Abstract
Profiles of small polar metabolites from aspen (Populus tremuloides Michx.) leaves spanning the sink-to-source transition zone were compared. Approximately 25% of 250 to 300 routinely resolved peaks were identified, with carbohydrates, organic acids, and amino acids being most abundant. Two-thirds of identified metabolites exhibited greater than 4-fold changes in abundance during leaf ontogeny. In the context of photosynthetic and respiratory measurements, profile data yielded information consistent with expected developmental trends in carbon-heterotrophic and carbon-autotrophic metabolism. Suc concentration increased throughout leaf expansion, while hexose sugar concentrations peaked at mid-expansion and decreased sharply thereafter. Amino acid contents generally decreased during leaf expansion, but an early increase in Phe and a later one in Gly and Ser reflected growing commitments to secondary metabolism and photorespiration, respectively. The assimilation of nitrate and utilization of stored Asn appeared to be marked by sequential changes in malate concentration and Asn transaminase activity. Principal component and hierarchical clustering analysis facilitated the grouping of cell wall maturation (pectins, hemicelluloses, and oxalate) and membrane biogenesis markers in relation to developmental changes in carbon and nitrogen assimilation. Metabolite profiling will facilitate investigation of nitrogen use and cellular development in Populus sp. varying widely in their growth and pattern of carbon allocation during sink-to-source development and in response to stress.
Quaking aspen (Populus tremuloides Michx.) is an ecological keystone species, an important component of the wood products industry, and a useful experimental model system for the study of woody plant development (Bradshaw et al., 2000). We are interested in using metabolic profiling to characterize the sink-to-source transition in developing aspen leaves. The approach is expected to provide a basis for more extended studies analyzing the effects of various carbon sinks on aspen development and growth. An illustrative example is the regulation of phenolic sinks in leaves of aspen and close relatives in the genera Populus and Salix (Orians and Fritz, 1995; Lindroth and Hwang, 1996; Kao et al., 2002). Phenolic deposition is very characteristic of these species, exhibits wide genetic variation, can begin early during aspen leaf expansion (Kleiner et al., 1999; Kao et al., 2002), and can reach levels that negatively affect aspen stem and branch growth (Lindroth and Hwang, 1996). The allocation of metabolic carbon into such sinks is influenced by plant nutrient status and photosynthesis rate (Bryant et al., 1983; Hemming and Lindroth, 1999; Mansfield et al., 1999) and in young leaves, therefore, may be linked to metabolic development.
Metabolic profiling of plants is generally geared toward the extraction of a broad spectrum of biochemical information from multiple sample types by relatively direct analytical means (for review, see Fiehn and Weckwerth, 2003; Sumner et al., 2003). The resolution and precision of gas chromatography-mass spectrometry (GC-MS)-based metabolic profiling for identifying uncommon metabolites and analyzing perturbations in plant metabolism have been demonstrated and promise to be widely accessible (Fiehn et al., 2000b; Roessner et al., 2000). In tandem with statistical data-mining tools, such as principal component analysis (PCA) and hierarchical clustering analysis (HCA; Eisen et al., 1998; Valafar, 2002), metabolic profiling has yielded informative biochemical phenotyping of Arabidopsis ecotypes and mutants (Fiehn et al., 2000a), as well as of experimentally and genetically manipulated potato (Solanum tuberosum) and tomato (Lycopersicon esculentum; Roessner et al., 2001; Roessner-Tunali et al., 2003). The application of profiling to whole-plant physiology was recently demonstrated in a study of the dynamics of pumpkin (Cucurbita maxima) phloem sap metabolism during transport (Fiehn, 2003).
Quaking aspen will likely be one of the first woody species to yield marker metabolic data that can be analyzed in a transcriptional context. Genome sequencing of Populus trichocarpa, a close aspen relative, is near completion (Wullschleger et al., 2002), and well over 150,000 Populus expressed sequence tags are available in public databases. To date, there are no reports describing the application of metabolic profiling and data-mining techniques to a woody species. As a first step, we have profiled extracts from a developmental gradient of aspen leaves undergoing transition from a sink-to-source organ. We monitored photosynthetic and respiratory development so that metabolic profile data could be interpreted in the context of the sink-to-source transition. Large fluctuations in malate and the malate-to-citrate ratio, as well as a switch in Asn homeostasis, were indications of broad changes in metabolism during rapid leaf expansion. Shifts in fatty acid-lipid intermediates, pectin residues, and carbohydrate composition marked membrane biogenesis/turnover and cell wall maturation in relation to the metabolic transients.
RESULTS
Photosynthesis and Respiration during Leaf Expansion
Leaves at leaf plastochron index (LPI) 0, 4, and 9 of the aspen used in this study, referred to as young (YL), expanding (EL), and mature leaves (ML), respectively, have typically expanded to 2% to 5%, 30% to 40%, and >90% of their mature size. YL are partially unfurled. During YL-to-ML expansion, aspen leaves undergo a number of structural changes consistent with the accumulation and export of photosynthate and the transition into source organs (Isebrands and Larson, 1973; Isebrands et al., 1976; Dickson and Larson, 1981; Dickson and Shive, 1982). The development of photosynthesis and respiration is integral to this process and should provide a framework for interpreting changes in metabolite profile during the sink-to-source transition (Table I). The rate of leaf disc O2 consumption in dark decreased by >50% from YL to ML and was considered to reflect a decline in resting respiration. Maximum photosynthetic rates were obtained in 10 mm sodium bicarbonate and were 1.7 and 2.4 times higher from EL and ML discs, respectively, than from YL discs (Table I). Dark respiration rates measured before and immediately after photosynthesis were highest, relative to photosynthesis, in YL (Table I). We consider this to indicate that a greater fraction of recent photosynthate was metabolized for oxidative phosphorylation in YL than in EL or ML. In sum, the physiological changes are consistent with the expected YL-to-EL-to-ML transition from net metabolic sink (low photosynthetic capacity and high respiratory flux) to net source or carbon exporter.
Table I.
Photosynthesis and dark respiration of aspen leaf discs
| Leaf Stage | Initial Respirationa | Post-Photosynthesis Respirationb | Maximum Photosynthesisc |
|---|---|---|---|
| μm O2 m−2 s−1 | |||
| YL | 2.2 ± 0.28 | 4.6 ± 0.50 | 2.4 ± 0.35 |
| EL | 1.6 ± 0.24 | 3.6 ± 0.42 | 4.1 ± 0.16 |
| ML | 0.8 ± 0.07 | 3.7 ± 0.58 | 5.9 ± 0.49 |
Photosynthetic O2 evolution and respiratory (dark) O2 consumption by low-light-adapted discs before and immediately following photosynthesis of YL, EL, and ML discs. Values represent the mean ± se (n = 6).
Dark respiration measured for a period of 10 min using discs from 2-h low-light-adapted plants.
Dark respiration measured for a period of 10 min immediately following a 30-min period of photosynthesis.
Maximum rate of photosynthesis during a 30-min period following the addition of 8 mm sodium bicarbonate.
The developmental course of leaf photorespiration has been well documented and provides another physiological marker by which to assess metabolic profile data (Lennon et al., 1995; Thompson et al., 1998). Photorespiration depends on assembly of the mitochondrial Gly decarboxylase complex, which normally occurs late relative to other mitochondrial components during monocot and dicot leaf development (Lennon et al., 1995; Thompson et al., 1998). In order to gauge photorespiratory activity, we monitored light-dependent O2 evolution as a function of increasing CO2 concentration (Table II). At very low CO2 concentration prior to sodium bicarbonate addition, light-dependent O2 evolution was 1.5 and 2 times higher from YL discs than from EL and ML discs, respectively. At 1.5 mm sodium bicarbonate, photosynthesis rates were 1.4 and 2 times higher from EL and ML, respectively, than from YL discs. This represented increases of 9-fold, 4.7-fold, and 1.3-fold by ML, EL, and YL discs, respectively. The trend continued when the concentration of sodium bicarbonate was doubled to 3.0 mm. The photosynthetic response of YL discs to added CO2 was therefore much weaker than that of EL or ML discs. The lower rates of O2 evolution by ML and EL discs in the absence of added CO2 could reflect greater stomatal resistance to CO2, or higher photorespiratory activity in those more mature tissues. Photorespiratory Gly supports high rates of mitochondrial O2 uptake by isolated mitochondria (Kromer and Heldt, 1991), and the stimulation of mitochondrial respiration by photorespiratory Gly has been measured in leaf discs (Dutilleul et al., 2003).
Table II.
Response of leaf disc photosynthetic O2 evolution to added bicarbonate
| Leaf Stage | 0 mm HCO3 | 1.5 mm HCO3 | 3.0 mm HCO3 |
|---|---|---|---|
| μm O2 m−2 s−1 | |||
| YL | 0.53 ± 0.03 | 1.2 ± 0.16 | 1.6 ± 0.12 |
| EL | 0.36 ± 0.04 | 1.7 ± 0.04 | 2.5 ± 0.16 |
| ML | 0.26 ± 0.05 | 2.4 ± 0.11 | 3.9 ± 0.19 |
Each measurement was obtained over a 15-min period beginning 10 min after addition of bicarbonate or equal volume of water. Values represent means ± se (n = 6).
Metabolite Abundance Trends during Leaf Expansion
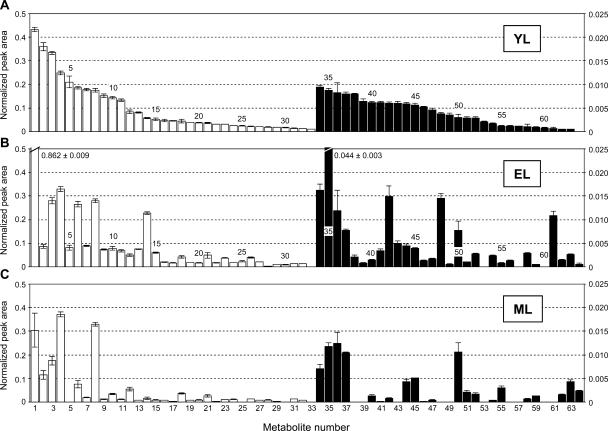
We next sought to determine whether methanol extracts would yield signature compositional changes consistent with, and supplemental to, the measured physiological parameters. More than 250 peaks were typically resolved from each extract. Sixty-four metabolites were identified with a high level of certainty (Table III), and these were arranged in a histogram according to decreasing normalized peak area in YL (Fig. 1). Overall, two-thirds of identified metabolites varied more than 4-fold between YL, EL, and ML tissues. Although the majority of metabolites decreased in abundance as leaves expanded, about one-third exhibited increased abundance in EL and/or ML compared to YL tissue.
Table III.
List of 64 metabolites identified in YL, EL, and ML
| No. | Metabolite | tR | No. | Metabolite | tR |
|---|---|---|---|---|---|
| min | min | ||||
| 1 | Malatea | 20.495 | 33 | Galactarate | 31.272 |
| 2 | Citratea | 27.859 | 34 | Propionate | 16.237 |
| 3 | Phosphorate | 14.864 | 35 | Gal | 29.418 |
| 4 | Inositol | 31.637 | 36 | Suberate | 25.447 |
| 5 | Asna | 24.847 | 37 | Palmitate | 31.042 |
| 6 | Trihydroxybutyrate | 22.238 | 38 | Glc-6-Pa | 34.334 |
| 7 | Glua | 23.666 | 39 | Anthraquinone derivativeb | 40.524 |
| 8 | Suc | 37.042 | 40 | Vala | 13.346 |
| 9 | Aspa | 21.293 | 41 | Valerate | 13.652 |
| 10 | Proa | 21.391 | 42 | Fumaratea | 16.921 |
| 11 | Methylphosphorate | 12.287 | 43 | Erythritol | 20.975 |
| 12 | Sera | 17.215 | 44 | Maltose | 35.510 |
| 13 | Shikimate | 27.642 | 45 | Phenylacetate | 15.459 |
| 14 | Glca | 29.127 | 46 | Glycolate | 9.534 |
| 15 | Glutarate | 18.158 | 47 | Catechin (flavan) | 38.897 |
| 16 | Thra | 17.902 | 48 | Malonate | 12.988 |
| 17 | Alaa | 10.266 | 49 | Oxalate | 10.985 |
| 18 | Ethanolamine | 14.758 | 50 | Azelate | 27.454 |
| 19 | Dimethylethanolamine | 5.671 | 51 | Glya | 15.838 |
| 20 | o-Hydroxyphenol | 16.154 | 52 | Arabinitol | 25.965 |
| 21 | Adipate | 20.929 | 53 | Nicotinamide | 6.833 |
| 22 | Glucosone | 20.261 | 54 | Phea | 23.890 |
| 23 | Succinatea | 15.980 | 55 | Glycerol galactopyranoside | 35.376 |
| 24 | Galactonate | 30.484 | 56 | Nicotinate | 15.556 |
| 25 | Xylitol | 25.450 | 57 | Noradrenaline | 26.316 |
| 26 | Frua | 28.732 | 58 | Glucarate | 30.756 |
| 27 | Hexadecenyl alcohol | 32.482 | 59 | Mannitola | 29.704 |
| 28 | Benzoate | 14.304 | 60 | Gluconate | 30.716 |
| 29 | Glna | 27.039 | 61 | Xyl | 24.444 |
| 30 | Norvaline | 21.999 | 62 | Cadaverine | 13.800 |
| 31 | Stearate | 33.113 | 63 | Man | 32.580 |
| 32 | α-Sitosterol | 45.904 | 64 | Putrescine | 26.192 |
Verified by authentic standards.
Refers to a compound with a MS signature most similar (match score of 865 out of 1,000) to that of 1,2,8-trihydroxy-6-methoxy-3-methylanthraquinone in the NIST database.
Figure 1.
Histogram of metabolites recovered in methanolic extracts from YL (A), EL (B), and ML (C) tissues of aspen. Metabolites are shown in order of decreasing normalized peak area in YL tissue (A). White histogram bars are scaled against the left axis, and black bars are scaled against the right axis. Numbers correspond to metabolites listed in Table III. Error bars represent se of means of n = 30 determinations.
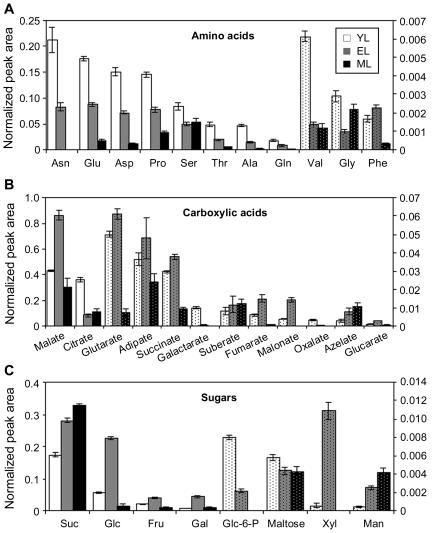
Amino acids, dicarboxylic organic acids, and sugars comprised more than 40% of the metabolites displayed in Figure 1. The 11 common amino acids we detected include all of the amino acids detected in light-adapted Arabidopsis, with the exception of Lys (Fig. 2A; Lam et al., 1995). Those we did not detect, such as His, Arg, Tyr, Trp, Met, Ile, and Leu, are only readily detected in dark-adapted Arabidopsis (Lam et al., 1995). The primary amino acids Asn, Glu, Asp, and Gln, decreased systematically to less than one-half their initial YL concentration by EL and to <15% by ML (Fig. 2A). Asn, a storage amino acid with low carbon-to-nitrogen ratio, exhibited the sharpest decrease and was not detected in ML. With the exception of Phe, concentrations of all detected amino acids decreased during YL-to-EL growth (Fig. 2A). Following this stage of leaf expansion, all amino acids except Gly and Ser exhibited a decrease in abundance. Gly and Ser are photorespiration intermediates, and their increase in leaves such as ML is consistent with the expected timing of photorespiratory development (Lennon et al., 1995; Thompson et al., 1998). The use of Gly-to-Ser ratios as markers of photorespiration in profiling studies has been described recently (Novitskaya et al., 2002).
Figure 2.
Histogram of highly represented metabolite classes in YL (white bars), EL (gray bars), and ML (black bars) aspen tissues. A, Amino acids; B, carboxylic acids; C, soluble carbohydrates. Solid-shade bars are scaled against the left axis and dotted histogram bars are scaled against the right axis. Error bars represent se of means of n = 30 determinations.
Dicarboxylic acids known to be by-products of fatty acid oxidation in animals (glutaric, adipic, suberic, and azelaic acids; Passi et al., 1983; Tserng and Jin, 1991) increased during the leaf expansion from YL to EL (Fig. 2B). TCA and glyoxylate cycle intermediates malate, succinate, and fumarate exhibited a similar trend, but citrate decreased quite sharply during YL-to-EL expansion, especially in relation to malate (Fig. 2B). Using our protocol, several common TCA cycle intermediates, including isocitrate and α-ketoglutarate, were not consistently detected in the aspen leaf extracts. There was a nearly 4-fold increase in malonate used for synthesis of flavonoids, membrane lipids, and lipoate cofactor for photorespiration (Gueguen et al., 2000). The synthesis of flavonoids is identified with membrane protection from the harmful effects of UV light (for review, see Winkel-Shirley, 2002). The correlative increase of malonate with dicarboxylate lipid oxidation products we observed in EL may reflect membrane repair and turnover specific to rapidly expanding leaves (Fig. 2B). The removal and processing of plastoglobuli rich in fatty acids is thought to prevent chloroplast membrane destabilization in active leaves (Ghosh et al., 1994; Smith et al., 2000). Turnover of phospholipids in plasma membrane vesicles of active plant tissues has also been reported (Madey et al., 2001).
Many of the nonstructural carbohydrates were most abundant in EL (Fig. 2C). At this early stage of analysis, we were interested in whether Glc, Fru, and Suc concentrations varied according to patterns expected during a sink-to-source transition. Glc levels were 4-fold higher in EL than YL tissues, consistent with increased photosynthetic capacity compared to YL and lower conversion to Suc compared to ML (Table I). In addition, the ratio of Glc to Fru was nearly 2-fold higher in EL than YL, a tendency that hinted at decreased provision of hexose sugars from Suc catabolism (Suc yields Glc and Fru in a 1:1 ratio) in EL and an increased reliance on photosynthesis. This trend in Suc utilization continued into ML, where Suc increased further, but Glc and Fru levels actually decreased sharply. It appeared that, as leaves expanded and gained photosynthetic capacity, an increasing fraction of their photosynthate was maintained as Suc for export.
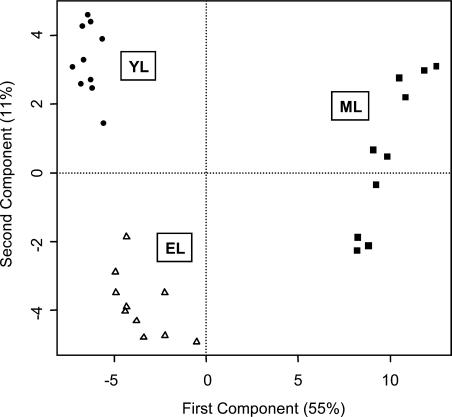
Tissue Clustering Analysis by PCA
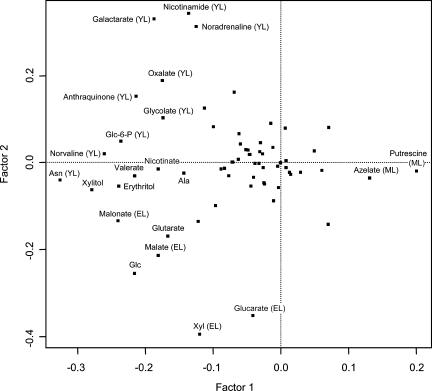
In general, the comparisons shown in Figure 2 point to differential trends in amino acid, fatty acid, TCA/glyoxylate cycle, and hexose metabolism in expanding aspen leaves undergoing photosynthetic development. The trends in mean metabolite levels provided the first indication that the aspen leaf profiles would offer a rational snapshot of changes in leaf chemistry during the sink-to-source transition. To determine whether the profiles contained sufficient information to metabolically distinguish YL, EL, and ML tissues, we analyzed the pattern of association among all metabolites using PCA (Fig. 3). YL, EL, and ML samples formed distinct clusters based on the first two components, which explained 55% (x axis) and 11% (y axis) of overall metabolite variance. YL and EL formed clusters that were much more closely related to each other than to the distantly clustered ML samples. PCA was used to calculate the loading (contribution) of individual metabolites to the clustering of the leaf tissues (Fig. 4). Metabolites plotting near the zero intercept of the axes contributed in a relatively small way, while a number of the more widely scattered metabolites, including Asn, oxalate, galactarate, nicotinamide, Glc-6-P, glycolate, malate, malonate, Xyl, and glucarate, contributed specifically to the separation of YL from EL tissue clusters as indicated (Fig. 4).
Figure 3.
Formation of YL, EL, and ML tissue sample clusters by PCA. Principal component vectors 1 (x axis) and 2 (y axis) describe 55% and 11%, respectively, of total sample variance.
Figure 4.
Scatter plot representation of the contribution of individual metabolites to principal component clustering of YL, EL, and ML tissue samples. Metabolites plotting farthest from the intersection of the principal component axes contributed most to tissue clustering and were identified by common name. Metabolites marked YL, EL, or ML were most abundant in those tissues, respectively.
Metabolite Clustering Based on Tissue Distribution
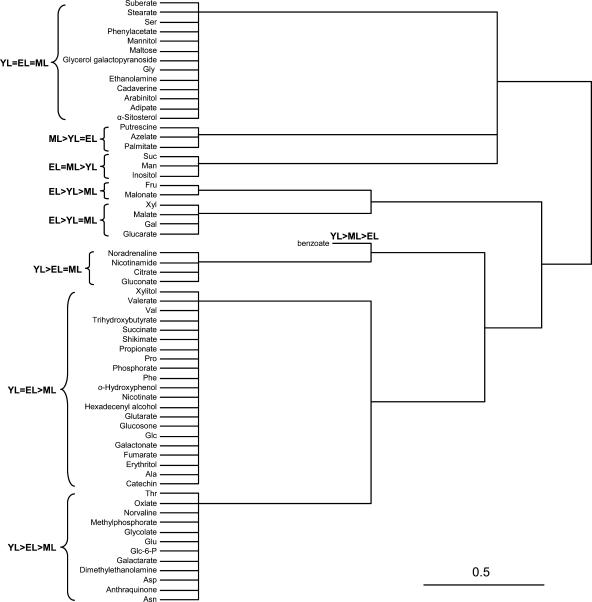
ANOVA was performed to determine whether mean normalized peak areas of individual metabolites differed significantly (P < 0.05) between leaf stages. By this criterion, 51 of the identified metabolites differed significantly in their abundance among YL, EL, and ML leaves, and 48 of those differed at P < 0.01. The metabolites were grouped according to their relative abundance in YL, EL, and ML tissues using Fisher's lsd mean comparison. HCA was then used to cluster the metabolites according to significant differences in distribution between YL, EL, and ML tissues (Fig. 5).
Figure 5.
HCA dendrogram grouping of metabolites based on significant differences in relative abundance in YL, EL, and ML tissues. Fisher's lsd mean comparison was used to rank the tissue abundance of each metabolite (see text). The average linkage method was used to group data by Euclidian distance relatedness.
Metabolites that clustered as (YL > EL > ML and YL = EL > ML) collectively exhibited a strong association with development of immature leaf tissues (Fig. 5). The decrease in methylphosphoric and phosphoric acids is consistent with a decrease in high-energy phosphorylated compounds during leaf expansion. Also decreasing with leaf age, nicotinamide (YL > EL = ML) and nicotinic acid (YL = EL > ML) are structural and precursor components, respectively, of cofactors NAD(P)H. The photorespiratory substrate glycolate (YL > EL > ML) decreased in accordance with increased photorespiratory activity (Table II) and anticipated the earlier noted trends in photorespiratory intermediates Gly and Ser (Fig. 2A). Interestingly, in this respect, Ser and Gly clustered separately from all other amino acids into the YL = EL = ML group.
A number of metabolites can be associated with development of cell structures. Dimethylethanolamine was present in the YL > EL > ML group and is an intermediate during biosynthesis of phosphatidylcholine from Ser (McNeil et al., 2000; Rontein et al., 2001). Phosphatidylcholine comprises 40% to 60% of nonplastid membrane lipid in plants (Moore, 1990). Oxalate and galactarate in the YL > EL > ML group are identified with juvenile cell wall development. Oxalate produced from ascorbate sequesters calcium and is thought to inhibit calcium-dependent cross-linking of pectins in young cell walls (for review, see Smirnoff, 1996). Oxalate, in turn, is degraded by an oxalate oxidase activity associated with increased cell wall cross-linking (Lane et al., 1993). Galactarate, derived from the pectin subunit galacturonate (Kessler et al., 1961), may indicate a higher percentage of pectinaceous cell wall material or greater extractability of pectins due to stronger oxalate inhibition of cross-linking in YL than older tissues (Lin and Varner, 1991). In addition, glucarate (YL = EL > ML) is derived from pectin units (Bean et al., 1961; Kessler et al., 1961). Consistent with increased cell wall integrity after YL growth, hemicellulose-forming sugars Xyl, Gal, and Man grouped into EL-associated clades.
Benzoate (YL > ML > EL), along with an anthroquinone derivative (metabolite no. 39 in Table III; YL > EL > ML), decreased sharply in abundance after YL and may signal a spike in certain shikimate (YL = EL > ML)-dependent pathways specific to YL development. The sustained abundance of shikimate, Phe, and catechin (YL = EL > ML) beyond the YL stage, along with the 4-fold increase in malonate (EL > YL > ML), is consistent with continued synthesis of flavonoid derivatives thought to be important for defense (Lindroth and Hwang, 1996) and protection from the harmful effects of UV light (for review, see Winkel-Shirley, 2002). It should be noted that Phe was the only consistently detected amino acid that trended higher during YL-to-EL growth (Fig. 2A).
In more mature leaves, the polyamines cadaverine (YL = EL = ML) and putrescine (ML > YL = EL) trended upward (Fig. 1). ML tissues were metabolically active and exhibited higher photosynthetic rates than YL or EL, but the only primary metabolites comparatively abundant with respect to YL and EL were Suc and inositol. The relative abundance of these export/storage forms in ML is consistent with an export-oriented metabolism.
DISCUSSION
The initial technical challenge was to determine whether reproducible profiles could be obtained from a clonal population of n = 10 trees, logistically a manageable experimental unit size. We were concerned that slight differences in early growth due to micropropagation and establishment protocols might have cumulative effects on source-to-sink and root-to-shoot ratios that could contribute to variation among large, albeit genetically identical, plants. Our target variance was based on RSDs (sd as a percentage of the mean) ranging from 20% to 50%, and averaging 40% in informative Arabidopsis, potato, and pumpkin profiling studies (Fiehn et al., 2000a; Roessner et al., 2000; Fiehn, 2003). Following protocol optimization steps in which the chromatogram data from triplicate 10-mg aliquots of each leaf powder were pooled, average RSDs were 38%, 43%, and 56% for YL, EL, and ML, respectively.
The second challenge was to construct a developmental scenario from the relatively small portion of the metabolome that was identified. In this study, LPI 0 (YL) represents a 2-d postemergent leaf. Since the interval between leaf emergences was approximately 40 h, LPI 4 (EL) was only 8 to 9 d postemergent, while LPI 9 (ML) was 16 to 17 d postemergent. Organelle biogenesis and the acquisition of functional maturity occur over a 7- to 14-d period in emerged leaves and cotyledons of various species (Titus and Becker, 1985; Lennon et al., 1995; Thompson et al., 1998) and could reasonably be expected to leave signature marks on cellular homeostasis during that period. As in the cases of other GC-MS profiling studies utilizing methanolic extracts (Fiehn et al., 2000a; Roessner et al., 2001; Roessner-Tunali et al., 2003), comparison of small assemblages of low-Mr polar metabolites yielded distinct biochemical phenotypes among the contrasted tissues (Fig. 3). In addition, we were able to detect leaf-stage-specific metabolite clusters indicative of transitions in cell membrane (dimethylethanolamine, malonate, fatty acid-derived dicarboxylic acids) and cell wall (pectin derivatives, oxalate, hemicellulose component sugars) biogenesis.
Malate as a Developmental Marker
Of particular interest were the oscillations in malate (EL > YL = ML) and malate-to-citrate ratio during leaf expansion (Fig. 2B). Malate can comprise the largest fraction of the leaf metabolite pool in leaves of C3 species (Gerhardt and Heldt, 1984; Muller et al., 2001), and its dynamic partitioning within the cell is essential for osmotic, turgor, charge, and pH control throughout the diurnal cycle (for review, see Martinoia and Rentsch, 1994). As reductant shuttle and counter anion, malate is important to the coordination of photosynthesis, glycolysis, TCA activity, glyoxysomal/peroxisomal activity, and nitrate assimilation (Hanning and Heldt, 1993; Martinoia and Rentsch, 1994; Champigny, 1995; Scheible et al., 2000; Muller et al., 2001).
Speculating about the causes of malate dynamics in developing tissues is made difficult by increasing vacuolization in expanding cells. Although cellular homeostasis of malate depends on large vacuolar stores (Martinoia and Rentsch, 1994), large transients in cellular malate content occur diurnally (Scheible et al., 2000) and, according to our data, during development (Fig. 2). Thus, shifts in malate content probably reflect changes in specific, malate-requiring cellular activities. Malate concentrations are known to rise in response to surplus photosynthetic electron transport (Backhausen et al., 1998) and as an outcome of anaplerotic CO2 fixation outside the chloroplast (Melzer and O'Leary, 1987), especially during periods of nitrate assimilation (Scheible et al., 2000). Anapleurotic NADH-dependent malate dehydrogenase (MDH) activity was at its peak in EL (Table IV), where leaf malate concentrations also peaked (Fig. 2). As cells, and presumably vacuoles, continued to expand, malate levels and MDH activity decreased. Chloroplast NADPH-dependent MDH activity exhibited a more sharply defined transient, increasing 2.4-fold during the period of YL-to-EL expansion (Table IV). However, NADPH-dependent MDH constituted a relatively minor activity and decreased rapidly, consistent with a protective role during surplus photosynthetic electron transport, as described in potato (Backhausen et al., 1998).
Table IV.
Comparison of MDH and Asn transaminase activities in developing aspen leaves
| LPI | Malate Dehydrogenase
|
Asn Transaminase | |
|---|---|---|---|
| NADH | NADPH | ||
| nkat g fresh weight−1 | |||
| 0 | 3.03 ± 0.06 | 0.11 ± 0.01 | <0.1 |
| 1 | 3.54 ± 0.18 | 0.19 ± 0.03 | 0.33 ± 0.03 |
| 3 | 3.95 ± 0.13 | 0.27 ± 0.06 | NDa |
| 5 | 3.46 ± 0.03 | 0.26 ± 0.02 | 0.91 ± 0.17 |
| 8 | 2.77 ± 0.12 | 0.17 ± 0.02 | 15.0 ± 1.70 |
Values represent the means ± se (n = 5).
ND, Not determined.
Malate Homeostasis and Nitrogen Assimilation
In Populus, nitrogen taken up by the roots is transported in the xylem to shoot tissues primarily in the nitrate- and amino-nitrogen forms (Siebrecht and Tischner, 1999; Black et al., 2002). The transported nitrate is then reduced and assimilated, primarily in young leaves (LPI 3), where nitrate reductase activity is most highly concentrated (Black et al., 2002). In C3 species like spinach (Spinacia oleracea) and tobacco (Nicotiana tabacum), nitrate reduction stimulates the anapleurotic production of malate to counter the imbalances in charge and pH caused by its assimilation (for review, see Martinoia and Rentsch, 1994; Chollet et al., 1996; Scheible et al., 2000; Muller et al., 2001). Consistent with this, we suggest from the correlation between MDH activity, malate content, malate-to-citrate ratio (Table IV; Fig. 2), and nitrate reductase activity (Black et al., 2002) that the peak in malate concentration in EL was associated with a peak in anapleurotic activity for nitrate assimilation. Based on the reported preferential expression of nitrate translocators in newly emerging leaves of Arabidopsis (Guo et al., 2002), nitrate was probably also delivered to YL. However, the 2-fold malate increase in EL compared to YL suggests that nitrate was more actively assimilated in EL, where chloroplasts were more mature.
Amino acids typically comprise more than 50% of xylem nitrogen in Populus (Siebrecht and Tischner, 1999), and their delivery in the transpiration stream probably contributed heavily to leaf nitrogen status. Asn, with its two amino groups and very low carbon-to-nitrogen ratio, was at its highest level in YL. This is consistent with a surplus of available nitrogen relative to photosynthetic capacity or available carbon in YL (for recent discussion of Asn regulation, see Thum et al., 2003). We determined that Asn was probably being synthesized in YL from xylem-delivered Gln. Reverse transcription-PCR of apex, YL, and EL tissues using primer sequences based on a conserved region of Arabidopsis class II Asn synthetase genes, and sequences (e.g. CA930978) from our aspen expressed sequence tag database (Ranjan et al., 2004), revealed that expression of Asn synthetase genes decreased 1 to 2 orders of magnitude between apex and EL tissues (data not shown). GC-MS of xylem tissue extracts confirmed (using authentic Gln and Asn standards for quantitative evaluation) that Gln was by far the major transport amino form delivered in the xylem stream. While Gln was readily detected at levels similar to Gln levels seen in YL (Fig. 2), the other major transport amino acid, Asn, was barely detected in xylem extracts (data not shown). This is in line with reports that Gln and Asn comprise nearly 100% of Populus xylem sap amino acids and occur in a Gln-to-Asn ratio of 3:1 to 10:1 (Dickson, 1979; Schneider et al., 1994; Siebrecht and Tischner, 1999). Thus, it would appear that nitrate and Asn accumulated in YL but that nitrate was much more actively assimilated in older leaves (EL).
The Metabolic and Developmental Fate of Asn
The catabolism of Asn has been characterized in legume species and is considered to proceed primarily by two routes (for review, see Ireland and Lea, 1999). Asparaginase yields Asp and ammonia-nitrogen while Asn transaminase (AT) yields hydroxysuccinamate and an amino acid (Lloyd and Joy, 1978). Substantial amounts of AT activity have also been reported in leaf extracts of the nonlegume species spinach and lettuce (Meister, 1952). We detected increasing AT activity with leaf age in aspen. Activity increased slowly in LPI 1 to 5 but increased rapidly in LPI 5 to 8 (Table IV). AT activity in ML was 15-fold greater than in EL and more than 100-fold greater than in YL. Several protocols were used to evaluate asparaginase activity (see “Materials and Methods”), but it was difficult to detect asparaginase-mediated catabolism of Asn at any leaf stage (data not shown). The contribution of AT to Asn turnover appears to correlate with the maturation of photosynthetic and, interestingly, photorespiratory activity. Increases in Ser and Gly abundance relative to other amino acids and in Gly-to-Ser ratio, like those we observed during the EL-to-ML transition (Fig. 2), are consistent with increased photorespiratory activity (Novitskaya et al., 2002). In this context it is perhaps noteworthy that the preferred substrate of AT is glyoxylate (Cooper, 1977), and its amination yields Gly.
The developmental pattern of Asn utilization we observed may represent a route for the attenuation of surplus Asn to lower steady-state levels via photorespiration, once nitrogen requirements for rapid growth are largely met. This pattern could vary, depending on the stress and nutritional status of expanding leaves. The aspen plants in this study were nitrogen sufficient and raised in a relatively stress-free environment. Had nutrient, moisture, or other stresses altered the timing or duration of nitrate assimilation relative to Asn stores and photorespiratory development, a different, and perhaps diagnostic, pattern of Asn, Gly, Ser, and malate might have developed.
Diurnal effects (Scheible et al., 2000) and phenolic sink formation would provide additional layers of complexity. In addition, phloem development substantially affects the movement of photosynthate within and between leaves in aspen (Isebrands and Larson, 1973; Isebrands et al., 1976; Dickson and Larson, 1981), and could certainly affect changes in malate accumulation and transport associated with nitrate uptake and assimilation (Martinoia and Rentsch, 1994). The extent to which Asn homeostasis in shoot tips senses and participates in the development of photosynthetic, photorespiratory, and nitrate-assimilating competence presents itself as an interesting topic with respect to metabolic development in aspen. Aspen leaf chemistry, as well as that of other Populus sp., varies widely by genotype and affects growth of individual trees (Lindroth and Hwang, 1996), as well as the productivity and carbon-sequestering capacity of temperate forest ecosystems. The ability to profile metabolic shifts that reflect photoassimilate utilization, transport, and storage in developing leaves will be an important step toward understanding the genetic basis for the wide pattern of carbon partitioning and growth in these species. In addition, markers of cell wall and membrane biogenesis can be integrated with profile data on carbon and nitrogen utilization to support analysis of secondary carbon sink effects on Populus growth.
MATERIALS AND METHODS
Plant Material
Quaking aspen (Populus tremuloides Michx.) plants were micropropagated from surface-sterilized stem sections, transferred to 3-inch pots filled with a 1:1 peat:perlite, and acclimated in a mist chamber for 1 week prior to greenhouse planting. Acclimated plantlets were potted in 2-L containers filled with 2:1:1 (v/v/v) topsoil:peat moss:perlite, watered daily, and fertilized biweekly with Miracle-Gro (15-30-15 with micronutrients; Marysville, OH) and Fe-chelate. Plants were grown to a height of approximately 1 m during late summer (August through October). Supplemental lighting (200 μE m−2 s−1 photosynthetically active radiation, 10 cm above shoot tips) was provided to maintain a 16/8-h photoperiod during October. These plants typically had a plastochron index (Larson and Isebrands, 1971) of 30 to 35. Emerged leaves at LPI positions 0 (YL, 2–3 cm in length and 75% unfurled), 4 (EL, 8–10 cm in length), and 9 (ML, 13–16 cm in length) were harvested at midday into aluminum foil packets, flash frozen, and ground under liquid nitrogen. Tissue powders (10.0 ± 0.1 mg) were weighed into liquid nitrogen-cooled microcentrifuge tubes and stored at −80°C for no more than 1 week prior to analysis.
Chemicals
Methanol was HPLC grade and was purchased from Fisher Scientific (Hanover Park, IL). Ribitol (99%) was obtained from Sigma (St. Louis). Methoxyamine hydrochloride was purchased from Acros (Hanover Park, IL) and N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA), N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA), and N-methyl-N-tert-butyldimethylsilyltrifluoroacetamide (MTBSTFA) were purchased from Aldrich (Milwaukee, WI). All other chemicals were from Acros.
Extraction and Derivatization
The extraction protocol was modified from Roessner et al. (2000). Briefly, frozen aspen leaf tissue powders (10 mg) were extracted in 1.4 mL of 80% (v/v) aqueous methanol in a 2.0-mL microcentrifuge tube for 2 h under 200 rpm at ambient temperature. Ten microliters of ribitol solution (2 mg mL−1 water) were added as an internal standard prior to incubation. Endogenous ribitol was not detected in the aspen leaf extracts. The mixture was centrifuged at 13,000 rpm for 3 min, and 1 mL of the supernatant was evaporated to dryness (5 h) in a vacuum concentrator (Eppendorf, Hamburg, Germany). The dry residue was modified for GC-MS analysis according to Fiehn et al. (2000a). Three derivatization reagents, MSTFA, BSTFA, and MTBSTFA, were compared in preliminary experiments using authentic standards Gly, Glc, mannitol, myoinositol, ribitol, malate, and salicylate. Overall, MSTFA was found to be superior to BSTFA and MTBSTFA. Carbonyl moieties were methoximated with methoxyamine hydrochloride at 30°C for 90 min and derivatized with MSTFA (37°C for 30 min). The mixture was left undisturbed for 2 h at room temperature to allow complete derivatization. Derivatized samples were diluted with 2 volumes of methylene chloride and analyzed within 24 h. Chromatographic changes indicative of deterioration were noted only after derivatized samples had been stored for more than 30 h.
GC-MS Analysis
GC with quadrupole mass spectrometric detection was performed using a 6890 GC/5973N system (Agilent Technologies, Wilmington, DE). Derivatized extract (1 μL) was injected using splitless mode and split/splitless single-tapered liners (4 mm i.d.) packed with deactivated glass wool (Superlco, Bellefonte, CA), onto a 30 m × 0.25 mm (i.d.), 0.25-μm film AT-5MS column fitted with a deactivated guard column (Alltech Associates, Deerfield, IL). The inlet temperature was set at 250°C. After a 5-min solvent delay, oven temperature was increased from 70°C to 200°C (5°C min−1), 200°C to 260°C (10°C min−1), and 260°C to 310°C (20°C min−1) and maintained at 310°C for 12 min. Helium was used as the carrier gas at a flow rate of 1 mL min−1. The interface temperature was maintained at 310°C. The ion source was adjusted to 230°C. Mass spectra were recorded at 2.69 scan s−1 with a scan range of 50 to 600 m/z. Chemstation software (Agilent Technologies) operated the system and validated chromatogram and spectrum output. Perfluorotributylamine (PFTBA), with m/z of 69, 219, and 502, was used for autotuning. The mass spectrum was deconvoluted, and peaks were assigned identities using the automated mass spectral deconvolution and identification system (AMDIS) and the National Institute of Standards and Technology (NIST) library (version 98). Peaks that were at least 70% pure were automatically assigned identities, depending on the presence of a corresponding mass signature (match) with a similarity score (match factor) of 80 (out of 100) or higher in the NIST library. Matches for peaks below 70% purity were verified by manual analysis of the mass spectrum, and peaks with signal-to-noise ratio smaller than 75 were discarded. Care was taken to minimize carry-over effects, and column performance was monitored in preliminary experiments to establish a schedule for guard column trimming during sample analysis. Liners were replaced every 10 to 15 injections, and 10 cm of guard column were trimmed every 30 injections. For background monitoring, method blanks were run every 20 injections, and for sample reproducibility, a sample blank was injected every 10 samples. Because we did not include the chloroform phase extraction step in our sample preparation, we compared column performance after 20 injections of phase-partitioned or nonpartitioned methanol extracts and observed no systematic difference with regard to the metabolites we report. Samples were prepared and loaded in random order with respect to YL, EL, and ML tissues, according to a schedule that allowed us to analyze an equal number of each tissue group per day.
Data Analysis
Metabolite peak areas were divided by peak area of the internal standard, ribitol, to correct for recovery differences, and then normalized on the basis of sample fresh weight. All data were log10 transformed for statistical calculations. One-way ANOVA was performed by SAS/STAT software (version 8.2; SAS Institute,Cary, NC). Multiple means comparison of normalized metabolite peak areas was performed using Fisher's lsd method. PCA and HCA were performed using the R statistical package (http://www.r-project.org). PCA was used to calculate the contribution, or loadings, of specific metabolites to the formation of leaf sample clustering. HCA separated metabolites into clusters on the basis of their abundance ranking by lsd in YL, EL, and ML tissue. Euclidean distance was used as the metric of data relatedness and the average linkage method was used for data clustering.
Photosynthesis and Respiration Assays
Plants were preconditioned under indoor lighting (20 μE m−2 s−1) for 2 h prior to leaf disc assays. O2 consumption (dark respiration) and CO2-dependent photosynthetic O2 evolution were monitored in a water-jacketed Hansatech (King's Lynn, UK) Clark-type oxygen electrode using 1-cm2 discs from leaves at LPI 0 (YL), 4 (EL), and 9 (ML). Discs were cored under buffer (0.3 m sorbitol containing 25 mm HEPES, pH 7.5) and suspended in 0.5 mL of the same buffer in the electrode chamber by use of a nylon basket affixed to the bottom of the chamber plunger. Actinic light (1,000 μE m−2 s−1) was supplied fiber-optically from a halogen lamp.
Enzyme Assays
Soluble protein for all assays was obtained by extracting liquid nitrogen leaf powders twice with 5 volumes of 50 mm potassium phosphate, pH 7.4, 0.5 mm DTT, 1 mm phenylmethylsulfonylfluoride, and 1 μg mL−1 leupeptin. Polyvinylpolypyrollidone (5% [w/v]) was included in the first extraction. Extraction and all subsequent operations were conducted at 4°C. The two extracts were pooled and debris removed by centrifugation at 24,000g for 10 min. Clarified extracts were desalted by low-speed centrifugation through 25 volumes of appropriately equilibrated and buffered Sephadex G25. Protein was quantified using the procedure of Bradford (1976), and extracts were then supplemented with 0.1% (w/v) BSA fraction V. Asparaginase activity in 1-mL assay mixtures (30 min at 30°C) containing 100 μg protein, 100 mm potassium phosphate, pH 8.0, and 20 mm l-Asn was estimated in three ways: (1) by colorimetric detection of chloroamine following the Bertholet reaction to measure ammonia released (Vorwerk et al., 2001); (2) by direct measurement of Asp produced from Asn following diode array detection (435 nm) of dabsylated (dimethylaminoazobenzene sulfonyl) amino acids eluting from an HPLC C-18 column (Stocchi et al.,1989); and (3) methoxymation and MSTFA derivatization of dried assay aliquots for GC-MS. Asp transaminase activity with glyoxylate was estimated following a 30-min incubation of 100 μg of protein in a 1-mL assay mixture containing 100 mm Tricine-KOH, pH 8.1, 2 mm DTT, 10 mm sodium-glyoxylate, 20 mm l-Asn, and 20 μm pyridoxal-5-phosphate. Assay aliquots (10 μL) were dried down under vacuum and derivatized in methoxamine hydrochloride and MSTFA, as above, for GC-MS detection of Gly. NADH-dependent MDH activity was measured following modifications of the procedure of Egli et al. (1989) in 1-mL assays containing 15 to 20 μg of crude protein, 100 mm potassium phosphate, pH 7.4, 1 mm EDTA, 10 mm DTT, 1 μg mL−1 leupeptin, 200 μm oxaloacetate, and 250 μm NADH. Activity was estimated at 25°C by the change in A340 during the first 2 min of the reaction. NADPH-dependent MDH activity was measured essentially according to the procedures and recommendations of Hatch and Agostino (1992). Protein (50 μg) was activated for 30 min at 25°C in 100 μL containing 50 mm Tricine-KOH, pH 8.8, 100 mm DTT, 0.4 mm EDTA, 100 mm KCl, 1 μg mL−1 pepstatin, 1 μg mL−1 leupeptin, and 0.0025% (v/v) Triton X-100. Assays were conducted by diluting the 100-μL aliquot of activated protein in a 1-mL reaction containing 25 mm Tricine-KOH, pH 8.3, 100 mm KCl, 10 mm DTT, 200 μm NADPH, and 2 mm oxaloacetate, and measuring the change in A340 over the first 2 min of the reaction. NADPH-dependent MDH activity was also measured in the absence of an activation step and found to yield rates that were 20% to 40% of those obtained from activated protein (data not shown).
This work was supported by the Michigan Life Sciences Corridor Fund of the Michigan Economic Development Corporation (grant no. 085P1000477 to C.-J.T. and S.A.H.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044776.
References
- Backhausen JE, Emmerlich A, Holtgrete S, Horton P, Nast G, Rogers JM, Muller-Rober B, Scheibe R (1998) Transgenic potato plants with altered expression levels of chloroplast NADP-malate dehydrogenase: interactions between photosynthetic electron transport and malate metabolism in leaves and in isolated intact chloroplasts. Planta 207: 105–114 [Google Scholar]
- Bean RC, Porter GG, Steinberg BM (1961) Carbohydrate metabolism of citrus fruits II. Oxidation of sugars by an aerodehydrogenase from young orange fruits. J Biol Chem 236: 1235–1240 [PubMed] [Google Scholar]
- Black BL, Fuchigami LH, Coleman GD (2002) Partitioning of nitrate assimilation among leaves, stems and roots of poplar. Tree Physiol 22: 717–724 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bradshaw HD, Reinhart C, Davis J, Stettler R (2000) Emerging model systems in plant biology: poplar (Populus) as a model forest tree. J Plant Growth Regul 19: 306–313 [Google Scholar]
- Bryant JP, Chapin FS III, Klein DR (1983) Carbon/nutrient balance of boreal plants in relation to vertebrate herbivory. Oikos 40: 357–368 [Google Scholar]
- Champigny ML (1995) Integrations of photosynthetic carbon and nitrogen metabolism in higher plants. Photosynth Res 46: 117–127 [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 40: 415–439 [DOI] [PubMed] [Google Scholar]
- Cooper AJL (1977) Asparagine transaminase from rat liver. J Biol Chem 252: 2032–2038 [PubMed] [Google Scholar]
- Dickson RE (1979) Xylem translocation of amino acids from roots to shoots in cottonwood plants. Can J For Res 9: 374–378 [Google Scholar]
- Dickson RE, Larson PR (1981) 14C fixation, metabolic labeling patterns, and translocation profiles during leaf development in Populus deltoids. Planta 152: 461–470 [DOI] [PubMed] [Google Scholar]
- Dickson RE, Shive JB (1982) 14CO2 fixation, translocation, and carbon metabolism in rapidly expanding leaves of Populus deltoides. Ann Bot (Lond) 50: 37–47 [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G (2003) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131: 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli MA, Griffith SM, Miller SS, Anderson MP, Vance CP (1989) Nitrogen assimilating enzyme activities and enzyme protein in effective and plant controlled ineffective alfalfa genotypes. Plant Physiol 91: 898–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O (2003) Metabolic networks of Cucurbita maxima phloem. Phytochemistry 62: 875–886 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Doermann P, Altmann T, Trethewey RN, Willmitzer L (2000. a) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Trethewey RN, Willmitzer L (2000. b) Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal Chem 72: 3573–3580 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Weckwerth W (2003) Deciphering metabolic networks. Eur J Biochem 270: 579–588 [DOI] [PubMed] [Google Scholar]
- Gerhardt R, Heldt HW (1984) Measurement of subcellular metabolite levels in leaves by fractionation of freeze-stopped material in non-aqueous media. Plant Physiol 75: 542–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Hudak KA, Dumbroff EB, Thompson JE (1994) Release of photosynthetic protein catabolites by blebbing from thylakoids. Plant Physiol 106: 1547–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen V, Macherel D, Jaquinod M, Douce R, Bourguignoni JC (2000) Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J Biol Chem 275: 5016–5025 [DOI] [PubMed] [Google Scholar]
- Guo F-Q, Wang R, Crawford N (2002) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL-1) is regulated by auxin in both shoots and roots. J Exp Bot 53: 835–844 [DOI] [PubMed] [Google Scholar]
- Hanning I, Heldt HW (1993) On the function of mitochondrial metabolism during photosynthesis in spinach (Spinacia oleracea L.) leaves. Partitioning between respiration and export of redox equivalents and precursors for nitrate assimilation products. Plant Physiol 103: 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD, Agostino A (1992) Bilevel disulfide group reduction in the activation of C4 leaf nicotinamide adenine dinucleotide phosphate-malate dehydrogenase. Plant Physiol 100: 360–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming JDC, Lindroth RL (1999) Effects of light and nutrient availability on aspen: growth, phytochemistry, and insect performance. J Chem Ecol 25: 1687–1714 [DOI] [PubMed] [Google Scholar]
- Ireland RJ, Lea PJ (1999) The enzymes of glutamine, glutamate, asparagine, and aspartate metabolism. In BK Singh, ed, Plant Amino Acids: Biochemistry and Biotechnology. Marcel Dekker, NY, pp 49–109
- Isebrands JG, Dickson RE, Larson PR (1976) Translocation and incorporation of 14C into the petiole from different regions within developing cottonwood leaves. Planta 128: 185–193 [DOI] [PubMed] [Google Scholar]
- Isebrands JG, Larson PR (1973) Anatomical changes during leaf ontogeny in Populus deltoides. Am J Bot 60: 199–208 [Google Scholar]
- Kao Y-Y, Harding SA, Tsai C-J (2002) Differential expression of two distinct phenylalanine ammonia-lyase genes in condensed tannin-accumulating and lignifying cells of quaking aspen. Plant Physiol 130: 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler G, Neufeld EF, Feingold DS, Hassid WZ (1961) Metabolism of D-glucuronic acid and D-galaturonic acid by Phaseolus aureus seedlings. J Biol Chem 236: 308–312 [PubMed] [Google Scholar]
- Kleiner KW, Raffa KF, Dickson RE (1999) Partitioning of 14C-labeled photosynthate to allelochemicals and primary metabolites in source and sink leaves of aspen: evidence for secondary metabolite turnover. Oecologia 119: 408–418 [DOI] [PubMed] [Google Scholar]
- Kromer S, Heldt HW (1991) Respiration of pea leaf mitochondria and redox transfer between the mitochondrial and extramitochondrial compartment. Biochim Biophys Acta 1057: 42–50 [Google Scholar]
- Lam H-M, Coshigano K, Schultz C, Meio-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh M-H, Coruzzi G (1995) Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane BG, Dunwell JM, Ray JA, Schmitt MR, Cuming AC (1993) Germin, a protein marker of early plant development, is an oxalate oxidase. J Biol Chem 268: 12239–12242 [PubMed] [Google Scholar]
- Larson PR, Isebrands JG (1971) The plastochron index as applied to developmental studies of cottonwood. Can J For Res 1: 1–11 [Google Scholar]
- Lennon AM, Pratt J, Leach G, Moore A (1995) Developmental regulation of respiratory activity in pea leaves. Plant Physiol 107: 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L-S, Varner JE (1991) Expression of ascorbic acid oxidase in zucchini squash (Cucurbita pepo L.). Plant Physiol 96: 159–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth RL, Hwang S-Y (1996) Diversity, redundancy and multiplicity in chemical defense systems of aspen. In JT Romeo, JA Saunders, P Barbosa, eds, Phytochemical Diversity and Redundancy in Ecological Interactions. Plenum Press, NY, pp 25–54
- Lloyd NDH, Joy KW (1978) Hydroxysuccinamic acid: a product of asparagine metabolism in plants. Biochem Biophys Res Commun 81: 186–192 [DOI] [PubMed] [Google Scholar]
- Madey E, Nowack LM, Su L, Hong Y, Hudak KA, Thompson JE (2001) Characterization of plasma membrane domains enriched in lipid metabolites. J Exp Bot 52: 669–679 [DOI] [PubMed] [Google Scholar]
- Mansfield JL, Curtis PS, Zak DR, Pregitzer KS (1999) Genotypic variation for condensed tannin production in trembling aspen (Populus tremuloides, salicaceae) under elevated CO(2) and in high- and low-fertility soil. Am J Bot 86: 1154–1159 [PubMed] [Google Scholar]
- Martinoia E, Rentsch D (1994) Malate compartmentation responses to a complex metabolism. Annu Rev Plant Physiol Plant Mol Biol 45: 447–467 [Google Scholar]
- McNeil SD, Nuccio ML, Rhodes D, Shachar-Hill Y, Hanson AD (2000) Radiotracer and computer modeling evidence that phospho-base methylation is the main route of choline synthesis in tobacco. Plant Physiol 123: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A (1952) Preparation and enzymatic reactions of the keto analogues of asparagine and glutamine. J Biol Chem 197: 571–589 [PubMed] [Google Scholar]
- Melzer E, O'Leary MH (1987) Anapleurotic CO2 fixation by phosphoenolpyruvate carboxylase in C3 plants. Plant Physiol 84: 58–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TS (1990) Biosynthesis of phosphatidylinositol. Inositol Metab Plant 9: 107–112 [Google Scholar]
- Muller C, Scheible WR, Stitt M, Krapp A (2001) Influence of malate and 2-oxoglutarate on the NIA transcript level and nitrate reductase activity in tobacco leaves. Plant Cell Environ 24: 191–203 [Google Scholar]
- Novitskaya L, Trevanion SJ, Driscoll S, Foyer CH, Noctor G (2002) How does photorespiration modulate leaf amino acid contents? A dual approach through modelling and metabolite analysis. Plant Cell Environ 25: 821–835 [Google Scholar]
- Orians CM, Fritz RS (1995) Secondary chemistry of hybrid and parental willows: phenolic glycosides and condensed tannins in Salix sericea, S. eriocephala and their hybrids. J Chem Ecol 21: 1245–1253 [DOI] [PubMed] [Google Scholar]
- Passi S, Nazzaro-Porro M, Picardo M, Mingrone G, Fasella P (1983) Metabolism of straight saturated medium chain length (C9 to C12) dicarboxylic acids. J Lipid Res 24: 1140–1147 [PubMed] [Google Scholar]
- Ranjan P, Kao Y-Y, Jiang H, Joshi CP, Harding SA, Tsai C-J (2004) Suppression subtractive hybridization-mediated transcriptome analysis from multiple tissues of aspen (Populus tremuloides) altered in phenylpropanoid metabolism. Planta 219: 694–704 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie HR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontein D, Nishida I, Tashiro G, Yoshioka K, Wu W-I, Voelker DR, Bassit G, Hanson AD (2001) Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J Biol Chem 276: 35523–35529 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Krapp A, Stitt M (2000) Reciprocal diurnal changes of phosphoenolpyruvate carboxylase expression and cytosolic pyruvate kinase, citrate synthase and NADP-isocitrate dehydrogenase expression regulate organic acid metabolism during nitrate assimilation in tobacco leaves. Plant Cell Environ 23: 1155–1167 [Google Scholar]
- Schneider H, Kreuzwieser J, Schums R, Sauter JJ, Rennenberg H (1994) Thiol and amino acid composition of the xylem sap of poplar trees. Can J Bot 72: 347–351 [Google Scholar]
- Siebrecht S, Tischner R (1999) Changes in the xylem exudate composition of poplar (Populus tremula x P. alba)-dependent on the nitrogen and potassium supply. J Exp Bot 50: 1797–1806 [Google Scholar]
- Smirnoff N (1996) The function and metabolism of ascorbic acid in plants. Ann Bot (Lond) 78: 661–669 [Google Scholar]
- Smith MD, Licatalosi DD, Thompson JE (2000) Co-association of cytochrome f catabolites and plastid-lipid-associated protein with chloroplast lipid particles. Plant Physiol 124: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocchi V, Piccoli G, Magnani M, Palma F, Biagiarelli B, Cucchiarini L (1989) Reversed-phase high-performance liquid chromatography separation of dimethylaminoazobenzene sulfonyl- and dimethylaminoazobenzene thiohydantoin-amino acid derivatives for amino acid analysis and microsequencing studies at the picomole level. Anal Biochem 178: 107–117 [DOI] [PubMed] [Google Scholar]
- Sumner LW, Mendes P, Dixon RA (2003) Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62: 817–836 [DOI] [PubMed] [Google Scholar]
- Thompson P, Bowsher CG, Tobin AK (1998) Heterogeneity of mitochondrial protein biogenesis during primary leaf development in barley. Plant Physiol 118: 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum KE, Shasha DE, Lejay LV, Coruzzi GM (2003) Light- and carbon-signaling pathways. Modeling circuits of interactions. Plant Physiol 132: 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus DE, Becker WM (1985) Investigation of the glyoxysome-peroxisome transition in germinating cucumber cotyledons using double-label immunoelectron microscopy. J Cell Biol 101: 1288–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tserng K-Y, Jin S-J (1991) Metabolic conversion of dicarboxylic acids to succinate in rat liver homogenates. J Biol Chem 266: 2924–2929 [PubMed] [Google Scholar]
- Valafar F (2002) Pattern recognition techniques in microarray data analysis. Ann N Y Acad Sci 980: 41–64 [DOI] [PubMed] [Google Scholar]
- Vorwerk S, Biernacki S, Hillebrand H, Janzik I, Müller A, Weiler EW, Piotrowski M (2001) Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 212: 508–516 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Wullschleger SD, Jansson S, Taylor G (2002) Genomics and forest biology—Populus emerges as the perennial favorite. Plant Cell 14: 2651–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]