Abstract
Background
The complex process of formation of storage roots (SRs) from adventitious roots affects sweetpotato yield. Identifying the genes that are uniquely expressed in the SR forming cultivated species, Ipomoea batatas (Ib), and its immediate ancestral species, Ipomoea trifida (It), which does not form SRs, may provide insights into the molecular mechanisms underlying SR formation in sweetpotato.
Results
Illumina paired-end sequencing generated ~208 and ~200 million reads for Ib and It, respectively. Trinity assembly of the reads resulted in 98,317 transcripts for Ib and 275,044 for It, after post-assembly removal of trans-chimeras. From these sequences, we identified 4,865 orthologous genes in both Ib and It, 60 paralogous genes in Ib and 2,286 paralogous genes in It. Among paralogous gene sets, transcripts encoding the transcription factor RKD, which may have a role in nitrogen regulation and starch formation, and rhamnogalacturonate lyase (RGL) family proteins, which produce the precursors of cell wall polysaccharides, were found only in Ib. In addition, transcripts encoding a K+ efflux antiporter (KEA5) and the ERECTA protein kinase, which function in phytohormonal regulation and root proliferation, respectively, were also found only in Ib. qRT-PCR indicated that starch and sucrose metabolism genes, such as those encoding ADP-glucose pyrophosphorylase and beta-amylase, showed lower expression in It than Ib, whereas lignin genes such as caffeoyl-CoA O-methyltransferase (CoMT) and cinnamyl alcohol dehydrogenase (CAD) showed higher expression in It than Ib. A total of 7,067 and 9,650 unique microsatellite markers, 1,037,396 and 495,931 single nucleotide polymorphisms (SNPs) and 103,439 and 69,194 InDels in Ib and It, respectively, were also identified from this study.
Conclusion
The detection of genes involved in the biosynthesis of RGL family proteins, the transcription factor RKD, and genes encoding a K+ efflux antiporter (KEA5) and the ERECTA protein kinase only in I. batatas indicate that these genes may have important functions in SR formation in sweetpotato. Potential molecular markers (SNPs, simple sequence repeats and InDels) and sequences identified in this study may represent a valuable resource for sweetpotato gene annotation and may serve as important tools for improving SR formation in sweetpotato through breeding.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0950-x) contains supplementary material, which is available to authorized users.
Keywords: Ipomoea batatas, Ipomoea trifida, Storage root, Fibrous root, Transcriptome, Molecular markers
Background
Sweetpotato (Ipomoea batatas [L.] Lam.) is a key food crop worldwide, with high levels of vitamin A and other essential nutrients; sweetpotatoes also produce large quantities of biomass suitable for conversion to bioethanol [1]. Sweetpotato storage roots (SRs) function in carbohydrate storage and vegetative propagation [2, 3] and form from adventitious roots. Adventitious roots develop from nodal primordia and cut ends or wounds of stem (slips) at 5–15 days after transplanting. These adventitious roots can then form SRs by a process that involves thickening of the vascular tissue, followed by the accumulation of starch and proteins [4]. Adventitious roots can also form fibrous roots (FRs), which undergo lignification of the stele; in contrast to FRs, SRs do not undergo stele lignification [4–6]. The conversion of adventitious roots to SRs involves the formation of new cambial cells, followed by the development of secondary cambium and thin-walled parenchyma cells. Despite its importance, key factors in SR development remain to be discovered.
Although the molecular mechanism underlying the transition from adventitious roots to SRs in sweetpotato is not yet clear, substantial prior work has implicated the plant hormones cytokinin, auxin, and abscisic acid (ABA) in the formation and thickening of SRs [7–11]. For example, ABA functions in the secondary thickening of vascular cambium during SR formation in sweetpotato [10]. Transcription factors from various families have also been implicated in SR formation. For example, the transcription factor gene IbMADS1 (Ipomoea batatas MADS-box 1) is expressed during the early stages of SR initiation [12]; also, cytokinins and jasmonic acid induce the expression of IbMADS1. Noh et al. [11] isolated a cDNA encoding the MADS-box protein SRD1, which plays an important role in the formation of SRs by activating the proliferation of cambium and metaxylem cells to induce the initial thickening of SRs. The expression of SRD1 is regulated by the auxin indole-3-acetic acid. Also, overexpression of the class I knotted1-like homeobox (KNOX1) genes, Ibkn1 and Ibkn2, results in increased cytokinin activity in sweetpotato, indicating that KNOX1 functions in controlling cytokinin levels in SRs [13].
Expression analysis during SR formation also identified a number of candidate genes [14–16]. For example, You et al. [14] identified 22 differentially expressed genes by comparing early SRs and fibrous roots. Several NAC family transcription factor genes are downregulated in SRs, and two NAM-like genes, as well as sporamin genes and genes involved in starch biosynthesis, are upregulated in SRs (compared to FRs) at six weeks after planting [15]. Noh et al. [16] used antisense RNA interference to demonstrate the negative role of an expansin gene (IbEXP1) in SR development; IbEXP1 suppresses the proliferation of metaxylem and cambium cells, and thus inhibits the initial thickening of SRs.
Recent work used microarray and next-generation sequencing technologies to examine the molecular mechanism of SR formation in sweetpotato. Wang et al. [17] used microarray analysis to identify transcription factors involved in SR development, such as DA1-related proteins, SHORT-ROOT, and BEL1-like proteins. Using Illumina sequencing, Tao et al. [18] identified genes that are differentially expressed at different stages of sweetpotato root formation. In particular, they found that a gene encoding sucrose phosphate synthase, which functions in sucrose metabolism, is highly expressed in SRs than in fibrous roots. Firon et al. [2] analyzed the root transcriptomes of sweetpotato SRs and non-storage/fibrous roots and demonstrated that phenylpropanoid pathway genes, such as those encoding coumaroyl CoA-synthase and phenylalanine ammonia lyase, are downregulated during the conversion of FRs to SRs, whereas starch metabolism genes, such as those encoding ADP-glucose pyrophosphorylase and starch synthase, are upregulated in SRs.
The cultivated sweetpotato likely evolved from the wild tetraploid I. trifida and diploid I. trifida/I. tabascana species [19–22]; these wild relatives do not form SRs. Previous transcriptome analyses investigating SR formation examined only the hexaploid cultivated species [2, 23, 24]. Therefore, comparative transcriptome analysis of the wild and cultivated species of sweetpotato may advance our knowledge on the mechanism underlying SR formation in this important crop. In this study, we performed transcriptome analysis of the roots from cultivated sweetpotato (Ib; Ipomoea batatas [L.] Lam) and its non-tuber forming relative (It; Ipomoea trifida [Kunth] G. Don) to elucidate possible pathways and candidate genes involved in SR formation.
Results and discussion
De novo assembly of root transcriptomes using Illumina sequencing
High-throughput sequencing of the root transcriptomes of cultivated Ipomoea batatas (Ib) and its wild ancestor Ipomoea trifida (It) generated 416 and 400 million reads, respectively. All reads were assembled using Trinity (Version r2012–10–15) with default parameters. The maximum contig length was 14,381 bp for Ib and 15,897 bp for It (Table 1). The contigs were grouped based on sequence length at an interval of 200 base pairs (bp). The majority of contigs were ranged from 200–399 bp in length (59.0% for Ib and 66.7% for It), followed by 400–599 bp (14.7% for Ib and 14.1% for It). In addition, 25.0 and 18.6% of the contigs were 600–3000 bp in length for Ib and It respectively, and only 1.3% of the contigs in Ib and 0.6% of the contigs in It were >3000 bp long (Figs. 1 and 2).
Table 1.
Summary of RNA-seq reads from Ib and It
| Genotype | No. of reads (PE, 2×100) | No. of contigs | N50 contig length | Min. length of contigs | Max. length of contigs | Filtered transcriptsa |
|---|---|---|---|---|---|---|
| I. batatas (Ib) | 208,213,047 | 240,915 | 1,791 | 201 | 14,381 | 98,317 |
| I. trifida (It) | 200,191,376 | 366,513 | 1,217 | 201 | 15,897 | 275,044 |
aSelected highly covered isoforms using RSEM and post-assembly trans-chimera cleanup using BLASTX results against the non-redundant protein database
Fig. 1.
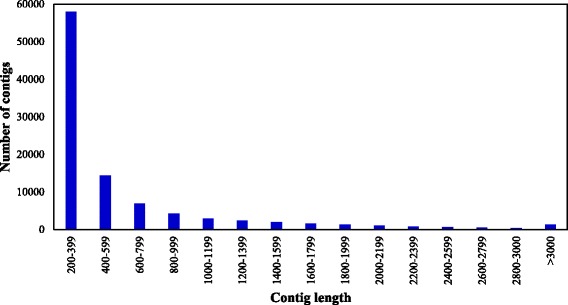
Length distribution of I. batatas contigs. Total reads were assembled using Trinity and grouped based on sequence length at 200-bp intervals
Fig. 2.
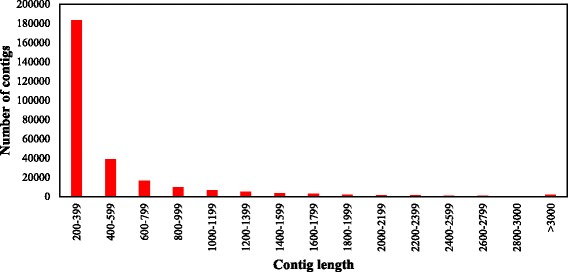
Length distribution of I. trifida contigs. Total reads were assembled using Trinity and grouped based on sequence length at 200-bp intervals
Functional annotation by sequence comparison with public databases
To perform functional annotation of the Ib and It contigs, we used BLASTX [25] with a cutoff e-value of 1.0E-3 to search public databases, such as the non-redundant (nr), Arabidopsis, cassava, and potato databases, and the sweetpotato gene index. This analysis showed that 44.1% of Ib transcripts and 63.8% of It transcripts matched the non-redundant (nr) database, but only 33.0% of Ib and 32.7% of It transcripts matched the Arabidopsis database. Similarity searches of the sweetpotato gene index revealed matches of only 25.7% and 8.7% of transcripts for Ib and It, respectively. The low percentage of Ib and It transcripts with sequence similarity to sweetpotato gene index may be attributed to the limited number of annotated genes in the database. Searches of the potato and cassava databases also showed the same degree of similarity as found for searches of the Arabidopsis database (Table 2). Overall, 51.7% of the transcripts from Ib and 66.4% of the transcripts from It showed homology with at least one database. The detailed functional annotation for each species is presented in Additional files 1 and 2.
Table 2.
BLAST annotation of Ib and It transcripts
| Number of transcripts with matches in: | Ib | It |
|---|---|---|
| Sweetpotato gene index | 25,258 | 23,909 |
| NCBI nr database | 43,369 | 175,495 |
| Arabidopsis database | 32,427 | 89,972 |
| Cassava database | 33,006 | 90,308 |
| Potato database | 33,335 | 87,150 |
| At least one database | 50,860 | 182,692 |
| No matches in any database | 47,457 | 92,352 |
| Information in Trinotate | 11,237 | 25,068 |
Comparative analysis of gene sets between Ib and It
To allow direct comparison of loci between Ib and It, we used the annotated transcripts to identify orthologous and paralogous genes in Ib and It. Among the 60 paralogous gene sets with 148 contigs identified in Ib, 18 contigs had no match in the Arabidopsis database, whereas the remaining 130 transcripts that matched with the Arabidopsis database were found to be involved in cellular organization, membrane transport, glucose metabolic activity, and carbohydrate metabolism. However, none of the 60 paralogous gene sets identified in Ib had matches in the sweetpotato gene index, most likely due to the small number of annotated genes available in this database (Additional file 3). In It, we identified 2,286 paralogous gene sets with 9,585 contigs. Among 9,585 contigs, 4,890 contigs had no match in the Arabidopsis database; the remaining 4,695 transcripts, which matched with the Arabidopsis database, were involved in transporter activity, stress-related functions, and ribosomal proteins involved in translation. In contrast, only 72 transcripts, which matched with sweetpotato gene index, showed annotated gene functions (Additional file 4). We also identified 5,695 and 6,289 transcripts in Ib and It, respectively, from 4,865 orthologous gene sets. Examples of orthologous genes include calcium-dependent lipid binding (CaLB) domain and the porcino tubulin-binding cofactor, which are involved in stress and defense responses. CaLB-domain genes are upregulated in drought-stressed sweetpotato [26]. The transcription factors observed among the orthologs include KNOX, BEL-1 like homeodomain, and NAC domain-containing proteins that are involved in DNA binding and transcription activities. In sweetpotato, KNOX1 is involved in secondary thickening of the SRs through enhanced cytokinin activity [13] and BEL-1 like homeodomain involved in SR development [17]. The NAC domain-containing proteins are upregulated in sweetpotato SRs compared to fibrous roots [15]. Overall, the orthologous genes and transcription factors in both Ib and It are involved in activities such as nucleic acid binding, defense responses, cell division and differentiation, transport, root gravitropism, hormonal control, and glucose metabolism (Additional file 5).
Among paralogous gene sets, genes encoding the rhamnogalacturonate lyase (RGL) family proteins, K+ efflux antiporter (KEA5), and ERECTA protein kinases were found only in Ib. RGL proteins comprise the major components of pectin polysaccharides in the cell wall [27]. RGL proteins also serve as signaling molecules for pectin polysaccharides [28], and are involved in cell wall modifications such as cell wall expansion, porosity, and textural changes during fruit ripening [29–32]. In Arabidopsis, the expression of RGL genes, which are involved in lateral root and root hair formation, is altered in response to the inhibition of primary root growth [33]. Also, in potato (Solanum tuberosum), overexpression of RGL genes leads to distinct morphological changes in the cortex and periderm [34]. The expression of RGL genes (with FPKM (Fragments per Kilobase of Exon per Million Fragments Mapped) value of 181.1) only in Ib suggests that pectin polysaccharides may play a role in cell expansion leading to the accumulation of storage proteins in developing SRs and that RGL proteins may have important roles in the secondary thickening of cell walls.
K+ (KEA5) transporter genes (with FPKM value of 14.9) are another group of genes that are expressed specifically in Ib. These transporters are involved in stomatal activity, leaf movements, ion transport, and the regulation of phytohormones such as auxin, ethylene, and jasmonic acid [35, 36]. K+ transporters also play an important role in cell expansion associated with turgor pressure in Arabidopsis [37, 38]. The expression of KEA5 transporter genes in Ib suggests that they may play a role in SR formation through the regulation of phytohormones such as auxin. In plant roots, auxin regulates lateral root development and gravitropism [39]. In sweetpotato, auxin regulates the expression of the transcription factor SRD1, which is involved in SR formation [11]. Therefore, the coordinated regulation of transporter genes such as KEA5 and transcription factors such as SRD1 in auxin regulation may be a possibility during SR formation in sweetpotato. In the present study, in addition to RGL family genes and KEA5, ERECTA protein kinase (with FPKM value of 2.2) was also identified in the paralogous genes of Ib but not in It. ERECTA protein kinase is a leucine-rich repeat receptor-like kinase (LRR-RLK) involved in the proliferation of organelles. ERECTA protein kinases regulate organ shape and inflorescence development in Arabidopsis [40, 41]. Interestingly, receptor-like kinases are involved in lateral root development in Arabidopsis [42] and cell wall-bound kinases are associated with pectin binding in Arabidopsis [43]. ERECTA, along with RGL family proteins, might represent an important link between the regulation of cell wall structure and SR development in sweetpotato. Among the transcription factors in paralogous genes, RKD (RWP-RK domain) belongs to the RWP-RK domain-containing proteins, which are involved in DNA binding and regulation of transcription activity, were expressed in Ib but not in It. The RWP-RK protein domain is required for embryonic pattern formation [44] and plays a key role in regulating responses to nitrogen availability [45]. Recent work reported that NIT2, a member of the RWP-RK family, influences starch and lipid storage in Chlamydomonas [46]. In Arabidopsis thaliana, NLP7, another member of the RWP-RK family, is an early regulator of cellular response to nitrogen assimilation [47]. The unique expression of RWP-RK proteins (with FPKM value of 18.4) only in Ib indicate that these proteins may have a role in nitrogen regulation and starch formation during the development of sweetpotato SRs.
Expression of genes involved in SR formation
We compared the expression of genes in Ib versus It based on their respective FPKM values (Table 3). Although two species were compared to estimate up— or down-regulation of gene expression, a clear limitation in our study is the lack of biological replications. Therefore, conclusions reached in this study, may change in future experiments with biological replications. While hundreds or thousands of mapped reads of differentially expressed genes are likely to be reliable, careful reading is required for the contigs/transcripts with few mapped hits. Some evidence for this differential gene expression is supported by the qRT-PCR results (Fig. 3). In the present study, the class-I knotted1-like homeobox gene KNOX1 was highly expressed in Ib (136.9) compared to It (50.3). Similarly, Firon et al. [2] observed the increased expression of KNOX1 in developing sweetpotato SRs versus FRs. KNOX1 is also associated with higher cytokinin levels, in addition to secondary thickening, in sweetpotato SRs [13, 14]. The cell wall loosening protein expansin was highly represented in Ib (70.5) compared to It (10.9). Firon et al. [2] demonstrated the involvement of this expansin gene in the initiating SRs of sweetpotato. Also, the gene encoding the sporamin was highly expressed in Ib (56,838.3) than in It (15.2) (Table 3). Sporamin, a key storage protein in sweetpotato SRs [15], forms in the endoplasmic reticulum, and the mature sporamin moves to the vacuoles of SRs [48]. In sweetpotato, sporamin was accounted for over 80% of the total protein in the SRs [49] and highly expressed in SRs than in FRs [2]. The high expression of sporamin in Ib confirmed its role in the synthesis of storage proteins. In the present study, starch and sucrose metabolic genes, such as β-amylase (381.7), glucose-1-phosphate adenylyltransferase (1745.2), phosphoglucomutase (170.2), starch synthase (110.0), ADP-glucose pyrophosphorylase (1752.6), and alpha 1–4 glucan phosphorylase (1305.1) were highly expressed in Ib. Similar to our study, the high expression of the gene encoding β-amylase is observed in the initiating SRs compared to fibrous roots in sweetpotato [2]. Also, the increased expression of ADP-glucose pyrophosphorylase is associated with starch accumulation in sweetpotato SRs [18]. Clearly, the high expression of both β-amylase, the second most abundant storage protein in sweetpotato after sporamin [48], and ADP-glucose pyrophosphorylase is positively correlated to their role in starch biosynthesis in sweetpotato [50]. In the present study, we observed higher expression of the gene encoding phosphoglucomutase in Ib than in It, indicating that the enhanced activity of this enzyme may provide abundant substrate for ADP-glucose pyrophosphorylase, the first step in starch biosynthesis pathway. Indeed, enhanced expression of phosphoglucomutase was previously observed in the SRs of sweetpotato [2]. Another highly expressed gene in Ib encodes alpha 1–4 glucan phosphorylase, an enzyme involved in starch phosphorylation in Arabidopsis [51]. The phosphorylation of starch promotes the accumulation of starch granules in the SRs of sweetpotato [18]. The high expression of genes encoding phosphoglucomutase and alpha 1–4 glucan phosphorylase in Ib reflects the high activity of starch metabolic genes in SRs.
Table 3.
Comparison of annotated genes involved in storage root formation between Ib and It using FPKM values
| Annotation | Ib | It | Ratio |
|---|---|---|---|
| Sporamin | 56,838.3 | 15.2 | 3739.4 |
| Expansin | 70.5 | 10.9 | 6.5 |
| Glucose-1-phosphate adenylyltransferase | 1745.2 | 178.7 | 9.8 |
| Alpha-1,4 glucan phosphorylase | 1305.1 | 0.5 | 2610.2 |
| Beta-amylase | 381.7 | 28.6 | 13.3 |
| Phosphoglucomutase | 170.2 | 76.7 | 2.2 |
| Class-I knotted1-like homeobox protein IbKN2 (KNOX) | 136.9 | 50.3 | 2.7 |
| ADP-glucose pyrophosphorylase | 1752.6 | 178.7 | 9.8 |
| Starch synthase | 110.0 | 15.9 | 6.9 |
Fig. 3.
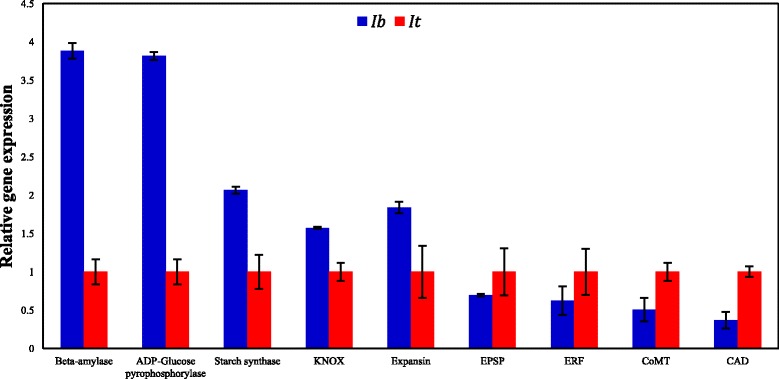
Validation of relative expression levels of selected genes (qRT-PCR). Expression levels were compared between I. batatas (Ib; indicated in blue) and I. trifida (It; indicated in red). Quantitative RT-PCR was performed with three biological replicates and two technical replicates for both Ib and It. The sweetpotato β-tubulin gene was used as an endogenous control and the gene expression levels were determined using the ΔΔCt method. Genes are shown with respective TAIR locus IDs: β-amylase (AT4G15210); ADP-glucose pyrophosphorylase (AT4G39210); Starch synthase (AT4G18240); KNOX: class I Knotted-like homeobox (AT4G08150); Expansin (AT2G39700); EPSP: 5- enolpyruvylshikimate-3-phosphate synthase (AT2G45300); ERF: Ethylene-responsive transcription factor (AT1G50640); CoMT: caffeoyl-CoA O-methyltransferase (AT4G34050); CAD: cinnamyl alcohol dehydrogenase (AT1G72680)
Expression of genes involved in non-storage/fibrous root formation
We compared the expression of genes in It versus Ib based on their respective FPKM values (Table 4). The results indicate that genes encoding cysteine protease (1709.6), cysteine proteinase (744.9), ethylene responsive transcription factor (ERF) (211.8), osmotin-like proteins (787.7), and peroxidase (689.3) were highly expressed in It. Cysteine protease is involved in the degradation of sporamin in sprouting sweetpotato SRs [52]. In addition, the cysteine protease gene is downregulated in sweetpotato SRs [2]. In our study, a cysteine protease gene was highly expressed in It, indicating that this enzyme may prevent the accumulation of sporamin in the roots and helps them remain as fibrous roots. Cysteine proteinases also exhibit protein degradation activity [53]. The cysteine proteinase gene is upregulated in FRs compared to the SRs in sweetpotato [2]. We observed similar high expression of the cysteine proteinase gene in It, indicating that this enzyme may be involved in proteolytic activity and promotes the development of FRs. Genes encoding osmotin-like protein were highly expressed in It compared to Ib (Table 4). The osmotin-like proteins are plant defense proteins [54] that regulate the production of jasmonic acid and ethylene in Arabidopsis [55]. In sweetpotato, the osmotic-like stress response genes are downregulated in the SRs compared to FRs [2]. The ethylene responsive transcription factor (ERF) was highly expressed in It than Ib, indicating the activity of this stress-responsive gene in the fibrous roots. Likewise, genes encoding peroxidase showed decreased expression in Ib versus It, indicating that peroxidase genes may be involved in phenylpropanoid and lignin (polymerization) biosynthesis pathways [2]. Another stress-response gene, which encodes succinate dehydrogenase, was minimally increased in It versus Ib; succinate dehydrogenase functions in the production of reactive-oxygen species during stress-related activities [56]. Overall, the higher expression of proteolytic enzymes such as cysteine protease and cysteine proteinase in It indicates that these genes may prevent the accumulation of storage proteins in the developing FRs. Moreover, the higher expression of stress-response genes such as those encoding peroxidases promotes the expression of phenylpropanoid pathway genes and the accumulation of lignin in FRs in sweetpotato [2].
Table 4.
Comparison of annotated genes involved in fibrous root formation between Ib and It using FPKM values
| Annotation | Ib | It | Ratio |
|---|---|---|---|
| Cysteine protease | 44.9 | 1709.6 | 38.1 |
| Cysteine proteinase | 145.4 | 744.9 | 5.1 |
| Osmotin-like protein | 20.4 | 787.7 | 38.6 |
| Succinate dehydrogenase | 41.9 | 49.0 | 1.2 |
| Caffeoyl-CoA O-methyltransferase (CoMT) | 410.7 | 975.2 | 2.4 |
| Phenylalanine ammonia-lyase (PAL) | 87.3 | 443.0 | 5.1 |
| Peroxidase | 105.5 | 689.3 | 6.5 |
| 4-coumarate--CoA ligase (4-CL) | 30.3 | 198.7 | 6.6 |
| Cinnamyl alcoholdehydrogenase (CAD) | 67.7 | 762.9 | 11.3 |
| 5-enolpyruvylshikimate-3-phosphate synthase (EPSP) | 29.1 | 144.3 | 5.0 |
| Ethylene responsive transcription factor (ERF) | 77.9 | 211.8 | 2.7 |
In addition, lignification helps roots remain as non-storage/fibrous roots and prevents the conversion of fibrous roots to SRs. In the current study, we observed higher expression of genes encoding phenylpropanoid and lignin biosynthetic enzymes, such as 5-enolpyruvylshikimate-3 phosphate (EPSP) synthase (144.3), phenylalanine ammonia-lyase (PAL) (443.0), 4-coumarate-CoA ligase (4-CL) (198.7), cinnamyl alcohol dehydrogenase (CAD) (762.9), peroxidase (689.3), and caffeoyl-CoA O-methyltransferase (CoMT) (975.2) in It (Table 4). The enzyme EPSP synthase produces an important precursor in the shikimate pathway, promotes the synthesis of lignin and phenylalanine [57]. Consistent with our observations, other studies showed that in sweetpotato, genes encoding cinnamyl alcohol dehydrogenase, coumaroyl-CoA synthase, and caffeoyl-CoA O-methyltransferase are downregulated during SR initiation [2]. Moreover, lignin biosynthesis genes are upregulated in the early stages of fibrous root formation in sweetpotato [17]. The increased expression of the phenylpropanoid and lignin genes in It provides evidence for the accumulation of lignins during FR development.
Validation of gene expression through quantitative reverse-transcription PCR (qRT-PCR)
We profiled different genes, based on FPKM values, for expression in Ib and It with the RNA samples used for Illumina sequencing. Genes involved in starch and sucrose metabolism, and lignin biosynthesis, such as those encoding ADP-glucose pyrophosphorylase, β-amylase, starch synthase, cinnamyl alcohol dehydrogenase (CAD), and caffeoyl-CoA O-methyltransferase (CoMT) were selected for qRT-PCR. The genes and their respective primers are presented in Additional file 6. The qRT-PCR results indicated the starch metabolic genes such as ADP-glucose pyrophosphorylase, beta-amylase, and starch synthase were highly expressed in Ib than in It (Fig. 3). The high expression of starch metabolism genes in Ib reflects the movement of storage proteins during SR development. A similar finding for starch metabolism genes was previously reported in sweetpotato [2]. The qRT-PCR results showed that the gene encoding expansin was more highly expressed in Ib than in It. By contrast, Noh et al. [16] found that the expression of the expansin gene is inhibited during SR formation in sweetpotato. However, Firon et al. [2] demonstrated that the expansin gene is involved in SR formation in sweetpotato. In the present study, the meristematic regulatory gene KNOX1 was highly expressed in Ib, which was previously demonstrated in the development of sweetpotato SRs [13]. The qRT-PCR results showed that the gene ethylene response factor (ERF) was expressed at higher levels in It than in Ib. By contrast, Firon et al. [2] showed high expression of ERF in sweetpotato SRs compared to FRs of the cultivar Georgia Jet. The shikimate pathway gene encoding EPSP synthase was highly expressed in It. This enzyme forms an important precursor in the shikimate pathway and promotes the biosynthesis of lignin [57]. The qRT-PCR results showed that the lignin biosynthetic genes CAD and CoMT were highly expressed in It compared to Ib. In summary, the high expression of starch and sucrose metabolism genes in Ib promotes SR formation, whereas the high expression of lignin biosynthetic genes in It promotes the development of FRs/non-storage roots.
Functional classification of transcripts using Gene Ontology (GO) and pathway analysis
The Ib and It annotated transcripts were grouped into three biological functions such as biological process, molecular function, and cellular component using the GO Slim database [58]. The majority of GO annotations (47.7% in Ib and 46.6% in It) were grouped into the biological process category, followed by molecular function (27.8% in Ib and 25.6% in It). In addition, 24.5% of Ib and 27.8% of It transcripts were grouped in the cellular component category (Fig. 4; Additional files 1 and 2). Most transcripts in the biological process category are involved in oxidation-reduction processes, protein phosphorylation, regulation of transcription, metabolic processes, salt stress, and signal transduction and translation. The transcripts in the molecular function category are involved in ATP binding, protein binding, DNA binding, phosphorus transferase, and catalytic activity. The transcripts grouped within the cellular component category were based on their predicted sub-cellular locations in the nucleus, plasma membrane, chloroplast, cytoplasm, extracellular space, and vacuole. Similar groupings of annotated genes have been reported in previous sweetpotato root transcriptome studies [2, 24].
Fig. 4.
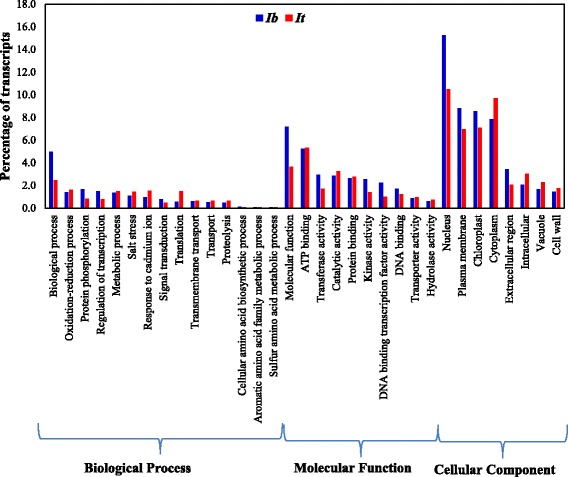
Functional classification of transcripts using Gene Ontology. I. batatas (Ib) transcripts are indicated in blue, and I. trifida (It) transcripts are indicated in red. The non-redundant transcripts were subjected to functional classification using GOSlim
The annotated transcripts between Ib and It were assigned to various biological pathways using DAVID [59, 60]. A total of 32,427 (33.0%) and 89,972 (32.7%) transcripts were assigned to 365 and 371 pathways in Ib and It respectively. Further, we also examined the pathways that are differentially enriched in Ib and It. The enriched transcripts in Ib and It (percentage of expressed transcripts) showed 16 major pathways that are involved in storage/fibrous roots (Fig. 5). The list of transcripts and KEGG-enriched pathways from DAVID are presented in Additional files 7 and 8. In Ib, we observed a higher percentage of differentially expressed transcripts in the starch (77 transcripts, 0.002% of the expressed transcripts) and sucrose biosynthetic pathways (10 transcripts, 0.001%) compared to It (177 transcripts, 0.001% in starch, and 24 transcripts, 0.0002% in sucrose biosynthetic pathways). The increased expression of sucrose and starch metabolic genes indicated the synthesis of sucrose and starch in the storage roots [2]. We also detected a large percentage of transcripts in Ib (13 transcripts, 0.0004%) than in It (20 transcripts, 0.0002%) for UDP-glucose biosynthesis, which is involved in the synthesis of UDP glucose. The increased expression of transcripts for UDP-glucose biosynthesis in Ib indicates the increased activity of UDP sugars in the storage roots. In Ib, enrichment of transcripts involved in the homogalacturonan pathway (33 transcripts, 0.001%) was observed in comparison with It (28 transcripts, 0.0003%), which indicates the accumulation of pectins in the storage roots. The pectin in the primary cell walls forms from homogalacturonans, rhamnogalacturonan-I, and rhamnogalacturonan-II [28], and strengthens the cell wall of the developing SRs.
Fig. 5.
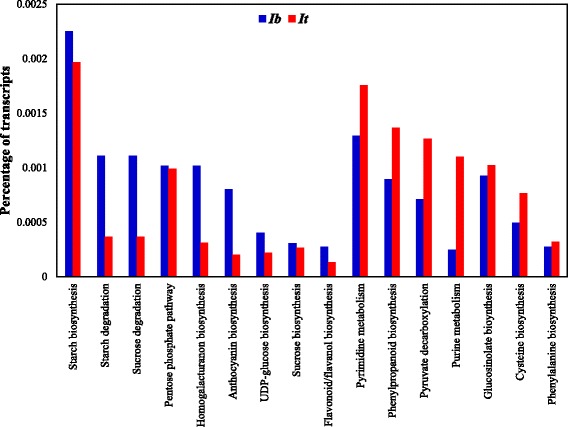
Functional classification of transcripts using DAVID. I. batatas (Ib) transcripts are indicated in blue, and I. trifida (It) transcripts are indicated in red. The functional grouping was based on KEGG pathway names associated with transcripts from DAVID analysis
The percentage of transcripts of secondary metabolic pathways such as the anthocyanin (26 transcripts, 0.0008%) and flavonoid biosynthetic pathways (9 transcripts, 0.0003%) were more represented in Ib than It (18 transcripts, 0.0002%, and 12 transcripts, 0.0001%). This is in contrast to the results of Firon et al. [2] who reported higher expression of anthocyanin and flavonoids in FRs than in SRs of sweetpotato. The more representation of anthocyanin and flavonoids in our study could be due to the use of orange-fleshed Beauregard variety. Earlier transcriptome studies showed that genes related to production of anthocyanin and flavonoid compounds were highly expressed in purple and yellow-colored sweetpotato SRs [61].
The transcripts representing the phenylpropanoid pathway (123 transcripts, 0.001%) and phenylalanine biosynthesis (29 transcripts, 0.0003%) were more represented in It than Ib (29 transcripts in It, 0.0008%, and 9 transcripts in Ib, 0.0002%) indicating the synthesis of lignin in the fibrous/non-storage roots. Similar results of higher phenylalanine biosynthesis were observed in the FRs of sweetpotato [2]. Transcripts of the pyruvate decarboxylation pathway were also more represented in It (114 transcripts, 0.001%) than Ib (23 transcripts, 0.0007%). The pyruvate decarboxylation pathway participates in carbohydrate metabolism and down-regulates the synthesis of glucose. In sweetpotato, Firon et al. [2] demonstrated the downregulation of pyruvate decarboxylase in the SRs in comparison with the FRs. We also observed more representation of transcripts related to biosynthesis of glucosinolate (92 transcripts, 0.001%) and cysteine (69 transcripts, 0.0007%), which are involved in stress and defense responses, in It compared to Ib (30 transcripts, 0.0009%, and 16 transcripts, 0.0004%). In Arabidopsis, the expression of glucosinolate occurs in the antifungal defense response [62]. In sweetpotato, Firon et al. [2] showed the high representation of cysteine biosynthesis genes, consistent with the results of our study, in the non-storage/fibrous roots. In addition, transcripts of purine and pyrimidine metabolism, which are involved in the synthesis of nucleic acids and energy carriers in the cell [63], were more represented in It (99 transcripts, 0.001%, and 158 transcripts, 0.002%) than Ib (8 transcripts, 0.0002%, and 42 transcripts, 0.001%).
Identification of Simple Sequence Repeat (SSR) markers
We predicted potential microsatellite markers based on the assembled transcripts generated from Ib and It using SSRfinder [64] and MISA (Version 1.0) [65]. We mined for new microsatellite markers in Ib and It using 98,317 and 275,044 transcripts, respectively. A total of 20,065 and 26,158 microsatellite markers were identified in Ib and It, respectively (Table 5). The list of identified SSRs, along with the associated forward and reverse primers are presented in Additional file 9 (Ib) and Additional file 10 (It). Overall, the results indicate that di-nucleotide repeats (GA/TA) were most abundant in both Ib and It. The percentage difference in the number of SSRs identified with di-, tri-, and tetra-nucleotide repeats was higher in It than in Ib (30.8%, 43.2%, and 52.5% higher, respectively). Functional genetic markers such as SSRs are useful in understanding the genetic variation in plants [66, 67].
Table 5.
Number of SSRs predicted in Ib and It
| Number of SSRs | Ib | It |
|---|---|---|
| SSRs predicted by MISA | 20,065 | 26,158 |
| SSRs with unique sets of designed primers | 7,067 | 9,650 |
Prediction of Single Nucleotide Polymorphisms (SNPs) and insertions and deletions (InDels) between Ib and It
The reads of Ib were mapped to the transcripts of It to identify the unique SNPs for Ib and vice versa for It. A total of 1,037,396 (Ib) and 495,931 (It) unique SNPs were identified for each species (Table 6). Within orthologous genes, 254,120 and 89,809 SNPs were identified in Ib and It, respectively. Also, 5,669 (Ib) and 883 (It) SNPs were identified within paralogous genes. A list of SNPs is provided in Additional file 11 for Ib and Additional file 12 for It. SNPs and InDels, the most abundant genetic variations in the genome, are often exploited for high-throughput genotyping and marker-assisted selection in plants [68]. Recently, Hirakawa et al. [69] reported a list of SNPs in the It genome. In the current study, we identified 103,439 and 69,194 InDels between Ib and It. In addition, 18,655 and 11,559 InDels were identified within orthologs between Ib and It, respectively. Within paralogs, 197 and 32 InDels were identified in Ib and It, respectively (Table 6). A list of InDels is provided in Additional file 13 for Ib and Additional file 14 for It.
Table 6.
Number of SNPs and InDels predicted between Ib and It
| Number of SNPs and InDels | Ib | It |
|---|---|---|
| Total SNPs | 1,037,396 | 495,931 |
| SNPs within orthologs | 254,120 | 89,809 |
| SNPs within paralogs | 5,669 | 883 |
| Total InDels | 103,439 | 69,194 |
| InDels within orthologs | 18,655 | 11,559 |
| InDels within paralogs | 197 | 32 |
Conclusion
We compared the root transcriptomes of SR forming cultivated I. batatas with its non-tuber-forming wild ancestor, I. trifida. Among paralogous gene sets, genes encoding RGL proteins were identified only in Ib; we speculate that RGL family proteins may play a role in SR formation in sweetpotato. In addition to the expression of the transcription factor RWP-RK domain-containing protein in Ib, other genes that are expressed in Ib, such as those encoding K+ transporters and ERECTA protein kinases, may also play a role in SR formation. qRT-PCR indicated that starch and sucrose metabolism genes such as those encoding ADP-glucose pyrophosphorylase, beta-amylase, and starch synthase, showed enhanced expression in Ib. By contrast, lignin biosynthetic genes, such as CAD and CoMT, were highly expressed in It than Ib. The root transcriptome data obtained in this study may serve as a resource for the development of molecular markers in sweetpotato and may facilitate annotation of the sweetpotato genome.
Methods
Plant materials and growth conditions
Five different I. batatas (L.) Lam cv. Beauregard (Ib) plants were grown in pots under greenhouse conditions at the University of Arkansas at Pine Bluff (UAPB), Pine Bluff, AR. Seeds from the non-tuber-forming ancestor I. trifida (Kunth) G. Don (It) (PI 540715) were obtained from the USDA-GRIN germplasm center (Plant Genetic Resources Conservation Unit, Griffin, GA). It seeds were scarified using 50% sulfuric acid and rinsed for five minutes in sterile water. The scarified seeds were germinated on sterile paper and the plants were transplanted in pots and grown under greenhouse conditions. Total roots were collected from 100-day-old plants of Ib and It. The FRs and SRs from five Ib plants were cleaned, pooled and frozen in liquid nitrogen. Similarly, the total roots of five It plants were cleaned, pooled and frozen for RNA isolation.
RNA extraction
Total RNA was extracted from roots of Ib (designated as Sp1) and It (designated as Sp2) using Qiagen RNeasy kit (Qiagen, Valencia, CA). RNA samples were subjected to DNase I (1 U/μl) treatment for 15 min., followed by heat inactivation at 65 °C for 10 min. The extracted RNA was quantified using NanoDrop 2000c (Thermo Fisher Scientific, Wilmington, DE), and the quality was checked by RNA gel electrophoresis (Additional file 15).
cDNA library construction and transcriptome sequencing
The cDNA library construction and sequencing of RNA from Ib and It (without biological controls) were carried out at the UC Davis Genome Center, San Diego, CA. The cDNA libraries were constructed following the manufacturer’s protocol (Illumina, Inc., San Diego, CA). Transcriptome sequencing was carried out using the HiSeq 2500 platform (Illumina, Inc., San Diego, CA).
Processing of RNA-seq reads
The quality of reads was assessed using FastQC (Version 0.10.0). All reads, after removing the adapter, were used for assembly without quality filtering, since at least 90% of reads passed the minimum quality score of Q30. Reads from Ib (Sp1) and It (Sp2) were assembled separately with Trinity (Version r2012– 10– 15) using default parameters [70]. Trinity assembly of Ib reads resulted in 240,915 contigs while It reads resulted in 366,513 contigs. The reads were mapped back to the assembled transcripts and isoform percentage was calculated using Trinity-RNA-seq by Expectation Maximization (RSEM) [70]. Redundant contigs were filtered from Ib and It using an optimized assembly method [71] in two steps: i) selecting a highly covered isoform for each unique component-subcomponent from Trinity output based on the RSEM results and ii) post-assembly trans-chimera cleanup using the BLASTX results against non-redundant (nr) protein database. These steps reduced the number of contigs from 240,915 to 98,317 transcripts for Ib and from 366,513 contigs to 275,044 transcripts for It. The percentage of redundant contigs was 40.8% and 75.0% for Ib and It, respectively. The filtered transcripts were used for further analysis. Likely coding sequences (ORFs) were predicted from filtered Trinity transcripts using TransDecoder located within trinity-plugins. This helped to predict longest putative ORFs from set of transcripts, out of which best scoring ORFs were selected based on Markov model (log likelihood ratio based on coding/non-coding) in each of six possible reading frames. Thus, ORFs were predicted from the filtered transcripts of both Ib and It. We also extracted FPKM values generated by RSEM for the genes of interest.
Identification of paralogous and orthologous groups
The best coding ORFs were used for identification of orthologs using OrthoMCL [72]. All proteins derived from best coding ORFs were first compared against each other by all-vs-all BLASTP search. BLAST was performed using BLOSUM62 matrix, e-value cutoff of 1.0E-5, and masked for low complexity regions. For all matching pairs of sequences “percent match length” score was calculated and all pairs with less than 50% scores were eliminated. A network of such sequence pairs across and within species was used to determine orthologous (similar set of sequences between Ib and It) and paralogous (similar set of sequences within Ib or It) sequences. All ortholog, paralog pairs identified were then clustered using Markov Clustering (MCL) program (orthoMCL). Cluster group IDs for both paralogs and orthologs are shown in Additional files 3, 4 and 5.
Functional annotation of transcripts
The filtered transcripts were classified into functional categories using different databases and tools. BLASTX was carried out of Ib and It transcripts against non-redundant database (nr), Arabidopsis protein database (The Arabidopsis Information Resource, TAIR10), Cassava protein database (ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Mesculenta) and potato protein database (ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Stuberosum); and BLASTN against the sweet potato gene index database (https://research.cip.cgiar.org/confluence/display/SPGI/Home). Both BLASTX and BLASTN were carried out with e-value cutoff of 10–3 and other default parameters. Transcripts with the best coding ORFs were predicted and additional annotation from trinotate was generated using databases such as Swissprot, PFAM, Protein signal prediction as well as EMBL Uniprot eggNOG and gene ontology (GO). Protein related information was added from PANTHER database based on BLAST hits to the Arabidopsis database. GO classification analysis was carried out using the GO Slim database [58] again based on Arabidopsis database. Based on level 3 category of GO classification, the transcripts were grouped into biological process, molecular function, and cellular component. The percentage of transcripts involved in each category was calculated based on the number of transcripts observed for individual functions versus the total number of transcripts (Fig. 4). Pathway information was added using information from TAIR10 database and DAVID (Fig. 5) [59, 60].
Quantitative Reverse-transcriptase PCR (qRT-PCR)
qRT-PCR analysis was conducted using StepOnePlus (Applied Biosystems, Carlsbad, CA); cDNA was synthesized from the total RNA used for sequencing analysis. Two-step cDNA conversion was carried out using a high-capacity cDNA reverse transcription kit according to the manufacturer’s instructions (Life Technologies, Foster City, CA). First-strand cDNA was synthesized using 10x reverse transcription buffer (2.0 μl), 100 mM dNTP mix (0.8 μl), 10x RT random primers (2.0 μl), MultiScribe reverse transcriptase-50 U/μl (1.0 μl), and RNA (500 ng). The reaction was carried out at 25 °C for 10 min., followed by 37 °C for 120 min. and 85 °C for 5 min. Subsequently, second-strand cDNA synthesis was carried out in a 10-μl reaction mixture consisting of 1.0 μl 1:1 diluted first-strand cDNA, 2x Fast SYBR Green mix (5.0 μl), 900 nM of each primer (0.8 μl), and 2.4 μl of nuclease-free water using MicroAmp optical 96-well reaction plates. The experiment was conducted with three biological replicates and two technical replicates for both Ib and It. The sweetpotato β-tubulin gene was used as an endogenous control, and gene expression levels were determined using the ΔΔCt method [73]; ΔΔCt was calculated by comparing the ΔCt values of Ib and It.
Simple sequence repeats discovery
SSRs were predicted from filtered transcripts of Ib (Sp1) and It (Sp2) using MISA software (Version 1.0) with default parameters (Table 5). Primers were designed for the predicted SSRs using Primer3 [74]. The forward and reverse primers were parsed using custom Perl script followed by filtering low-complexity primer sequences using SSRfinder to obtain a unique set of primers for predicted SSRs. A custom Perl script was used to extract repeat information such as SSR type, SSR size, and SSR start and end positions for the predicted SSRs with unique sets of primers and associated annotation information for Ib (Additional file 9) and It (Additional file 10).
Discovery of single nucleotide polymorphisms and InDels
Ib and It reads were mapped against both It and Ib genes using Burrows-Wheeler Aligner (BWA) [75]. As the read coverage was very high and SNP calling programs failed to call SNPs for very high density, Picard (Version 1.107) ‘MarkDuplicates’ was used to remove the duplicate reads. About 60% of reads were marked as duplicate by Picard and were not considered for SNP calling. SNP and InDel calling was carried out with GATK (Version 3.1.1) [76] and Samtools (Version 0.1.18) [77] with all default parameters except for the following options (--baq, --mbq=20, mmq=10, stand_call_conf=50, stand_emit_conf=30 for GATK and -Q=20, -q=10 and –C=50 for Samtools). Custom perl scripts were used for further processing to denote specifically unique and common SNPs predicted by GATK and Samtools. Additional filtering of GATK predicted SNPs were filtered with GATK filters such as FisherStrand (>60), Mapping Quality (MQ <40), HaplotypeScore (>13), MQRankSum (<−12.5) and ReadPosRankSum (<−8). InDels were filtered with additional filters such as ReadPosRankSum < −20.0 and FS > 200.0. Filtered SNPs and InDels were not removed from final set, however SNPs and InDels which passed the filters were labelled as “PASS” in “FILTER” columns while SNPs and InDels that failed to pass filters have blank “FILTER” column. Annotation and ortholog/paralog information were also added to the SNPs (Additional file 11 for Ib and Additional file 12 for It) and InDels (Additional file 13 for Ib and Additional file 14 for It).
Acknowledgements
Authors thank USDA-GRIN for the supply of I. trifida seeds (PI 540715).
Funding
Funding support for this work was provided to MM by The Plant Powered Production (P3) Center, which was funded wholly or in part by the National Science Foundation (NSF) EPSCoR Program and the Arkansas Science & Technology Authority (NSF EPSCoR award number: EPS-1003970), and USDA-NIFA Capacity Building Grant (2014–38821–22460).
Availability of data and material
Seeds from non-tuber-forming ancestor I. trifida (Kunth) G. Don (It) (PI 540715) may be obtained from the USDA-GRIN germplasm center (Plant Genetic Resources Conservation Unit, Griffin, GA, U.S.A.); and plant material for the cultivated I. batatas (L.) Lam cv. Beauregard (Ib) can be obtained from the corresponding author. Data presented in the manuscript are provided in Additional files 1-15. All sequence data of Ib (designated as Sp1) and It (designated as Sp2) from this article have been deposited in the Sequence Read Archive (SRA) at the NCBI database under the accession number: SRP090387 (https://www.ncbi.nlm.nih.gov/sra/?term=SRP090387).
Author’s contributions
MM, SKP and VK conceived and designed the experiments; SKP performed the experiments; JT, KB and SKP analyzed the data; SKP, MM, VK, JT and KB wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- 4-CL
4-coumarate-CoA ligase
- ABA
Abscisic acid
- CAD
Cinnamyl alcohol dehydrogenase
- CaLB
Calcium-dependent lipid binding protein
- CoMT
Caffeoyl-CoA O-methyltransferase
- EPSP
5-enolpyruvylshikimate-3 phosphate synthase
- ERF
Ethylene responsive transcription factor
- FR
Fibrous root
- Ib
Ipomoea batatas
- IbMADS1
Ipomoea batatas MADS-box 1
- InDels
Insertions and deletions
- It
Ipomoea trifida
- KEA5
K+ efflux antiporter
- KNOX1
Class I knotted1-like homeobox
- PAL
Phenylalanine ammonia-lyase
- RGL
Rhamnogalacturonate lyase family protein
- SNP
Single nucleotide polymorphism
- SR
Storage root
- SSR
Simple sequence repeat
Additional files
BLAST annotation and Gene Ontology (GO) classification of I. batatas (Sp1) contigs. (XLSX 20 MB)
BLAST annotation and Gene Ontology (GO) classification of I. trifida contigs (Sp2). (ZIP 42.9 MB)
Paralogous genes identified in I. batatas (Sp1). (XLSX 16 KB)
Paralogous genes identified in I. trifida (Sp2). (XLSX 285 KB)
Orthologous genes identified between I. batatas (Sp1) and I. trifida (Sp2). (XLSX 394 KB)
Primer sequences used for quantitative reverse-transcription PCR (qRT-PCR). (XLSX 12.5 KB)
Functional annotation of I. batatas (Sp1) transcripts using DAVID. (XLSX 5.85 MB)
Functional annotation of I. trifida (Sp2) transcripts using DAVID. (XLSX 12.4 MB)
SSRs and primers predicted in I. batatas (Sp1). (XLSX 5.29 MB)
SSRs and primers predicted in I. trifida (Sp2). (XLSX 7.21 KB)
SNPs identified in I. batatas (Sp1). (ZIP 190 MB)
SNPs identified in I. trifida (Sp2). (ZIP 89.7 MB)
InDels identified in I. batatas (Sp1). (ZIP 34.3 MB)
InDels identified in I. trifida (Sp2). (ZIP 22.5 MB)
Gel containing RNA samples from I. batatas and I. trifida. (DOCX 1.27 MB)
Contributor Information
Sathish K. Ponniah, Email: ponniahs@uapb.edu
Jyothi Thimmapuram, Email: jyothit@purdue.edu.
Ketaki Bhide, Email: bhide@purdue.edu.
Venu (Kal) Kalavacharla, Email: vkalavacharla@desu.edu.
Muthusamy Manoharan, Email: manoharanm@uapb.edu.
References
- 1.Ziska LH, Runion GB, Tomecek M, Prior SA, Torbet HA, Sicher R. An evaluation of cassava, sweet potato and field corn as potential carbohydrate sources for bioethanol production in Alabama and Maryland. Biomass Bioenergy. 2009;33:1503–1508. doi: 10.1016/j.biombioe.2009.07.014. [DOI] [Google Scholar]
- 2.Firon N, LaBonte D, Villordon A, Kfir Y, Solis Y, Lapis E, Perlman TS, Doron-Faigenboim A, Hetzroni A, Althan L, Nadir LA. Transcriptional profiling of sweetpotato (Ipomoea batatas) roots indicates down-regulation of lignin biosynthesis and up-regulation of starch biosynthesis at an early stage of storage root formation. BMC Genomics. 2013;14:460. doi: 10.1186/1471-2164-14-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villordon AQ, Ginzberg I, Firon N. Root architecture and root and tuber crop productivity. Trends Pl Sci. 2014;19:419–425. doi: 10.1016/j.tplants.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Villordon AQ, LaBonte DR, Firon N, Kfir Y, Schwartz A, Pressman E. Characterization of adventitious root development in sweetpotato. Hortsci. 2009;44:651–655. [Google Scholar]
- 5.Togari Y. A study of tuberous root formation in sweet potato. Bul Nat Agr Expt Sta Tokyo. 1950;68:1–96. [Google Scholar]
- 6.Wilson LA, Lowe SB. The anatomy of the root system in West Indian sweet potato [Ipomoea batatas (L.) Lam.] cultivars. Ann Bot. 1973;37:633–643. [Google Scholar]
- 7.Matsuo T, Yoneda T, Itoo S. Variations in the levels of major free cytokinins and free abscisic acid during tuber development of sweet potato. J Plant Growth Regul. 1988;7:249–258. doi: 10.1007/BF02025267. [DOI] [Google Scholar]
- 8.Matsuo T, Yoneda T, Itoo S. Identification of free cytokinins and the changes in endogenous levels during tuber development of sweet potato (Ipomoea batatas Lam.) Plant Cell Physiol. 1983;24:1305–1312. [Google Scholar]
- 9.Nakatani M, Komeichi M. Changes in endogenous indole acetic acid level during development of roots in sweet potato. Jap J Crop Sci. 1992;61:683–684. doi: 10.1626/jcs.61.683. [DOI] [Google Scholar]
- 10.Wang Q, Zhang L, Guan Y, Wang Z. Endogenous hormone concentration in developing tuberous roots of different sweet potato genotypes. Agrl Sci China. 2006;5:919–927. doi: 10.1016/S1671-2927(07)60005-4. [DOI] [Google Scholar]
- 11.Noh SA, Lee HS, Huh EJ, Huh GH, Paek KH, Shin JS, Bae JM. SRD1 is involved in the auxin-mediated thickening growth of storage root by enhancing proliferation of metaxylem and cambium cells in sweetpotato (Ipomoea batatas) J Exp Bot. 2010;61:1337–1349. doi: 10.1093/jxb/erp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ku AT, Huang YS, Wang YS, Ma D, Yeh KW. IbMADS1 (Ipomoea batatas) MADS-box 1 gene) is involved in tuberous root initiation in sweet potato (Ipomoea batatas) Ann Bot. 2008;102:57–67. doi: 10.1093/aob/mcn067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka M, Kato N, Nakayama H, Nakatani M, Takahata Y. Expression of class 1 Knotted1-like homeobox genes in the storage roots of sweetpotato (Ipomoea batatas) J Plant Physiol. 2008;165:1726–1735. doi: 10.1016/j.jplph.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 14.You MK, Hur CG, Ahn YS, Suh MC, Jeong BC, Shin JS, Bae JM. Identification of genes possibly related to storage root induction in sweetpotato. FEBS Lett. 2003;53:101–105. doi: 10.1016/S0014-5793(03)00035-8. [DOI] [PubMed] [Google Scholar]
- 15.McGregor C. Differential expression and detection of transcripts in sweetpotato (Ipomoea batatas (L.) LAM.) using cDNA microarrays. PhD thesis. Louisiana State University and Agricultural and Mechanical College; 2006. http://etd.lsu.edu/docs/available/etd-05092006-145816/
- 16.Noh SA, Lee H-S, Kim Y-S, Paek K-H, Shin JS, Bae JM. Down-regulation of the IbEXP1 gene enhanced storage root development in sweetpotato. J Exp Bot. 2013;64:129–142. doi: 10.1093/jxb/ers236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Fang B, Chen X, Liao M, Chen J, Zhang X, Huang L, Luo Z, Yao Z, Li Y. Temporal patterns of gene expression associated with tuberous root formation and development in sweetpotato. BMC Plant Biol. 2015;15:180. doi: 10.1186/s12870-015-0567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao X, Gu Y-H, Wang H-Y, Zheng W, Li X, Zhao C-W, Zhang YZ. Digital gene expression analysis based on integrated de novo transcriptome assembly of sweetpotato [Ipomoea batatas (L.) Lam.] PLoS One. 2012;7:e36234. doi: 10.1371/journal.pone.0036234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin FW, Jones A, Ruberté RM. A wild Ipomoea species closely related to the sweet potato. Eco Bot. 1974;28:287–292. doi: 10.1007/BF02861425. [DOI] [Google Scholar]
- 20.Oración MZ, Niwa K, Shiotani I. Cytological analysis of tetraploid hybrids between sweet potato and diploid Ipomoea trifida (H.B.K.). Don. Theor Appl Genet. 1990;80:617–624. doi: 10.1007/BF00224220. [DOI] [PubMed] [Google Scholar]
- 21.Srisuwan S, Sihachakr D, Yakovled SS. The origin and evolution of sweet potato (Ipomoea batatas Lam.) and its wild relatives through the cytogenic approaches. Plant Sci. 2006;17:424–433. doi: 10.1016/j.plantsci.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya T, et al. Self-incompatibility system of Ipomoea trifida, a wild-type sweet potato. In: Sawada H, et al., editors. Sexual reproduction in animals and plants. 2014. pp. 305–325. [Google Scholar]
- 23.Schafleitner R, Tincopa LR, Palomino O, Rossel G, Robles RF, Alagon R, Rivera C, Quispe C, Rojas L, Pacheco JA, Solis J, Cerna D, Kim JY, Hou J, Simon R. A sweetpotato gene index established by de novo assembly of pyrosequencing and Sanger sequences and mining for gene-based microsatellite markers. BMC Genomics. 2010;11:604. doi: 10.1186/1471-2164-11-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Fang BP, Chen J, Zhang X, Luo Z, Huang L, Chen X, Li Y. De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweetpotato (Ipomoea batatas) BMC Genomics. 2010;11:726. doi: 10.1186/1471-2164-11-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Solis J, Villordon A, Baishakh N, LaBonte D, Firon N. Effect of drought on storage root development and gene expression profile of sweetpotato under greenhouse and field conditions. J Amer Soc Hort Sci. 2014;139:317–324. [Google Scholar]
- 27.Jensen MH, Otten H, Christensen U, Borchert TV, Christensen LLH, Larsen S, Leggio LL. Structural and biochemical studies elucidate the mechanism of rhamnogalacturonan lyase from aspergillus aculeatus. J Mol Biol. 2010;404:100–111. doi: 10.1016/j.jmb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Willats WGT, McCartney L, Mackie W, Knox JP. Pectin: cell biology and prospects for functional analysis. Plant Mol Biol. 2001;47:9–27. doi: 10.1023/A:1010662911148. [DOI] [PubMed] [Google Scholar]
- 29.Wolf S, Hematy K, Hofte H. Growth control and cell wall signaling in plants. Annu Rev Pl Biol. 2012;63:381–407. doi: 10.1146/annurev-arplant-042811-105449. [DOI] [PubMed] [Google Scholar]
- 30.Aldington S, Fry SC. Oligosaccharins. Adv Bot Res. 1993;19:1–101. doi: 10.1016/S0065-2296(08)60203-7. [DOI] [Google Scholar]
- 31.Darvill AG, Augur C, Bergmann C, Carlson RW, Cheong JJ, Eberhard S, Hahn MG, Ló VM, Marfá V, Meyer B, Mohnen D, O’Neill MA, Spiro MD, van Halbeek H, York WS, Albersheim P. Oligosaccharins-oligosaccharides that regulate growth, development and defence responses in plants. Glycobiology. 1992;2:181–198. doi: 10.1093/glycob/2.3.181. [DOI] [PubMed] [Google Scholar]
- 32.Vicente AR, Saladié M, Rose JKC, Labavitch JM. The linkage between cell wall metabolism and fruit softening: looking to the future. J Sci Food Agric. 2007;87:1435–1448. doi: 10.1002/jsfa.2837. [DOI] [Google Scholar]
- 33.Hernández GM, Elena M, Rojas M, Lewis AR, Bucio JL, Pena EB, Bello ELS. Plant immunity induced by oligogalacturonides alters root growth in a process involving flavonoid accumulation in Arabidopsis thaliana. J Plant Growth Regul. 2010;29:441–454. doi: 10.1007/s00344-010-9156-x. [DOI] [Google Scholar]
- 34.Oomen RJ, Doeswijk-Voragen CH, Bush MS, Vincken JP, Borkhardt B, van den Broek LA, Corsar J, Ulvskov P, Voragen AG, McCann MC, Visser RG. In muro fragmentation of the rhamnogalacturonan I backbone in potato (Solanum tuberosum L.) results in a reduction and altered location of the galactan and arabinan side-chains and abnormal periderm development. Plant J. 2002;30:403–413. doi: 10.1046/j.1365-313X.2002.01296.x. [DOI] [PubMed] [Google Scholar]
- 35.Ashley MK, Grant M, Grabov A. Plant responses to potassium deficiencies: a role for potassium transport proteins. J Exp Bot. 2006;57:425–436. doi: 10.1093/jxb/erj034. [DOI] [PubMed] [Google Scholar]
- 36.Adams E, Shin R. Transport, signaling, and homeostasis of potassium and sodium in plants. J Int Plant Biol. 2014;56:231–249. doi: 10.1111/jipb.12159. [DOI] [PubMed] [Google Scholar]
- 37.Elumalai RP, Nagpal P, Reed JW. A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell. 2002;14:119–131. doi: 10.1105/tpc.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan YL, Llewellyn DJ, Furbank RT. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell. 2001;13:47–60. doi: 10.1105/tpc.13.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci U S A. 2003;100:2987–2991. doi: 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shpak ED, Lakeman MB, Torii KU. Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell. 2003;15:1095–1110. doi: 10.1105/tpc.010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poustka F, Irani NG, Feller A, Lu Y, Pourcel L, Frame K, Grotewold E. A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol. 2007;145:1323–1335. doi: 10.1104/pp.107.105064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hattan J, Kanamoto H, Takemura M, Yokota A, Kohchi T. Molecular characterization of the cytoplasmic interacting protein of the receptor kinase IRK expressed in the inflorescence and root apices of Arabidopsis. Biosci Biotech Biochem. 2004;68:2598–2606. doi: 10.1271/bbb.68.2598. [DOI] [PubMed] [Google Scholar]
- 43.Wagner TA, Kohorn BD. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 2001;13:303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waki T, Hiki T, Watanabe R, Hashimoto T, Nakajima K. The Arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr Biol. 2011;21:1277–1281. doi: 10.1016/j.cub.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Chardin C, Girin T, Roudier F, Meyer C, Knapp A. The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J Exp Bot. 2014;65:5577–5587. doi: 10.1093/jxb/eru261. [DOI] [PubMed] [Google Scholar]
- 46.Remacle C, Eppe G, Coosemans N, Fernandez E, Vigeolas H. Combined intracellular nitrate and NIT2 effects on storage carbohydrate metabolism in Chlamydomonas. J Exp Bot. 2014;65:23–33. doi: 10.1093/jxb/ert339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, Meyer C, Krapp A. The nodule inception- like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009;57:426–435. doi: 10.1111/j.1365-313X.2008.03695.x. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura K, Ohto M, Yoshida N, Nakamura K. Sucrose induced accumulation of β-amylase occurs concomitant with the accumulation of starch and sporamin in leaf-petiole cuttings of sweet potato. Plant Physiol. 1991;96:902–909. doi: 10.1104/pp.96.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senthilkumar R, Yeh KW. Multiple biological functions of sporamin related to stress tolerance in sweet potato (Ipomoea batatas Lam) Biotechnol Adv. 2012;30:1309–1317. doi: 10.1016/j.biotechadv.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 50.Wang QM, Zhang LM, Wang ZL. Formation and thickening of tuberous roots in relation to the endogenous hormone concentrations in sweetpotato. Sci Agric Sin. 2005;38:2414–2420. [Google Scholar]
- 51.Baunsgaard L, Lütken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005;41:595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- 52.Chen H-J, Liang S-H, Huang G-J, Lin Y-H. Sweet potato cysteine proteases SPAE and SPCP2 participate in sporamin degradation during storage root sprouting. J Plant Physiol. 2015;186:39–49. doi: 10.1016/j.jplph.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Grudkowska M, Zagdańska B. Multifunctional role of plant cysteine proteinases. Acta Biochim Pol. 2004;51:609–624. [PubMed] [Google Scholar]
- 54.Edreva A. Pathogenesis-related proteins: Research progress in the last 15 years. Gen Appl Plant Physiol. 2005;31:105–124. [Google Scholar]
- 55.Lorenza O, Piqueras R, Jose J. Sánchez-Serrano, Solano R. Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jardim-Messeder D, Caverzan A, Rauber R, Ferreira ES, Margis-Pinheiro M. Galina. Succinate dehydrogenase (mitochondrial complex II) is a source of reactive oxygen species in plants and regulates development and stress responses. New Phytol. 2015;208:776–789. doi: 10.1111/nph.13515. [DOI] [PubMed] [Google Scholar]
- 57.Mottiar Y, Vanholme R, Boerjan W, Ralph J, Mansfield SD. Designer lignins: harnessing the plasticity of lignification. Curr Opin Biotechnol. 2016;37:190–200. doi: 10.1016/j.copbio.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 58.Gene ontology consortium Gene ontology consortium: moving forward. Nucleic Acids Res. 2015;43:1049–1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie F, Burklew CE, Yang Y, Liu M, Xiao P, Zhang B, Qiu D. De novo sequencing and a comprehensive analysis of purple sweet potato (Ipomoea batatas L.) transcriptome. Planta. 2012;236:101–113. doi: 10.1007/s00425-012-1591-4. [DOI] [PubMed] [Google Scholar]
- 62.Bednarek P, Piślewska-Bednarek M, Svatoś A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, Molina A, Schulze-Lefert P. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- 63.Moffatt BA, Ashihara H. Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis book. 2002;1:0018. doi: 10.1199/tab.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao L, Tang J, Li H, Jia J. Analysis of microsatellites in major crops assessed by computational and experimental approaches. Mol Breed. 2003;12:245–261. doi: 10.1023/A:1026346121217. [DOI] [Google Scholar]
- 65.Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theo Appl Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 66.Varshney RK, Graner A, Sorrells ME. Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 2005;23:48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 67.Varshney RK, Sigmund R, Borner A, Korzun V, Stein N, Sorrells ME, Langridge P, Graner A. Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci. 2005;168:195–202. doi: 10.1016/j.plantsci.2004.08.001. [DOI] [Google Scholar]
- 68.Rafalski JA. Novel genetic mapping tools in plants: SNPs and LD-based approaches. Plant Sci. 2002;162:329–333. doi: 10.1016/S0168-9452(01)00587-8. [DOI] [Google Scholar]
- 69.Hirakawa H, Okada Y, Tabuchi H, Shirasawa K, Watanabe A, Tsuruoka H, Minami C, Nakayama S, Sasamoto S, Kohara M, Kishida Y, Fujishiro T, Kato M, Nanri K, Komaki A, Yoshinaga M, Takahata Y, Tanaka M, Tabata S, Isobe SN. Survey of genome sequences in a wild sweet potato, Ipomoea trifida (H. B. K.) G. Don. DNA Res. 2015;22:171–9. doi: 10.1093/dnares/dsv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, Palma FD, Birren BW, Nusbaum C, Toh KL, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Y, Smith SA. Optimizing de novo assembly of short-read RNA-seq data for phylogenomics. BMC Genomics. 2013;14:328. doi: 10.1186/1471-2164-14-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ. Using orthoMCL to assign proteins to orthoMCL-DB groups to cluster proteomics into new ortholog groups. Curr Protoc Bioinformatics. 2011;6:1211–19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Livak KJ, Schmittgen TD. Analysis of relative expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 74.Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 2012;40:115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 genome project data processing subgroup. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2010;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]


