Abstract
Ten years ago, a method to reprogram cells to a pluripotent state changed biomedical research
Ten years ago, Shinya Yamanaka and his student Kazutoshi Takahashi did an experiment of exquisite simplicity and elegance that changed biomedical research forever (1). By showing that a set of transcription factors could reprogram somatic cells to acquire a pluripotent stem cell state, they ushered in the era of induced pluripotent stem cells (iPSCs). The discovery made it crystal clear that cell identity is much more malleable than previously thought, and provided an invaluable tool for disease-oriented and translational researchers, bridging reductionism with patient-derived relevance. Combined with other maturing technologies, most notably genome editing and three-dimensional (3D) cell culture systems, iPSC technology has enabled investigation that was hitherto only possible in model organisms (2, 3).
THE PAST
The Takahashi and Yamanaka report took the stem cell community by storm. The first years were marked by studies published at a remarkable pace by researchers primarily from the stem cell, developmental biology, and genetic engineering fields who jointly contributed to milestones and controversies that have ultimately shaped our current—and still very incomplete—understanding of what iPSCs are and how reprogramming works.
Milestones of the first decade
Technical achievements were reached, proofs of principle in disease models were established, and biological insights into reprogramming with transcription factors were gained precipitously over the past decade. The methods of reprogramming cells to pluripotency evolved from using integrating retroviral vectors and excisable lentiviral and transposon-based vehicles, to nonintegrating episomes and RNA-based systems (Sendai virus vectors and messenger RNA transfection). Thus far, methods based solely on protein transduction or chemical compounds have been only partially successful. The stoichiometric and kinetic requirements for expressing the key transcription factors [octamer-binding transcription factor 4 (OCT4), sex determining region Y-box 2 (SOX2), Kruppel-like factor 4 (KLF4), and cellular myelocytomatosis (cMYC)] were determined, as were alternative combinations of these factors and other factors or compounds as reprogramming “enhancers” (4). As the cell types that could be reprogrammed quickly expanded, it became clear that transcription factor reprogramming to pluripotency is a robust and universal process that works with nearly any cell type from any species. An array of disease-specific iPSCs were derived from patients, and proof-of-concept for their use in disease modeling, drug testing, and regenerative medicine by means of cell therapy (which can also be combined with gene therapy) was achieved (5, 6).
How reprogramming works at the molecular level has been, not surprisingly, a harder problem to crack. Models of transcription factor reprogramming provided some evidence for both stochastic and deterministic components, potentially occurring at distinct phases of the process. Intermediate steps were described that may involve the “pioneer” binding of transcription factors to closed chromatin, a balancing act between opposing actions of lineage-specifying factors and/or a reversal of normal development, ultimately mediating the silencing of somatic genes and the activation of an endogenous network of pluripotency regulators, thereafter establishing a remarkably stable new cell identity (4). However, a clear picture of the critical events and a consensus universal model of transcription factor reprogramming remain largely elusive.
Controversies of the first decade
Along with important milestones came a series of controversial issues that quickly captured the imagination of the scientific and lay communities, as they provided grounds for questioning the value of iPSCs as sources for cell therapies and disease modeling. Fortunately, most of the initial concerns turned out to be based either on artifacts or on much more nuanced phenomena, rather than problems intrinsic to iPSCs (7). The idea of epigenetic memory arose from the finding that at an early stage of cell cultivation, iPSCs harbor DNA methylation remnants and exhibit differentiation propensities biased towards their cell of origin. This was later proven to be due mostly to incomplete reprogramming as a result of poor technique or early passage in cell culture. The idea that iPSCs may have more unstable genomes than other cells arose from the discovery of copy-number and single-nucleotide variants in iPSC lines (which are typically clonal) at a higher frequency compared to the donor tissue (which is typically highly polyclonal). These initial observations were later largely attributed to detection limits, as most genetic aberrations found in iPSCs could be shown, with sensitive methods, to preexist in the cell of origin (most human somatic tissues are highly genetically mosaic), and to have been merely captured and clonally amplified. Additional genomic variation was found to occur during cell expansion, as happens with all cultured cells. Concerns that iPSCs have immunogenic properties that could lead to their rejection, even in an autologous setting, turned out to be based on heavily context-dependent observations, while immune tolerance of iPSCs and their derivatives turned out to be, not surprisingly, dependent on multiple parameters related to the quality of the starting cell and the final cell product. Concerns that variability between iPSC lines—epigenetic, transcriptional, or in differentiation potential—could pose a serious impediment to their use were eased by conclusive evidence that most of the variability is neither random nor intractable, but stems from differences in genetic background (as iPSC lines are as genetically diverse as people and certainly much less so than inbred mice). The iPSC line-to-line variability is also not higher than that observed among the gold-standard embryonic stem cell (ESC) lines, derived either from blastocysts or by somatic cell nuclear transfer (SCNT).
THE FUTURE
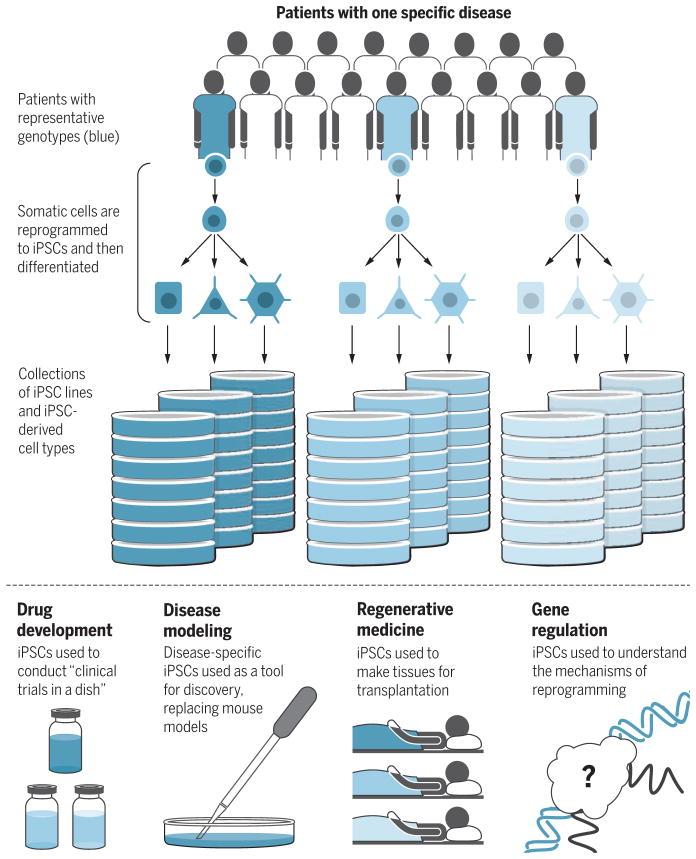
Whereas iPSCs can contribute to breakthroughs in all fields, some areas in which iPSCs can be uniquely useful over other models include disease modeling, drug development, regenerative medicine, and understanding gene regulation (see the figure).
Figure 1. New frontiers in iPSC research.
Collections of iPSC lines and diverse differentiated cell types derived from them can be assembled in repositories and used for multiple purposes.
Disease modeling
The first disease modeling studies mostly focused on well-characterized, monogenic diseases and established the principle that iPSCs derived from patients could recapitulate disease manifestations already suspected or known from previous studies using other disease models. A next wave of studies started tackling polygenic and more complex disorders, including psychiatric diseases, infectious diseases, and cancer, and also started to examine the role of human genetic variation (8). The next frontier is turning disease-specific iPSCs into a true tool for primary discovery. Because of the compelling strengths and new opportunities they provide, iPSCs are expected to become what mouse models have been in the past two to three decades. A collection of iPSC models for practically any disease and any disease-related gene mutation—also leveraging genome editing technologies, mainly the clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 system—will be steadily assembled over the next few years. These cell models will also be increasingly used in conjunction with other emerging technologies, including organoids and other 3D culture systems, organs-on-chips, and xenografts. These approaches will also likely leverage advances from fields such as synthetic biology and bioengineering to increasingly afford multicellularity, and more physiological modeling of healthy and diseased human tissue at a higher level of organization.
Building collective experience in the scientific community in the strengths and weaknesses of iPSC disease models, and increasing confidence in their readouts, will benefit from lessons in the mouse modeling field. iPSC disease models will only be as good as the iPSC lines and the differentiated cells made from them, and readouts will only be as relevant and predictive as the surrogate cellular phenotypes. Careful characterization of disease iPSC lines is therefore imperative, as is their maintenance, authentication, and distribution—which would be best handled by dedicated repositories. It will also be important to strike the right balance between studying a few well-validated lines and still capturing enough of the genetic diversity of the disease under study. Although the need for high-quality well-characterized iPSC lines and standardized differentiation procedures is broadly recognized, a less widely discussed issue is what constitutes a disease-relevant phenotype and how one can determine for what organismal phenotype a cellular phenotype observed in an iPSC disease model can serve as a surrogate. This is of utmost importance as human diseases typically manifest with diverse clinical symptoms and signs that are linked to the underlying pathogenic processes with varying degrees of relatedness and causality.
Drug development
Apart from serving as tools to discover new mechanisms of disease and new therapeutic targets, iPSCs will occupy increasingly more central roles in drug development, including candidate drug testing, compound screening, drug repurposing, and toxicity testing. Collections of iPSCs that capture a range of diseases and genotypes can be used to conduct “clinical trials in a dish” to discover biomarkers of response, while phenotype-based screens empowered by isogenic controls can provide a road to drug discovery in diseases with no known molecular targets. Candidate drug testing and high-throughput screening in iPSC-derived cells from patients with neurological disorders, such as amyotrophic lateral sclerosis (ALS) and familial dysautonomia, have already produced promising compounds, which are making their way to clinical trials (9, 10). In parallel, the ability to generate multiple normal tissues from the same initial cells offers well-defined conditions for toxicology studies (11).
Regenerative medicine
The idea of regenerating humans by replacing damaged organs and tissues with brand-new ones has fascinated scientists and the public alike for years and is feeding into most of the hype around stem cells. Initial aspirations that stem, progenitor, and precursor cells administered to an animal or human will find the cues needed to home to the right place, integrate harmoniously with the environment, and functionally benefit the host organism have proved to be overly optimistic [with the sole exception of hematopoietic stem cells (HSCs)], and it is now obvious that all of these requirements are major obstacles that stand in the way of translating stem cell research into cell therapies. The diseases that will be in line for clinical trials using cells derived from human pluripotent stem cells (hPSCs, including iPSCs and ESCs) is by and large determined by two questions: (i) What cells can be made from hPSCs in vitro with sufficient yield and functionality; and (ii) is there a disease that can be treated with these hPSC-derived cells with a reasonable chance of success and acceptable risk to patients? By these criteria, eye diseases gained priority and the first clinical trial using iPSC-derived retinal pigment epithelial cells to treat macular degeneration started patient enrollment in 2014 in Japan. The next candidates approaching the clinic include Parkinson’s disease, spinal cord trauma, diabetes, and myocardial infarction. Although hPSC-derived HSC therapies are not currently on the horizon because of the inability to generate transplantable HSCs from hPSCs, a single breakthrough may catapult them to the finish line, owing to the ease of administration and the long clinical experience with HSC transplantation.
Apart from scientific advances in cell and tissue engineering, a paramount task for moving the field forward is establishing guidelines on what tests and thresholds should be used to determine quality and safety of iPSC-derived cell products. The first iPSC clinical trial was halted 1 year later, after the safety of a patient-derived iPSC line could not be firmly established because the effects of two mutations found by next-generation sequencing could not be determined.
A major decision is whether to pursue iPSC-derived cell therapies in an autologous or allogeneic setting. As both approaches have obvious pros and cons, it is likely that this decision will be made on a case-by-case basis. Different tissues have less or more stringent requirements for tissue matching, as experience from solid organ transplantation has shown. The acceptable level of immunosuppression and the alignment of cell preparation with treatment time-lines depend on the underlying condition. Cell therapies will also be influenced by the present and future status of good manufacturing practice capabilities that will largely determine the financial sustainability, scalability, and time scales of cell manufacturing. For acute life-threatening conditions, an immediately available off-the-shelf treatment may be a more viable option. The development of banks of iPSC lines homozygous for major histocompatibility complex antigens or even universal donor lines that can potentially be engineered through genome editing could provide histocompatible cells that a large number of patients can accept without mounting an immune response. For patients requiring repeated cell administration, such as blood product transfusions or enzyme replacement therapies, and who often become sensitized to major and minor antigens, autologous therapies might be both more efficacious and cost-effective. iPSC products may be transplanted, at increasing levels of complexity, as cells in suspension, 2D sheets, 3D tissues, or even entire organs. The latter could well be generated entirely in vitro or in xenogeneic conditions in large animal chimeras.
Gene regulation
The success of transcription factor reprogramming could not have been predicted and cannot be explained with current models of gene regulation, which is why the mere fact that it worked ten years ago was originally met with so much disbelief and awe. The “Yamanaka experiment” required a leap of faith, as the only evidence at the time supporting the rewiring of a somatic cell nucleus to pluripotency were nuclear transplantation and cell fusion experiments and a few transcription factor–mediated cell fate conversions (notably the conversion of fibroblasts to muscle by the transcription factor MyoD). These were viewed as isolated oddities. Although it is now a well-established phenomenon that combinations of only few key transcription factors can induce dramatic cell fate changes (including transitions across somatic cell lineages without going through a pluripotent state), how and why this happens remains as much of a mystery as it was 10 years ago.
It is entirely possible that current models of gene regulation are missing some fundamental functionality of the genome and/or action of transcription factors. Revised models that are built upon new insights into how gene expression programs and cell identity are established may be needed to explain cell fate transitions. New experimental systems that accommodate reprogramming with high efficiency and similar kinetics will be needed to address these questions, preferably in human cells. Interdisciplinary approaches that combine single-cell analysis and systems biology methodologies are also part of this next frontier. Breakthroughs in understanding the molecular mechanisms of transcription factor reprogramming may ultimately lead to processes that generate iPSCs of the highest possible quality.
Two highly synergistic technologies in the past decade, iPSCs and the CRISPR-Cas9 system “democratized” stem cell modeling and genome editing, respectively, by making them widely accessible to the scientific community. Their convergence, together with the recent avalanche of human genome sequencing data, is revolutionizing the study and treatment of human disease. iPSCs are poised to play a central role in a shift that is already underway in the use of primary human patient material from late-stage validation studies to the front line of biomedical discovery. As iPSCs will be put to use in increasingly diverse scientific questions and applications, it will be important for all stakeholders to manage the expectations of scientists, patients, and the public, and to provide safeguards on the one hand without impeding progress on the other. As iPSC-based therapies move to the clinic, lessons from other related fields should be remembered: the boldness of the HSC transplantation field (in severe cases where this is justified) that pushed the boundaries of clinical practice beyond scientific knowledge of the time; the caution of the chimeric antigen receptor (CAR) T cell field that, through engineering of T cells and adoptive cell transfer achieved so much in treating cancers with few fatalities; and the lesson from the gene therapy field that safety cannot be ascertained until therapeutic levels of the administered cell product are reached.
References
- 1.Takahashi K, Yamanaka S. Cell. 2006;126:663. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Hockemeyer D, Jaenisch R. Cell Stem Cell. 2016;18:573. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lancaster MA, Knoblich JA. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Nat Rev Mol Cell Biol. 2016;17:183. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 5.Hanna J, et al. Science. 2007;318:1920. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 6.Papapetrou EP, et al. Nat Biotechnol. 2011;29:73. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tapia N, Schöler HR. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Zeltner N, Studer L. Curr Opin Cell Biol. 2015;37:102. doi: 10.1016/j.ceb.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 9.McNeish J, et al. Cell Stem Cell. 2015;17:8. doi: 10.1016/j.stem.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Lee G, et al. Nat Biotechnol. 2012;30:1244. doi: 10.1038/nbt.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burridge PW, et al. Nat Med. 2016;22:547. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]