Abstract
Sleep is a highly conserved and essential behaviour in many species, including the fruit fly Drosophila melanogaster. In the wild, sensory signalling encoding environmental information must be integrated with sleep drive to ensure that sleep is not initiated during detrimental conditions. However, the molecular and circuit mechanisms by which sleep timing is modulated by the environment are unclear. Here we introduce a novel behavioural paradigm to study this issue. We show that in male fruit flies, onset of the daytime siesta is delayed by ambient temperatures above 29 °C. We term this effect Prolonged Morning Wakefulness (PMW). We show that signalling through the TrpA1 thermo-sensor is required for PMW, and that TrpA1 specifically impacts siesta onset, but not night sleep onset, in response to elevated temperatures. We identify two critical TrpA1-expressing circuits and show that both contact DN1p clock neurons, the output of which is also required for PMW. Finally, we identify the circadian blue-light photoreceptor CRYPTOCHROME as a molecular regulator of PMW, and propose a model in which the Drosophila nervous system integrates information encoding temperature, light, and time to dynamically control when sleep is initiated. Our results provide a platform to investigate how environmental inputs co-ordinately regulate sleep plasticity.
Sleep is controlled by circadian and homeostatic mechanisms1, and is observed in organisms as divergent as humans and insects2. Such deep conservation suggests a fundamental requirement for sleep in maintaining organismal fitness. Indeed, recent work has demonstrated a key role for sleep in regulating several aspects of nervous system function in vertebrates and invertebrates, including synaptic plasticity, neuronal development and metabolite clearance3,4,5,6.
The quiescent and high arousal-threshold state that is the primary characteristic of sleep has obvious fitness costs in an environment containing predators and a limited supply of food and mates. Thus, the timing of sleep must be tightly regulated by an array of sensory modalities to match sleep onset to current external conditions. In other words, sleep must be a plastic phenotype that is sensitive to environmental alterations. However, the mechanisms by which ethologically-relevant environmental cues modulate sleep timing are unclear. We have used the fruit fly, Drosophila melanogaster, to address this issue.
Drosophila has emerged as an important model system for investigating genes and circuits that influence the levels, timing, and homeostatic regulation of sleep7,8,9,10,11,12,13,14, as well as its neurobiological functions3,4,5,15. Drosophila exhibits a daytime siesta and long bouts of consolidated sleep during the night. Components of this sleep pattern are sexually dimorphic, with female flies exhibiting reduced siesta sleep relative to males7. As poikilotherms, Drosophila physiology is temperature-sensitive, and fruit flies possess an array of thermo-sensory signalling pathways that facilitate adaptive behavioral responses to the surrounding ambient temperature, enabling Drosophila to sense attractive or noxious temperatures and initiate appropriate locomotor programs16,17. Recent work has also shown that increasing ambient temperature modifies sleep architecture18,19,20. However, the thermo-sensory molecules and circuits that transmit temperature information to sleep regulatory centers remain unclear.
We performed an in-depth analysis of how sleep in Drosophila is modified by changes in ambient temperature. Interestingly, we found that the male siesta exhibits a complex response to temperature increases, with the onset advanced during mild increases and delayed by further thermal increases (≥30 °C). We term this delay in siesta onset due to elevated temperature Prolonged Morning Wakefulness (PMW). Through mutational analysis and circuit mapping approaches, we identify the thermo-receptor, thermo-sensory circuits and downstream sleep-regulatory neurons involved in PMW. We show that temperature increases are encoded by two distinct sub-circuits expressing the TrpA1 thermo-sensor, both of which contact DN1p clock neurons. These, in turn, promote wakefulness during hot mornings. We also demonstrate that loss of the circadian photoreceptor CRYPTOCHROME suppresses PMW. Our work suggests that the Drosophila nervous system integrates light, temperature, and temporal information to drive changes in daytime sleep architecture.
Results
Increased ambient temperature alters sleep architecture in Drosophila
To study how sleep architecture dynamically adapts to environmental changes, we measured sleep levels and timing in adult male flies during two consecutive days. On Day 1 of the experimental paradigm, flies were housed at 22 °C, in 12 h light: 12 h dark (LD) conditions. On Day 2, flies were exposed to a range of warm temperatures (27–31 °C) for 24 h, beginning at Zeitgeber Time 0 (ZT0, lights-on). In these and all subsequent experiments, sleep was defined as 5 min of inactivity, as measured by the Drosophila Activity Monitoring (DAM) system, a well-described standard in the field21.
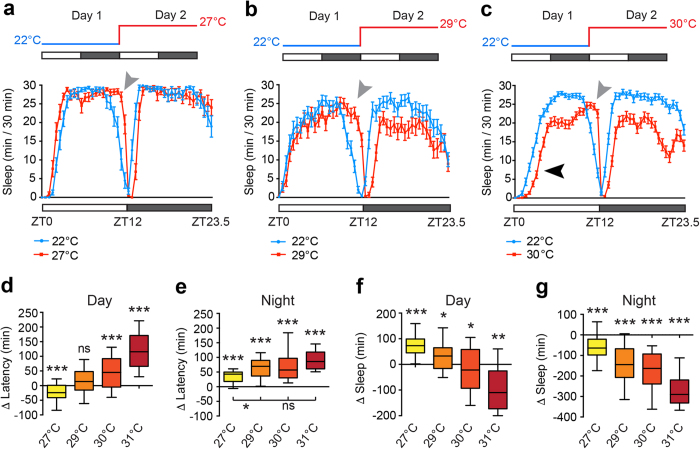
Temperature increases resulted in complex changes to the architecture of siesta sleep. Shifting male flies from 22 °C to either 27 °C or 29 °C prolonged siesta sleep towards lights-off (ZT12), yielding a net increase in siesta sleep at 27 °C and 29 °C compared to 22 °C (Fig. 1a,b,f). This may be due to a delay in the initiation of locomotor increases before lights-off (evening anticipation; Fig. S1), an output of the circadian clock previously shown to be temperature-sensitive22,23. At 27 °C, siesta onset was slightly advanced relative to 22 °C (Fig. 1a,d), quantified as the change in latency to initiate sleep between cold and hot days (Δ Latency). However, further increases in temperature shifted siesta onset to later time periods. In particular, at ≥30 °C we observed a robust delay in siesta onset that was not observed at 29 °C (Fig. 1b–d), contributing to a net reduction in siesta sleep at ≥30 °C (Fig. 1f). With respect to nighttime sleep, we found that heightened ambient temperatures induced a delay in night sleep onset (Fig. 1a–c,e). In addition, we observed a roughly linear decrease in night sleep levels in response to increasing temperature levels (Fig. 1g). Thus, temperature increases differentially affect the onset of day versus night sleep, with siesta sleep onset both advanced and delayed by temperature increases within a relatively narrow range, and night sleep onset consistently delayed. To our knowledge, the response of siesta onset to elevated temperatures has not previously been characterized, and for simplicity, we refer to the temperature-induced delay in siesta onset as PMW - Prolonged Morning Wakefulness.
Figure 1. Warm temperatures prolong morning wakefulness in male Drosophila.
(a–c) Average sleep patterns of adult male flies shifted from 22 °C to either 27 °C (n = 20) (a), 29 °C (n = 32) (b) or 30 °C (n = 69) (c). Sleep traces are presented as mean ± SEM for each time point in these and all subsequent figures. Temperature-shift paradigms are indicated above. Sleep was measured under 12 h light: 12 h dark conditions (white/grey bars) with Zeitgeber Times (ZT) shown below. Black arrowhead indicates the delay of the sleep onset observed at 30 °C (PMW). Grey arrowheads indicate the delay of sleep offset induced at 27 °C or above. (d,e) Change in time taken to the first day sleep episode (d) or night sleep episode (e) (Δ Latency) between consecutive 24 h periods at 22 °C and 27–31 °C. (f,g) Difference in total sleep during the day (f) or night (g) between consecutive 24 h periods at 22 °C and 27–31 °C. n-values: 27 °C, n = 39; 29 °C, n = 32; 30 °C, n = 79; 31 °C, n = 18. In this and all subsequent figures, box plots show the 10th, 25th, median, 75th and 90th percentiles, and p-values are indicated as follows: *p < 0.05, **p < 0.005, ***p < 0.0005, ns – p > 0.05. Statistical comparisons: (d–g) Wilcoxon signed rank test compared to a theoretical median of zero and (e) Kruskal-Wallis test with Dunn’s post-hoc test.
We also tested for the presence of PMW in adult female Drosophila. At 22 °C, female Drosophila initiate the siesta later during the day compared to males (Fig. S1), and we did not observe PMW in females when shifted from 22 °C to 30 °C (Fig. S1). Thus, PMW is sexually dimorphic at 30 °C and correlates with a siesta onset occurring earlier in males than in females at 22 °C.
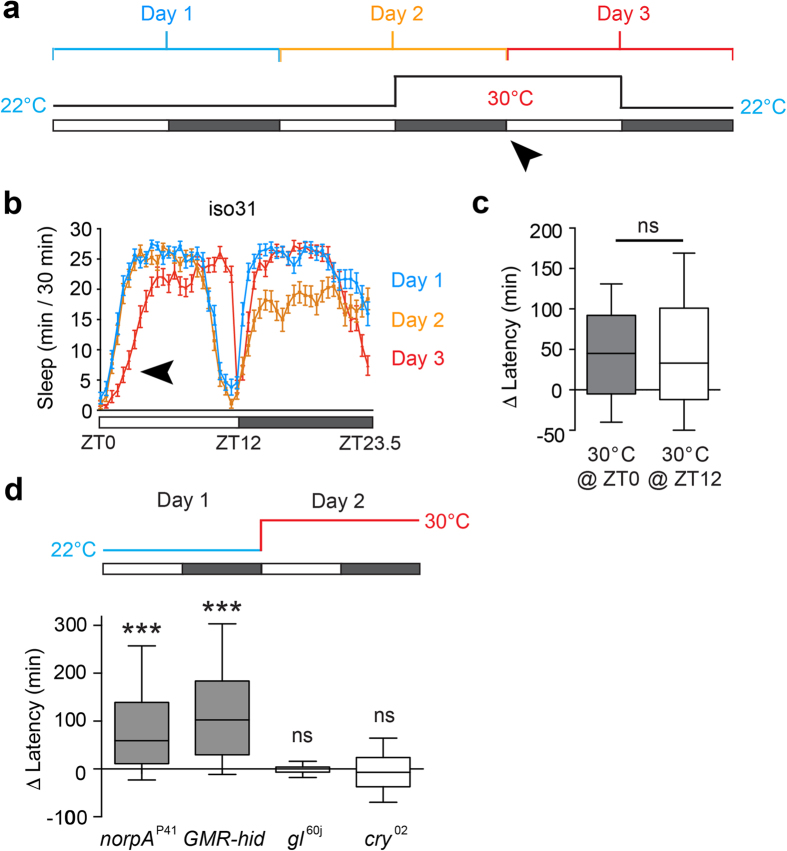
Does PMW simply represent an acute avoidance response to rapidly increased ambient temperature? To rule out this possibility, we shifted male flies from 22 °C to 30 °C at ZT12 rather than ZT0 and measured sleep the following morning, after 12 h at elevated temperature (Fig. 2a). Indeed, under these conditions we still observed robust PMW, and the magnitude of PMW was equivalent to that caused by a shift from 22 °C to 30 °C at ZT0 (Fig. 2b,c). Thus, PMW is not solely a reaction to a rapid environmental change, but is a behavioral response linked to high temperatures during the morning.
Figure 2. PMW is CRYPTOCHROME- and GLASS-dependent.
(a) Three-day temperature-shift paradigm to test whether PMW is an acute avoidance response. Ambient temperature is raised from 22 °C to 30 °C at ZT12 on Day 2 for 24 h. Sleep latency was subsequently measured on Day 3 (black arrowhead). (b) Average sleep patterns of control adult male flies during the above temperature-shift paradigm. Subsequent days are juxtaposed to allow direct comparison. Day 1: blue, Day 2: orange, Day 3: red. Black arrowhead indicates PMW during Day 3. n = 43. (c) Comparison of the PMW when ambient temperature is increased at either ZT0 on the experimental day, or at ZT12 – the beginning of the previous night. ns – p > 0.05, Mann-Whitney U-test. ZT0: n = 79 (data also presented in Fig. 1); ZT12: n = 45. (d) PMW in light-pathway mutants. Statistical comparison: Wilcoxon signed rank test compared to a theoretical median of zero. norpAP41: n = 31, GMR-hid: n = 38, gl60j: n = 44, cry02: n = 58.
PMW is GLASS- and CRYPTOCHROME-dependent
Since PMW occurs shortly after lights-on, we tested whether PMW could be modified by mutations that impact light-sensing pathways, the circadian clock, or both. We examined PMW in three photoreceptor mutants where signaling through the compound eye is abolished. norpAP41 is a loss of function allele in the phospholipase C-β-encoding gene norpA, a critical component in the canonical light transduction pathway24,25; GMR-hid flies express the pro-apoptotic gene hid in all photoreceptor cells26; and gl60j mutants are developmentally blind due to loss of GLASS, a transcription factor required for photoreceptor cell development27. Surprisingly, PMW was still observed in norpAP41 and GMR-hid males, yet was suppressed in gl60j homozygotes (Fig. 2d). GLASS is also required for the development of a subset of clock cells termed DN1p neurons26,28, providing a possible explanation for this discrepancy (see below). Interestingly, we also found that PMW was suppressed by loss of CRYPTOCHROME (CRY), a circadian blue-light photoreceptor29 (Fig. 2d; see Discussion). Finally, to test for a direct role of the clock in gating PMW, we generated a new timeless (tim) null allele by replacing the tim coding sequence with a mini-white+ reporter gene using homologous recombination (timko; see Materials and Methods). TIM is an essential component of the negative arm of the circadian transcription-translation feedback loop30. We confirmed that clock-driven morning and evening anticipation are lost in timko homozygote males (Fig. S2), and that timko homozygotes are arrhythmic in constant-dark conditions (data not shown). PMW was reduced in timko homozygotes, but not fully suppressed (Fig. S2). These results suggest that light- and temperature information collectively drive PMW, with the light-sensing pathway involving the CRY photoreceptor. Furthermore, the circadian clock may play a modulatory role in this process.
PMW requires the TrpA1 thermo-receptor
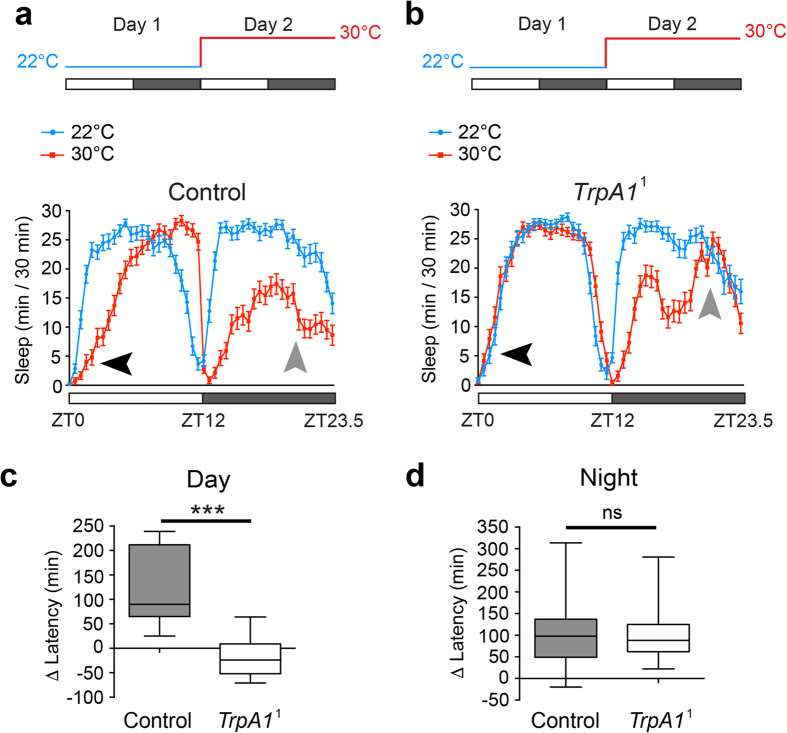
What molecular pathways signal sleep-relevant temperature information to regulate siesta onset? The Drosophila genome encodes several thermo-sensory proteins31. Of these, TrpA1, a cation-conducting channel active at 30 °C, is required for temperature-induced changes in the phase of morning and evening anticipation23. We therefore tested whether TrpA1 also impacted PMW. Indeed, a loss of function mutation in TrpA1 (TrpA11) suppressed PMW, while PMW was still robustly observed in a paired genetic control (Fig. 3a–c). These results were confirmed using a previously validated TrpA1-RNAi32 transgene to knock down TrpA1 expression throughout the Drosophila nervous system using the pan-neuronal driver elav-GAL4. In the TrpA1 knockdown background, PMW was suppressed (Fig. S3), similarly to TrpA11 homozygotes (Fig. 3a–c). In contrast, null or strongly hypomorphic mutations affecting the Gr28b33 and Pyrexia34,35 thermo-receptors did not suppress PMW (Fig. S3). From the above data, we conclude that TrpA1 is the critical thermo-sensor that mediates PMW.
Figure 3. The TrpA1 thermo-sensor is required for PMW.
(a,b) Average sleep patterns of adult male control or TrpA11 homozygotes shifted from 22 °C to 30 °C at ZT0. Temperature-shift paradigms are indicated above. Black arrowheads: presence/absence of PMW. Grey arrowheads: presence/absence of enhanced wakefulness prior to lights-on during a warm night. (c,d) Comparison of change in latency to the first sleep episode between control and TrpA11 homozygote males during the day (c) and night (d) following a shift from 22 °C to 30 °C. Statistical comparison: Mann-Whitney U-test. Control: n = 43; TrpA11: n = 62.
As shown previously23, loss of TrpA1 also suppressed temperature-induced changes in morning and evening anticipation (Fig. 3a,b), but had no effect on temperature-induced delay of sleep onset during the night (Fig. 3d). Furthermore, sleep loss at 30 °C was still strongly observed in the early-middle of the night in TrpA11 mutants (Fig. 3b). Thus, temperature-dependent modulation of day sleep, but not night sleep, appears strongly dependent on TrpA1.
Two populations of TrpA1-expressing neurons are necessary for PMW
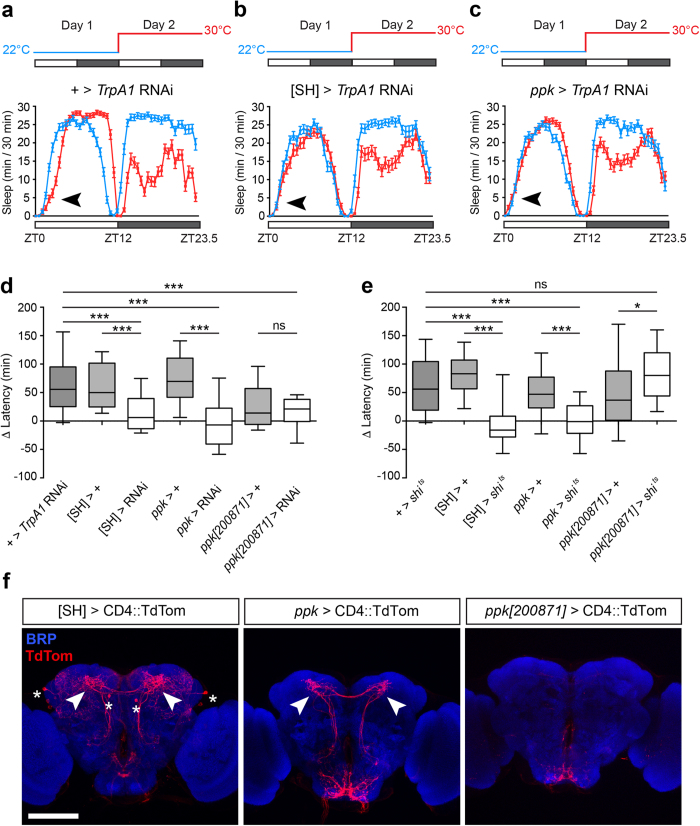
We next sought to identify subpopulations of TrpA1-expressing neurons that transduce thermo-sensory information to drive PMW. Recent work has shown that a group of TrpA1-expressing neurons defined by the TrpA1[SH]-GAL4 driver modulates the phase of morning anticipation in response to temperature changes23. To test if these neurons also play a role in PMW, we used TrpA1-RNAi to knockdown TrpA1 in TrpA1[SH]-neurons. Indeed, PMW was reduced when TrpA1 expression was inhibited in TrpA1[SH]-neurons (Fig. 4a,b,d). To test whether additional TrpA1-expressing neurons also influenced PMW we screened several driver lines shown, or predicted, to label TrpA1-positive neurons. From this mini-screen, we found that expression of TrpA1-RNAi using pickpocket-GAL4 (ppk-GAL4), also robustly suppressed PMW (Fig. 4a,c,d). We further found that acute inhibition of TrpA1[SH]- and ppk-neuron output using temperature-sensitive dominant-negative shibire (UAS-shits), which blocks endocytosis of synaptic vesicles at 30 °C but not 22 °C36, also suppressed PMW (Fig. 4e). However, neither TrpA1 knockdown nor inhibition of synaptic transmission in TrpA1[SH]- and ppk-neurons suppressed temperature-induced delays in nighttime sleep onset (Fig. S4). Thus, TrpA1 expression in, and neurotransmitter release from, TrpA1[SH]- and ppk-neurons are required for PMW, and these circuits primarily impact daytime, as opposed to nighttime, sleep onset.
Figure 4. TrpA1-expressing TrpA1[SH]- and ppk-neurons are necessary for PMW.
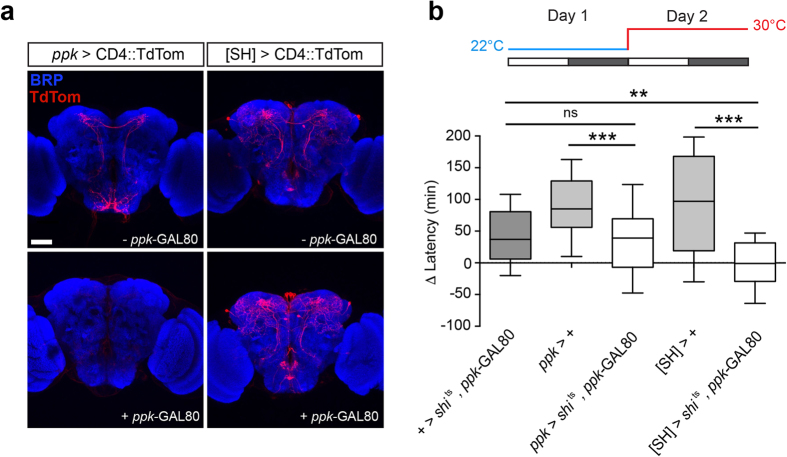
(a–c) Average sleep patterns of adult males containing a UAS-TrpA1 RNAi transgene but lacking a promoter-GAL4 driver (a), or expressing UAS-TrpA1 RNAi under control of the TrpA1[SH]- and ppk-GAL4 drivers (b,c respectively). Temperature-shift paradigms are indicated above. Black arrowheads: presence/absence of PMW. (d) Comparison of PMW in males expressing UAS-TrpA1 RNAi under GAL4 drivers and their corresponding controls. n = 32–120. Statistical comparison: Kruskal-Wallis test with Dunn’s post-hoc test. All controls are ***p < 0.0001. [SH] > RNAi: p = 0.0481; ppk > RNAi: p = 0.2420; ppk[200871] > RNAi: p = 0.0054, Wilcoxon signed rank test compared to a theoretical median of zero. (e) Effect of acute inhibition of synaptic output (using UAS-shits) from TrpA1[SH]-, ppk-, and ppk[200871]-neurons on PMW. Statistical comparison: Kruskal-Wallis test with Dunn’s post-hoc test. n = 24–77. All controls except ppk[200871] > + (**p = 0.0008) are ***p < 0.0001. [SH] > shits: ns (p = 0.06); ppk > shits: ns (p = 0.89); ppk[200871] > shits: ***p < 0.0001, Wilcoxon signed rank test compared to a theoretical median of zero. (f) Expression patterns of TrpA1[SH]-, ppk- and ppk[200871]-GAL4 in the adult Drosophila brain. CD4::TdTom is labeled with anti DsRed. Synaptic neuropil (BRP) is labeled using an anti-nc82 antibody. Stars: cell bodies labeled by TrpA1[SH]-GAL4. Arrows: projections to the dorsal posterior protocerebrum from subsets of TrpA1[SH]- and ppk-neurons. Similar projection were not observed in ppk[200871]-positive neurons. Scale bar: 100 μm.
What are the neuro-anatomical correlates of PMW suppression through blocking TrpA1-signaling? ppk-GAL4 is widely used for labeling TrpA1-expressing sensory class IV multi-dendritic (mdIV) neurons in the larval and adult body wall37,38, but is also expressed in additional neurons in the adult legs, wings and antennae (data not shown). Interestingly, comparison of the projection patterns of TrpA1[SH]- and ppk-neurons in the adult brain suggested a potential commonality: both drivers encompass neurons that project to the dorsal posterior protocerebrum (DPP: Fig. 4f, arrows). As part of our mini-screen we also tested eight promoter fragments of the ppk-promoter fused to GAL4 (see Materials and Methods). Of these, the ppk[200871]-GAL4 driver labeled mdIV neurons on the adult body wall and exhibited a similar projection pattern to ppk-GAL4 in the suboesophageal ganglion (SOG) region of the brain (Fig. 4f and Fig. S4). However, projections to the DPP were absent in ppk[200871]-neurons, and both knockdown of TrpA1 and acute inhibition of synaptic output in ppk[200871]-neurons did not impact PMW (Fig. 4d,e). We note that while the ppk[200871]/+ flies exhibited a trend towards lower levels of PMW compared to other transgene or driver controls, they still showed significant PMW at 30 °C (Fig. 4d,e; p ≤ 0.0008, Wilcoxon signed rank test). Thus, while we are yet to identify the precise cells within the TrpA1[SH]- and ppk-neuron populations that mediate PMW, the above results suggest that TrpA1[SH]- and non-mdIV ppk-positive neurons that project to the DPP may be critical mediators of PMW.
Dorsal-projecting TrpA1[SH]- and ppk-neurons are distinct cell-types
Since both the TrpA1[SH]- and ppk-positive populations include neurons that send projections to the DPP and regulate PMW, we wondered whether the TrpA1[SH]- and ppk-GAL4 drivers label a common set of sensory neurons. TrpA1[SH]-GAL4 labels internal thermo-sensory AC neurons, whose axons project to the DPP from cell bodies located close to the antennal lobes39. In support of the above premise, we stochastically observed AC cell bodies when examining fluorescently labeled TrpA1[SH]- and ppk-neurons (Fig. S5). Thus, we used an intersectional strategy to provide more definitive evidence for common, or distinct, circuits labeled by TrpA1[SH]- and ppk-GAL4. We drove expression of the GAL4-inhibitory protein GAL80 under control of the ppk-promoter (ppk-GAL8040). In the presence of ppk-GAL80, we observed a strongly penetrant loss of ppk-GAL4 expression using CD4::TdTomato (CD4::TdTom) as a fluorescent reporter (Fig. 5a). We confirmed suppression of ppk-GAL4 by ppk-GAL80 at the behavioral level by driving UAS-shits in ppk-GAL4/ppk-GAL80 males. In this background we still observed robust PMW (Fig. 5b), in contrast to the effect of driving UAS-shits with ppk-GAL4 in the absence of ppk-GAL80 (Fig. 4e). Thus, ppk-GAL80 robustly suppresses ppk-GAL4 activity. Is the same true for TrpA1[SH]-GAL4? Unlike ppk-GAL4, TrpA1[SH]-GAL4-driven CD4::TdTom fluorescence was clearly observed in the presence of ppk-GAL80 (Fig. 5a), and expression of UAS-shits in a TrpA1[SH]-GAL4/ppk-GAL80 background still inhibited PMW at 30 °C (Fig. 5b). These data demonstrate that the critical TrpA1[SH]- and ppk-positive neurons required for PMW are distinct populations.
Figure 5. TrpA1[SH]- and ppk-neurons are distinct cellular populations.
(a) Expression patterns of the ppk- and TrpA1[SH]-GAL4 drivers in adult male brains in the absence (top) or presence (bottom) of ppk-GAL80 - a GAL4-inhibitory protein under control of the ppk-promoter. Scale bar: 50 μm. (b) Effect of acute inhibition of synaptic output from ppk-and TrpA1[SH]-neurons on PMW using UAS-shits in the presence of ppk-GAL80. n = 34–62, Kruskal-Wallis test with Dunn’s post-hoc test. All controls p < 0.0001; ppk > shits, ppk- GAL80: p = 0.0005; [SH] > shits, ppk- GAL80: p = 0.91. Statistical comparison: Wilcoxon signed rank test compared to a theoretical median of zero.
DN1p clock neurons are required for PMW and contact thermo-sensory neurons
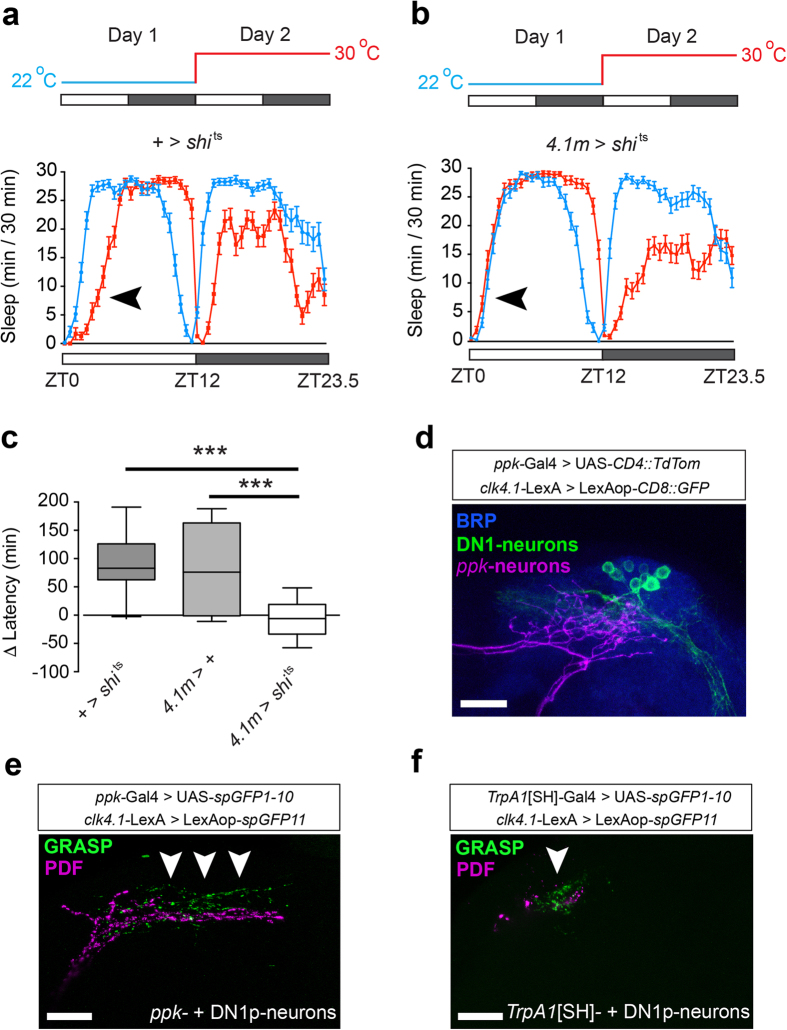
Loss of the circadian photoreceptor CRY suppresses PMW (Fig. 2d). Therefore, we wondered whether subsets of clock neurons in the brain might regulate PMW. Since both TrpA1[SH]- and ppk-neurons send projections to the DPP, we focused on clock neurons with cell bodies and/or projections in this region. Within the network of clock neurons, CRY-positive s-LNvs, LNds, and DN1p neurons send projections to the DPP41. DPP-projecting s-LNvs express the neuropeptide Pigment Dispersing Factor (PDF), which acts as a critical mediator of morning anticipation and rhythmicity in constant-dark conditions42, and both PDF-expressing s-LNvs, and DN1p neurons drive clock-dependent morning anticipation43,44. Interestingly, the output of DN1p neurons during the morning has also been shown to be temperature-dependent44, and the excitability of DN1p neurons peaks around dawn45, the time period in which PMW occurs (Fig. 1c). Furthermore, the development of DN1p neurons, but not s-LNvs, is GLASS-dependent26,28, and as shown above, loss of GLASS suppresses PMW (Fig. 2d). Therefore, we tested for a direct role for DN1p neurons by acutely inhibiting DN1p synaptic output at 30 °C, accomplished by driving UAS-shits with the driver clk4.1M-GAL4 (4.1 M), which labels both CRY-positive and -negative DN1p neurons in the adult brain44. When shifted to 30 °C, inhibiting DN1p output suppressed PMW, whereas PMW was intact in control lines (Fig. 6a–c). In contrast, inhibiting DN1p output did not alter the delay in night sleep onset at 30 °C (Fig. S6). Similar expression of UAS-shits in the CRY-positive s-LNvs and LNds using mai179-GAL443 did not suppress PMW (Fig. S6). Since blocking classical neurotransmitter release does not inhibit PDF exocytosis46, we also tested for PMW in pdf null males (pdf 01)42. In this background, PMW was present, albeit slightly reduced (Fig. S6). These results suggest that DN1p clock neurons are wake-promoting in the early morning at elevated temperatures and undertake a privileged role within the circadian CRY-positive network in regulating PMW.
Figure 6. DN1p clock-neurons are necessary for PMW.
(a,b) Average sleep patterns of adult males with synaptic output of DN1p neurons inhibited using UAS-shits (4.1 M > shits, a), and + > shits control (b). Temperature-shift paradigms are indicated above. Black arrowheads: presence/absence of PMW. (c) Comparison of PMW in 4.1 M > shits males and associated controls. n = 37–53, Kruskal-Wallis test with Dunn’s post-hoc test. All controls p < 0.0001; 4.1 M > shits: p = 0.24, using Wilcoxon signed rank test compared to a theoretical median of zero. (d) Co-localization of projections from ppk-neurons (magenta) and DN1p neurons (green) in the dorsal posterior protocerebrum of the adult Drosophila brain. BRP-positive neuropil is labeled with an anti-nc82 antibody. (e,f) GRASP between DN1p neurons and ppk-neurons (e) or TrpA1[SH]-neurons (f). Arrows indicate regions of punctate GRASP signal. Scale bars: 20 μm.
DN1p cell bodies are located in the DPP, potentially in close proximity to projections from thermo-sensory ppk- and TrpA1[SH]-neurons. To confirm this, we used orthogonal binary systems to drive distinct fluorophores in both ppk- and DN1p neurons. Indeed, we observed a clear overlap between ppk- and DN1p-projections (Fig. 6d). To study potential connectivity between thermo-sensory and DN1p neurons, we used GRASP47 to test for physical interactions between ppk- and DN1p neurons, and TrpA1[SH]- and DN1p neurons, using PDF immuno-reactivity of s-LNv axons as a marker for the location of DN1p projections48. In both cases, expression of complementary split-GFP fragments in either ppk- and DN1p-neurons (Fig. 6e), or TrpA1[SH]- and DN1p-neurons (Fig. 6f) resulted in GRASP fluorescence. In contrast, we only observed minimal GRASP fluorescence between ppk- and PDF-neurons (Fig. S6). Collectively, these results suggest that DN1p-neurons receive dual input from two distinct populations of thermo-sensory neurons to drive temperature-induced increases in morning arousal.
Discussion
How plasticity of distinct sleep periods is regulated at the molecular and circuit levels is unclear. Here we show that the TrpA1 thermo-sensor imparts temperature-sensitivity to siesta sleep in Drosophila, but modulates night sleep to a more subtle degree. Furthermore, we define a novel circuit linking thermo-sensory cells to clock neurons that, in turn, delay sleep onset in response to elevated temperatures.
Modulation of Drosophila sleep by temperature has recently been examined, but only up to an ambient temperature of 29 °C19,20. At this temperature, siesta sleep was shown to increase relative to 25 °C in both male and female flies20, and our results are consistent with this finding (Fig. 1f and Fig. S1). However, we find that in male flies, this effect is specific to the 27–29 °C range. At higher temperatures that would nonetheless be common during summer months (≥30 °C), siesta sleep is reduced. In particular, we observed a clear delay in siesta onset at ≥30 °C that we term PMW (Fig. 1c,d). The timing and magnitude of siesta sleep is sexually dimorphic7, with male flies initiating sleep earlier in the morning at 22 °C when females are still active. While the causes of this sex-specific sleep pattern are still being elucidated, it is noticeable that females do not show PMW at 30 °C. We suggest that the relative hyperactivity of females in the morning masks the effect of temperature on arousal, and later in the afternoon, circadian and/or homeostatic mechanisms act to initiate sleep, whether at mild or higher ambient temperatures. These results imply that arousal during the early morning is particularly sensitive to temperature increases. The circuits we have identified suggest an explanatory basis for this effect.
We found that TrpA1 acts in two distinct thermo-sensory subpopulations defined by the TrpA1[SH]- and ppk-GAL4 drivers to drive PMW, and that both DPP-projecting TrpA1[SH]- and ppk-neurons make physical contact with DN1p neurons that promote arousal in the early morning at 30 °C (Figs 4 and 6). When ectopically expressed, enhanced synaptic transmission induced by TrpA1 can be detected at 26 °C and is further increased at 29 °C39. We hypothesize that DN1p neurons receive weak excitatory drive from DPP-projecting TrpA1[SH]- and ppk-neurons, perhaps due to low TrpA1 expression or intrinsic excitability in each cell-type. In this model, excitatory drive scales with temperature39, and simultaneous input from TrpA1[SH]- and ppk-neurons, in combination with strong TrpA1-dependent activation of both circuits, is required to cause robust DN1p firing. This, in turn, prolongs arousal during morning periods.
Our model, combined with prior literature, suggests a mechanism for the relatively specific effect of TrpA1 signalling and DN1p activation on the onset of siesta, rather than night sleep. Under LD conditions, DN1p neurons promote morning anticipation, i.e increased locomotion before lights-on, and this output is reduced at low temperatures44,49. Recent work has also shown that thermo-genetic activation of CRY-positive DN1p neurons with a distinct driver (R18H11-GAL450) induces a PMW-like phenotype (Fig. 4 of ref. 51), further supporting a role for DN1p neurons in this process. Importantly, the intrinsic excitability of DN1p neurons is under circadian control, peaking between ZT20-ZT4 and reaching a minimum between ZT8-16 due to clock-dependent oscillations in the resting membrane potential45 (RMP). PDF signalling from s-LNvs further enhances DN1p excitability in the late night/early morning52,53. Thus, DN1p neurons are ‘primed’ to receive excitatory input from thermo-sensory neurons during the early morning.
Consistent with clock- and PDF-dependent increases in DN1p excitability during the morning, we observed that loss of both the clock protein TIM, and PDF, reduce the magnitude of PMW (Figs S2 and S6). We further demonstrate a role for the blue-light photoreceptor CRY as an essential molecular regulator of PMW (Fig. 2d). CRY is expressed in s-LNv, LNd and DN1p clock neurons41,54, and cry transcription is clock-controlled55. Inhibiting neurotransmitter or neuropeptide release from the s-LNvs and LNds does not phenocopy the effect of loss of CRY on PMW (Fig. 2 and Fig. S6). In contrast, inhibiting DN1p output fully suppresses PMW (Fig. 6), as observed in cry null males (Fig. 2). Thus, the most parsimonious hypothesis is that CRY is acting in DN1p neurons.
How might CRY influence PMW, and since cry transcription is under clock control, why is PMW not fully suppressed in tim mutants? While CRY undertakes several roles in the Drosophila nervous system29,56,57,58,59, recent work has shown that CRY additionally mediates acute light-dependent increases in clock cell excitability via interaction with the potassium β-subunit Hyperkinetic60,61. CRY stability is light-dependent, and thus CRY protein levels increase during the night55. We hypothesize that in the early morning, strongly-expressed CRY confers light-dependent excitation to DN1p neurons, enhancing the effect of excitatory drive from TrpA1-expressing neurons. Loss of the negative feedback loop of the circadian clock results in constant low-level transcription of cry55. However, CRY protein may still accumulate during the night and promote PMW in the early morning. This may explain why loss of TIM reduces, but does not fully suppress, PMW (Fig. S2). Further experiments are required to test the predictions outlined above, and to identify the critical TrpA1-expressing cells that contact DN1p neurons.
In summary, we propose that DN1p neurons integrate both TrpA1-dependent temperature- and CRY-dependent light-information with clock-driven changes in intrinsic excitability to time sleep onset during the early day. In the wild, signalling from a wide array of sensory modalities must be computed in parallel to match sleep onset with environmental conditions. Our work provides a framework to unravel how multi-sensory processing in the Drosophila nervous system facilitates dynamic control of sleep timing.
Materials and Methods
Fly Strains and Husbandry
Fly strains and crosses were reared on standard yeast-containing fly flood at constant temperature 25 °C in 12 h: 12 h Light-Dark cycles (LD). The ppk-Gal4 (BL32078), UAS-shits (BL44222), UAS-TrpA1 (BL26263), TrpA11 (BL26504), and gl60j (BL509) were obtained from Bloomington stock center. The TrpA1[SH]-Gal4, pyx3, cry02, GMR-hid, norpAP41, pdf01, Clk4.1M-LexA, UAS-spGFP1-10 and LexAop-spGFP11 fly lines were described previously24,26,34,39,42,47,48,62. Apart from gl60j, norpAP41 and pdf01, all Drosophila lines used for sleep studies were outcrossed at least 5 times into an isogenic (iso31) background. This stock also served as a wild type control line. For the gl60j and pdf01 stocks, chromosomes X and II were replaced with iso31 counterparts, while the original chromosome III (containing the glass or the pdf allele) was retained. The Vienna Tile (VT) lines were obtained from the Vienna Drosophila Resource Centre (VDRC). VDRC ID: 200748, 200871, 200905, 201215, 207296, 213670, 205646, 200782. VT insertions were also outcrossed 5 times into an iso31 background. 200871 and 207296 exhibit the most similar projection patterns in brain compared to ppk-Gal4 (see Fig. 4 and S4 for description of 200871). However, 207296 exhibits expression in Giant Commisural Interneurons in addition to the ppk-like SOG projections and mdIV neurons.
Generation of the timeless knockout allele
The timeless knock-out fly line was generated using homologous recombination63. Homologous regions flanking the target gene locus were amplified via PCR, using w1118 genomic DNA as a template and primers containing appropriate restriction sites for cloning into the pGX-attP vector64. Primer sequences were as follows. (5′ homologous region (4124 bp): 5′-ATAGCGGCCGCGAAGATTGTATACTCTAGAAG-3′/5′-CGATGGTACCATACCCTAATCGAAGTTGGTT-3′, NotI/KpnI. 3′ homologous region (3666 bp): 5′-ACGTACTAGTAACTGTGCAGGATATACGAATC-3′/5′-ATCGCTCGAGGGTCAAGATCTATTGGGAGTT-3′, SpeI/XhoI). pGX-attP was incorporated into the genome of w1118 flies via P-element insertion (BestGene Inc., and mapped to the second chromosome (BestGene Inc., California, USA). Ends-out targeting was performed as described previously63,64,65,66. Successful targeting events generated a deletion of ~17 kb, including putative 5′ promoter and 3′ UTR sequences. Fly lines required to initiate homologous recombination were obtained from the Bloomington Drosophila Stock Center (BL#25679, BL#26258, BL#1092). After screening for potential targeting events (using a mini-white+ reporter as a visual marker), knockout was verified via genomic, proteomic and behavioural investigations (data not shown and Fig. S2).
Behavioral assays
Individual 2–4 day old males were loaded into glass tubes (Trikinetics) containing 2% agar and 4% sucrose. For all experiments shown in this manuscript, Trikinetics monitors were housed in temperature- and light-controlled incubators (LMS, UK). Light intensity was measured to be between 700–1000 Lux using an environmental monitor (Trikinetics). Locomotor activity was recorded in 1 min bins using the Drosophila Activity Monitoring (DAM) system (Trikinetics). During temperature-shift experiments, flies were left for 22 °C prior to recording for 24 h. Activity counts were subsequently measured for 24 h at 22 °C, then for 24 h at elevated temperature (27 °C, 29–31 °C). The time taken for ambient temperature to increase from 22 °C to 30 °C was approximately 25 min. Sleep was defined as a period of inactivity of at least 5 min67. A modified version of a previously described Microsoft Excel script68 was used to measure all sleep parameters detailed in this article. Siesta sleep onset is defined as the latency of the first sleep bout and siesta offset as the end of the last sleep episode. We note that, using incubators from LMS (UK) in the lab of J.E.C.J, consistent and significant delays in siesta onset were observed in response to increasing ambient temperature from 22 °C to 30 °C in control lines. A delayed onset using Percival DR36VLC8 incubators (IA, USA) was also observed in the lab of K.K, although the magnitude of the effect was marginally smaller. Thus, at 30 °C, differences in incubator model may contribute to slightly altered effect sizes. When shifting flies from 22 °C to 31 °C, substantial and highly significant delays in siesta onset of iso31 flies were observed using both incubator models. Sleep graphs were generated using GraphPad Prism 6.
Immunohistochemistry and confocal microscopy
Adult male Drosophila brains were immuno-stained as described previously10. Briefly, brains were fixed in 4% paraformaldehyde for 20 min at RT, and blocked in 5% goat serum for 1 h at RT. Primary antibodies used were as follows: rabbit anti-DsRed (Clontech) – 1:2000; mouse anti-PDF (Developmental Studies Hybridoma Bank, DSHB) – 1:2000; mouse anti-Bruchpilot (nc82, DSHB) – 1:200; chicken anti-GFP (Invitrogen) – 1:1000. Alexa-fluor secondary antibodies (goat anti-rabbit 555, goat anti-chicken 488 and goat anti-mouse 647; Invitrogen) were used at 1:2000 except for labeling anti-BRP where goat anti-mouse 647 where a dilution of 1:500 was used. Confocal images were taken using an inverted Zeiss LSM 710.
Statistical analysis
Since many of the datasets derived from sleep experiments exhibited a non-normal distribution, the following statistical tests were used. Firstly, for binary analysis of whether temperature increases did or did not alter a given sleep parameter, Wilcoxon signed rank test was used, with experimental medians compared to a theoretical median of zero. When simultaneously comparing multiple genotypes, Kruskal-Wallis tests were used, followed by Dunn’s post-hoc tests. All statistical analysis was performed using GraphPad Prism 6.
Additional Information
How to cite this article: Lamaze, A. et al. Regulation of sleep plasticity by a thermo-sensitive circuit in Drosophila. Sci. Rep. 7, 40304; doi: 10.1038/srep40304 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Dr. Ko-Fan Chen for generating the Excel script used to measure sleep onset. We thank Drs. Ralf Stanewsky, Nirao Shah, Yuh Nung Jan, Francois Rouyer and the Bloomington Stock Center for providing fly stocks. Dr. Ralf Stanewsky and Dr. Ko-Fan Chen gave helpful comments on the manuscript, and Abhishek Chatterjee gave helpful suggestions during the research process. Dr. Ralf Stanewsky also gave infrastructure support to J.E.C.J. when initially constructing his lab. Huihui Pan provided technical support, and Alexandra Kenny performed pilot experiments. This work was supported by a grant from the National Institutes of Health (R01NS086887) to K.K. and a UCL start-up grant to J.E.C.J.
Footnotes
Author Contributions Conceptualization, A.L. and J.E.C.J.; Methodology, A.L. and J.E.C.J.; Formal Analysis, A.L.; Investigation, A.L. and A.O.-C.; Resources, R.F. and N.P.; Writing – Original Draft, J.E.C.J. and A.L.; Writing – Review & Editing; A.O.-C., N.P. and K.K.; Visualization, A.L., A.O.-C., K.K. and J.E.C.J.; Funding Acquisition, K.K. and J.E.C.J.; Supervision, K.K. and J.E.C.J.
References
- Borbély A. A. A two process model of sleep regulation. Hum Neurobiol 1, 195–204 (1982). [PubMed] [Google Scholar]
- Sehgal A. & Mignot E. Genetics of sleep and sleep disorders. Cell 146, 194–207, doi: 10.1016/j.cell.2011.07.004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D., Tononi G. & Cirelli C. Sleep and synaptic homeostasis: structural evidence in Drosophila. Science 332, 1576–1581, doi: 10.1126/science.1202839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilestro G. F., Tononi G. & Cirelli C. Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324, 109–112, doi: 10.1126/science.1166673 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M. S., Yue Z. & Sehgal A. A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science 344, 269–274, doi: 10.1126/science.1250553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L. et al. Sleep drives metabolite clearance from the adult brain. Science 342, 373–377, doi: 10.1126/science.1241224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C. et al. Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092, doi: 10.1038/nature03486 (2005). [DOI] [PubMed] [Google Scholar]
- Koh K. et al. Identification of SLEEPLESS, a sleep-promoting factor. Science 321, 372–376, doi: 10.1126/science.1155942 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Liu S., Kodama L., Driscoll M. R. & Wu M. N. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol 22, 2114–2123, doi: 10.1016/j.cub.2012.09.008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. WIDE AWAKE mediates the circadian timing of sleep onset. Neuron 82, 151–166, doi: 10.1016/j.neuron.2014.01.040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Liu Q., Tabuchi M. & Wu M. N. Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell 165, 1347–1360, doi: 10.1016/j.cell.2016.04.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T. et al. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci 15, 1516–1523, doi: 10.1038/nn.3238 (2012). [DOI] [PubMed] [Google Scholar]
- Donlea J. M., Pimentel D. & Miesenbock G. Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81, 860–872, doi: 10.1016/j.neuron.2013.12.013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D. et al. Operation of a homeostatic sleep switch. Nature 536, 333–337, doi: 10.1038/nature19055 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J. A., Cervantes-Sandoval I., Chakraborty M. & Davis R. L. Sleep Facilitates Memory by Blocking Dopamine Neuron-Mediated Forgetting. Cell 161, 1656–1667, doi: 10.1016/j.cell.2015.05.027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler M. A. & Montell C. Drosophila TRP channels and animal behavior. Life Sci 92, 394–403, doi: 10.1016/j.lfs.2012.07.029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity P. A., Goodman M. B., Samuel A. D. & Sengupta P. Running hot and cold: behavioral strategies, neural circuits, and the molecular machinery for thermotaxis in C. elegans and Drosophila. Genes Dev 24, 2365–2382, doi: 10.1101/gad.1953710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto H., Lark A. R. & Kitamoto T. Factors that differentially affect daytime and nighttime sleep in Drosophila melanogaster. Front Neurol 3, 24, doi: 10.3389/fneur.2012.00024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W. & Edery I. A novel pathway for sensory-mediated arousal involves splicing of an intron in the period clock gene. Sleep 38, 41, doi: 10.5665/sleep.4322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky K. M., Rivera J. L. A., Donelson N. C., Kotecha S. & Griffith L. C. Reorganization of Sleep by Temperature in Drosophila Requires Light, the Homeostat, and the Circadian Clock. Curr Biol 26, 882–892, doi: 10.1016/j.cub.2016.02.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C., Lear B. C., Keegan K. P. & Allada R. Processing sleep data created with the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc 2010, pdb prot5520, doi: 10.1101/pdb.prot5520 (2010). [DOI] [PubMed] [Google Scholar]
- Majercak J., Sidote D., Hardin P. E. & Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230 (1999). [DOI] [PubMed] [Google Scholar]
- Das A., Holmes T. C. & Sheeba V. dTRPA1 in Non-circadian Neurons Modulates Temperature-dependent Rhythmic Activity in Drosophila melanogaster. J Biol Rhythms 31, 272–288, doi: 10.1177/0748730415627037 (2016). [DOI] [PubMed] [Google Scholar]
- Szular J. et al. Rhodopsin 5–and Rhodopsin 6–Mediated Clock. Synchronization in Drosophila melanogaster Is Independent of Retinal Phospholipase C-β Signaling. J Biol Rhythms 27, 25–36, doi: 10.1177/0748730411431673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C. Drosophila visual transduction. Trends Neurosci 35, 356–363, doi: 10.1016/j.tins.2012.03.004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarsfeld A. et al. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci 24, 1468–1477, doi: 10.1523/JNEUROSCI.3661-03.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses K., Ellis M. C. & Rubin G. M. The glass gene encodes a zinc: finger protein required by Drosophila photoreceptor cells. Nature 340, 531–536, doi: 10.1038/340531a0 (1989). [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C., Winter C., Hofbauer A., Hall J. C. & Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30, 249–261 (2001). [DOI] [PubMed] [Google Scholar]
- Stanewsky R. et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692 (1998). [DOI] [PubMed] [Google Scholar]
- Sehgal A., Price J. L., Man B. & Young M. W. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263, 1603–1605 (1994). [DOI] [PubMed] [Google Scholar]
- Barbagallo B. & Garrity P. A. Temperature sensation in Drosophila. Curr Opin Neurobiol 34, 8–13, doi: 10.1016/j.conb.2015.01.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E. W. et al. Drosophila circadian rhythms in seminatural environments: summer afternoon component is not an artifact and requires TrpA1 channels. Proc Natl Acad Sci 112, 8702–8707, doi: 10.1073/pnas.1506093112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L. et al. A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature 500, 580–584, doi: 10.1038/nature12390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang W., Simoni A., Gentile C. & Stanewsky R. The Pyrexia transient receptor potential channel mediates circadian clock synchronization to low temperature cycles In Drosophila melanogaster. Proc R Soc Lond B Bio Sci 280, 20130959, doi: 10.1098/rspb.2013.0959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. et al. Pyrexia is a new thermal transient receptor potential channel endowing tolerance to high temperatures in Drosophila melanogaster. Nat Genet 37, 305–310, doi: 10.1038/ng1513 (2005). [DOI] [PubMed] [Google Scholar]
- Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol 47, 81–92 (2001). [DOI] [PubMed] [Google Scholar]
- Grueber W. B. et al. Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development 134, 55–64, doi: 10.1242/dev.02666 (2007). [DOI] [PubMed] [Google Scholar]
- Xiang Y. et al. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 468, 921–926, doi: 10.1038/nature09576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F. N. et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220, doi: 10.1038/nature07001 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-h. et al. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron 61, 519–526, doi: 10.1016/j.neuron.2008.12.021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T., Todo T., Wülbeck C., Stanewsky R. & Helfrich‐Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol 508, 952–966, doi: 10.1002/cne.21702 (2008). [DOI] [PubMed] [Google Scholar]
- Renn S. C., Park J. H., Rosbash M., Hall J. C. & Taghert P. H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802 (1999). [DOI] [PubMed] [Google Scholar]
- Grima B., Chélot E., Xia R. & Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873, doi: 10.1038/nature02935 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu Y., Bilodeau-Wentworth D., Hardin P. E. & Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol 20, 600–605, doi: 10.1016/j.cub.2010.02.044 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourakis M. et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell 162, 836–848, doi: 10.1016/j.cell.2015.07.036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard H. A. et al. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol 516, 59–73, doi: 10.1002/cne.22099 (2009). [DOI] [PubMed] [Google Scholar]
- Feinberg E. H. et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363, doi: 10.1016/j.neuron.2007.11.030 (2008). [DOI] [PubMed] [Google Scholar]
- Cavanaugh D. J. et al. Identification of a circadian output circuit for rest: activity rhythms in Drosophila. Cell 157, 689–701, doi: 10.1016/j.cell.2014.02.024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol 20, 591–599, doi: 10.1016/j.cub.2010.02.056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F. et al. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature 536, 292–297, doi: 10.1038/nature19097 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst M. et al. Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr Biol 24, 2652–2664, doi: 10.1016/j.cub.2014.09.077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A. et al. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol 12, e1001810, doi: 10.1371/journal.pbio.1001810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H. et al. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci 97, 3608–3613, doi: 10.1073/pnas.070036197 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J., Houl J. H., Roman G. W. & Hardin P. E. The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J Biol Rhythms 23, 296–307, doi: 10.1177/0748730408318588 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P., So W. V., Kaneko M., Hall J. C. & Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679 (1998). [DOI] [PubMed] [Google Scholar]
- Kumar S., Chen D. & Sehgal A. Dopamine acts through Cryptochrome to promote acute arousal in Drosophila. Genes Dev 26, 1224–1234, doi: 10.1101/gad.186338.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani M. F. et al. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285, 553–556 (1999). [DOI] [PubMed] [Google Scholar]
- Gegear R. J., Casselman A., Waddell S. & Reppert S. M. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature 454, 1014–1018, doi: 10.1038/nature07183 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzotta G. et al. Fly cryptochrome and the visual system. Proc Natl Acad Sci 110, 6163–6168, doi: 10.1073/pnas.1212317110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle K. J. et al. CRYPTOCHROME-mediated phototransduction by modulation of the potassium ion channel beta-subunit redox sensor. Proc Natl Acad Sci 112, 2245–2250, doi: 10.1073/pnas.1416586112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle K. J., Parson K. G., Dahm N. A. & Holmes T. C. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science 331, 1409–1413, doi: 10.1126/science.1199702 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezelova E., Dolezel D. & Hall J. C. Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics 177, 329–345, doi: 10.1534/genetics.107.076513 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A. M. & Hong Y. From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci 106, 8284–8289, doi: 10.1073/pnas.0900641106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Watson A. M., Jan Y. N. & Hong Y. Efficient ends-out gene targeting in Drosophila. Genetics 180, 703–707, doi: 10.1534/genetics.108.090563 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W. J. & Golic K. G. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci 100, 2556–2561, doi: 10.1073/pnas.0535280100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert K. A., Gong W. J. & Golic K. G. Methods for homologous recombination in Drosophila. Methods Mol Biol 420, 155–174, doi: 10.1007/978-1-59745-583-1_9 (2008). [DOI] [PubMed] [Google Scholar]
- Shaw P. J., Cirelli C., Greenspan R. J. & Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837 (2000). [DOI] [PubMed] [Google Scholar]
- Roessingh S., Wolfgang W. & Stanewsky R. Loss of Drosophila melanogaster TRPA1 function affects “siesta” behavior but not synchronization to temperature cycles. J Biol Rhythms 30, 492–505, doi: 10.1177/0748730415605633 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.