Abstract
Whereas Glc is stored in small-sized hydrosoluble glycogen particles in archaea, eubacteria, fungi, and animal cells, photosynthetic eukaryotes have resorted to building starch, which is composed of several distinct polysaccharide fractions packed into a highly organized semicrystalline granule. In plants, both the initiation of polysaccharide synthesis and the nucleation mechanism leading to formation of new starch granules are currently not understood. Ostreococcus tauri, a unicellular green alga of the Prasinophyceae family, defines the tiniest eukaryote with one of the smallest genomes. We show that it accumulates a single starch granule at the chloroplast center by using the same pathway as higher plants. At the time of plastid division, we observe elongation of the starch and division into two daughter structures that are partitioned in each newly formed chloroplast. These observations suggest that in this system the information required to initiate crystalline polysaccharide growth of a new granule is contained within the preexisting polysaccharide structure and the design of the plastid division machinery.
Starch and glycogen define the most widespread form of Glc storage in living cells and consist of α-1,4 linked glucan chains with α-1,6 branches (Buléon et al., 1998; Ball and Morell, 2003). Hydrosoluble glycogen particles cannot grow greater than 40 nm in diameter because the structure becomes too crowded with Glc at the periphery of the particle to accommodate the presence of enzymes of glycogen synthesis or degradation (Meléndez et al., 1998). This limitation is due to both the higher branching level of the polymer and to its rather uniform branching pattern. There presently seems to be no other limit to the size of a starch granule than that afforded by the availability of substrate for continuing synthesis. Amylopectin, the major polysaccharide of starch, aggregates into insoluble semicrystalline material because of its asymmetrical distribution of α-1,6 linkages generating clusters of branches responsible for formation of arrays of parallel double helical structures (Buléon et al., 1998). The appearance of starch coincides with the acquisition of photosynthesis by the eukaryotic cell. No such polymer can presently be found in bacteria, archea, fungi, or animal cells. Although starch is clearly associated with the acquisition of photoautotrophy by eukaryotes, glycogen seems to be the predominant form of Glc storage within cyanobacteria and purple nonsulfur bacteria (for review, see Preiss and Romeo, 1989). In addition, nonphotosynthetic eukaryotes accumulating amylopectin-like polymers such as apicomplexa parasite heterotrophic dinoflagellates or others were always subsequently inferred to be derived from a photosynthetic eukaryote ancestor (McFadden et al., 1996). Starch has been found to accumulate in plastids in green algae and land plants, while red algae, glaucophytes, dinoflagellates, apicomplexa parasites, and cryptophytes accumulate an extraplastidial form of so-called floridean starch (Viola et al., 2001). Green plants and algae are known to use the bacterial ADP-Glc-based pathway of storage polysaccharide synthesis (Recondo and Leloir, 1961; Greenberg and Preiss, 1964). Although chloroplasts and cyanobacteria share a common ancestor, the latter appear to synthesize glycogen, while starch as such may have appeared after endosymbiosis of the chloroplast. Plants and Chlorophycean algae such as Chlamydomonas reinhardtii have acquired multiple enzyme forms for each step of the metabolic pathway.
Ostreococcus tauri is a picophytoplanktonic species that belongs to the Prasinophyceae, a group of green algae thought to have diverged very early from the ancestor of all chloroplast-containing green plants and algae. Ostreococcus, with a 0.8-μm diameter, presently defines the tiniest eukaryotic cell and the smallest currently described genome for a photosynthetic eukaryotic organism (Chrétiennot-Dinet et al., 1995; Courties et al., 1998). A DNA-sequencing effort has recently yielded the full genome of this organism (H. Moreau and Y. Van de Peer, unpublished data). Because of its relative simplicity and because of its particular position with respect to the evolution of green plants, we have undertaken a detailed study of storage polysaccharide synthesis. We report that despite the genome simplification that took place in picophytoplanktonic species, a unique starch granule is synthesized at the center of the tiny plastid by using a pathway of similar complexity as that of higher plants or Chlamydomonas. We observe, however, that this granule elongates and divides into two daughter structures at the time of plastid division. We propose that localized synthesis and degradation regulate starch granule partitioning and maintenance in this species. The implications for the priming of starch granules in higher plants and algae are discussed.
RESULTS
Structural Characterization of O. tauri Storage Polysaccharide
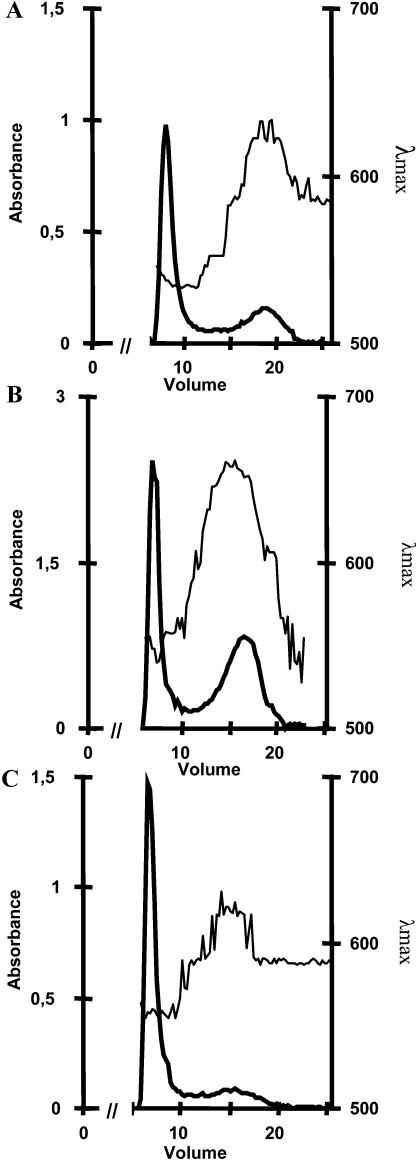
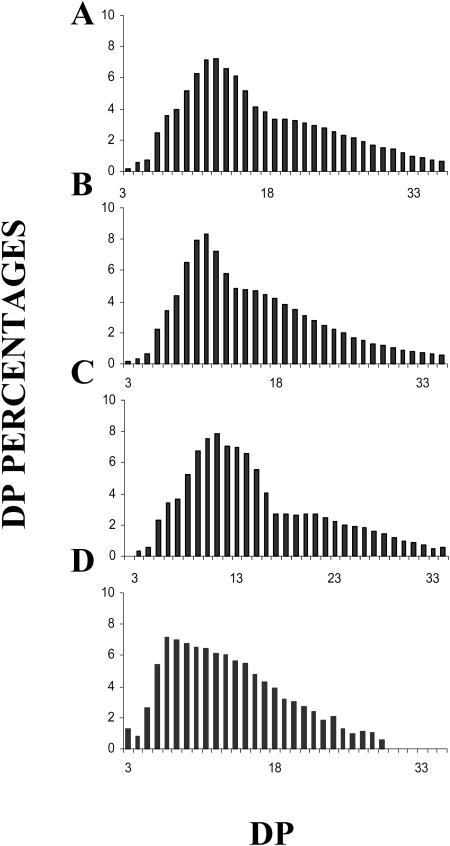
The granule present at the center of each unique plastid was purified and shown to be starch by the following criteria. It contained an amyloglucosidase-sensitive polysaccharide that was shown by NMR to be composed of approximately 96% and 4% α-1,4 and α-1,6 glucosidic linkages, respectively. The polysaccharide dispersed in 10 mm NaOH could be fractionated by gel permeation chromatography (Fig. 1) on a sepharose CL2B column. A major polysaccharide fraction with an iodine interaction identical to that of plant amylopectin was always witnessed. A minor fraction whose relative importance depended on the growing conditions was purified with an average mass distribution and iodine interaction identical to those of cereal amyloses. Proton NMR performed on the amylopectin-like and amylose-like fractions yielded 6% and less than 1% α-1,6 glucosidic linkages, respectively, and thereby confirmed the identity of both fractions (see supplemental material, available at www.plantphysiol.org). The chain-length (CL) distribution of the O. tauri amylopectin fraction was compared to that of rabbit liver glycogen and of Arabidopsis and Chlamydomonas amylopectin fractions (Fig. 2). The CL distribution of the material purified from Ostreococcus was typically trimodal and similar to those of the plant standards and differed noticeably from the unimodal CL distribution of the animal glycogens. The semicrystalline nature of the native starch granule was further checked by x-ray diffraction. Plant starches contain parallel double helical glucan chains that display either the A or B type of crystalline packing. The polysaccharide granules from Ostreococcus were shown to be predominantly of the B type, which further establishes this material as starch.
Figure 1.
Separation of amylose and amylopectin CL2B gel permeation chromatography of starch from O. tauri (1 mg) dispersed in 10 mm NaOH (A) in comparison with C. reinhardtii (2 mg; B) and Arabidopsis (1 mg; C). The λmax (wavelength of the maximal absorbance of the iodine-polysaccharide complex in nanometers) is scaled on the right axis. The absorbency of the complex at λmax measured for each fraction is indicated on the left axis. The low λmax fraction excluded from the column defines amylopectin, while the high λmax amylose is separated throughout the column. The x axis shows the elution volume in milliliters.
Figure 2.
Polysaccharide CL distributions. Histograms of CL distributions obtained after isoamylase-mediated enzymatic debranching through capillary electrophoresis of 8-amino-1,3,6-pyrenetrisulfonic acid-labeled fluorescent glucans. The x axis displays degree of polymerization scales (DP 3–35) and the y axis represents the relative frequency of chains expressed as percentages. A, CL distribution of purified amylopectin from O. tauri. B and C, CL distribution of purified amylopectin from Arabidopsis (B) and C. reinhardtii (C) wild-type references. D, Bovine liver glycogen CL distribution.
O. tauri Synthesizes Starch through the ADP-Glc Pathway
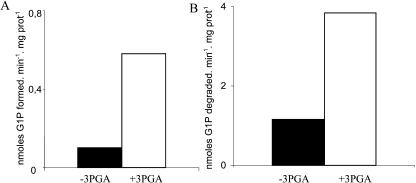
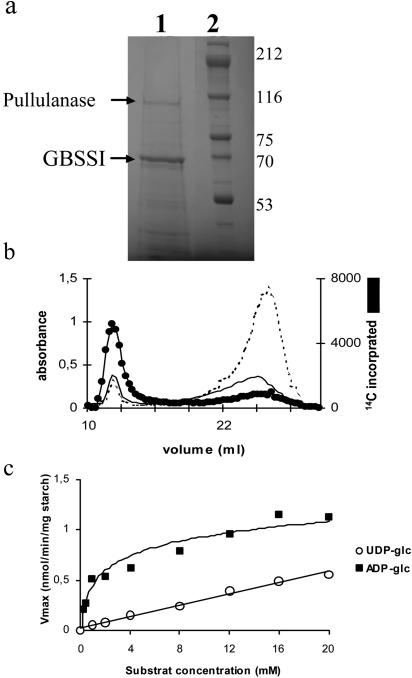
To identify the biochemical pathway of starch biosynthesis, we concentrated our efforts on two key activities. Bacteria and plants both synthesize their storage polysaccharides through the use of ADP-Glc (Recondo and Leloir, 1961; Greenberg and Preiss, 1964). ADP-Glc pyrophosphorylase defines an enzyme devoted to glycogen and starch biosynthesis (Ballicora et al., 2003). It has not been found in organisms such as yeast (Saccharomyces cerevisiae) or humans, which use the UDP-Glc pathway of storage polysaccharide synthesis. We found an ADP-Glc pyrophosphorylase activity in O. tauri crude extracts through the use of two distinct enzyme assays (Fig. 3). The enzyme was activated most efficiently by 3-phosphoglycerate (PGA) and responded to inhibition by orthophosphate (see supplemental material) in a fashion similar to that described for land plants and cyanobacteria (Ballicora et al., 2003). To further confirm that Ostreococcus was using the ADP-Glc pathway, we turned our attention to granule-bound starch synthase (GBSSI), the most abundant enzyme bound to the starch granule in plants (Delrue et al., 1992). This enzyme has been shown to be selectively responsible for amylose synthesis. The major protein bound to starch was analyzed by matrix-assisted laser-desorption ionization-mass spectrometry (MALDI-MS) following trypsic digestion and shown to be identical to an expressed Ostreococcus genome sequence bearing strong resemblance to the plant GBSSI class of enzymes (Fig. 4A). As was demonstrated in Chlamydomonas, we were able to synthesize a significant amount of amylose in vitro by labeled ADP-Glc (Fig. 4B). In addition, the Ostreococcus GBSSI was shown to prefer markedly ADP-Glc (Km, 1.3 mm), while the enzyme could not be saturated at 10 mm with UDP-Glc in a fashion similar to that which was originally shown in maize (Zea mays; Fig. 4C). Taken together, these results suggest strongly that prasinophytes synthesize starch through the ADP-Glc pathway.
Figure 3.
Histograms of ADP-Glc pyrophosphorylase activity. A displays the assay in the nonphysiological direction of pyrophophorolysis in the presence (white box) or absence (black box) of 1 mm 3PGA. Units are expressed in nanomoles of Glc-1-P formed per minute and per milligram of protein. B represents the activity of ADP-Glc pyrophosphorylase in the direction of ADP-Glc synthesis in the presence or absence of 1 mm activator. Units are expressed in nanomoles of Glc-1-P degraded per minute and per milligram of protein.
Figure 4.
Analysis of GBSSI activity and protein from O. tauri starch granules. A, Coomassie Brilliant Blue R-250 stained 7.5% SDS-acrylamide gels of starch-bound proteins. Lane 1 represents starch-bound proteins extracted from 2 mg of polysaccharides purified after nitrogen starvation. Lane 2 displays the molecular-mass marker with the corresponding masses expressed in kilodaltons. B, Kinetics of in vitro synthesis of amylose. Starch from O. tauri was subjected to in vitro synthesis in the presence of 14C-labeled ADP-Glc. After 16 h (black line) and 40 h (doted line) of in vitro synthesis, the amylopectin and amylose were separated by CL2B-sepharose chromatography. C, ADP-Glc kinetics of the GBSSI from O. tauri. A total of 100 μg of polysaccharides were incubated with different concentrations of ADP-Glc (▪) or UDP-Glc (○).
Probing the Ostreococcus Genome for Enzymes of Starch Metabolism
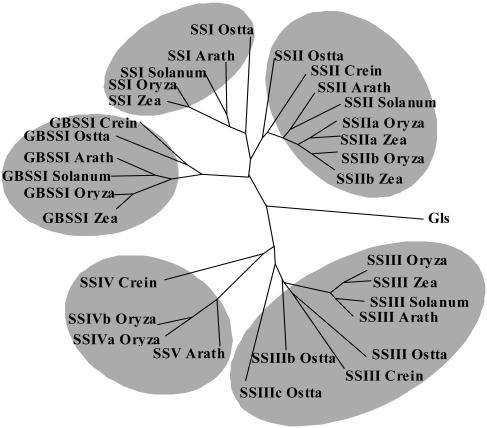
We probed the full Ostreococcus genome sequence for genes of storage polysaccharide metabolism (see supplemental material for Ostreococcus accession numbers list). Each sequence was checked for expression by reverse transcription (RT)-PCR and was found to be expressed and to yield the expected RT-PCR fragment sizes. The results are displayed in Table I, and an example of the phylogenetic tree is given in Figure 5 for the starch synthases. The Ostreococcus small-genome size had initially raised our hopes that this organism may have streamlined its metabolic pathways and reduced the number of enzyme forms required at each step of the pathway, thereby facilitating functional studies dealing with starch metabolism. Table I demonstrates that this is not the case and that Ostreococcus displays the same level of complexity as that of vascular plants with respect to starch biosynthesis and degradation. It is evident from the tree displayed in Figure 5 that the ancestor of Chlorophyceae, Prasinophyceae, and plants contained a minimum of four sequence families of starch synthases and that these have been conserved throughout evolution. The same observation holds for most other enzymes of starch metabolism despite the obvious general simplification of the Ostreococcus genome.
Table I.
Storage polysaccharide metabolism in plants and bacteria
| Genes in E. coli Genome | Genes in O. taurii Genome | Genes in Arabidopsis Genome | Genes in O. sativa Genome |
|---|---|---|---|
| AGPase | AGPase small subunit | AGPase small subunit | AGPase small subunit 1 and 2 |
| AGPase large subunit A and B | AGPase large subunit 1, 2, 3, and 4 | AGPase large subunit 1 and 2 | |
| α-Amylase A and B | α-Amylase A, B, and C | α-Amylase 1, 2, and 3 | α-Amylase 1 to 9 |
| 0 | β-Amylase A and B | β-Amylase 1 to 8 | β-Amylase 1 to 7 |
| GBE | SBE I, II-1, and II-2 | SBE, 2-1, and 2-2 | SBE 1 to 4 |
| 4-α-Glucanotransferase | 4-α-Glucanotransferase 1 and 2 | 4-α-Glucanotransferase 1 and 2 | 4-α-Glucanotransferase 1 and 2 |
| Glycogen debranching enzyme | Isoamylase A and B | Isoamylase 1, 2, and 3 | Isoamylase 1, 2, and 3 |
| Pullulanase | Pullulanase | Pullulanase | |
| GPHO and MPHO | PHO A, B, and C | PHO1 and PHO2 | PHO1 and PHO2 |
| 0 | R1 protein A and B | R1 protein | R1 protein |
| GS | GBSS I | GBSS I | GBSS I |
| SS I, II, IIIa, IIIb, and IIIC | SS I, II, III, IV, and V | SSI, Iia, IIb, III, III-2, IVa, and IVb | |
| 0 | 0 | Glycogenin 1 to 7 | Glycogenin 1 to 4 |
The protein-deduced from four different species was generated using Bioedit (Hall, 1999) and Forcon (Van de Peer et al., 1998). The species included were E. coli, O. tauri, Arabidopsis, and O. sativa. Amino acid sequence of O. tauri genome provided from the Ostreococcus blast research Web site (http://194.167.19.196/). Ostreococcus accession numbers for all reported sequences are given as supplemental material. Expression of all Ostreococcus sequences involved in starch metabolism was checked by RT-PCR and double checked by analysis of the Expression Sequence Tag Database. Amino acid sequences from other organisms were provided from the Arabidopsis Information Resource Web site (www.arabidopsis.org) and the RiceBlastDB Web site (http://www.tigr.org/tdb/e2k1/osa1/osa1.shtml; GenBank accession numbers are given as supplemental material). The following abbreviations were used: AGPase, ADP-Glc pyrophosphorylase; GS, glycogen synthase; SS, soluble starch synthase; GBE and SBE, respectively, glycogen and starch branching enzyme; GPHO and MPHO, glycogen and maltodextrin phosphorylase.
Figure 5.
Dendogram of starch synthases in plants and algae and glycogen synthases from Synechocystis. Sequences were aligned with ClustalW (Thompson et al., 1994). The sequence alignment was manually improved using BioEdit (Hall, 1999) and ForCon (Van de Peer et al., 1998). TreeCon (Van de Peer and De Wachter, 1997a, 1997b) was used for constructing the neighbor-joining (Saitou and Nei, 1987) tree based on Poisson-corrected distances, only taking into account unambiguously aligned positions (600 amino acids). Bootstrap analysis with 500 replicates was performed to test the significance of the nodes. The amino acid sequences used were as follows: C. reinhardtii (Crein) GBSSI (AF26420), SSII (AAC17970), SSIII, SSIV (TO7926); maize (Zea) GBSSI (M24258), SSI (AF036891), SSIIa (Harn et al., 1998), SSIIb (Harn et al., 1998); rice (Oryza) GBSSI (X62134), SSI (D16202), SSIIa (AF419099), SSIIb (Harn et al., 1998), SSIII (Gao et al., 1998), SSIVa (AY1OO470), SSIVb (AY100471); Solanum tuberosum (Solanum) GBSSI (X58453), SSI (Y10416), SSII (X87988), SSIII (X94400); Arabidopsis (Arath) GBSSI (AC006424), SSI (AF121673), SSII (AC008261), SSIII (AC007296), SSV (A021713), and Synechocystis glycogen-synthase Gls (NP441947). Accession numbers for Ostreococcus sequences are given as supplemental material.
It is worth noting that the Ostreococcus, unlike Arabidopsis and other plants, do not appear to contain sequences related to glycogenins of the yeast or mammalian type. Glycogenin, a protein capable of autoglucosylation from UDP-Glc, was proven to be involved in the priming of yeast glycogen synthesis (Cheng et al., 1995).
Starch Granules Elongate and Divide during the Ostreococcus Cell Cycle
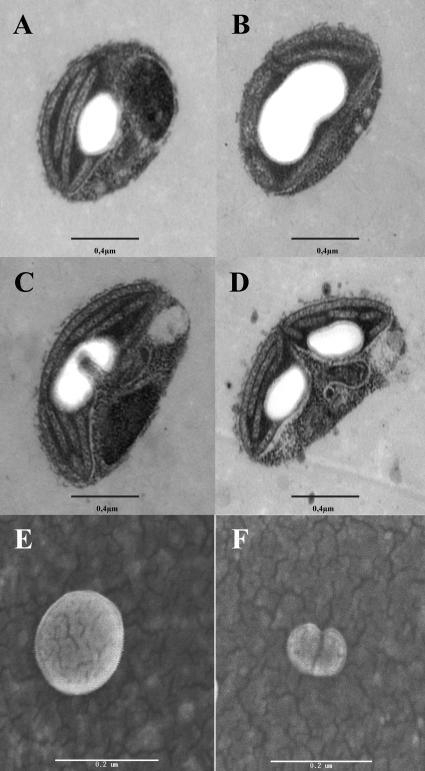
We proceeded to examine cytologically the pattern of starch synthesis and degradation by using transmission electron microscopy (TEM) of cells harvested at various stages of their diurnal rhythm of cell division and starch metabolism. We were surprised to see by TEM that cells were apparently engaged in a process of elongation, division, and partitioning of their unique starch granule into two daughter structures at the time of plastid division at the end of the light phase (Fig. 6, A–D). To check that the granule morphology was indeed modified and not only the subject of constriction through the plastid division machinery, starch granules purified from synchronized cultures engaged in plastid division and were subjected to scanning electron microscopy (SEM). The purified granules still displayed these division figures (Fig. 6, E and F), demonstrating that the starch itself was subjected to these modifications. The TEM observations show that in O. tauri, the number of starch granules is restricted to one and suggest that most if not all granules derive from a parental granule by a localized process of elongation and degradation. Interestingly, attempts to cure O. tauri of starch through prolonged incubation in darkness failed as viability was lost before the granules were entirely degraded. In an effort to understand the basis of this mechanism, we subjected minor proteins associated to starch to trypsic digestion followed by MALDI-MS. We were thus able to find pullulanase, the most abundant of the minor (non-GBSS) proteins associated to starch. Pullulanase has not been reported to date as associated to starch granules, and its presence may reflect a particular function of this enzyme in the partitioning process.
Figure 6.
Micrographs of dividing starch granule in O. tauri. A to D, TEM of dividing O. tauri cells. Poorly synchronized cultures contain a mixture of cells at different stages of plastid division (bar = 0.4 μm). E and F, Field emission type SEM images of dividing starch granule purified from synchronized cultures. Starch granules isolated from G1 cells (E) and G2/M-phase (F). Cultures were synchronized by 12-h-day/12-h-night growth cycles. G1 granules always appear spherical with a somewhat smoother surface, while G2/M starch granules are small often compounded with a rough surface (bar = 0.2 μm).
DISCUSSION
Prasinophytes Display the Full Complexity of Higher Plant Starch Metabolism
The results reported in this paper suggest that starch metabolism has appeared at the earliest stage in the green lineage with the full complexity that characterizes storage polysaccharide metabolism in vascular plants. This complexity seems to be a requisite to build starch through the ADP-Glc pathway. Indeed, it has not been subjected to any simplification to accommodate small-genome size despite the obvious reduction of complexity that has occurred in Ostreococcus concerning a variety of important processes such as those exemplified by cell cycle control (Khadaroo et al., 2004). It is, however, not obvious that this multiplicity of enzyme forms is required to build crystalline polysaccharides, as apicomplexan parasites are able to synthesize starch from UDP-Glc through the use of one enzyme form for each biochemical step defined in this pathway (Coppin et al., 2004).
Prasinophytes Lack Glycogenin-Like Proteins
Glycogenins were found as proteins associated to glycogen particles that are able to glucosylate themselves through the transfer of Glc from UDP-Glc to specific Tyr residues present on the protein. Several Glc residues are then elongated to form a protein-bound oligosaccharide. They have been suspected to be involved in the priming of glycogen synthesis in yeast and mammalian cells by supplying the primer for glycogen synthesis in these systems (for review, see Alonso et al., 1995). That this glycogenin function is indeed required for glycogen accumulation was demonstrated by selectively disrupting both glycogenin genes present in the yeast genome (Cheng et al., 1995). The yeast double mutants lost their ability to store glycogen. In plants, several proteins capable of autoglucosylation were reported. However, in most instances, these were proven to be quite different with respect to the nature both of the sugars transferred and of glycosyl-linkage formed (Singh et al., 1995; Langeveld et al., 2002). The extraplastidial localization of such proteins also argues that they may not be involved in the priming of starch synthesis. Despite this, other genes encoding glycogenin proteins much more similar to the fungal-mammalian type of protein can be detected through analysis of the Arabidopsis and rice (Oryza sativa) genomes. Some of these contain predicted transit peptide sequences and may be involved in both polysaccharide and granule priming. We were unable to find, using standard bioinformatic tools, Ostreococcus genes that are related to the yeast or Arabidopsis glycogenin or glycogenin-like sequences. It remains possible, however, that proteins with similar functions but with divergent primary structures have escaped detection.
The Apparent Elongation and Division of Ostreococcus Starch Granules
The most surprising aspect revealed by Ostreococcus is no doubt defined by the apparent elongation and division of the unique starch granule at the center of the plastid. Elongation could be explained by a modification of the geometry of the photosynthetic membranes prior to plastid division. In fact, in this system, starch is centrally located with respect to the algal thylakoids and could result from the appearance of novel thylakoids supplying additional substrate during the cycle of plastid division. Division of the starch could be explained by a localized starch degradation process targeted at the plastid constriction site. The net result would be the creation of two starch granules of more or less equal size ready for another round of synthesis elongation and division. Such a behavior would render granule and possibly polysaccharide priming unnecessary. However if starch is completely degraded, one has to envision the existence of a mechanism regenerating novel granules. Interestingly, we have never been able to have Ostreococcus degrade its starch to completion through prolonged incubation in darkness. Along similar lines and at variance with results reported for Arabidopsis (Critchley et al., 2001), we have never been able to get conditions where Chlamydomonas cells degrade its starch completely. Interestingly, both of these systems seem to lack glycogenin.
We do not know if our results in Ostreococcus relate to the propagation of starch granules in vascular plants. In addition, we do not know if glycogenin-like sequences have any relevance to the mechanisms of granule seeding and polysaccharide priming in higher plants. It is indeed evident that the volume occupied by starch in the tiny Ostreococcus chloroplast could present a serious physical problem and challenge to the plastid division machinery in this alga only. This may have prompted the evolution of a sophisticated polysaccharide partitioning machinery and could offer an explanation for the absence of starch in prokaryotes in general and cyanobacteria in particular. Alternatively, it remains possible that localized starch degradation may be at work in other species to provide the seeds of new starch granules. That starch catabolic enzymes are involved in controlling the number of seeds for novel granules in dividing plant cells remains an interesting possibility.
MATERIALS AND METHODS
Strains and Growth Conditions
The Ostreococcus tauri strain OTTHO595 (Chrétiennot-Dinet et al., 1995) was grown in K medium prepared on the base of 36% seawater and sterilized by 0.2 μm filtration (Keller and Selvin, 1987). The nitrogen-limited medium defines K medium where NH4Cl was substituted by equivalent concentration of NaCl. Experiments were performed under 12-h day/12-h night growth cycles or continuous light (100 μE m−2s−1). The wild-type Arabidopsis Wassilewskija reference strain was grown in the greenhouse (Zeeman et al., 1998). Media and culture conditions for growing the wild-type Chlamydomonas 137C strain were previously detailed (Delrue et al., 1992).
Starch Purification
Pure native starch from O. tauri was prepared from nitrogen-limited culture and harvested after 4 d of growth under continuous light. Cells were centrifuged (10,000g for 20 min) with 0.2% Pluronic. The pellet was resuspended in 300 μL of 10 mm Tris acetate, pH 7.5, 1 mm EDTA. Algal suspensions were disrupted by sonication. A crude starch extract was obtained by spinning down the lysate at 10,000g for 15 min. The pellet obtained from 1-L cultures was resuspended in 1 mL of 90% Percoll. The gradient was self-formed by centrifugation at 10,000g for 30 min. The starch pellet was collected and resuspended in 1 mL of 90% Percoll. After a 30-min spin at 10,000g, the purified starch pellet was rinsed in sterile distilled water, centrifuged at 10,000g, and kept dry at 4°C. Starch yields through this purification procedure were greater than 80%. Starch amounts were measured by the amyloglucosidase assay (Delrue et al., 1992). Starch granule purification from Chlamydomonas reinhardtii and Arabidopsis were previously described (Ball et al., 1991; Zeeman et al., 1998).
Separation of Starch Polysaccharide by Gel Permeation Chromatography
A total of 1 to 2.5 mg of starch dissolved in 500 μL of 10 mm NaOH was applied to a Sepharose CL2B column (0.5 cm [i.d.] × 65 cm) equilibrated in 10 mm NaOH. Fractions of 250 to 300 μL were collected at a rate of 1 fraction/1.5 min. Glucans in each fraction were detected through the iodine-polysaccharide interaction.
CL Distribution Analysis
A total of 500 μg of dialyzed and lyophilized amylopectin purified after gel permeation chromatography were suspended in 55 mm sodium acetate, pH 3.5, and debranched by 10 units of Pseudomonas amylodermosa Isoamylase (Hayashibara Biochemical Laboratory, Okayama, Japan) at 45°C during 4 h. The reaction was stopped by boiling 10 min. After neutralization with 10 m NaOH, the samples were lyophilized and analyzed by fluorophore-assisted carbohydrate electrophoresis using the procedure previously described (Morell et al., 1998).
Enzyme Assays
GBSSI was assayed as described previously (Delrue et al., 1992). In vitro synthesis of amylose was performed by using the method detailed by Van de Wal et al. (1998).
ADP-Glc pyrophosphorylase was assayed both in the direction of pyrophophorolysis and in the direction of ADP-Glc synthesis by using the protocols set up for C. reinhardtii (Ball et al., 1991). Activation by 3-PGA was measured at a concentration of 1 mm activator.
Km Determination of GBSSI
The GBSSI was measured from freshly purified starch granules after 15 min of incubation at 30°C in the presence of 50 mm Gly, pH 9.0, 100 mm (NH4)2SO4, 0.4% β-mercaptoethanol, 5 mm MgCL2, and 0.05% bovine serum albumin with various concentrations of ADP- or UDP-Glc containing 62 nCi of glycosyl-nucleotide at 10.5 GBq/mmol (for both UDP-Glc and ADP-Glc). Km constants were measured through the Hanes and Woolf procedure from a series of two datasets.
In Vitro Synthesis of Amylose
A total of 500 μg of starch was incubated with 3.2 mm ADP-Glc in the presence of 22 mm Tris-HCl, pH 8.0, 0.47% β-mercaptoethanol, 5.5 mm MgCL2, and 2.2 μm ADP[U-14C]Glc at 10.5 GBq/mmol in a total volume of 2 mL at 30°C for 16 and 40 h. The reaction was terminated by adding 3 volumes of 96% ethanol. The ethanol-rinsed pellet was resuspended in 1 mL of 90% dimethyl sulfoxide and dispersed through boiling. The dispersed polysaccharide was precipitated by 3 volumes of 96% ethanol and stored overnight at −20°C.
After centrifugation at 15,000g for 20 min, the polysaccharide was resuspended in 500 μL of 10 mm NaOH and subjected to gel permeation on a Sepharose CL2B column (Sigma-Aldrich, Steinhem, Germany) as described above.
X-Ray Diffraction, NMR, SDS-PAGE, and Matrix-Assisted Laser-Desorption Ionization Time of Flight Analysis of GBSSI
Powder x-ray diffractograms were collected from Percoll purified starches as described previously (Buléon et al., 1997). Proton NMR spectra of dispersed polysaccharides were produced according to the protocol developed for C. reinhardtii (Fontaine et al., 1993). To analyze granule-bound proteins, purified starches (2 mg) were boiled for 10 min in 80 μL of 2% SDS and 5% β-mercaptoethanol and centrifuged for 20 min at 10,000g. Supernatants were loaded onto 7.5% SDS-PAGE and stained with Coomassie Brilliant Blue R250. Isolated proteins were extracted from the gel and bleached using acetonitrile/100 mm ammonium bicarbonate (v/v) at 30°C. After acetonitrile dehydration, dried proteins were subjected to trypsic digestion in 50 mm ammonium bicarbonate buffer for 8 h. Peptides were eluted twice by a 45% acetonitrile/10% formic acid bath at 30°C and a 95% acetonitrile/5% formic acid at 30°C. Samples were analyzed by matrix-assisted laser-desorption ionization time of flight.
TEM and SEM
O. tauri cells were collected by centrifugation and fixed in paraformaldehyde/glutaraldehyde/PIPES buffer, then postfixed in osmium tetroxyde in PIPES buffer and dehydrated and embedded in Epon (Soyer, 1977). Sections were stained with uranyl acetate and lead citrate and examined in a Hitachi (Tokyo) H-600 transmission electron microscope. O. tauri starch granules were concentrated during centrifugation on a plastic Thermanox (Naperville, IL) coverslip. Coverslips were then mounted on a stub and coated with gold. Starch granules were viewed with a field emission scanning electron microscope Hitachi S-4500.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY570699 to AY570722.
Supplementary Material
Acknowledgments
We thank Frédéric Chirat and Emmanuel Maes for their excellent technical assistance.
This work was supported by the French Ministry of Education, by the Centre National de la Recherche Scientifique, and by the Génopole Languedoc-Roussillon.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044131.
References
- Alonso MD, Lomako J, Lomako WM, Whelan WJ (1995) A new look at the biogenesis of glycogen. FASEB J 12: 1126–1137 [DOI] [PubMed] [Google Scholar]
- Ball S, Marianne T, Dirick L, Fresnoy M, Delrue B, Decq AA (1991) Chlamydomonas reinhardtii low-starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta 185: 17–26 [DOI] [PubMed] [Google Scholar]
- Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54: 207–233 [DOI] [PubMed] [Google Scholar]
- Ballicora MA, Iglesias AA, Preiss J (2003) ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol Mol Biol Rev 67: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buléon A, Colonna P, Planchot V, Ball S (1998) Starch granules: structure and biosynthesis. Int J Biol Macromol 23: 85–112 [DOI] [PubMed] [Google Scholar]
- Buléon A, Gallant D-J, Bouchet B, Mouille G, D'Hulst C, Kossman J, Ball SG (1997) Starches from A to C. Chlamydomonas reinhardtii as a model microbial system to investigate the biosynthesis of the plant amylopectin crystal. Plant Physiol 115: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Mu J, Farkas I, Huang D, Goebl MG, Roach PJ (1995) Requirement of the self-glucosylating initiator proteins Glg1p and Glg2p for glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol 12: 6632–6640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétiennot-Dinet MJ, Courties C, Vaquer A, Neveux J, Claustre H, Lautier J, Machado MC (1995) A new marine picoeucaryote: Ostreococcus tauri gen. Et sp. Nov. (Chlorophyta, Prasinophyceae). Phycologia 34: 285–292 [Google Scholar]
- Coppin A, Varré J-S, Liénard L, Dauvillée D, Guérardel Y, Soyer-Gobillard M-O, Buléon A, Ball S, Tomavo S (2004) Evolution of plant-like crystalline storage polysaccharide in the protozoan parasite Toxoplasma gondii argues for red alga ancestry. J Mol Evol (in press) [DOI] [PubMed]
- Courties C, Perasso R, Chrétiennot-Dinet MJ, Gouy M, Guillou L, Troussellier M (1998) Phylogenetic analysis and genome size of Ostreococcus tauri (Chlorophyta, Prasinophyceae). J Phycol 34: 844–849 [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM (2001) A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J 26: 89–100 [DOI] [PubMed] [Google Scholar]
- Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski J-M, Van den Koornhuyse N, Maddelein ML, Fournet B, Ball S (1992) Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J Bacteriol 174: 3612–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine T, D'Hulst C, Maddelein M-L, Routier F, Marianne-Pepin T, Decq A, Wieruszeski JM, Delrue B, Van Den Koornhuyse N, Bossu JP, et al (1993) Toward an understanding of the biogenesis of the starch granule. Evidence that Chlamydomonas soluble starch synthase II controls the synthesis of intermediate size glucans of amylopectin. J Biol Chem 268: 16223–16230 [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM (1998) Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell 10: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg E, Preiss J (1964) The occurrence of adenosine diphosphate glucose:glycogen transglucosylase in bacteria. J Biol Chem 239: 4314–4315 [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Harn C, Knight M, Ramakrishnan A, Guan H, Keeling PL, Wasserman BP (1998) Isolation and characterization of the zSSIIa and zSSIIb starch synthase cDNA clones from maize endosperm. Plant Mol Biol 37: 639–649 [DOI] [PubMed] [Google Scholar]
- Keller MD, Selvin RC (1987) Media for the culture of oceanic ultraphytoplankton. J Phycol 23: 633–638 [Google Scholar]
- Khadaroo B, Robbens S, Ferraz C, Derelle E, Eychenie S, Cooke R, Peaucellier G, Delseny M, Demaille J, Van De Peer Y, et al (2004) The first green lineage cdc25 dual-specificity phosphatase. Cell Cycle 3: 513–518 [PubMed] [Google Scholar]
- Langeveld SMJ, Vennik M, Kottenhagen M, van Wijk R, Buijk A, Kijne JW, de Pater S (2002) Glucosylation activity and complex formation of two classes of reversibly glycosylated polypeptides. Plant Physiol 129: 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI, Reith ME, Munholland J, Lang-Unnasch N (1996) Plastid in human parasites. Nature 381: 482. [DOI] [PubMed] [Google Scholar]
- Meléndez R, Meléndez-Hevia E, Mas F, Mach J, Cascante M (1998) Physical constraints in the synthesis of glycogen that influence its structural homogeneity: a two-dimensional approach. Biophys J 75: 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell MK, Samuel MS, O'Shea MG (1998) Analysis of starch structure using fluorophore-assisted carbohydrate electrophoresis. Electrophoresis 19: 2603–2611 [DOI] [PubMed] [Google Scholar]
- Preiss J, Romeo T (1989) Physiology, biochemistry and genetics of bacterial glycogen synthesis. Adv Microb Physiol 30: 183–238 [DOI] [PubMed] [Google Scholar]
- Recondo E, Leloir L (1961) Adenosine diphosphate glucose and starch biosynthesis. Biochem Biophys Res Commun 6: 85–88 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Singh DG, Lomako J, Lomako WM, Whelan WJ, Meyer HE, Serwe M, Metzger JW (1995) Beta-Glucosylarginine: a new glucose-protein bond in a self-glucosylating protein from sweet corn. FEBS Lett 376: 61–64 [DOI] [PubMed] [Google Scholar]
- Soyer MO (1977) Une modification de la technique de Karnosky pour la préservation optimale des structures nucléaires chez les dinoflagellés. Biol Cell 30: 297–300 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Caers A, De Rijk P, De Wachter R (1998) Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res 26: 179–182 [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R (1997. a) Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci 13: 227–230 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R (1997. b) Evolutionary relationships among the eukaryotic crown taxa taking into account site-to-site rate variation in 18S rRNA. J Mol Evol 45: 619–630 [DOI] [PubMed] [Google Scholar]
- Van de Wal M, D'Hulst C, Vincken J-P, Buléon A, Visser R, Ball S (1998) Amylose is synthesized in vitro by extension of and cleavage from amylopectin. J Biol Chem 273: 22232–22240 [DOI] [PubMed] [Google Scholar]
- Viola R, Nyvall P, Pedersen M (2001) The unique features of starch metabolism in red algae. Proc R Soc Lond B Biol Sci 268: 1417–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman S, Northrop F, Smith AM, ap Rees T (1998) A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch hydrolyzing enzyme. Plant J 15: 357–365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.