Abstract
Demand-driven signaling will contribute to regulation of sulfur acquisition and distribution within the plant. To investigate the regulatory mechanisms pedospheric sulfate and atmospheric H2S supply were manipulated in Brassica oleracea. Sulfate deprivation of B. oleracea seedlings induced a rapid increase of the sulfate uptake capacity by the roots, accompanied by an increased expression of genes encoding specific sulfate transporters in roots and other plant parts. More prolonged sulfate deprivation resulted in an altered shoot-root partitioning of biomass in favor of the root. B. oleracea was able to utilize atmospheric H2S as S-source; however, root proliferation and increased sulfate transporter expression occurred as in S-deficient plants. It was evident that in B. oleracea there was a poor shoot to root signaling for the regulation of sulfate uptake and expression of the sulfate transporters. cDNAs corresponding to 12 different sulfate transporter genes representing the complete gene family were isolated from Brassica napus and B. oleracea species. The sequence analysis classified the Brassica sulfate transporter genes into four different groups. The expression of the different sulfate transporters showed a complex pattern of tissue specificity and regulation by sulfur nutritional status. The sulfate transporter genes of Groups 1, 2, and 4 were induced or up-regulated under sulfate deprivation, although the expression of Group 3 sulfate transporters was not affected by the sulfate status. The significance of sulfate, thiols, and O-acetylserine as possible signal compounds in the regulation of the sulfate uptake and expression of the transporter genes is evaluated.
In general plants utilize sulfate taken up by the roots as the main S source for growth (Cram, 1990; Clarkson et al., 1993) and sulfate is reduced to sulfide before it is assimilated into organic S compounds. Under normal conditions the rate of uptake and assimilation of S will depend on the requirement for growth, which can be defined as the rate of S uptake and assimilation required per gram plant biomass produced with time (De Kok et al., 2002). The S requirement will fluctuate during plant development and may vary between species differing in S need for growth and the potential sink capacity of secondary S compounds.
The uptake of sulfate by the roots and its transport to the shoot appear to be major sites of regulation of S assimilation (Hawkesford and Wray, 2000; De Kok et al., 2002). In addition to sulfate uptake from the soil by the roots, there are further requirements for transmembrane transport of sulfate in the process of long distance transport of sulfate from the root to the shoot, for intracellular transport of sulfate into the vacuole as the main storage pool of sulfate, and into plastids where sulfate reduction takes place.
Sulfate transporter genes have been identified from Arabidopsis and other plant species (Smith et al., 1995, 1997; Takahashi et al., 1996, 1997, 1999a, 1999b, 2000; Yoshimoto et al., 2002; 2003, Howarth et al., 2003). In the Arabidopsis genome (The Arabidopsis Genome Initiative, 2000), 12 sulfate transporter genes have been identified in the gene family, which may be subdivided into four different groups. Group 1 sulfate transporters are assumed to be principally responsible for the primary uptake of sulfate by the root (Hawkesford, 2003). All Group 1 members tested to date have a high affinity for sulfate (Kms in the range 1.5–10 μm) and expression analysis indicated that these transporters were derepressed in roots upon sulfate deprivation, and repressed when sulfate supply was restored (Smith et al., 1997; Hawkesford and Wray, 2000). The expression of Group 1 transporters in the root epidermis, in the cortex, and other plant tissues suggested additional functions for these sulfate transporters within the plant (Yoshimoto et al., 2002). A specialized role is suggested for Arabidopsis Sultr1;3, which is localized solely in the phloem and which may mediate the redistribution of sulfate between source and sink organs (Yoshimoto et al., 2003). The Group 2 sulfate transporters have a lower affinity for sulfate compared to Group 1 sulfate transporters (Smith et al., 1995; Takahashi et al., 2000). Both Arabidopsis Group 2 transporters are expressed in the vascular tissue and may mediate the transport of sulfate within the stele for xylem loading and uploading in roots and leaves (Sultr2;1) or for loading and transport via root phloem and leaf bundle sheath cells (Sultr2;2) (Takahashi et al., 2000). The Group 3 sulfate transporters are less well characterized. Studies in Arabidopsis have shown leaf specific expression for three Group 3 transporters, which was not affected by the S status of the plant (Takahashi et al., 1999b). The N-terminal region of the both Arabidopsis Group 4 proteins contains a putative plastidal targeting sequence and combined with GFP, the N-terminal region is able to target GFP to chloroplasts (Takahashi et al., 1999b). However, more recent experiments using the full protein combined with a GFP reporter gene indicated that this isoform may be targeted to the tonoplast and is responsible for sulfate efflux from the vacuole (Kataoka et al., 2004).
There is little information on shoot-root coordination of the uptake of sulfate versus its assimilation and the signal transduction pathways involved. There may be fast responses to environmental change, for instance S supply, via metabolite activation and deactivation of enzymes involved in S assimilation (Hell et al., 2002) or metabolite-induced expression and derepression of genes encoding the sulfate transporters (Hawkesford and Wray, 2000). In addition, regulation may occur via altered growth patterns, e.g. changes in root development and/or shoot to root ratio. It is still largely unclear to what extent sulfate itself or other metabolic products of S assimilation are directly involved in sensing or act as regulatory signals.
Plants are able to utilize forms of S other than sulfate taken up from the pedosphere such as foliarly absorbed atmospheric H2S, which is directly metabolized into Cys and subsequently into other organic S compounds, including glutathione and proteins (De Kok, 1990; De Kok et al., 1997, 1998, 2000, 2002; De Kok and Tausz, 2001; Stuiver and De Kok, 2001). In the absence of pedospheric sulfate, plants may grow with H2S as the sole S source for growth (De Kok et al., 1997, 1998, 2000, 2002; Westerman et al., 2000a). H2S exposure of B. oleracea resulted in a negative feedback regulation of the uptake and assimilation of pedospheric sulfate (Westerman et al., 2000a, 2000b, 2001a, 2001b).
In this paper the modulation of S nutrition of B. oleracea was achieved by changing the levels of atmospheric H2S and pedospheric sulfate in order to probe the regulation of sulfate uptake and transport as indicated by the expression of the respective genes in relation to the S status of the plant. The expression patterns of the complete Brassica sulfate transporter gene family are presented for roots, stem, and leaves. In addition, analysis of sulfate, thiols, total S, and O-acetylserine (OAS) was examined to determine the relationship of these metabolite pools to the expression of the sulfate transporter gene family in uptake and movement of sulfate within the plant.
RESULTS AND DISCUSSION
Impact of Sulfate Deprivation and H2S Exposure on Growth, Sulfur, and Nitrogen Metabolites and Sulfate Uptake Capacity
H2S is a potentially phytotoxic gas; however, B. oleracea is not very susceptible to H2S and its growth is only significantly reduced at atmospheric levels higher than 400 nL L−1 (De Kok et al., 1997, 2000, 2002). B. oleracea is able to use atmospherically supplied H2S taken up by the foliage as the sole S source for growth (De Kok et al., 1997, 2000, 2002). From this study it was evident that an atmospheric H2S level as low as 75 nL L−1 was almost sufficient to meet the S requirements to maintain shoot growth during the seedling stage of the cultivar used here (Arsis).
As has been observed previously, the transfer of B. oleracea seedlings to sulfate-deprived conditions resulted in a rapid development of S deficiency symptoms from approximately 6 d onwards (Table I; De Kok et al., 1997, 2000; Stuiver et al., 1997). The young developing leaves started to turn yellow and growth rate declined, and after 10 d the plants became severely S deficient. Biomass production of leaves and stem plus petioles was affected more rapidly by sulfate deprivation compared to roots (Table I). This was also reflected by a decrease in the shoot to root ratio, which was already significantly decreased after 6 d of deprivation (Table II). Shoot biomass production of plants, which were cultivated under sulfate-deprived conditions and simultaneously exposed to 75 nL L−1 H2S, was comparable to that of sulfate-sufficient plants (Table I). Here, plants started to develop visible S deficiency symptoms only from day 10 onwards. However, the shoot to root ratio remained equally low as the S-deprived, non-H2S-exposed plants due to a stimulated root growth of sulfate-deprived plants upon H2S exposure (Table II).
Table I.
Growth of Brassica oleracea as affected by pedospheric and atmospheric sulfur nutrition
| Growth Rate
|
Biomass Production
|
||
|---|---|---|---|
| t6 − t2 | t10 − t6 | t10 − t0 | |
| g g−1 day−1 | g | ||
| Leaves | |||
| −S | 0.201 ± 0.040 | 0.106 ± 0.006* | 2.42 ± 0.46 (63)* |
| −S, H2S | 0.205 ± 0.019 | 0.212 ± 0.016 | 3.99 ± 0.72 (104) |
| +S | 0.211 ± 0.008 | 0.197 ± 0.009 | 3.84 ± 0.82 (100) |
| +S, H2S | 0.203 ± 0.037 | 0.210 ± 0.005 | 4.26 ± 0.95 (111) |
| Stem Plus Petioles | |||
| −S | 0.279 ± 0.043 | 0.129 ± 0.017* | 0.76 ± 0.15 (65)* |
| −S, H2S | 0.257 ± 0.023 | 0.252 ± 0.044 | 1.17 ± 0.17 (100) |
| +S | 0.234 ± 0.012 | 0.257 ± 0.011 | 1.17 ± 0.21 (100) |
| +S, H2S | 0.282 ± 0.071 | 0.258 ± 0.028 | 1.44 ± 0.30 (123) |
| Roots | |||
| −S | 0.248 ± 0.072 | 0.128 ± 0.029* | 1.09 ± 0.22 (89) |
| −S, H2S | 0.243 ± 0.067 | 0.242 ± 0.026 | 1.76 ± 0.50 (143)* |
| +S | 0.186 ± 0.026 | 0.200 ± 0.021 | 1.23 ± 0.41 (100) |
| +S, H2S | 0.189 ± 0.048 | 0.195 ± 0.019 | 1.18 ± 0.38 (96) |
B. oleracea was grown on a 25% Hoagland nutrient solution (0.5 mm sulfate) in a climate controlled room for 7 d and subsequently transferred to a fresh nutrient solution at 0 mm sulfate (−S) or 0.5 mm sulfate (+S) and simultaneously exposed to 0 or 75 nL L−1 H2S for 2, 6, and 10 d. Growth rates were measured over the period from day 2 to day 6 (t6 − t2) and from day 6 to day 10 (t10 − t6), and biomass production (fresh weight) was measured over the entire 10 d period (t10 − t0). Growth rate was calculated using the natural log transformed fresh weight. Data represent the mean of 3 independent experiments on 3 to 4 (4–12 for t0) measurements per experiment with 3 plants in each (± sd). Data between brackets represent values as percent of the control (+S). Values marked with an asterisk are significantly different from the control value (+S; P < 0.01).
Table II.
Shoot to root ratio of B. oleracea as affected by pedospheric and atmospheric sulfur nutrition
| Shoot to Root Ratio
|
||||
|---|---|---|---|---|
| t0 | t2 | t6 | t10 | |
| −S | 3.42 ± 0.64 | 3.70 ± 0.47 | 3.23 ± 0.43* | 2.98 ± 0.23* |
| −S, H2S | 3.42 ± 0.64 | 3.78 ± 0.31 | 3.34 ± 0.46* | 3.07 ± 0.51* |
| +S | 3.42 ± 0.64 | 3.57 ± 0.42 | 4.00 ± 0.53 | 4.15 ± 0.66 |
| +S, H2S | 3.42 ± 0.64 | 3.88 ± 0.26 | 4.35 ± 0.34 | 4.77 ± 0.62 |
Shoot to root ratio, based on fresh weight, represents the mean of three independent experiments on 3 to 4 (4–12 for t0) measurements per experiment on three plants in each (± sd). Data derived from Table I. Values marked with an asterisk are significantly different from the control value (+S; P < 0.01).
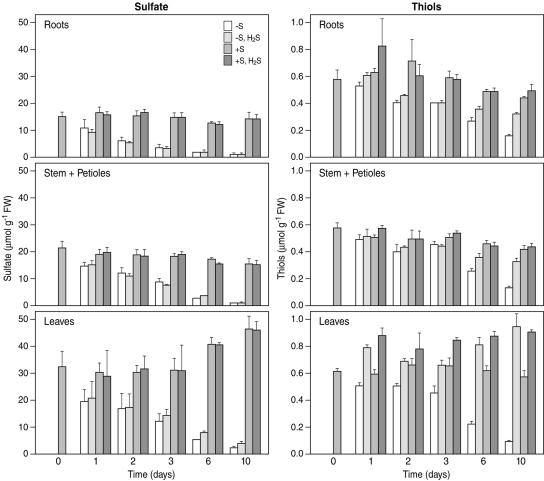
Upon sulfate deprivation, the appearance of S deficiency symptoms was preceded by a rapid decrease in the sulfate content in all plant parts, which was already significant after 1 d (Fig. 1). However, after 6 d, when plants started to show visible symptoms of sulfate deficiency, all plant parts still contained detectable amounts of sulfate (Fig. 1). In sulfate-deprived, H2S-exposed plants, the sulfate contents in leaves, stems and roots were as low as in sulfate-deprived nonexposed plants.
Figure 1.
Sulfate and total water-soluble nonprotein thiol content of B. oleracea as affected by pedospheric and atmospheric sulfur nutrition. B. oleracea was grown on a 25% Hoagland nutrient solution (0.5 mm sulfate) in a climate controlled room for 7 d and subsequently transferred to a fresh nutrient solution at 0 (−S) or 0.5 mm sulfate (+S) and simultaneously exposed to 0 or 75 nL L−1 H2S for 1, 2, 3, 6, and 10 d. Sulfate contents represent the means of two experiments with three to five measurements on one (day 10) to three plants in each (± sd). Thiol content represents the means of three measurements with three or six plants in each (± sd).
Sulfate deprivation resulted in a decrease in the water-soluble nonprotein thiol content, however to a lesser extent than for sulfate (Fig. 1). H2S exposure generally results in an increased size of the water-soluble nonprotein thiol pool (predominantly glutathione) in the shoot, since the absorbed H2S is metabolized into Cys with high affinity and subsequently into glutathione and other S metabolites (De Kok et al., 1997, 2000, 2002). Indeed, in sulfate-sufficient, H2S-exposed plants the thiol content in leaves was enhanced up to 1.4-fold after 1 d of exposure and it was hardly affected during further exposure. The thiol contents of the roots and stem plus petioles of sulfate-sufficient plants were not affected by H2S exposure (Fig. 1). However, in sulfate-deprived, H2S-exposed plants the total water-soluble nonprotein thiol content in leaves was increased 1.5-fold after 1 d of H2S exposure and further increased with time reaching a level similar to that in leaves of sulfate-sufficient, H2S-exposed plants (Fig. 1). From day 1 to 3, the thiol content in the stem and roots of sulfate-deprived, H2S-exposed plants decreased by nearly the same extent as for sulfate-deprived plants. However, from day 6 onwards the thiol content of both, stem and roots of sulfate-deprived, H2S-exposed plants remained unaltered (Fig. 1).
Ten days of sulfate deprivation resulted in a 4- and 8-fold decrease in total S content of roots and shoot, respectively; it did not affect the total N content, which resulted in a strongly increased N:S ratio (Table III). It was striking that the total S content was hardly affected in sulfate-deprived, H2S-exposed plants as compared to that of sulfate-deprived nonexposed plants (Table III). The total N content was not affected by H2S exposure and as a consequence the total N/S ratio remained extremely high as compared to that of sulfate-sufficient plants (Table III).
Table III.
Total sulfur and nitrogen content of B. oleracea as affected by pedospheric and atmospheric sulfur nutrition
| Total S | Total N | N:S Ratio (on a molar basis) | |
|---|---|---|---|
| mmol g−1 DW | |||
| Shoot | |||
| Initial | 0.426 ± 0.021 | 4.29 ± 0.16 | 10.1 |
| −S | 0.059 ± 0.010* | 3.87 ± 0.25 | 65.6* |
| −S, H2S | 0.063 ± 0.002* | 4.12 ± 0.12 | 65.4* |
| +S | 0.481 ± 0.005 | 4.16 ± 0.04 | 8.6 |
| +S, H2S | 0.503 ± 0.025 | 4.06 ± 0.08 | 8.1 |
| Roots | |||
| Initial | 0.385 ± 0.014 | 3.94 ± 0.03* | 10.2 |
| −S | 0.093 ± 0.029* | 3.53 ± 0.09 | 38.0* |
| −S, H2S | 0.097 ± 0.023* | 3.68 ± 0.06 | 37.9* |
| +S | 0.394 ± 0.018 | 3.61 ± 0.10 | 9.2 |
| +S, H2S | 0.352 ± 0.026 | 3.66 ± 0.02 | 10.4 |
B. oleracea was grown on a 25% Hoagland nutrient solution at 0.5 mm sulfate in a climate controlled room for 7 d and subsequently transferred to a fresh nutrient solution at 0 mm sulfate (−S) or 0.5 mm sulfate (+S) and simultaneously exposed to 0 or 75 nL L−1 H2S for 10 d. Total S and N are expressed as mmol g−1 DW and represent the mean of 3 measurements with 27 (initial) and 3 plants in each (± sd). Values marked with an asterisk are significantly different from the control value (+S; P < 0.01).
Brassica originates from saline and S enriched environments and is considered to have a high S requirement for growth (Westerman et al., 2001a). However, in Brassica seedlings a large proportion of total S (up to 90%) may be present as sulfate, presumably located in the vacuole and here the concept of “the S need for structural growth” may have to be redefined (Castro et al., 2003). From the present data it is evident that sulfate-deprived, H2S-exposed plants were able to grow nearly optimally and quite efficiently at a minimum S level (Table III).
The depletion of S-containing amino acids and a subsequently restricted protein synthesis is likely the primary cause of the disturbance of physiological functioning of the plant and the appearance of S deficiency symptoms. The development of S deficiency symptoms is generally accompanied by accumulation of nitrate, free amino acids (especially in the shoots), and an increase in dry matter content due to an accumulation of nonstructural carbohydrates such as starch (De Kok et al., 1997, 2000; Stuiver et al., 1997).
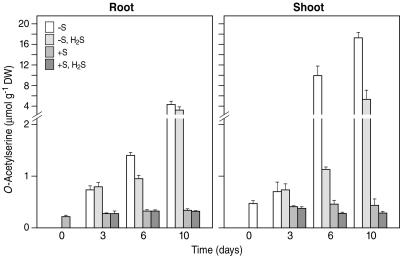
Despite the fact that sulfate deprivation did not affect total N content (Table III), it had a decisive impact on the content of several N metabolites. For instance, the content of OAS, which is the direct carbon/N precursor for Cys synthesis, is assumed to have significance in the regulation of sulfate uptake and reduction (Smith et al., 1997; Clarkson et al., 1999; Hawkesford and Wray, 2000). It increased both in roots and shoot by 24% and 54%, respectively, after 1 d of sulfate deprivation (data not shown). After an approximately 2- to 3-fold increase upon 3 d of sulfate deprivation, the OAS concentration increased further dramatically up to 13- and 40-fold in roots and shoots, respectively, after 10 d of sulfate deprivation (Fig. 2). Furthermore, in S-deficient shoots (from 6 d onwards) there was also an increase in nitrate and free amino acid content (data not shown). The OAS content in the shoot of sulfate-deprived, H2S-exposed plants was similar to that of the sulfate-deprived plants after 3 d; however, it increased substantially from day 6 onwards, though to a lower extent than in sulfate-deprived plants (Fig. 2). Its content in the roots showed a similar pattern to that observed in the shoot but its increment upon sulfate deprivation was lower (Fig. 2). In sulfate-sufficient plants, growth, the contents of sulfate, total S, and total N in leaves, stem, and roots were not affected upon H2S exposure (Fig. 1; Table III). There was a decrease in OAS content of the shoots during prolonged H2S exposure (Fig. 2).
Figure 2.
OAS content of B. oleracea as affected by pedospheric and atmospheric sulfur nutrition. B. oleracea was grown on a 25% Hoagland nutrient solution (0.5 mm sulfate) in a climate controlled room for 7 d and subsequently transferred to a fresh nutrient solution at 0 (−S) or 0.5 mm sulfate (+S) and simultaneously exposed to 0 or 75 nL L−1 H2S for 3, 6, and 10 d. Data represent the means of three measurements with three plants in each (± sd). Breaks in the chart indicate two different scales of OAS concentrations.
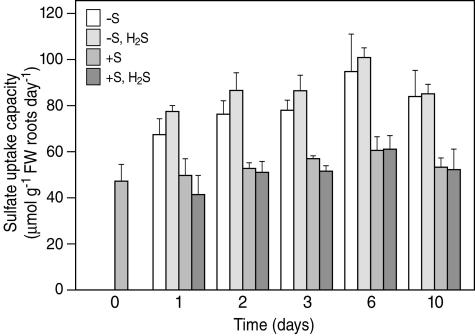
Similarly to previous observations, sulfate deprivation resulted in a rapid increase in the sulfate uptake capacity (Fig. 3; Clarkson et al., 1993; Hawkesford and Smith, 1997). The sulfate uptake capacity increased 1.4-fold after 1 d of sulfate deprivation and it slightly further increased with time upon deprivation up to 1.6-fold after 10 d. The nitrate uptake capacity, however, was decreased from 6 d onwards down to 0.5-fold (to 0.23 mmol g−1 root fresh weight d−1) after 10 d of sulfate deprivation (data not shown). Even when H2S was used as the S source and growth of sulfate-deprived plants was (except root growth) quite similar to that of sulfate-sufficient plants, the enhanced sulfate uptake capacity upon sulfate deprivation was not affected by H2S exposure (Fig. 3). The sulfate uptake capacity of sulfate sufficient plants was hardly affected by H2S exposure.
Figure 3.
Sulfate uptake capacity of B. oleracea as affected by pedospheric and atmospheric sulfur nutrition. B. oleracea was grown on a 25% Hoagland nutrient solution (0.5 mm sulfate) in a climate controlled room for 7 d and subsequently transferred to a fresh nutrient solution at 0 mm sulfate (−S) or 0.5 mm sulfate (+S) and simultaneously exposed to 0 or 75 nL L−1 H2S for 1, 2, 3, 6, and 10 d. Sulfate uptake was measured over a 24-h period (see “Materials and Methods”) and represents the mean of three to six measurements with three plants in each (± sd).
Isolation and Identification of the Brassica Sulfate Transporter Family
Plants contain a variety of sulfate transporters with specific functions for the uptake of sulfate by the roots, transport to the shoot, and subcellular distribution (Hawkesford and Smith, 1997; Hawkesford, 2000; Hawkesford and Wray, 2000; Hawkesford, 2003).
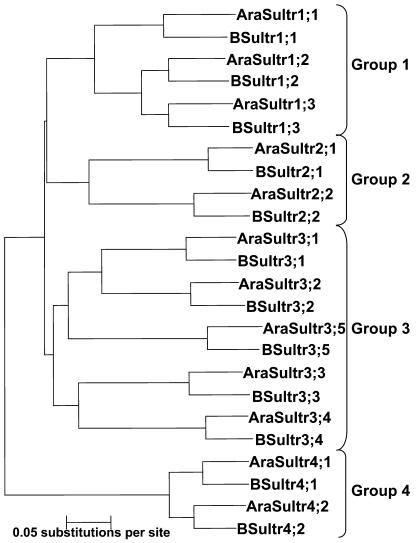
The use of degenerate primers based on the high identity sequence regions of known plant sulfate transporter groups (Table IV) as well as Brassica genomic sequence information enabled the isolation by PCR and the identification of 11 sulfate transporter cDNAs from B. napus and B. oleracea. Sequence comparison and phylogenetic analysis of the coding regions of the isolated Brassica sulfate transporter mRNAs to the known sulfate transporter family from Arabidopsis indicated that three transporters, BSultr1;1, 1;2, 1;3, belong to Group 1, one, BSultr 2;1, belongs to Group 2, five, BSultr 3;1, 3;2, 3;3, 3;4 and 3;5 belong to group 3 and two, BSultr4;1 and 4;2, belong to the Group 4 sulfate transporter family (Fig. 4). The sequence similarity of the coding mRNA region of the Brassica sulfate transporters to the homologous Arabidopsis genes was in the range of 84% to 89%.
Table IV.
Oligonucleotide primers used for Brassica sulfate transporter cDNA iolation
| Group and Name | Degenerated Oligo Nucleotide Primer Sequences | Orientation |
|---|---|---|
| Group 1 | ||
| BST sense 1 | 5′-CTCACCATYGCHAGYCTYTGYAT-3′ | Sense |
| BST sense 2 | 5′-CCTCAGGAYMTYGSWTATGC-3′ | Sense |
| BST as 1 | 5′-GCATTBGGMGTGTACTTRAA-3′ | Antisense |
| BST as 2 | 5′-TTCAWKGCMGCAAATGTTCT-3′ | Antisense |
| Bo1.3sense1 | 5′-AGAGCTCATCCTGTGGACG-3′ | Sense |
| Bo1.3as1 | 5′-TCAGACCTCGTYGGASAGTTT-3′ | Antisense |
| Group 2 | ||
| STL sense1 | 5′-AAAGARATGDTGGCHATKGG-3′ | Sense |
| STL sense2 | 5′-TCMAACATWGTDATGGCGRT-3′ | Sense |
| STL as 1 | 5′-AGAAGWCCRATCTCDAYGGA-3′ | Antisense |
| STL as2 | 5′-TTATCRASYTTCCAWATGTG-3′ | Antisense |
| Group 3 | ||
| STN sense1B | 5′-CCTCAGGRSATYRGYTAYGC-3′ | Sense |
| STN sense2 | 5′-GGDTTYATGGSNGGWGCNGC-3′ | Sense |
| STN as 1 | 5′-GADCCWASGAYGTTCATSR-3′ | Antisense |
| STN as 2 | 5′-GCBABCATYTCTTTGTTYCCRTC-3′ | Antisense |
| Bo3.2sense1 | 5′-CAATGTATAAGAATTACAACATAG-3′ | Sense |
| Bo3.2sense2 | 5′-GAGATGATTGCATTTGGGATG-3′ | Sense |
| Bo3.3sense1 | 5′-GTTGCAGGAACTAATCGATCC-3′ | Sense |
| Bo3.3sense2 | 5′-CTTAGGCTACACGAAACTCGT-3′ | Sense |
| Bo3.4sense1 | 5′-CAAATGGGGTTCTCAGTACG-3′ | Sense |
| Bo3.4sense2 | 5′-TAGGTGTGATGTTGTCTCTGG-3′ | Sense |
| Bo3.5sense1 | 5′-CGAGGAGATGTACCACCTCTT-3′ | Sense |
| Bo3.5sense2 | 5′-TCCTTCATTAGCATGGACTACG-3′ | Sense |
| Group 4 | ||
| STPas1 | 5′-ATCCCATYACRAAWGGTGGCC-3′ | Antisense |
| STPas2 | 5′-TGGYAYAATYTTGCTGCTYC-3′ | Antisense |
| STPsense1 | 5′-AGGCTYGGWTGGCTTATTCG-3′ | Sense |
| Bo4.2sense1 | 5′-TATAGTGTTTTGGGCCGTC-3′ | Sense |
| Bo4.2sense2 | 5′-TCGAATAGACGCTCCCATAT-3′ | Sense |
Figure 4.
Phylogenetic analysis. Neighbor-joining tree (MEGA V.2.1; Kumar et al., 2001) from the multiple alignment (ClustalX V.1.81; Thompson et al., 1997) of the coding cDNAs of the Arabidopsis: AB018695, AB042322, AB049624, AB003591, D85416, D89631, AB004060, AB023423, AB054645, AB061739, AB008782, AB052775, AC018848, and AC006053; and Brassica oleracea sulfate transporter family: AJ416460, AJ311388, AJ633707, AJ633705, AJ581745, AJ601439, AJ704373, AJ704374, AJ633706, AJ416461, AJ555124 (this report), and AJ223495 (Heiss et al., 1999).
There is a close phylogenetic relationship between Arabidopsis and Brassica species, which was evident from the high sequence identities between the Brassica and the homologous Arabidopsis sulfate transporter mRNAs including the subdivision into 4 groups (Fig. 4). To date proton-coupled sulfate transport activity has been demonstrated only for Group 1 and 2 transporters; however, because of the high homology between the members of the family, it is reasonable to expect that most are involved in sulfate transport (Hawkesford, 2000). The close phylogenetic relationship between Arabidopsis and Brassica species, and the high sequence similarities of the sulfate transporters suggest a similar if not identical function of the Brassica sulfate transporter homologs.
Expression of Sulfate Transporters in Sulfur-Sufficient Conditions
The expression pattern in B. oleracea roots indicated that there was no need for expression of all sulfate transporters under sufficient sulfate supply. As in Arabidopsis, the abundant expression of BSultr1;2 (Fig. 5) suggested that the 1;2 transporter is mainly responsible for the initial uptake of sulfate (Takahashi et al., 2000; Shibagaki et al., 2002). The expression of the Group 2 transporters differed from Arabidopsis. In Arabidopsis both Group 2 transporters were expressed in the vascular tissue (Takahashi et al., 2000). In Brassica the sole expression of BSultr2;2 suggested the requirement of only one low affinity transporter for transport in vascular tissue in B. oleracea. The expression of Sultr1;3 in Brassica roots was low, which may be due to companion-cell specificity as was found in Arabidopsis (Yoshimoto et al., 2003).
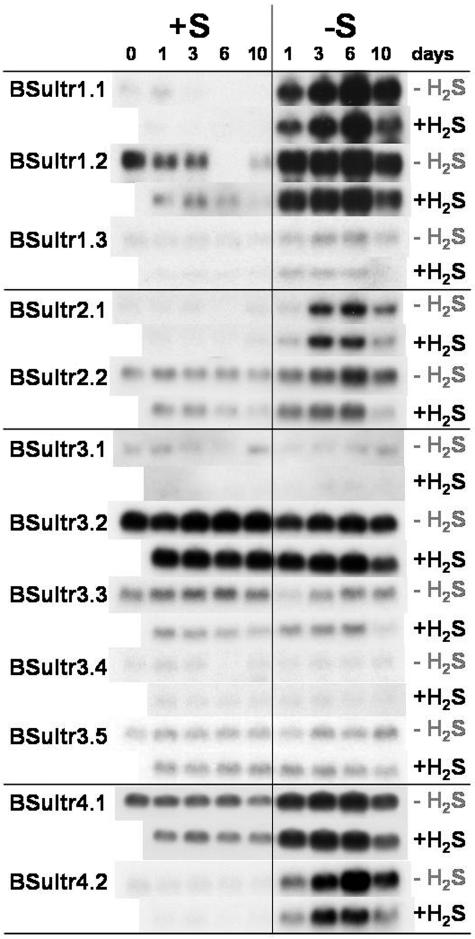
Figure 5.
Expression analysis by northern hybridization of the Brassica sulfate transporter gene family in root tissues of B. oleracea as affected by pedospheric and atmospheric sulfur nutrition. B. oleracea was grown on a 25% Hoagland nutrient solution (0.5 mm sulfate) in a climate controlled room for 7 d and subsequently transferred to a fresh nutrient solution at 0 (−S) or 0.5 mm sulfate (+S) and simultaneously exposed to 0 or 75 nL L−1 H2S for 1, 2, 3, 6, and 10 d.
To our knowledge, this is the first study reporting Group 3 sulfate transporter expression in roots. The highest transcript abundances were seen for BSultr3;2, 3;3, and 3;5 (Fig. 5). In Arabidopsis transcripts of Sultr3;1, 3;2, and 3;3 were detected in leaves but not in roots (Takahashi et al., 2000). Whether the Brassica Group 3 sulfate transporters fulfill a novel function is speculative without any cellular localization data of the different members of Group 3.
There was also a high expression of the Group 4 Sultr4;1 in B. oleracea roots. The expression of a sulfate efflux transporter in the tonoplast, as attributed to this group, would limit accumulation of sulfate in the vacuole. Although the nature of the vacuole influx transporter is not known yet, the expression of a vacuole efflux transporter in roots would favor transport of sulfate in the direction of vascular long distance transport rather than vacuole accumulation for storage.
The stem is the major aerial support system in most plants and provides a pathway for transport between the shoot and the root. The exclusive Sultr2;1 expression in the stem contrasted with the expression of Sultr2;2 in the root (Fig. 6). The role of a vascular sulfate transporter in the stem under sufficient S supply would be to support sulfate distribution from the vascular tissue to the surrounding parenchyma tissue for growth and maintenance compared to the role in the root which would be to take up sulfate for vascular transport. The weak expression of the Group 4 transporter in the stems (Fig. 6) might suggest a similar role as for BSultr4;1 in roots. There was no change in the sulfate content in stems visible under sufficient sulfate indicating reduced or no vacuolar sulfate accumulation. Compared to the roots a change of expression pattern was also found for the Group 3 transporters. In stems, BSultr3;1, 3;3, and 3;4 were the prominent Group 3 transporter transcripts (Fig. 6), which might indicate tissue or organ specific function of the different Group 3 transporters.
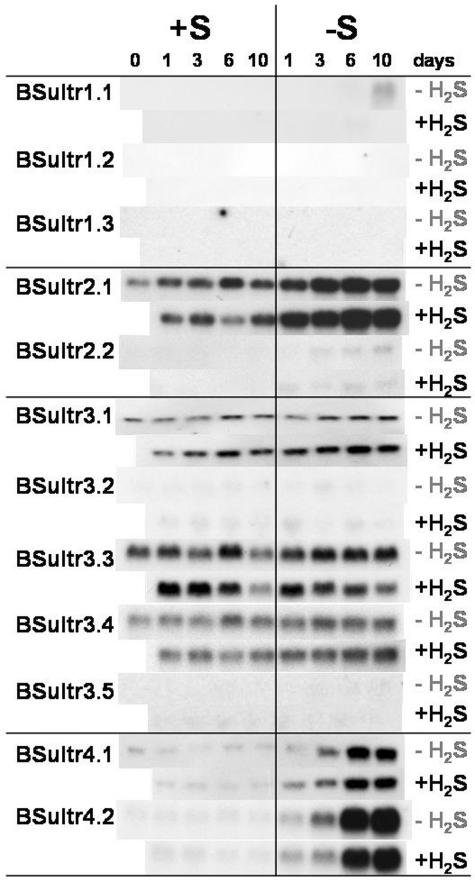
Figure 6.
Expression analysis by northern hybridization of the Brassica sulfate transporter gene family in stem tissues including petioles of B. oleracea as affected by pedospheric and atmospheric sulfur nutrition. For details see legend Figure 5.
Leaves are the major site of sulfate assimilation. If sufficient sulfate was supplied to B. oleracea, sulfate accumulation was found in leaves, which apparently was not needed for assimilation and growth. Apart from the expression of the Group 2 Sultr 2;1, there was only clear expression of the Group 3 transporter, Sultr3;3, and a weak expression of Sultr3;1 in the leaves (Fig. 7). This result would indicate the need for high expression of only three transporters for plasma membrane proton/sulfate cotransport in the entire leaf: BSultr2;1 for the vascular cellular uptake of sulfate delivered via xylem in vascular tissues and BSultr3;3 and 3;1 for cellular uptake of sulfate into nonvascular parenchyma cells. These results differed from the expression pattern found in Arabidopsis leaves, in which expression of both Group 2 transporters as well as of Sultr4,1 and the Group 3 transporters Sultr3;1, 3;2, and 3;3 were detectable by reverse transcription (RT)-PCR (Takahashi et al., 2000).
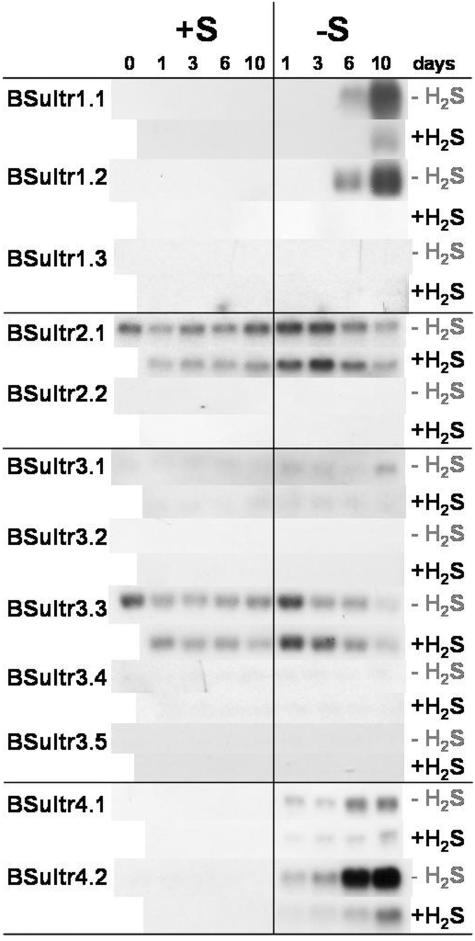
Figure 7.
Expression analysis by northern hybridization of the Brassica sulfate transporter gene family in leaf tissues of B. oleracea as affected by pedospheric and atmospheric sulfur nutrition. For details see legend Figure 5.
Expression of Sulfate Transporters upon Sulfate Deprivation and H2S Exposure
The transfer of the plants to sulfate-deprived conditions resulted in a drastic change of the expression pattern of the Group 1, 2, and 4 sulfate transporters in the roots (Fig. 5). After 1 d of sulfate deprivation an up-regulation and induction, respectively, of expression was found for Group 1 BSultr1;1, 1;2, and both Group 4 transporters. An induction of expression was also noticeable for BSultr1;3 but with a lower intensity than for BSultr1;1 and 1;2 (Fig. 5). The increase of the Group 2 transporter mRNA abundance occurred only at day 3 of deprivation, with a highest peak of transcript abundance for all Group 1, 2, and 4 transporters detected at day 6 of sulfate deprivation. In parallel to the reduction of uptake capacity with further sulfate depletion, all transcript abundances were diminished by day 10 (Fig. 5). H2S exposure did not alter the sulfate deprivation induced coordinated expression of the Group 1, 2, and 4 transporters in roots. A slightly reduced transcript signal was seen for BSultr1;1 and 4;2 under H2S treatment, which did not result in a reduction of the sulfate uptake capacity (Figs. 3 and 5). In contrast, the expression of the Group 3 sulfate transporters in the roots was not influenced by the S status of the plants, either by H2S exposure or by sulfate deprivation (Fig. 5).
A differential expression pattern of the sulfate transporter genes was also detectable in stem tissues (Fig. 6). The Group 1 sulfate transporters were not expressed in the stem except after 10 d of sulfate deprivation, when there was a small induction of BSultr1;1 transporter expression. In contrast to the expression in the roots, the Group 2 BSultr2;1 transporter expression was up-regulated; however, no induction of BSultr2;2 expression occurred in stems of plants by sulfate deprivation (Fig. 6). Sulfate deprivation resulted in an increase of BSultr4;1 and BSultr4;2 transcript with the duration of deprivation (Fig. 6). H2S exposure repressed BSultr1;1 expression, but otherwise had no effect on BSultr4;1 and 4;2 expression (Fig. 6).
In leaves, from day 1 of sulfate deprivation onwards, an induction of both Group 4 transporters occurred and transcript abundance increased with the duration of the sulfate deprivation. In addition, after 6 d of sulfate deprivation, expression of BSultr1;1 and BSultr1;2 was induced (Fig. 7). H2S exposure reduced the expression pattern of the Group 1 and 4 transporters in sulfate-deprived leaves (Fig. 7).
Both in roots and stems, the transcript abundances of the leaf expressed transporters BSultr3;1 and BSultr3;3 were not altered by changing the S status (Fig. 7).
Regulatory Aspects of Expression of Sulfate Transporters
The expression analysis of all of the Brassica sulfate transporters indicated that the sulfate transport system can be split into nutritionally regulated and nonregulated parts. The regulated system is represented by the Group 1, 2, and 4 transporters and the nonregulated system by the Group 3 transporters.
When the sulfate supply was removed, the reduction of the root sulfate concentration was concomitant with an increase of the total root sulfate uptake capacity (Figs. 1 and 3). The increased transporter activity in plasma membrane vesicles isolated from sulfate-deprived roots (Hawkesford et al., 1993) was confirmed by increased sulfate transporter protein found by western blotting (Hawkesford and Wray, 2000). Further studies have shown that the regulation occurs predominantly at the level of transcription (Smith et al., 1995, 1997). This sulfate deprivation related derepression of expression was found for all Group 1, 2, and 3 sulfate transporter genes in the roots of sulfate-deprived B. oleracea (Fig. 5).
Although large increases of both Group 1 sulfate transporter transcripts were observed, the total root sulfate uptake capacity was only approximately doubled during sulfate deprivation. This might be an indication of further posttranscriptional regulation of Group 1 transporters. Although the signal intensities differed drastically, all genes were coordinately controlled. After 1 d of sulfate deprivation, derepression occurred with increasing transcript abundance to day 6, followed by a coordinated reduction of transcript at day 10. This coordinated reduction of expression may be a consequence of alteration of the root structure indicated by the reduced shoot to root ratio in S deprived H2S nonfumigated as well as fumigated plants.
Based on the putative function of the Group 1, 2, and 4 transporters, this derepression under S stress will maximize uptake, vacuole efflux, and vascular transport of sulfate to the growing shoot. Additionally the induction and/or up-regulation of Sultr1;3 and the Group 2 transporters may fulfill a dual function for translocation of sulfate from the phloem to the root derived from upper source organs by remobilization.
Group 3 sulfate transporter expression was not regulated by the S nutritional status of the plant. As there was no expression of Group 3 transporters found in Arabidopsis roots, an involvement of the Group 3 transporters in the initial uptake of sulfate is unlikely. The contrasting expression pattern in B. oleracea roots compared to Arabidopsis suggests a need for further analysis of the Group 3 sulfate transporters to elucidate the importance of this nonregulated system in sulfate transport.
In the upper parts of the plant, the nutritional regulated sulfate transport system followed a dual pattern: an early and a late response on sulfate deficiency. One of the early responses in stems and leaves is the up-regulation in stems, and induction in leaves, of both Group 4 transporters. Increased expression of Group 4 sulfate transporters would favor efflux of sulfate from the vacuoles to maintain the supply of sulfate for assimilation and remobilization of sulfate from leaves for transport to other organs. Up-regulated expression of Sultr2;1 would strengthen the cellular uptake of xylem derived sulfate in the vascular tissue. The export of sulfate by source leaves, especially under sulfate stress, would require changes in the expression of transporters involved in vascular transport especially for phloem loading and/or retransportation into xylem. Interestingly, none of the regulated Arabidopsis genes expressed in the vascular tissue were influenced by sulfate deprivation in the B. oleracea leaves. In Arabidopsis Sultr2;1 expression was down-regulated, but in B. oleracea no change of expression of Sultr2;1 was visible. The expression of AraSultr2;2 and 1;3 was up-regulated in Arabidopsis leaves under sulfate deficiency (Takahashi et al., 2000; Yoshimoto et al., 2003) but in B. oleracea no induction of Sultr2;2 and 1;3 occurred. The function of AraSultr2;2 is thought to strengthen uptake by the bundle sheath cells in order to maintain a supply of sulfate to leaf cells for assimilation (Takahashi et al., 2000). In contrast AraSultr1;3 seems to be involved in phloem export of sulfate from sink organs (Yoshimoto et al. 2003).
In oilseed rape an export of sulfate from older leaves was found after 6 d of sulfate deprivation (Blake-Kalff et al., 1998). The late response on sulfate deficiency was represented in the leaves of B. oleracea by induction and increased expression of Sultr1;1 and 1;2 after 6 d of sulfate deprivation. At this time the sulfate concentration in leaves was reduced to less that 15% of the initial concentration before the onset of deprivation. This may be required to maintain plasmamembrane transport of sulfate when only low concentrations of sulfate are available.
The expression of Group 3 transporters in the stem and leaves suggested a need for the constitutive, nonregulated sulfate transport system. Differences in the expression pattern of the individual Group 3 transporters suggest specific functions in the respective tissues.
Regulatory Aspects of Sulfate Uptake and Transport: Signals and Shoot-Root Interactions
Sulfate uptake and transport is dependent on the S-nutritional status of the plant. In addition activities of enzymes involved in sulfate reduction and assimilation are regulated (Leustek and Saito, 1999). This regulation is embedded in the overall control of S, N, and C assimilation and metabolism. This study showed that sulfate deprivation of B. oleracea seedlings induced multiple physiological responses, with the obvious aim to increase efficiency of sulfate uptake and distribution in the plants. First, sulfate deprivation resulted in a rapid increase in the sulfate uptake capacity of the roots, accompanied with increased expression of sulfate transporters. Second, upon more prolonged sulfate deprivation, there was a change in shoot and root biomass partitioning in favor of root production. When the pedospheric sulfate was replaced by atmospheric H2S as the S source for growth, there was no repression of sulfate uptake capacity or expression of the sulfate transporters in roots and it did not affect the low shoot to root ratio response to sulfate deprivation. Apparently, there was no direct shoot to root signaling in the coordination between the S status of the shoot and the sulfate uptake potential of the roots, via modulation of the sulfate transporter activity and root growth. Atmospheric H2S did however repress leaf sulfate transporter expression.
The up-regulation of the transporters is much reduced in response to S deficiency when N is limiting (Clarkson et al., 1989, 1999). Mimicking N-surplus by supplying external OAS, the C/N precursor skeleton of Cys, under adequate S nutrition, resulted in an induction of high affinity sulfate transporter expression (Smith et al., 1997). It has been proposed also that thiols such as glutathione may act as a regulatory signal to decrease sulfate uptake in the roots (Rennenberg et al., 1989; Lappartient and Touraine, 1996). The generally accepted model involves a combination of a negative feedback by a reduced S-containing compound such as Cys or glutathione and a positive effector OAS (Hawkesford, 2003).
From the present data it is obvious that the shoot to root signaling of the regulation of the sulfate uptake capacity in B. oleracea roots is unlikely to be mediated by the size of the thiol pool (Westerman et al., 2000a). The initial decrease in thiol content of the root and stems including petioles upon sulfate deprivation was relatively low, its content only decreased substantially after 6 d of deprivation. It is more likely that the concentration of sulfate in the root itself has a determining influence as a direct or an indirect signal in the regulation of the sulfate uptake potential, by altering the sulfate uptake capacity and the shoot to root biomass partitioning. Its content decreased rapidly upon sulfate deprivation; however, more information is needed on the intracellular distribution of the sulfate. The major proportion of sulfate may be present in the vacuole and the cytoplasmic concentration would be very low upon deprivation. The possible significance of sulfate as a direct or an indirect signal in the regulation of the sulfate uptake potential, at least in the root, is also supported by the absence of an effect of H2S exposure on the root sulfate uptake capacity and the shoot to root ratio upon sulfate-deprivation. It remains to be resolved to what extent an increase of the OAS content upon S deprivation is a primary (signaling) response or the consequence of a disturbed balance between S and N assimilation. The lack of correlation of thiols, OAS, and sulfate contents to the repression of the sulfate deficiency up-regulated sulfate transporter under H2S treatment in leaves is an indication of a complex regulatory network behind the environmental response, which may be only partly explained from measurements of the related metabolites or nutrient factors. Moreover, the cross-talk and communication in regulation of the different sulfate transporters at a cellular, intracellular and tissue level needs to be investigated in detail.
CONCLUSION
This study confirmed that sulfate deprivation of B. oleracea seedlings induced multiple responses, which may facilitate increased sulfate uptake efficiency. Initially, there was a rapid induction of the sulfate uptake capacity by the roots, accompanied by an expression of genes encoding sulfate transporters in the roots and in the rest of the plant. Second, upon more prolonged sulfate deprivation there was a change in shoot and root biomass partitioning in favor of root production. Indirect evidence suggested that sulfate itself has the potential to be involved in sensing or as a regulatory signal in the roots, while the role of OAS and reduced thiols such as glutathione remains unclear. However, it is evident that in B. oleracea there was a poor shoot to root signaling in the regulation of sulfate uptake and expression of the sulfate transporters.
MATERIALS AND METHODS
Plant Material
Brassica oleracea L. cv Arsis (Royal Sluis, Enkhuizen, The Netherlands) was germinated in vermiculite in a climate-controlled room. Day and night temperatures were 21°C and 17°C, respectively, relative humidity was 60% to 70%, and the photoperiod was 14 h at a photon fluence rate of 250 to 300 μmol m−2 s−1 (within the 400–700 nm range).
Ten-day-old seedlings were transferred to a 25% Hoagland nutrient solution (pH 6) at 0.5 mmol L−1  (+S) and grown for 7 d. Subsequently, plants were transferred to stainless steel containers containing a 25% Hoagland solution at 0 mmol L−1
(+S) and grown for 7 d. Subsequently, plants were transferred to stainless steel containers containing a 25% Hoagland solution at 0 mmol L−1 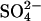 (−S; all
(−S; all 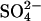 salts were replaced by Cl− salts) or 0.5 mmol L−1
salts were replaced by Cl− salts) or 0.5 mmol L−1 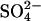 (+S). The containers were sealed with terostat (Henkel Teroson, Heidelberg) in order to prevent absorption of atmospheric H2S into the nutrition solution. Plants were harvested after 0, 1, 2, 3, 6, and 10 d of exposure. Roots were rinsed in ice-cold demineralized water (for 3 × 20 s) and plants were separated into leaves, stem plus petioles, and roots.
(+S). The containers were sealed with terostat (Henkel Teroson, Heidelberg) in order to prevent absorption of atmospheric H2S into the nutrition solution. Plants were harvested after 0, 1, 2, 3, 6, and 10 d of exposure. Roots were rinsed in ice-cold demineralized water (for 3 × 20 s) and plants were separated into leaves, stem plus petioles, and roots.
For analysis of the expression of sulfate transporters, plant material was frozen in liquid N2 and stored at −80°C until further use.
H2S Exposure
Plants were exposed in 150-L cylindrical double-walled stainless steel exposure cabinets (0.6 m diameter) with a polycarbonate top. Day and night temperatures were 20°C and 16°C (±1°C), respectively, relative humidity was 40% to 50%, and the photoperiod was 14 h at a photon fluence rate of 320 to 350 μmol m−2 s−1 at plant height. The air exchange in the cabinets was 40 L min−1 and the air inside was stirred continuously by two ventilators (air exchange capacity of 59 m3 h−1 each) placed on the bottom. Pressurized H2S diluted with N2 (1 mL L−1) was injected into the incoming air stream and adjusted to the desired concentration by AMS electronic mass flow controllers (Bilthoven, The Netherlands). The H2S concentration in the cabinets was controlled by an SO2 analyzer (model 9850) equipped with an H2S converter (model 8770, Monitor Labs, Measurement Controls, Englewood, CO).
Total Sulfur
Analysis of total S content was performed according to a modification of the method of Jones (1995). Plant tissue was dried at 100°C for 24 h, pulverized by a Retsch Mixer-Mill (type MM2; Haan, Germany). Drop by drop a 50% Mg(NO3) solution was added to 100 to 250 mg dry material just to saturate the tissue and the samples were dried in an oven at 100°C overnight. Subsequently, the trays were covered with a lid and the samples were ashed in an oven (Carbolite BWF 11/13, Hope Valley, United Kingdom) at 650°C for 12 h or longer, until the residue had become completely white. The residue was dissolved in 10 mL 20% aqua regia (50 mL concentrated HNO3 and 150 mL concentrated HCL in 1 L distilled water), quantitatively transferred to a measuring flask, and added up to 100 mL with distilled water. To 25 mL extract one SulfaVer 4 Reagent Powder Pillow (HACH, Permachem Reagents, Loveland, CO) containing BaCl2 was added and the turbidity was measured with a spectrophotometer (HACH DR/400V) at 450 nm.
Total Nitrogen
The total N content was determined with a modified Kjeldahl method. Plant tissue was dried at 100°C for 24 h, pulverized by a Retsch Mixer-Mill (type MM2; Haan, Germany), and 50 mg was weighed on a small ash free filter. To the filter 0.33 g Na2S2O3.5H2O and a small spoon of catalyst (K2SO4:CuSO4:Na2SeO3, 15:5:0.085 [w/w/w]) were added; the filter was folded and transferred into a destruction tube. To each tube 2 mL of destruction solution (33.3 g Na salicylate in 1 L H2SO4 [95%–97%]) was added and incubated overnight. The tubes were placed in a Kjeldatherm-Digestion unit (type KB 40 S, Gerhardt, Bonn) coupled with a Variostat temperature-time-programmer (Gerhardt) and the temperature was very slowly increased up to 250°C, then small funnels were placed on top, and then the temperature was further increased to 360°C. At this temperature the samples were boiled for 3 h until the solution was clear and subsequently cooled down overnight. The tubes were heated up to 100°C, the nondestructed particles were washed down from the wall, the temperature was raised to 360°C again, and the samples were boiled for a short time. After cooling down, the samples were quantitatively transferred to 50-mL flasks with demineralized water and made up to 50 mL. NH4+ was determined colorimetrically at 410 nm after reaction with Nessler's reagent.
Sulfate
For measurement of the 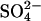 content, frozen material was homogenized in demineralized water at 0°C with an Ultra Turrax for 30 s (10 mL/g fresh weight). The homogenate was filtered over one layer of Miracloth, incubated at 100°C in a boiling water bath for 10 min, and centrifuged at 30 × 103 g for 15 min (0°C). The anions were separated by HPLC on a IonoSpher A anion-exchange column (250 × 4.6 mm; Varian/Chrompack Benelux, Bergen op Zoom, The Netherlands) and the anions were determined refractometrically according to Maas et al. (1986) using a Knauer differential refractometer (model 98.00, Bad Homburg, Germany). As a mobile phase 25 mm potassium biphthalate (pH 4.3) containing 0.02% (w/v) NaN3 was used. The HPLC apparatus consisted of a Separations high precision pump (model 300; H.I. Ambacht, The Netherlands) provided with a Rheodyne sample injector (model 7175; loop volume 20 μL; Cotati, CA). The flow rate was 1 mL min−1 and the detector temperature was kept at 25°C by a water bath. Peak analysis was performed with a Shimadzu Chromatopac C-R8A data processor (Kyoto).
content, frozen material was homogenized in demineralized water at 0°C with an Ultra Turrax for 30 s (10 mL/g fresh weight). The homogenate was filtered over one layer of Miracloth, incubated at 100°C in a boiling water bath for 10 min, and centrifuged at 30 × 103 g for 15 min (0°C). The anions were separated by HPLC on a IonoSpher A anion-exchange column (250 × 4.6 mm; Varian/Chrompack Benelux, Bergen op Zoom, The Netherlands) and the anions were determined refractometrically according to Maas et al. (1986) using a Knauer differential refractometer (model 98.00, Bad Homburg, Germany). As a mobile phase 25 mm potassium biphthalate (pH 4.3) containing 0.02% (w/v) NaN3 was used. The HPLC apparatus consisted of a Separations high precision pump (model 300; H.I. Ambacht, The Netherlands) provided with a Rheodyne sample injector (model 7175; loop volume 20 μL; Cotati, CA). The flow rate was 1 mL min−1 and the detector temperature was kept at 25°C by a water bath. Peak analysis was performed with a Shimadzu Chromatopac C-R8A data processor (Kyoto).
Total Water-Soluble Nonprotein Thiols
Plant tissue was homogenized in 80 mmol L−1 sulfosalicylic acid, 1 mmol L−1 EDTA, and 0.15% (w/v) ascorbic acid with an Ultra Turrax (Polytron pt 3000, Kinematica AG, Littau, Switzerland) at 0°C (1 g fresh weight in 10 mL). Oxygen was removed from the solution by N2 bubbling. The homogenate was filtered through one layer of Miracloth (Calbiochem Corporation, LaJolla, CA) and the filtrate was centrifuged at 30 × 103 g for 15 min (0°C). Total water-soluble nonprotein thiol content was determined colorimetrically at 412 nm after reaction with 5,5′-dithiobis[2-nitrobenzoic acid] according to De Kok et al. (1988).
OAS
Plant tissue was freeze-dried in a Heto LyoLaB 3000 freeze dryer (Heto-Holten A/S, Allerød, Denmark) and 100 mg tissue material was ground in liquid N2 to a fine powder and extracted with 1 mL 0.1 m HCl for 15 min at 4°C while shaking. Homogenates were centrifuged twice at 15 400g, 4°C for 5 min. The resulting supernatants were used for further analysis. Determination of OAS was based upon derivatization with the fluorescence dye AccQ-Tag (Waters, Milford, MA). An appropriate volume of supernatant (5–20 μL) was derivatized according to the manufacturer (Manual WAT052874TP). The resulting OAS derivative was separated from other amino acid derivatives by reverse phase HPLC as described in Wirtz et al., 2004, with the exception that the pH of buffer A was optimized to pH 6.3. Data acquisition and processing were performed with Millenium32 software (Waters). Recovery rates for OAS were higher than 90% in all tested tissues as determined by spiking of samples with internal standards.
Sulfate Uptake Capacity
For measurement of the 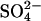 uptake capacity three sets of plants (three plants per set) per treatment were transferred to beakers with 250 mL 25% Hoagland solution at 0.5 mmol L−1
uptake capacity three sets of plants (three plants per set) per treatment were transferred to beakers with 250 mL 25% Hoagland solution at 0.5 mmol L−1 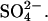 At the beginning of the uptake measurements and after 24 h, the nutrient solution was adjusted to the original volume by weighing. Aliquots were taken from the nutrient solution and the
At the beginning of the uptake measurements and after 24 h, the nutrient solution was adjusted to the original volume by weighing. Aliquots were taken from the nutrient solution and the 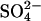 contents were analyzed by HPLC as described above. The
contents were analyzed by HPLC as described above. The 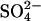 uptake capacity was calculated according to Westerman et al. (2001a).
uptake capacity was calculated according to Westerman et al. (2001a).
Total RNA Isolation
Total RNA from roots, stems, and leaves was isolated by a method based on Verwoerd et al. (1989), which involved an additional phenol-chloroform-isoamyl alcohol extraction of the aqueous phase after the first centrifugation. The final air dried pellet was dissolved in an appropriate volume of diethyl pyrocarbonate treated water. The quality of the RNA preparations was checked by electrophoresis of a 1-μg aliquot on a Tris-acetate/agarose gel. The concentration was calculated from the A260 in water.
cDNA Isolation, Sequencing, and Sequence Analysis
Partial cDNAs corresponding to the 11 sulfate transporter genes were isolated by RT-PCR from RNA from Brassica napus and B. oleracea. Degenerate primers were designed based on highly homologous regions identified in sequence alignments of published sulfate transporter genes of the different sulfate transporter subfamilies 1 to 4 (Table IV). For the Group 1 BSultr 1;3, Group 3 BSultr 3;2, 3;3, 3;4, 3;5, and the Group 4 BSultr 4;2 sulfate transporter partial gene sequence information was obtained via BLAST search using Arabidopsis sulfate transporter sequence information (data not shown) on The Brassica oleracea Genome Database at the Institute for Genomic Research in Rockville, Maryland. Oligonucleotide primers were generated (Table IV) to isolate the corresponding cDNAs via 3′-RACE approach.
First strand cDNA synthesis was performed according the Invitrogene Superscript II Reverse Transcriptase manual with 1-μg aliquots of total root and leaf RNA and antisense primer. Following PCR was performed according the Promega Taq-DNA polymerase standard protocol for 50 μL reaction by using a 1-μL aliquot of the each first strand cDNA solutions and specific sense/antisense primer combinations. 5′- and 3′-region of the sulfate transporter transcripts were isolated by 5′- and 3′-RACE approach according the Invitrogene 5′-RACE and 3′-RACE kit manual and sequence specific oligonucleotide primers based on the sequence results from the partial cDNA fragment isolation (data not shown). Finally the full-length cDNA containing the full coding region were generated via RT-PCR and sequence specific primer obtained from the 5′- and 3′-RACE fragments (data not shown) using proofreading Pfu-DNA polymerase in the PCR reaction. All PCR fragments derived from RT-PCR or RACE were verified by electrophoresis, gel-eluted according the Qiagen QIAquick gel extraction kit manual, ligated into the Promega pGEM-Teasy vector system, and sequenced in both directions.
DNA sequences were determined by PCR based the dideoxy method on an Applied Biosystems 377 DNA sequencer or a Perkin Elmer ABI Prism 310 genetic analyzer. ClustalX V.1.81 (Thompson et al., 1997) was used for multiple sequence alignment of the coding cDNA sequences.
Northern Analysis
Northern hybridization of the described Brassica sulfate transporter transcripts was performed according Church and Gilbert (1984), with prehybridization/hybridization at 65°C. Ten micrograms of total RNA per slot was separated on an agarose/formaldehyde gel and blotted onto positively charged nylon membrane. Sequence diversity especially in the 3′ noncoding region allowed the use of partial cDNA fragments for gene specific hybridization to the respective Brassica sulfate transporter mRNA. cDNA fragments were labeled with 32P-dCTP and used as hybridization probes. For hybridization of BST2.2 a partial cDNA fragment was provided by Prof. T. Rausch, Institute of Botany, University of Heidelberg (Heiss et al., 1999). After hybridization the membrane was washed under high-stringency conditions and exposed to Kodak BioMax MS film.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ416460, AJ311388, AJ633707, AJ633705, AJ581745; AJ601439, AJ704373, AJ704374, AJ633706, AJ416461, AJ555124.
This work was supported by the Biotechnology and Biological Science Research Council (BBSRC), UK.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046441.
References
- Blake-Kalff MA, Harrison KR, Hawkesford MJ, Zhao FJ, McGrath SP (1998) Distribution of sulfur within oilseed rape leaves in response to sulfur deficiency during vegetative growth. Plant Physiol 118: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A, Stulen I, De Kok LJ (2003) Nitrogen and sulfur requirement of Brassica oleracea L. cultivars. In J-C Davidian, D Grill, LJ de Kok, I Stulen, MJ Hawkesford, E Schnug, H Rennenberg, eds, Sulfur Transport and Assimilation in Plants. Backhuys Publishers, Leiden, The Netherlands, pp 181–183
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson DT, Saker LR, Purves JV (1989) Depression of nitrate and ammonium transport in barley plants with diminished sulphate status. Evidence for co-regulation of nitrogen and sulphate uptake. J Exp Bot 40: 953–963 [Google Scholar]
- Clarkson DT, Hawkesford MJ, Davidian J-C (1993) Membrane and long-distance transport of sulfate. In LJ De Kok, I Stulen, H Rennenberg, C Brunold, WE Rauser, eds, Sulfur Nutrition and Sulfur Assimilation in Higher Plants: Fundamental, Environmental and Agricultural Aspects. SPB Academic Publishing, The Hague, pp 3–19
- Clarkson DT, Diogo E, Amancio S (1999) Uptake and assimilation of sulphate by sulphur deficient Zea mays cells: The role of O-acetyl-L-serine in the interaction between nitrogen and sulphur assimilatory pathways. Plant Physiol Biochem 37: 283–290 [Google Scholar]
- Cram WJ (1990) Uptake and transport of sulfate. In H Rennenberg, C Brunold, LJ De Kok, I Stulen, eds, Sulfur Nutrition and Sulfur Assimilation in Higher Plants: Fundamental, Environmental and Agricultural Aspects. SPB Academic Publishing, The Hague, pp 3–11
- De Kok LJ (1990) Sulfur metabolism in plants exposed to atmospheric sulfur. In H Rennenberg, C Brunold, LJ De Kok, I Stulen, eds, Sulfur Nutrition and Sulfur Assimilation in Higher Plants; Fundamental, Environmental and Agricultural Aspects. SPB Academic Publishing, The Hague, pp 111–130
- De Kok LJ, Tausz M (2001) The role of glutathione in plant reaction and adaptation to air pollutants. In D Grill, M Tausz, LJ De Kok, eds, Significance of Glutathione to Plant Adaptation to the Environment. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 185–201
- De Kok LJ, Buwalda F, Bosma W (1988) Determination of cysteine and its accumulation in spinach leaf tissue upon exposure to excess sulfur. J Plant Physiol 133: 502–505 [Google Scholar]
- De Kok LJ, Stuiver CEE, Rubinigg M, Westerman S, Grill D (1997) Impact of atmospheric sulfur deposition on sulfur metabolism in plants: H2S as sulfur source for sulfur deprived Brassica oleracea L. Bot Acta 110: 411–419 [Google Scholar]
- De Kok LJ, Stuiver CEE, Stulen I (1998) The impact of elevated levels of atmospheric H2S on plants. In LJ De Kok, I Stulen, eds, Responses of Plant Metabolism to Air Pollution and Global Change. Backhuys Publishers, Leiden, pp 51–63
- De Kok LJ, Castro A, Durenkamp M, Stuiver CEE, Westerman S, Yang L, Stulen I (2002) Sulphur in plant physiology. In Proceedings No. 500. The International Fertiliser Society, York, UK, pp 1–26
- De Kok LJ, Westerman S, Stuiver CEE, Stulen I (2000) Atmospheric H2S as plant sulfur source: interaction with pedospheric sulfur nutrition—a case study with Brassica oleracea L. In C Brunold, H Rennenberg, LJ De Kok, I Stulen, J-C Davidian, eds, Sulfur Nutrition and Sulfur assimilation in Higher Plants: Molecular, Biochemical and Physiological Aspects. Paul Haupt, Bern, pp 41–55
- Hawkesford MJ (2000) Plant response to sulphur deficiency and the genetic manipulation of sulphate transporters to improve S-utilisation efficiency. J Exp Bot 51: 131–138 [PubMed] [Google Scholar]
- Hawkesford MJ (2003) Transporter gene families in plants: the sulfate transporter gene family– redundancy or specialization? Physiol Plant 117: 155–163 [Google Scholar]
- Hawkesford MJ, Wray JL (2000) Molecular genetics of sulphur assimilation. Adv Bot Res 33: 159–223 [Google Scholar]
- Hawkesford MJ, Smith FW (1997) Molecular biology of higher plant sulphate transporters. In WJ Cram, LJ De Kok, I Stulen, C Brunold, H Rennenberg, eds, Sulphur Metabolism in Higher Plants, Backhuys Publishers, Leiden, pp 13–25
- Hawkesford MJ, Davidian J-C, Grignon C (1993) Sulfate proton cotransport in plasma vesicles isolated from roots of Brassica napus L. Increased transport in membranes isolated from sulphur deprived plants. Planta 190: 297–304 [Google Scholar]
- Heiss S, Schafer HJ, Haag-Kerwer A, Rausch T (1999) Cloning sulfur assimilation genes from Brassica juncea L.: cadmium differentially affects the expression of a putative low-affinity sulfate transporter and isoforms of ATP sulfurylase and APS reductase. Plant Mol Biol 39: 847–857 [DOI] [PubMed] [Google Scholar]
- Hell R, Jost R, Berkowitz O, Wirtz M (2002) Molecular and biochemical analysis of the enzymes of cysteine biosynthesis in the plant Arabidopsis thaliana. Amino Acids 22: 245–257 [DOI] [PubMed] [Google Scholar]
- Howarth J, Fourcroy P, Davidian J-C, Smith FW, Hawkesford MJ (2003) Cloning of two contrasting high-affinity sulphate transporters from tomato induced by low sulphate and infection by the vascular pathogen Verticillium dahlia. Planta 218: 58–64 [DOI] [PubMed] [Google Scholar]
- Jones JB Jr (1995) Determining total sulphur in plant tissue using the HACH kit spectrophotometer technique. Sulphur Agric 19: 58–62 [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashia H (2004) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell, in press [DOI] [PMC free article] [PubMed]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245 [DOI] [PubMed] [Google Scholar]
- Lappartient AG, Touraine B (1996) Demand-driven control of root ATP sulfurylase activity and sulfate uptake in intact canola. Plant Physiol 111: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Saito K (1999) Sulfate transport and assimilation in plants. Plant Physiol 120: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas FM, Hoffmann I, Van Harmelen MJ, De Kok LJ (1986) Refractometric determination of sulfate and other anions in plants separated by High Performance Liquid Chromatography. Plant Soil 91: 129–132 [Google Scholar]
- Rennenberg H, Kemper O, Thoene B (1989) Recovery of sulfate transport into heterotrophic tobacco cells from inhibition by reduced glutathione. Physiol Plant 76: 271–276 [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP (2002) Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J 29: 475–486 [DOI] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT (1995) Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Hawkesford MJ, Ealing PM, Clarkson DT, van den Berg PJ, Belcher AR, Warrilow AGS (1997) Regulation of expression of a cDNA from barley roots encoding a high affinity sulphate transporter. Plant J 12: 875–884 [DOI] [PubMed] [Google Scholar]
- Stuiver CEE, De Kok LJ, Westerman S (1997) Sulfur deficiency in Brassica oleracea L.: Development, biochemical characterization, and sulfur/nitrogen interactions. Russ J Plant Physiol 44: 505–513 [Google Scholar]
- Stuiver CEE, De Kok LJ (2001) Atmospheric H2S as sulfur source for plant growth: Kinetics of H2S uptake and activity of O-acetylserine(thiol)lyase as affected by sulfur nutrition. Environ Exp Bot 46: 29–36 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Sasakura N, Noji M, Saito K (1996) Isolation and characterization of a cDNA encoding a sulfate transporter from Arabidopsis thaliana. FEBS Lett 392: 95–99 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, de Almeida-Engler J, Engler G, van Montagu M, Saito K (1997) Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate deprived roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Asanuma W, Saito K (1999. a) Cloning of an Arabidopsis cDNA encoding a chloroplast localizing sulphate transporter isoform. J Exp Bot 50: 1713–1714 [Google Scholar]
- Takahashi H, Sasakura N, Kimura A, Watanabe A, Saito K (1999. b) Identification of two leaf-specific sulfate transporters in Arabidopsis thaliana (Accession, AB012048 and AB004060). (Pgr.99-154). Plant Physiol 121: 686 [Google Scholar]
- Takahashi H, Watanabe-Takahasi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23: 171–182 [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TT, Dekker BMM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman S, De Kok LJ, Stulen I (2000. a) Interaction between metabolism of atmospheric H2S in the shoot and sulfate uptake by the roots of curly kale (Brassica oleracea L.). Physiol Plant 109: 443–449 [Google Scholar]
- Westerman S, Weidner W, De Kok LJ, Stulen I (2000. b) Effect of H2S exposure on 35S-sulfate uptake, transport and utilization in curly kale. Phyton 40: 293–302 [Google Scholar]
- Westerman S, Blake-Kalff MMA, De Kok LJ, Stuiver CEE, Stulen I (2001. a) Sulfate uptake and utilization by two varieties of Brassica oleracea with different sulfur need as affected by atmospheric H2S. Phyton 41: 49–62 [Google Scholar]
- Westerman S, Stulen I, Suter M, Brunold C, De Kok LJ (2001. b) Atmospheric H2S as sulfur source for Brassica oleracea: consequences for the activity of the enzymes of the assimilatory sulfate reduction pathway. Plant Physiol Biochem 39: 425–432 [Google Scholar]
- Wirtz M, Droux M, Hell R (2004) O-acetylserine (thiol) lyase – an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot 55: 1785–1798 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H (2003) Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol 131: 1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K (2002) Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J 29: 465–473 [DOI] [PubMed] [Google Scholar]