Carbonyl blots were performed in 2–4 replicates and quantified using Image J.
Proteasome Activity Assays
N2 and lax120 animals were treated at Day 3 and Day 10 with ±1 or 10 µM of H2O2 for 20 minutes and allowed to adapt for 16 hours on seeded NGM plates. Animals were collected in M9 and sonicated in proteolysis buffer (50-mM Tris, 20-mM KCL, 5-mM MgAc, 0.5-mM dithiothreitol, pH 7.5) until obliterated. Samples were normalized to 5 µg and diluted in 100 µL of proteolysis buffer in a 96-well plate. Either 20-µM Suc-LLVY-AMC substrate or 1-µg oxidized [3H] hemoglobin were added, and samples were incubated in a plate shaker at 37°C for 4 and 2 hours, respectively. Degradation of Suc-LLVY-AMC substrate was measured at 355nm/444nm ex/em, and degradation of oxidized [3H] hemoglobin substrate was measured using a scintillation counter.
Results
Adaptation to Oxidative Stress Declines During Aging and Is Dependent on SKN-1
Adaptation occurs for a broad spectrum of stressors at both mild and moderate intensity. For our aging studies, we chose Day 10 as an old age time point, at which more than 90% of the population has survived, thus eliminating selection bias by only examining an older, geriatric population. In support of this, microarray analysis has shown that postreproductive C. elegans at this age undergo considerable transcriptional changes and therefore represent a fundamentally different life stage than young adult animals (22).
In order to determine the amount of H2O2 to induce 50% survival, we performed a dose curve for Day 3 and Day 10 animals (Figure 1A). Interestingly, older animals tolerated a slightly higher dose of H2O2 compared with younger animals. Therefore, a median lethal dose (LD50) of 6- and 7-mM H2O2 was used for Day 3 and Day 10 animals, respectively. Next, we evaluated stress resistance, both before and after H2O2 preconditioning. As expected, adaptation to oxidative stress resulted in cytoprotection for Day 3 animals exposed to a median lethal dose of H2O2 (Figure 1B). Interestingly, low (1 µM) and moderate (2mM) doses of acute H2O2 were equally sufficient to induce cytoprotection, indicating that survival after adaptation is not dose dependent (Supplementary Figure 2). In contrast, Day 10 animals were unable to adapt after pretreatment with any dose of H2O2. These data suggest that adaptation to acute oxidative stress declines with age.
In order to determine if adaptation that occurs early in life has a lasting effect on organismal life span or if moderate doses of H2O2 would have a negative effect, we conducted life-span assays for both conditions. We found no significant impact on longevity for animals treated with 1- or 2-mM adaptive doses of H2O2 at Day 3 (Figure 1C).
Figure 1.
Adaptation to H2O2 diminishes during aging and is dependent on SKN-1. (A) Dose survival curve for N2 animals exposed to H2O2. The LD50 dose was determined to be 6 and 7mM for Day 3 and Day 10 animals, respectively. (B) Oxidative stress tolerance for N2 animals adapted to 1- to 10-µM H2O2 preconditioning at Day 3 and Day 10 and challenged with LD50 determined in (A). (C) Survival assay for N2 animals exposed to 1-µM or 2-mM H2O2 for 20 minutes at Day 3. Significance determined by log-rank (Mantel-Cox) test. p = .2054, not significant. (D) Oxidative stress tolerance at 6mM for untreated versus preconditioned (1-µM H2O2) skn-1(zu67), hsf-1(sy441), hif-1 (ia4), and daf-16(m26) animals. Error bars denote standard error of the mean values. *p < .05, **p < .01, ***p < .001 relative to control using Student’s t test.
Preconditioning with a stressor may engage multiple adaptive mechanisms that could result in the activation of more than one transcription factor. To test this idea, we examined the requirement of key stress response regulators for adaptation to oxidative stress. Using the skn-1 (zu67) deletion mutant, we verified our model that SKN-1 is necessary for adaptation to H2O2 (Figure 1D). In addition, neither HSF-1, which mediates intrinsic thermotolerance (23), nor DAF-16, which mediates stress resistance induced by the downregulation of insulin-like signaling (24), is required for H2O2 adaptation (Figure 1D). Furthermore, HIF-1, which regulates the hypoxic stress response, is also not necessary for adaptation to H2O2 (Figure 1D). Based on these findings, we conclude that SKN-1 specifically promotes survival after preconditioning with H2O2.
Aging Results in a Loss of SKN-1 Recruitment to the Pbs-5 Promoter During Aging
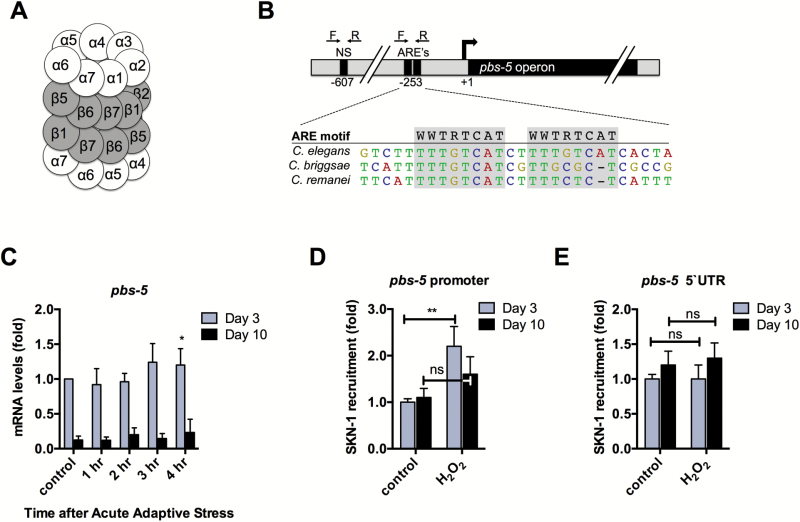
We next characterized changes in regulation of the proteasome during the adaptive response in young compared with old animals. The proteasome is composed of two β-rings that make up the internal enzyme complex of the proteasome, flanked by α-rings on either end that manage substrate entry. Each ring is composed of seven subunits known as proteasome beta or alpha subunits (PBS or PAS), which share significant homology to their mammalian counterparts (Figure 2A).
Figure 2.
SKN-1 recruitment to the pbs-5 promoter declines during aging. (A) Schematic representation of the α and β subunits of the 20S proteasome. (B) Representation of the pbs-5 putative promoter and the ARE-specific and nonspecific (NS) primers used for qPCR of SKN-1::GFP ChIP samples. This region is closely conserved for two other nematode species. (C) qPCR analysis of pbs-5 for Day 3 and Day 10 N2 animals in response to 2-mM H2O2. Expression levels were adjusted to N2 Day 3 control animals. (D) ChIP showing that SKN-1 is recruited to the pbs-5 promoter in response to H2O2 for Day 3, but not for Day 10 animals. (E) SKN-1 occupancy does not change in response to stress at a nonspecific site upstream of the pbs-5 promoter. Error bars denote standard error of the mean values. *p < .05, **p < .01, ***p < .001 relative to control using Student’s t test.
Several proteasome subunits contain AREs within their promoters, which recruit the transcription factor Nrf2/SKN-1 (10). There are two SKN-1 binding consensus sites at position –253 in the promoter of the pbs-5 operon. The sequences and positions of these sites are closely conserved between C. elegans and other nematode species (Figure 2B). Both of these sites have the sequence TTTGTCAT, which matches the consensus ARE motif (WWTRTCAT) (25,26). Due to the close proximity of these conserved ARE sites, we chose to focus on the pbs-5 subunit to further investigate regulation of the proteasome.
We performed qPCR and found that adaptive H2O2 treatment resulted in a significant increase in the levels of pbs-5 transcripts at Day 3, but no change in levels for Day 10 (Figure 2C). The proteasome comprises 1% of total cellular proteins and has an abundance of transcripts similar to that of a housekeeping gene (27). However, at Day 10, we found a marked decrease in pbs-5 transcripts compared with Day 3 (Figure 2C), indicating that induction of pbs-5 declines with age.
To evaluate recruitment of SKN-1 to the pbs-5 promoter in response to stress during aging, we performed ChIP of SKN-1 from SKN-1::GFP-expressing animals. We monitored SKN-1 occupancy by PCR using primers that flank the ARE sites within the promoter (Figure 2B). For Day 3 animals, SKN-1 occupancy at the pbs-5 AREs increased approximately twofold after treatment with H2O2 (Figure 2D). This increase is similar to that observed with other SKN-1 targets (28). However, there was no increase in SKN-1 recruitment in response to stress in Day 10 animals. Furthermore, we found no significant change in the association of SKN-1 to a nearby nonspecific region within the pbs-5 promoter in response to stress (Figure 2E). These findings suggest that the decline of adaptation during aging may be due to a reduction in proteasome gene transcription by SKN-1.
SKN-1 Induction of gst-4 Decreases With Age
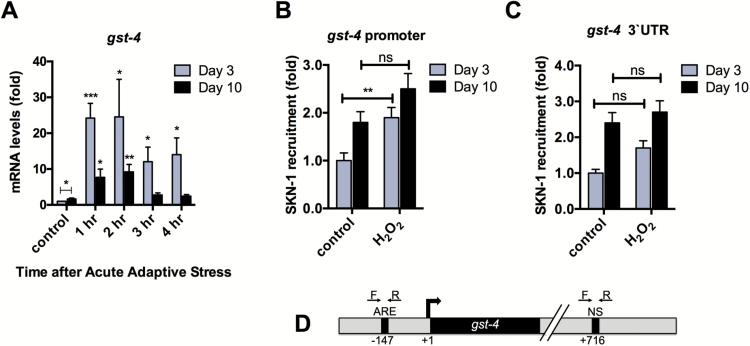
Phase II detoxification is an element of glutathione metabolism and is a cellular response to a multitude of stressors, including xenobiotics, electrophiles, and reactive oxygen species (29–31). Because this response is also mediated by SKN-1, we next investigated whether induction of gst-4 by H2O2 also declines during aging. We found that an acute adaptive dose of 2-mM H2O2 induces the transcription of gst-4 (Figure 3A). Whereas Day 3 animals exhibited an increase in transcription following oxidative stress, induction of gst-4 in Day 10 animals was reduced (Figure 3A). Furthermore, induction of phase II genes gcs-1 and gst-10 in response to H2O2 was also reduced during aging (Supplementary Figure 3). Interestingly, basal gst-4 levels are higher at Day 10 suggesting that the increased tolerance for H2O2 observed in older animals (Figure 1A) may be due to elevated detoxification enzymes during aging (Figure 3A).
Figure 3.
Induction of phase II detoxification gene gst-4 declines in response to H2O2 in aged animals. (A) qPCR analysis of gst-4 for Day 3 and Day 10 N2 animals in response to 2-mM H2O2. Expression levels were adjusted to N2 Day 3 control animals for each gene. (B) SKN-1 is recruited to the gst-4 promoter in response to 2-mM H2O2 for Day 3, but not for Day 10 animals. (C) SKN-1 occupancy does not change in response to stress at a nonspecific site downstream of the gst-4 promoter. Error bars denote standard error of the mean values. *p < .05, **p < .01, ***p < .001 relative to control using Student’s t test. (D) Representation of the gst-4 putative promoter and the ARE-specific and nonspecific (NS) primers used for qPCR of SKN-1::GFP ChIP samples.
Similar to Figure 2D, we also examined promoter occupancy of gst-4 by SKN-1. We found that upon treatment with H2O2, SKN-1 is significantly recruited to the gst-4 promoter for Day 3 animals, but not for Day 10 animals (Figure 3B). To verify that an increase in SKN-1 binding to DNA was specifically due to recruitment to an ARE motif, we also performed qPCR using primers to a nonspecific site in the downstream untranslated region for gst-4 and found no significant change between the control and H2O2-treated animals (Figure 3C and D). Together, the results shown in Figures 2 and 3 suggest a defect in SKN-1 signaling sensitivity in response to oxidative stress during aging.
Proteasome Expression Increases During Aging, But Is Not Induced During Adaption to Oxidative Stress
To further evaluate declining SKN-1 signaling during aging on the 20S proteasome, we examined the impact of adaptive H2O2 pretreatment on pbs-5 expression. We generated a P.pbs-5::GFP chromosomal array that expresses GFP driven by 557bp of the pbs-5 promoter, which contains the entire 5′ intergenic region between pbs-5 and the neighboring gene (Supplementary Figure 4). We found that P.pbs-5::GFP expression increased after pretreatment with oxidative stress for Day 3 animals (Figure 4A and Supplementary Figure 7A).. We also found that Day 10 animals exhibited increased basal levels of P.pbs::GFP, but did not show an increase in expression after pretreatment with H2O2 (Figure 4A and Supplementary Figure 7A).
Figure 4.
Defective SKN-1 signaling impacts proteasome expression in response to stress. (A) Quantification of fluorescence for animals in Supplementary Figure 7A. AFU normalized to 100 in comparison with each control. N = 65–91 animals. (B) Densitometry of Western blots described in (A). Error bars denote standard error of the mean values. *p < .05, **p < .01, ***p < .001 relative to control using Student’s t test.
The pbs-5 gene is part of an operon (CEOP1752), which includes the jagunal 1 (K05C4.2) and sol-2 (K05C4.11) genes. We mutated the two ARE sites within the pbs-5 operon promoter by scrambling six of the nucleotides within the motif and generated transgenic animals expressing these mutated AREs driving expression of GFP (P.pbs-5(ARE)::GFP, Supplementary Figure 4). We found that mutating these ARE sites completely blocked the increase in expression of P.pbs-5(ARE)::GFP after treatment with H2O2 (Figure 4A and Supplementary Figure 7A). In addition, we found that the expression of P.pbs-5(ARE)::GFP did not change despite age or stress. These results indicate that the aging-dependent increase in proteasome expression is also dependent on SKN-1. It should be noted that the P.pbs-5::GFP and P.pbs-5(ARE)::GFP strains were generated by microinjection of DNA encoding the desired transgene, which may have been incorporated unequally and thus cannot be directly compared. Therefore, we compared the fold change in fluorescence after exposure to H2O2.
Although there is no commercially available antibody for the PBS-5 subunit of the proteasome, there is one available for PAS-7. We found that H2O2 adaptation results in an increase in PAS-7 expression for Day 3 animals (Supplementary Figure 7B). However, consistent with our data for P.pbs::GFP (Figure 4A and Supplementary Figure 7A), Day 10 animals exhibited increased basal levels of PAS-7, but no adaptive increase in expression. Because proteasome transcript levels are at least four times lower in Day 10 animals compared with Day 3 animals (Figure 2C), the increase in PAS-7 expression during aging may indicate a passive accumulation of proteasomes that do not become degraded. Accumulation of proteasomes during aging may also account for the slight increase in median lethality observed for Day 10 animals (Figure 1A).
Proteolytic Activity Increases During Aging, But Is Not Induced During Adaptation to Oxidative Stress
The decrease in the adaptive expression of proteasome subunits with age suggested that there might be a corresponding defect in proteasome function. Therefore, we measured proteasome function with two in vitro proteolytic activity assays. First, we characterized the capacity for the 20S proteasome to degrade a Suc-LLV-AMC fluorogenic proteasome substrate in vitro (Figure 5A). For Day 10 animals, we found that basal proteolytic capacity increased significantly, but adaptation to H2O2 was lost with age. An increase in the in vitro capacity for proteolytic activity at Day 10 is consistent with the increase in basal proteasome expression observed in Figure 4 and Supplementary Figure 7.
Figure 5.
Proteasome adaptation is lost during aging, despite an increase in proteolytic activity. (A) Pretreatment with 1- to 10-µM H2O2 results in an increase in adaptive proteolytic activity for Day 3 animals, but not for Day 10 animals. Proteasome peptidase activity was measured based on the degradation of a Suc-LLVY-AMC fluorogenic proteasome substrate measured at 355nm/444nm ex/em. (B) Pretreatment with 1-µM H2O2 results in an increase in adaptive proteolytic activity for Day 3 animals, but not for Day 10 animals. Proteolytic capacity to degrade oxidized [3H] hemoglobin was measured by release of acid-soluble counts (liquid scintillation). Error bars denote standard error of the mean values. *p < .05, **p < .01, ***p < .001 relative to control using Student’s t test.
Second, we measured the proteolytic capacity to degrade oxidized [H3] hemoglobin in vitro (Figure 5B). As in Figure 5A, we found that basal proteasome activity increased with age, but Day 10 animals lost the capacity to adapt to oxidative stress.
For both small peptide and hemoglobin degradation assays, inhibition of proteasomal catalytic activity using epoxomicin, lactacystin, or MG132 resulted in nearly complete inhibition of proteolysis, indicating that other cellular proteases did not significantly contribute to substrate degradation (Supplementary Figure 5).
Increased Activity of SKN-1 Does Not Rescue Adaptation During Aging
We next asked if increased activity of SKN-1 could rescue proteasome function during aging. In order to evaluate increased SKN-1 activity, we utilized a mutant expressing skn-1 gain-of-function (gf) allele lax120 (32), which encodes S245L, to determine if increased activation of SKN-1 can rescue adaptation during aging.
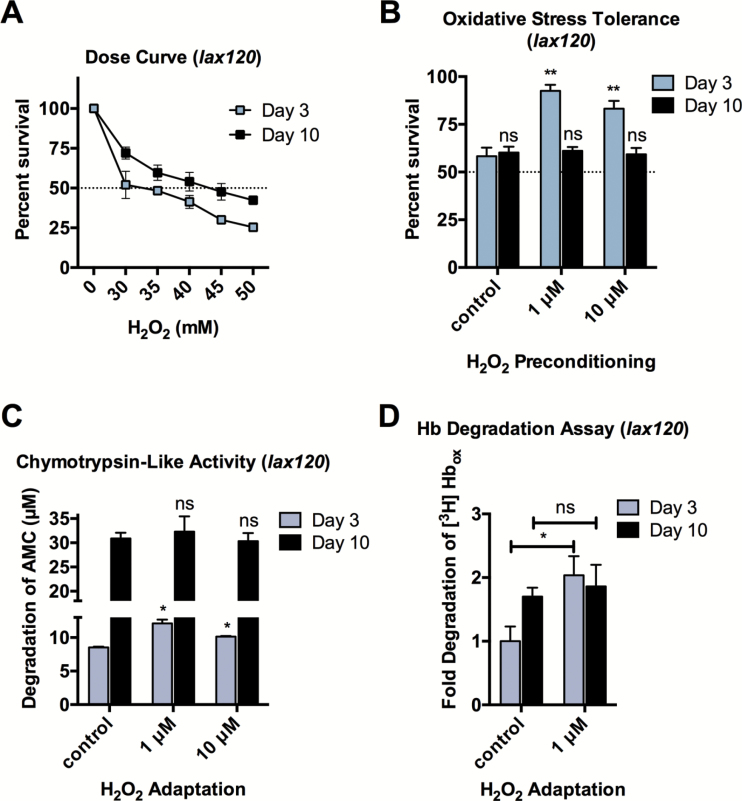
We performed a survival assay and found that increased activation of SKN-1 results in a 5-fold increase in stress resistance compared to N2 animals (Figures 1A and 6A). While young lax120 animals showed adaptation similar to wild-type, increased SKN-1 activation did not rescue adaptation for Day 10 animals at the adjusted median lethal dose (Figure 6B). Therefore, while increased SKN-1 activity directly impacts SKN-1 gene target levels and oxidative stress tolerance, it was not able to rescue the aging-related decline of adaptation.
Figure 6.
Increased SKN-1 activation does not rescue adaptive proteolytic capacity during aging. (A) Animals with increased SKN-1 activity (lax120) have an increased tolerance for H2O2 compared with N2. The LD50 dose was determined to be 30 and 50mM for Day 3 and Day 10 animals, respectively. (B) Oxidative stress tolerance for lax120 animals adapted to 1- to 10-μM H2O2 preconditioning at Day 3 and Day 10 and challenged with LD50 determined in (A). (C) Increased activity of SKN-1 (lax120) does not rescue proteolytic capacity to degrade Suc-LLVY-AMC for Day 10 animals. (D) Increased activity of SKN-1 (lax120) does not rescue proteolytic capacity to degrade oxidized [3H] hemoglobin for Day 10 animals. Error bars denote standard error of the mean values. *p < .05, **p < .01, ***p < .001 relative to control using Student’s t test.
We next asked if increased activation of SKN-1 could rescue proteasome functional adaptation during aging. We performed a chymotrypsin-like activity assay and found that SKN-1 gain-of-function does not rescue 20S proteasome-dependent adaptation during aging (Figure 6C). In addition, increased SKN-1 activity did not rescue adaption of proteolytic capacity to degrade oxidized [H3] hemoglobin in Day 10 animals (Figure 6D).
Adaptive Proteolytic Degradation Declines During Aging In Vivo
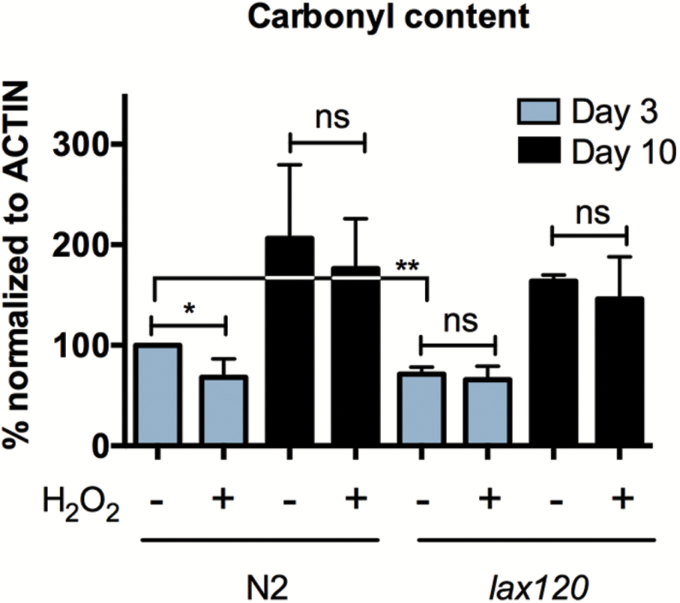
In order to evaluate proteasome capacity in vivo, we measured total protein carbonylation, an irreversible consequence of oxidative damage that often leads to loss of protein function (33). In accordance with other studies (34–36), we found that protein carbonylation increases with age. We also found that C. elegans exhibit an increase in carbonylation as early as Day 5 (Supplementary Figure 6).
We next sought to evaluate the impact of H2O2 pretreatment and SKN-1 gain-of-function on protein carbonylation during aging. Pretreatment with H2O2, followed by a 16-hour recovery, resulted in a significant decrease in total protein carbonylation for young animals (Figure 7 and Supplementary Figure 8). SKN-1 gain-of-function also resulted in a significant decrease in carbonylation compared with the untreated N2 control, though pretreatment did not impact carbonylation for lax120 animals. Moreover, for both Day 10 N2 and lax120 animals, protein carbonylation did not decrease after pretreatment indicating that SKN-1 activation is beneficial for young animals, but does not alleviate detrimental phenotypes associated with decreased proteasome utility in older animals. Therefore, these results demonstrate that while young lax120 mutants were able to increase proteolytic function in response to the adaptive H2O2 treatment, old animals did not exhibit adaptation in any of these assays.
Figure 7.
Adaptation to oxidative stress reduces total protein carbonylation for Day 3, but not for Day 10 animals. Densitometry of total protein carbonylation for N2 and lax120 animals ± adaptation to H2O2. Error bars denote standard error of the mean values. *p < .05, **p < .01, ***p < .001 relative to control using Student’s t test.
Discussion
The overall decline of adaptive stress responses is thought to be a major factor in organismal aging and aging-related disease. Acute stress leads to the induction of transcriptional programs evolved to promote cellular homeostasis. Several studies have indicated that aging correlates with a defect for induction of these adaptive mechanisms (37). In parallel, another key to adaptation is the immediate response to stress marked by changes in affinity of existing cytoprotective proteins that play a role in proteome quality control. Our results demonstrate dynamic changes for adaptive signaling and proteolytic capacity during aging. These findings highlight the complexity of the proteasome system and the decline of transcriptional signaling in aging animals.
Many cell culture studies have found that proteasome function declines with replicative senescence (38–42) and that inhibition of the proteasome can induce senescence (43). These findings rely on in vitro proteolytic activity assays and Western blot analysis to evaluate ubiquitylation and expression of proteasome subunits. In addition to immortalized cell culture studies, primary cells cultured from old tissues have also exhibited a decrease in proteasome function for cells propagated from both human patients (44–46) and mammalian models (47). A caveat for these findings is that cells grown in culture become progressively closer to replicative senescence with each passage, which may not be the best parallel to model normal progressive physiological aging in an organism.
Interestingly, mammalian animal studies have indicated tissue-specific variability in proteasome function during aging. For instance, it was found that proteasome expression and proteolytic activity decline in the spinal cord of aged rats (48). However, despite increased oxidative stress, there was no change in proteolytic activity found in the cortex, cerebellum, or hippocampus during aging (49). In contrast, several studies have found aging-related increases in 20S expression and activity in skeletal muscle (50–54). Furthermore, Altun and colleagues found that expression of ubiquitin ligases and deubiquitylating enzymes also increase during aging, indicating a role for both the 20S and 26S conformations of the proteasome in muscle wasting (51). In addition to muscle tissue, increased 20S proteasome function has been noted in the liver (55) and spleen (56), whereas decreased function has been noted in the heart (47) and lung (57). Therefore, the regulation of proteasome activity is likely tissue dependent and varies depending on the aging continuum. Because our study used whole animal lysate for all biochemistry analyses, it is possible that the major proteasome pool is representative of muscle and intestine, which constitute the largest group of tissues in C. elegans. Our findings add to the field of proteasome research and help to further elucidate the dynamic regulation of 20S proteasome regulation during aging.
However, despite an increase in 20S proteasome activity, protein oxidation also increased with age. It should be noted that assays for proteolytic capacity are in vitro and it is not yet feasible to measure 20S proteasome activity in vivo. Therefore, it is possible that, although 20S proteasomes from aged cells have the capacity for proteolytic degradation, they may not actively degrade proteins in vivo. Proteins that are heavily oxidized have been shown to inhibit the proteasome (41,58–60). In addition, a variety of proteome quality control machinery are sequestered at protein aggregates, which increase during aging (61–63). Consistent with our data, a recent study in yeast found that enhancing protein disaggregation restores proteasome activity in replicatively aged cells, indicating that in vivo inactivation of proteasome function is reversible (64). Thus, increased protein oxidation during aging may be attributed to the accumulating, yet reversible, inactivation of the 20S proteasome.
We propose that the aging-related decline in SKN-1 signaling sensitivity in response to increasing stress contributes to the accrual of dysfunctional 20S proteasomes and the subsequent loss of adaptation during aging. Although enhanced SKN-1 activity increases stress tolerance, it does not rescue adaptation or result in an increase in life span. However, recent work has shown that direct overexpression of proteasome expression has a positive impact on health span and life span in C. elegans (65).
Our results indicate that understanding the balance between proteasome function and oxidatively damaged aggregated proteins is important in managing the collapse of protein homeostasis during aging. Further work is necessary to identify methods to alleviate declining adaptation of the 20S proteasome to resolve aging-dependent protein oxidation and aggregation.
Supplementary Material
Supplementary material can be found at: http://biomedgerontology.oxfordjournals.org/
Funding
This work was supported by National Institutes of Health grants ES03598-25 (PI: K.J.A.D.) and NS071085 (PI: D.S.).
Supplementary Material
Acknowledgments
We thank Sean Curran for helpful advice and for the skn-1(gf) allele mutant (lax120). We also thank the Pinchas Cohen lab for shared equipment and undergraduate researcher Sara Bilimoria for technical assistance. Some strains were provided by the Caenorhabditis Genetics Center.
References
- 1. Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011;3:a004440 doi:10.1101/cshperspect.a004440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi:10.1161/01.RES.0000252342.61447.a2 [DOI] [PubMed] [Google Scholar]
- 3. Chen H, Yoshioka H, Kim GS, et al. Oxidative stress in ischemic brain damage: mechanisms of cell death and potential molecular targets for neuroprotection. Antioxid Redox Signal. 2011;14:1505–1517. doi:10.1089/ars.2010.3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi:10.1101/gad.1657108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacob KD, Noren Hooten N, Trzeciak AR, Evans MK. Markers of oxidant stress that are clinically relevant in aging and age-related disease. Mech Ageing Dev. 2013;134:139–157. doi:10.1016/j.mad.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barber SC, Mead RJ, Shaw PJ. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochim Biophys Acta. 2006;1762:1051–1067. doi:10.1016/j.bbadis.2006.03.008 [DOI] [PubMed] [Google Scholar]
- 7. Rattan SI. Hormesis in aging. Ageing Res Rev. 2008;7:63–78. doi:10.1016/ j.arr.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 8. Calderwood SK, Murshid A, Prince T. The shock of aging: molecular chaperones and the heat shock response in longevity and aging–a mini-review. Gerontology. 2009;55:550–558. doi:10.1159/000225957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Texel SJ, Mattson MP. Impaired adaptive cellular responses to oxidative stress and the pathogenesis of Alzheimer’s disease. Antioxid Redox Signal. 2011;14:1519–1534. doi:10.1089/ars.2010.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pickering AM, Linder RA, Zhang H, Forman HJ, Davies KJ. Nrf2-dependent induction of proteasome and Pa28alphabeta regulator are required for adaptation to oxidative stress. J Biol Chem. 2012;287:10021–10031. doi:10.1074/jbc.M111.277145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pickering AM, Staab TA, Tower J, Sieburth D, Davies KJ. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative stress adaptation in mammals, Caenorhabditis elegans and Drosophila melanogaster . J Exp Biol. 2013;216:543–553. doi:10.1242/jeb.074757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brignull HR, Morley JF, Garcia SM, Morimoto RI. Modeling polyglutamine pathogenesis in C. elegans . Methods Enzymol. 2006;412:256–282. doi:10.1016/S0076-6879(06)12016-9 [DOI] [PubMed] [Google Scholar]
- 13. Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans . Free Radic Biol Med. 2015;88:290–301. doi:10.1016/j.freeradbiomed.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feldman N, Kosolapov L, Ben-Zvi A. Fluorodeoxyuridine improves Caenorhabditis elegans proteostasis independent of reproduction onset. PLoS One. 2014;9:e85964. doi:10.1371/journal.pone.0085964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brunquell J, Bowers P, Westerheide SD. Fluorodeoxyuridine enhances the heat shock response and decreases polyglutamine aggregation in an HSF-1-dependent manner in Caenorhabditis elegans . Mech Ageing Dev. 2014;141–142:1–4. doi:10.1016/j.mad.2014.08.002 [DOI] [PubMed] [Google Scholar]
- 17. Andersen NK, Anderson BA, Wengel J, Hrdlicka PJ. Synthesis and characterization of oligodeoxyribonucleotides modified with 2’-amino-alpha-L-LNA adenine monomers: high-affinity targeting of single-stranded DNA. J Org Chem. 2013;78:12690–12702. doi:10.1021/jo4022937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shemesh N, Shai N, Ben-Zvi A. Germline stem cell arrest inhibits the collapse of somatic proteostasis early in Caenorhabditis elegans adulthood. Aging Cell. 2013;12:814–822. doi:10.1111/acel.12110 [DOI] [PubMed] [Google Scholar]
- 19. Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ch’ng Q, Sieburth D, Kaplan JM. Profiling synaptic proteins identifies regulators of insulin secretion and lifespan. PLoS Genet. 2008;4:e1000283. doi:10.1371/journal.pgen.1000283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans . Nat Protoc. 2008;3:698–709. doi:10.1038/nprot.2008.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Golden TR, Hubbard A, Dando C, Herren MA, Melov S. Age-related behaviors have distinct transcriptional profiles in Caenorhabditis elegans . Aging Cell. 2008;7:850–865. doi:10.1111/j.1474-9726.2008.00433.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nate Rev Mol Cell biol. 2010;11:545–555. doi:10.1038/nrm2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi:10.1038/88850 [DOI] [PubMed] [Google Scholar]
- 25. Blackwell TK, Bowerman B, Priess JR, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi:10.1126/science.7939715 [DOI] [PubMed] [Google Scholar]
- 26. Oliveira RP, Porter Abate J, Dilks K, et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi:10.1111/j.1474-9726.2009.00501.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kremmer E, Ohst K, Kiefer J, Brewis N, Walter G. Separation of PP2A core enzyme and holoenzyme with monoclonal antibodies against the regulatory A subunit: abundant expression of both forms in cells. Mol Cell Biol. 1997;17:1692–1701. doi:10.1128/MCB.17.3.1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robida-Stubbs S, Glover-Cutter K, Lamming DW, et al. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metabol. 2012;15:713–724. doi:10.1016/j.cmet.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi:10.1111/j.1474-9726.2009.00501.x [DOI] [PubMed] [Google Scholar]
- 30. Talalay P, Dinkova-Kostova AT, Holtzclaw WD. Importance of phase 2 gene regulation in protection against electrophile and reactive oxygen toxicity and carcinogenesis. Adv Enzyme Regul. 2003;43:121–134. doi:10.1016/S0065-2571(02)00038-9 [DOI] [PubMed] [Google Scholar]
- 31. An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi:10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paek J, Lo JY, Narasimhan SD, et al. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metabol. 2012;16:526–537. doi:10.1016/j.cmet.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi:10.1007/s00726-003-0011-2 [DOI] [PubMed] [Google Scholar]
- 34. Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi:10.1101/gad.439307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levine RL, Stadtman ER. Oxidative modification of proteins during aging. Exp Gerontol. 2001;36:1495–1502. doi:10.1016/S0531-5565(01)00135-8 [DOI] [PubMed] [Google Scholar]
- 36. Baraibar MA, Ladouce R, Friguet B. Proteomic quantification and identification of carbonylated proteins upon oxidative stress and during cellular aging. J Proteomic. 2013;92:63–70. doi:10.1016/j.jprot.2013.05.008 [DOI] [PubMed] [Google Scholar]
- 37. Haigis MC, Yankner BA. The aging stress response. Mol Cell. 2010;40:333–344. doi:10.1016/j.molcel.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chondrogianni N, Stratford FL, Trougakos IP, Friguet B, Rivett AJ, Gonos ES. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J Biol Chem. 2003;278:28026–28037. doi:10.1074/jbc.M301048200 [DOI] [PubMed] [Google Scholar]
- 39. Ahmed EK, Picot CR, Bulteau AL, Friguet B. Protein oxidative modifications and replicative senescence of WI-38 human embryonic fibroblasts. Ann NY Acad Sci. 2007;1119:88–96. doi:10.1196/annals.1404.020 [DOI] [PubMed] [Google Scholar]
- 40. Lu L, Song HF, Wei JL, et al. Ameliorating replicative senescence of human bone marrow stromal cells by PSMB5 overexpression. Biochem Biophys Res Commun. 2014;443:1182–1188. doi:10.1016/j.bbrc.2013.12.113 [DOI] [PubMed] [Google Scholar]
- 41. Sitte N, Huber M, Grune T, et al. Proteasome inhibition by lipofuscin/ceroid during postmitotic aging of fibroblasts. FASEB J. 2000;14:1490–1498. doi:10.1096/fj.99-0843com [DOI] [PubMed] [Google Scholar]
- 42. Sitte N, Merker K, von Zglinicki T, Grune T. Protein oxidation and degradation during proliferative senescence of human MRC-5 fibroblasts. Free Radic Biol Med. 2000;28:701–708. doi:10.1016/S0891-5849(99)00279-8 [DOI] [PubMed] [Google Scholar]
- 43. Chondrogianni N, Gonos ES. Proteasome inhibition induces a senescence-like phenotype in primary human fibroblasts cultures. Biogerontology. 2004;5:55–61. [DOI] [PubMed] [Google Scholar]
- 44. Hwang JS, Hwang JS, Chang I, Kim S. Age-associated decrease in proteasome content and activities in human dermal fibroblasts: restoration of normal level of proteasome subunits reduces aging markers in fibroblasts from elderly persons. J Gerontol A Biol Sci Med Sci. 2007;62:490–499. [DOI] [PubMed] [Google Scholar]
- 45. Petropoulos I, Conconi M, Wang X, et al. Increase of oxidatively modified protein is associated with a decrease of proteasome activity and content in aging epidermal cells. J Gerontol A Biol Sci Med Sci. 2000;55, B220–B227. doi:10.1093/gerona/55.5.B220 [DOI] [PubMed] [Google Scholar]
- 46. Boraldi F, Bini L, Liberatori S, et al. Proteome analysis of dermal fibroblasts cultured in vitro from human healthy subjects of different ages. Proteomics. 2003;3:917–929. doi:10.1002/pmic.200300386 [DOI] [PubMed] [Google Scholar]
- 47. Bulteau AL, Szweda LI, Friguet B. Age-dependent declines in proteasome activity in the heart. Arch Biochem Biophys. 2002;397:298–304. doi:10.1006/abbi.2001.2663 [DOI] [PubMed] [Google Scholar]
- 48. Keller JN, Huang FF, Markesbery WR. Decreased levels of proteasome activity and proteasome expression in aging spinal cord. Neuroscience. 2000;98:149–156. doi:10.1016/S0306-4522(00)00067-1 [DOI] [PubMed] [Google Scholar]
- 49. Abd El Mohsen MM, Iravani MM, Spencer JP, et al. Age-associated changes in protein oxidation and proteasome activities in rat brain: modulation by antioxidants. Biochem Biophys Res Commun. 2005;336:386–391. doi:10.1016/j.bbrc.2005.07.201 [DOI] [PubMed] [Google Scholar]
- 50. Ferrington DA, Husom AD, Thompson LV. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005;19:644–646. doi:10.1096/fj.04-2578fje [DOI] [PubMed] [Google Scholar]
- 51. Altun M, Besche HC, Overkleeft HS, et al. Muscle wasting in aged, sarcopenic rats is associated with enhanced activity of the ubiquitin proteasome pathway. J Biol Chem. 2010;285:39597–39608. doi:10.1074/jbc.M110.129718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bardag-Gorce F, Farout L, Veyrat-Durebex C, Briand Y, Briand M. Changes in 20S proteasome activity during ageing of the LOU rat. Mol Biol Rep. 1999;26:89–93. [DOI] [PubMed] [Google Scholar]
- 53. Hepple RT, Qin M, Nakamoto H, Goto S. Caloric restriction optimizes the proteasome pathway with aging in rat plantaris muscle: implications for sarcopenia. Am J Physiol Regul Integr Comp Physiol. 2008;295, R1231–R1237. doi:10.1152/ajpregu.90478.2008 [DOI] [PubMed] [Google Scholar]
- 54. Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci USA. 2009;106:20405–20410. doi:10.1073/pnas.0911570106 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55. Shibatani T, Nazir M, Ward WF. Alteration of rat liver 20S proteasome activities by age and food restriction. J Gerontol A Biol Sci Med Sci. 1996;51, B316–B322. doi:10.1093/gerona/51A.5.B316 [DOI] [PubMed] [Google Scholar]
- 56. Zhang L, Li F, Dimayuga E, Craddock J, Keller JN. Effects of aging and dietary restriction on ubiquitination, sumoylation, and the proteasome in the spleen. FEBS Lett. 2007;581:5543–5547. doi:10.1016/j.febslet.2007.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Breusing N, Arndt J, Voss P, et al. Inverse correlation of protein oxidation and proteasome activity in liver and lung. Mech Ageing Dev. 2009;130:748–753. doi:10.1016/j.mad.2009.09.004 [DOI] [PubMed] [Google Scholar]
- 58. Jung T, Grune T. The proteasome and the degradation of oxidized proteins: Part I-structure of proteasomes. Redox Biol. 2013;1:178–182. doi:10.1016/j.redox.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shringarpure R, Grune T, Sitte N, Davies KJ. 4-Hydroxynonenal-modified amyloid-beta peptide inhibits the proteasome: possible importance in Alzheimer’s disease. Cell Mol Life Sci. 2000;57:1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grune T, Jung T, Merker K, Davies KJ. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int J Biochem Cell Biol. 2004;36:2519–2530. doi:10.1016/j.biocel.2004.04.020 [DOI] [PubMed] [Google Scholar]
- 61. Stenoien DL, Cummings CJ, Adams HP, et al. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum Mol Genet. 1999;8:731–741. doi:10.1093/hmg/8.5.731 [DOI] [PubMed] [Google Scholar]
- 62. Verhoef LG, Lindsten K, Masucci MG, Dantuma NP. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum Mol Genet. 2002;11:2689–2700. doi:10.1093/hmg/11.22.2689 [DOI] [PubMed] [Google Scholar]
- 63. Shringarpure R, Davies KJ. Protein turnover by the proteasome in aging and disease. Free Radic Biol Med. 2002;32:1084–1089. doi:10.1016/S0891-5849(02)00824-9 [DOI] [PubMed] [Google Scholar]
- 64. Andersson V, Hanzen S, Liu B, Molin M, Nystrom T. Enhancing protein disaggregation restores proteasome activity in aged cells. Aging. 2013;5:802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chondrogianni N, Georgila K, Kourtis N, Tavernarakis N, Gonos ES. 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans . FASEB J. 2015;29:611–622. doi:10.1096/fj.14-252189 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.