Abstract
γ-Glutamylcysteine synthetase (γGCS), a rate-limiting enzyme in glutathione biosynthesis, plays a central role in glutathione homeostasis and is a target for development of potential therapeutic agents against parasites and cancer. We have determined the crystal structures of Escherichia coli γGCS unliganded and complexed with a sulfoximine-based transition-state analog inhibitor at resolutions of 2.5 and 2.1 Å, respectively. In the crystal structure of the complex, the bound inhibitor is phosphorylated at the sulfoximido nitrogen and is coordinated to three Mg2+ ions. The cysteine-binding site was identified; it is formed inductively at the transition state. In the unliganded structure, an open space exists around the representative cysteine-binding site and is probably responsible for the competitive binding of glutathione. Upon inhibitor binding, the side chains of Tyr-241 and Tyr-300 turn, forming a hydrogen-bonding triad with the carboxyl group of the inhibitor's cysteine moiety, allowing this moiety to fit tightly into the cysteine-binding site with concomitant accommodation of its side chain into a shallow pocket. This movement is caused by a conformational change of a switch loop (residues 240–249). Based on this crystal structure, the cysteine-binding sites of mammalian and parasitic γGCSs were predicted by multiple sequence alignment, although no significant sequence identity exists between the E. coli γGCS and its eukaryotic homologues. The identification of this cysteine-binding site provides important information for the rational design of novel γGCS inhibitors.
Glutathione homeostasis is critically important for maintaining both intracellular redox balance and defense against oxidative or chemical stress (1–3). γ-Glutamylcysteine synthetase (l-glutamate:l-cysteine γ-ligase, EC 6.3.2.2; γGCS) catalyzes the first and rate-limiting step of glutathione biosynthesis. The enzyme's activity is precisely controlled by nonallosteric feedback inhibition by glutathione, the limited availability of cellular l-Cys, and the transcriptional and posttranscriptional regulation of the enzyme's expression under various physiological conditions (4). Strategies to control the tissue level of glutathione pharmacologically have received considerable attention, because some parasitic and cancer cells rely on this tripeptide for protection against drugs or attacks from host defense systems (5, 6). In light of its central role in glutathione homeostasis, γGCS is an attractive target for drug design. For example, l-buthionine sulfoximine (BSO), a classical inhibitor of γGCS, was shown to prolong the survival of mice infected with the parasitic protozoan Trypanosoma brucei, which is the causative agent of African sleeping sickness in humans (7), and to inhibit the growth of Plasmodium falciparum, an agent of malaria tropica (8). Furthermore, pretreatment of cancer cells with BSO was reported to abolish the drug resistance caused by the multidrug resistance protein, MRP (6, 9, 10). In other investigations of the enzyme as a chemotherapeutic target, a number of biochemical studies have identified several residues involved in the glutamate- and nucleotide-binding sites (11–13), but none of those studies has been successful in identifying the binding site of l-Cys, the physiologically indispensable component of glutathione.
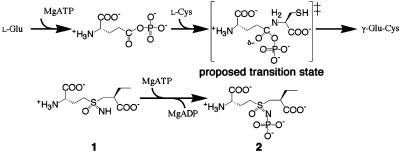
The catalytic mechanism of γGCS has been proposed to involve the initial activation of the γ-carboxyl group of l-Glu by ATP-phosphorylation to form a γ-glutamylphosphate intermediate, followed by the nucleophilic attack of the amino group of l-Cys on the carbonyl to generate a tetrahedral transition state (1, 11). The phosphate is then eliminated to yield the product γ-glutamylcysteine (γGC; Scheme 1). We have reported that a sulfoximine-based multisubstrate inhibitor, (2S)-2-amino-4-[(2S)-2-carboxybutyl-(R)-sulfonimidoyl]butanoic acid (1), served as an ATP-dependent, slow, and tight-binding inhibitor of the Escherichia coli enzyme, and that it showed ≈500 times higher affinity (K*i = 99 nM) than did BSO (K*i = 49 μM) to the enzyme (14, 15). This compound is thought to undergo mechanism-based phosphorylation to yield the corresponding N-phosphoryl sulfoximine (2) as a good transition-state analog (Scheme 1). Here, we present the 3-dimensional structure of γGCS unliganded and in complex with the transition state analog 2 at resolutions of 2.5 and 2.1 Å, respectively. By comparing these structures, we hope to elucidate the amino acid residues responsible for substrate recognition and catalysis, and thus gain structural insights into the catalytic mechanisms of this class of enzyme.
Scheme 1.
Experimental Procedures
Crystallization and Data Collection. The unliganded γGCS used in this study (518 amino acid residues; molecular mass, 58,205 Da) was mutated, expressed, purified, and crystallized as described (16). Selenomethionine (Se-Met)-substituted protein was obtained by the pathway inhibition method (17), and was purified and crystallized by using the same protocol as for the native protein. Both the unliganded and derivative crystals were dialyzed against mother liquor containing 20% glucose for flash cooling.
In preparing the complexed crystals, the purified enzyme was incubated with 20 mM of 1 in the presence of 5 mM ATP and 50 mM MgCl2 at 25°C until its activity was completely abolished, as described (14). The complex was crystallized by using the sitting-drop vapor diffusion method against 15% (wt/vol) polyethylene glycol (PEG) 2000 MME and 250 mM MgCl2 in 20 mM Tris·HCl buffer (pH 7.5) at 20°C. Diaminooctane 0.5% (vol/vol) was added to the protein solution to prevent twinning. The crystals obtained were momentarily soaked in a cryoprotective solution containing 25% PEG 400 and were cooled directly in liquid nitrogen for data collection.
X-ray diffraction data were collected at 100 K by using an MAR charge-coupled device detector at the SPring-8 beamline BL41XU. The XAFS spectrum of the selenomethionine derivative showed a clear peak and edge, and multiwavelength anomalous diffraction (MAD) data were collected. Intensity data were processed, merged, and scaled with mosflm and the ccp4 program suite (18) or crystalclear software (Rigaku/MSC, Tokyo). Data collection statistics are given in Table 1.
Table 1. Data collection and refinement statistics.
| γGCS | Se-Met-γGCS | γGCS-sulfoximine 2-MgADP | |||
|---|---|---|---|---|---|
| Data collection statistics | |||||
| Space group | R3 | R3 | P21 | ||
| Unit cell parameters | a = b = 326.82 Å | a = b = 327.03 Å | a = 70.45 Å, b = 97.36 Å | ||
| c = 104.73 Å | c = 104.45 Å | c = 102.18 Å, β = 109.63° | |||
| No. of monomers/asymmetric unit | 4 | 4 | 2 | ||
| Wavelength, Å | 0.9726 | 0.9792 (peak) | 0.9794 (edge) | 0.9839 (remote) | 1.0000 |
| Resolution, Å | 47.2-2.50 (2.59-2.50) | 55.0-2.60 (2.74-2.60) | 48.8-2.10 (2.21-2.10) | ||
| Total no. of reflections | 776,878 | 724,749 | 711,447 | 728,434 | 280,959 |
| No. of unique reflections | 144,279 | 127,496 | 127,456 | 127,571 | 73,644 |
| Completeness, % | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 97.2 (96.2) |
| Redundancy | 5.4 (5.3) | 5.7 (5.4) | 5.6 (5.3) | 5.7 (5.5) | 3.8 (3.8) |
| I/σ | 9.0 (3.4) | 9.1 (2.1) | 9.1 (1.7) | 7.3 (1.3) | 13.3 (3.2) |
| Rmerge (= ΣhΣi| Ih,I - 〈Ih〉|/ΣhΣi|Ih,i|) | 0.095 (0.346) | 0.065 (0.324) | 0.071 (0.431) | 0.087 (0.547) | 0.080 (0.371) |
| Refinement statistics | |||||
| Resolution, Å | 40.0-2.5 | 40.0-2.6 | 40.0-2.1 | ||
| Rcryst | 0.206 (0.288) | 0.199 (0.283) | |||
| Rfree | 0.236 (0.331) | 0.225 (0.298) | |||
| No. of protein atoms | 15,901 | 7,998 | |||
| No. of solvent molecules | 316 | 346 | |||
| rmsd bond length, Å | 0.021 | 0.017 | |||
| rmsd angles, ° | 1.8 | 1.4 | |||
| Ramachandran plot, % | |||||
| Most favored/allowed regions | 91.1/100.0 | 91.0/99.8 |
Values in parentheses are for the highest-resolution shell.
Structure Solution and Refinement. Multiwavelength anomalous diffraction data in the resolution range of 40 to 2.6 Å were of sufficient quality to identify the positions of all 52 Se atoms by using solve (19). The program resolve (20) was used to generate the initial solvent-flattened electron density map for the polypeptide chains with distinct solvent boundaries. The initial model was built with the program o (21) and refined with cns (22). After an initial round of refinement, the model was refined against the 2.5-Å data of the unliganded γGCS crystal by iterative maximum likelihood positional and translation, liberation, and screw-rotation displacement (TLS) refinement in refmac5 (23, 24). Each round of refinement was alternated with a round of manual rebuilding by using o, and the progress of the refinement was monitored by tracking decreases in Rcryst and Rfree. The space group of the unliganded enzyme crystal was R3, and there were four monomer molecules in the asymmetric unit. The refined overall structure was similar from monomer to monomer, because the monomer molecules in the asymmetric unit could be superimposed with <0.67-Å rms distance (rmsd) for the Cα atoms.
The crystal structure of γGCS in complex with the sulfoximine 2 and MgADP (see below) was solved by molecular replacement by using the program molrep (25). The search model used was one N-terminal half and one C-terminal half from the catalytic domain of the unliganded enzyme. The space group of this crystal was P21, and two copies of each model were searched for in the asymmetric unit. The rotation and translation functions were determined over the resolution range of 10 to 3 Å. After several rounds of iterative maximum likelihood positional and TLS refinement, the electron density from the Fo – Fc map depicted a clear density for MgADP and the sulfoximine 2. The structures of the two enzyme molecules in the asymmetric unit were essentially the same, because their Cα atoms exhibited a low rmsd of 0.30 Å. The quality of the final structures was assessed with the procheck (26) and whatcheck (27) programs. Statistics for refinement are given in Table 1.
Results and Discussion
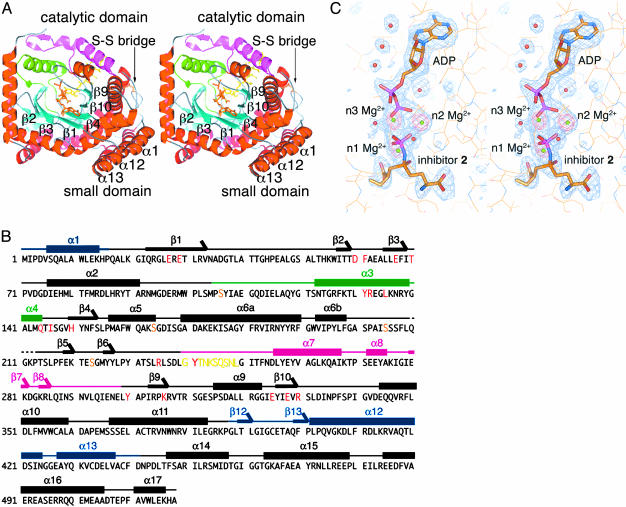
Overall Structure of the γGCS-Inhibitor Complex. Fig. 1A shows that the crystal structure comprises two structural domains: a catalytic domain (residues 18–387 and 442–518) and a small domain (residues 1–16 and 388–441). These domains are linked by swapping their terminal short chains (α1, α12, and α13) with each other and through a disulfide bridge (Cys-372–Cys-395), which is quite rare in cytosolic proteins. The catalytic domain contains six anti-parallel β-strands that form a curved partial barrel with a funnel-shaped internal cavity. The electron-density peak corresponding to the inhibitor appears near the tip of the funnel's cone, and that representing the nucleotide is found in the cone's interior. Two arm-like structures cover the open side of the β-barrel in the active site. The amino acid sequence of these arms varies widely among γGCS family members, and sequence homology analysis revealed no regions corresponding to these sequences among glutamine synthetase family members (12, 28). Thus, we termed these structures the N-terminal variable arm (residues 105–144) and the central variable arm (residues 240–298), respectively. The inhibitor model obtained is anchored by Tyr-131 and Leu-135 on the α3 helix of the N-terminal variable arm and by Tyr-241 at the N terminus of the central variable arm (see Fig. 3); all of these residues are highly conserved among prokaryotic γGCS family members. The adenine base of the nucleotide model is oriented anti to the ribose sugar, with the diphosphate moiety elongating toward the bound inhibitor (Fig. 1C).
Fig. 1.
Structures of γGCS in complex with MgADP and the sulfoximine-based transition-state analog 2. (A) Top view of a schematized model of the γGCS monomer. Helices are red and β-strands are cyan. Green and magenta indicate N-terminal and central variable arms, respectively. Each strand in the central β-barrel is labeled, and the ligands bound are shown as stick models. A switch loop (residues 240–249) is depicted as a stick model in yellow. Figures were prepared with pymol (40). (B) Amino acid sequence and secondary structure of E. coli γGCS. The residue number is indicated at the beginning of each line. Residues important for interaction with the transition-state analog are highlighted in red. Thick bars above the sequences indicate α helices (labeled α1to α17), and arrows indicate β strands (labeled β1to β12). Blue, green, and pink bars show the small domain, the N-terminal variable arm, and the central variable arm, respectively. Yellow and orange letters show the region corresponding to the switch loop and the serine residues substituted for crystallization (16), respectively. (C) SIGMAA-weighted Fo – Fc omit electron density maps for the inhibitor molecule 2, three Mg2+ ions, and ADP (blue) and for only the Mg2+ ions (red). These maps are contoured at 3 and 8 σ, respectively. Inhibitor and ADP are drawn as stick models, and the protein is drawn as a line model. Mg2+ ions and water molecules are shown as green and red spheres, respectively.
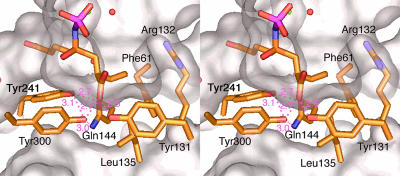
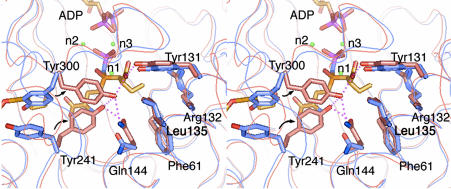
Fig. 3.
Stereoview of the residues surrounding the Cys-analog moiety of sulfoximine 2, showing the distances between the ligands. The molecular surface around the Cys-binding site is drawn in white.
The Nucleotide- and Metal-Binding Sites. The γ-phosphate of ATP has already been transferred to the NS sulfoximine nitrogen atom of the inhibitor 1 to form the N-phosphoryl sulfoximine 2. The phosphorylation of nitrogen, as noted first by Griffith, using BSO (29), mimics the phosphorylation of the γ-carboxyl group of l-Glu. The observed R-configuration around the sulfur atom in 2 is consistent with the previous finding that the sulfoximine-based compound 1, with its R-sulfur atom, is an extremely potent time- and ATP-dependent inhibitor of E. coli γGCS, whereas the corresponding (S)-sulfoximine- or sulfone-based compounds were weak and simple reversible inhibitors (14). The same relative configuration as that of the sulfur atom in 2 is thought to occur in a tetrahedral adduct, suggesting that l-Cys approaches from the si-face of the carbonyl of the γ-glutamylphosphate intermediate.
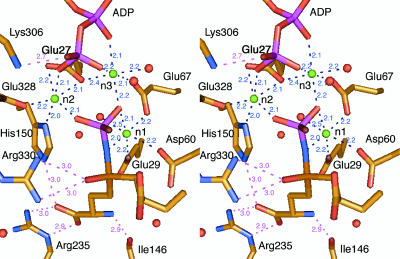
A constellation of three important Mg2+ ions is found around the phosphate-binding pocket; these ions are located at sites termed n1, n2, and n3 (12). The n1 Mg2+ is coordinated to Glu-29 OE2, Asp-60 OD1, Glu-67 OE1, a water molecule, and atoms of both the NS nitrogen and OP1 oxygen of the N-phosphoryl sulfoximido group of 2 (Fig. 2). The NS and OP1 atoms of 2 are located at equatorial positions in the octahedral coordination group, so that the OS oxygen atom of the sulfoximine is oriented toward Arg-330 to accept symmetrical bifurcated hydrogen bonds from its guanidino group. In addition to being coordinated with the n1 Mg2+, the carbonyl group of the γ-glutamylphosphate intermediate is also probably anchored by Arg-330, thus orienting its si-face toward the Cys-binding site (see below). Furthermore, the guanidino group of Arg-330 can polarize the carbonyl group and then stabilize the oxyanion formed in the transition state to promote the reaction.
Fig. 2.
Stereoview of the residues surrounding the Glu-analog moiety of the sulfoximine 2 and the organization of the trimetal cluster, showing the distances between the ligands.
The other two Mg2+ ions form a bridge between 2 and ADP. The n2 Mg2+-binding site consists of Glu-27 OE1 and OE2, His-150 ND1, Glu-328 OE2, the oxygen atom O3B of the β-phosphate group of ADP, and the oxygen atom OP3 of the transferred phosphate group. The n3 site is constructed from Glu-27 OE1, Glu-67 OE2, a water molecule, the oxygen atoms O2A and O3B, respectively, of the α- and β-phosphate groups of ADP, and the oxygen atom OP1 of the transferred phosphate. Although the presence of a third Mg2+ ion has not yet been reported for γGCS, this pattern of Mg2+ binding has been observed in pyruvate kinase (30), glutathione synthetase (31), d-Ala-d-Ala ligase (32), and d-Ala-d-lactate ligase (33). Therefore, the bridging coordination of the n2 and n3 Mg2+ ions may be a common catalytic motif found in the family of ATP-dependent ligases (ADP-forming), and is probably responsible for increasing the reactivity of the γ-phosphate group of ATP and for coordinating, and thus stabilizing, the γ-glutamylphosphate intermediate (34, 35). These Mg2+ ions also play a key role as Lewis acids in facilitating the elimination of ADP and phosphate.
The Glutamate-Binding Site. In contrast to its relatively loose fit of the cysteine-binding site, γGCSs bind l-Glu and its analogs with quite high specificity (11, 36). The glutamate moiety of 2 occurs in an extended conformation in the interior of the hydrophilic stem of the active-site funnel, which also contains part of a polyethylene glycol 400 molecule that was added as a cryoprotectant. The α-amino group of 2 is bound to the main-chain carbonyl group from Ile-146 located at the C terminus of the short α helix α4 in the N-terminal variable arm. Meanwhile, the α-carboxyl group of 2 is bound to both the guanidino group of Arg-235 and the imidazolyl group of His-150. Only small differences are found near this site between the unliganded and the complexed structures, except for the side chain of Tyr-241, which covers the glutamate moiety in the hydrophilic stem in the inhibitor-bound structure, as discussed below.
The Cysteine-Binding Site. Fig. 3 illustrates the residues around the moiety of 2 that mimic l-α-aminobutyrate (l-Abu), a substitute for l-Cys. Studies of the substrate specificity of the E. coli and mammalian enzymes have indicated that, whereas the E. coli enzyme exhibits high specificity for l-Cys and accepts only a few unnatural amino acids, whose side chains closely approximate the size and polarity of that of l-Cys (36), homologs from mammals such as rat and bovine accept a variety of natural and unnatural amino acids (11). The ethyl (-C2H5) side chain of 2 fills up a shallow pocket that is open to the cavity in the funnel cone and is built from the hydrophobic residues Phe-61, Tyr-131, and Leu-135, which make up its wall, and the polar side chain of Arg-132, which forms its floor. The terminal methyl of the -C2H5 side chain is 4.2 Å from the guanidino group of Arg-132; no significant polar interactions are expected between these groups. However, the side chain of l-Cys, which is larger, would be ≈0.3 Å closer to the bottom; polar interaction with this Arg residue would increase its binding affinity. In fact, the Km value for l-Cys (0.10 mM) is 40 times smaller than that for l-Abu (3.9 mM), and β-chloro- and β-cyanoalanine are good substrates for E. coli γGCS (36).
The O2 oxygen atom of the terminal carboxyl group of 2 is stabilized by hydrogen bonds from Tyr-131 and from Tyr-300, which in turn is involved in a hydrogen-bonding triad with Tyr-241 and Gln-144. This hydrogen-bond network accounts for a significant contribution (3.8 kcal·mol–1) of the carboxyl group of inhibitor 2 to the overall binding energy, and is evidenced by the lower affinity of BSO, a carboxyl-free version of 2 (15).
There are no specific residues around the -CH2-next to the sulfur, implying that the enzyme does not have a basic residue to abstract a proton from the α-NH+3 group of l-Cys. The lack of a catalytic base for this purpose has also been noted in the mechanistically related ATP-dependent peptide ligases, such as glutathione synthetase (37) and d-Ala-d-Ala ligase (32).
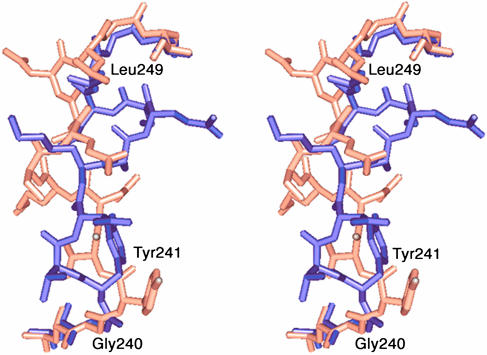
Induced Fit Essential for Intrinsic Binding of Cysteine at the Transition State. Comparison of the complexed and unliganded crystal structures reveals major conformational changes around the central variable arm induced by the binding of transition-state analog 2 (Fig. 4). The mechanisms by which γGCS properly recognizes its essential substrate, l-Cys, and the competitive inhibitor, glutathione (38), which regulates the synthesis of γGC, are unknown. In the unliganded structure, the side chains of Tyr-241 and Tyr-300 are located out of the active site and are stacked loosely on top of each other. In this state, an open space remains around the representative Cys-binding site. This space is large enough to accommodate the cysteinylglycine moiety of glutathione and is probably responsible for the feedback inhibition by glutathione. Upon binding of the inhibitor 2, an altered path of a switch loop (residues 240–249; Fig. 5) is facilitated, and this conformational change of the loop permits an inward shift of the central variable arm (α-carbon rmsd, 0.9 Å), thereby moving the key residues into the Cys-binding site. Thus, the side chain of Tyr-300 is rotated in a 137° χ-1 rotation from the stacking position into the Cys-binding site, allowing it to form a hydrogen bond with the terminal carboxyl group of 2. The side chain of Tyr-241 also turns by a 108° χ-1 rotation, forming a hydrogen bond triad with the side chains of Tyr-300 and Gln-144. As a result, the C-terminal carboxyl group of 2, which corresponds to the carboxyl group of l-Cys, is recognized by the hydrogen-bonding network involving Tyr-131, Gln-144, Tyr-241, and Tyr-300. This hydrogen-bonding network probably contributes in the orientation of l-Cys for nucleophilic attack toward the carbonyl of the γ-glutamylphosphate intermediate from its si-face. The cooperative binding of both the carboxyl group and the side chain of l-Cys by these induced-fit mechanisms (39) thus establishes the tetrahedral geometry at the transition state, which the inhibitor 2 mimics.
Fig. 4.
Conformational changes upon binding of the transition-state analog. Superimposition of the side (stick) and main (ribbon) chains of γGCS involved inl-Cys binding and recognition is shown. The binding region of the inhibitor's Cys-analog moiety of the free enzymes and the complex with the sulfoximine 2 are shown in blue and pink, respectively.
Fig. 5.
Superimposition of residues 238–251, including the switch loop. The loop's hinge residues, Gly-240 and Leu-249, are labeled.
These major conformational changes may be responsible for the slow-binding kinetics commonly observed for the inhibition of γGCS by sulfoximine-based inhibitors such as BSO and compound 1. This speculation is consistent with the view that the observed slow-binding kinetics are not due to a chemical process, such as the phosphorylation of the NS nitrogen, because they persisted when chemically synthesized N-phosphoryl sulfoximine 2 was used as an inhibitor (14).
Proposed Cysteine-Binding Sites of Other γGCS Members. In contrast with the highly conserved sequences around the MgATP- and Glu-binding sites, the sequences of the N-terminal and central variable arms of the E. coli γGCS are quite different from those of other γGCSs. This result has made it difficult to predict the Cys-binding site by homology search experiments. Using the crystal structure shown here, we obtained a set of predicted multiple alignments of the sequences of the variable arms from a number of eukaryotic and prokaryotic enzymes. The alignment predicts several conserved residues that form part of the putative Cys-binding site (Fig. 6). Tyr-241 is conserved in all of the species shown, whereas Leu-135 and Gln-144 are replaced by the functionally equivalent amino acids Ile and Asn, respectively. Among the eukaryotic enzymes, Tyr-131 and Tyr-300, which together bind the carboxyl group of l-Cys in the E. coli enzyme, are replaced with Thr and Trp, respectively. It is not clear to what degree these substitutions would preserve or weaken the hydrogen-bonding network seen in the E. coli enzyme, and the resulting impact on binding affinity. Interestingly, a well-conserved Arg-132 in the side-chain pocket of the cysteine-binding site is replaced by Asn and Gln in the enzymes from T. brucei and P. falciparum, respectively. These alignment results suggest that all γGCSs share the basic mechanism for the recognition of l-Cys seen in the E. coli enzyme, where the side chain and carboxyl group are bound cooperatively in association with a conformational change.
Fig. 6.
Multiple sequence alignment of N-terminal and central variable arm sequences for prokaryotic and eukaryotic γGCSs. Ec, E. coli K strain; Vc, Vibrio cholerae; Pa, Pseudomonas aeruginosa; Ba, Buchnera aphidicola; Hu, human γGCS heavy chain; Ra, rat γGCS heavy chain; Tc, Trypanosoma cruzi; Tb, T. brucei; Pf, P. falciparum. The first and last residue numbers of each sequence are indicated, and the total length of each protein is shown at the end. Residues that are inserted or that occur between the arms are not displayed, and the number of omitted residues is in parentheses. Residues important to the binding of the sulfoximine inhibitor in the E. coli enzyme are highlighted in red, and the hinge residues of the switch loop are in blue. The substitution of His-370 in the human γGCS (green), which has been found to cause low red-blood cell glutathione levels and hereditary hemolytic anemia (41), is proposed to be included in the central variable arm.
However, the differences found among these active-site residues may underlie their respective differences in binding of l-Cys and its analogs. In fact, the mammalian enzymes show a broader specificity for the side chain of cysteine substrates and are inhibited potently by BSO and its higher homologs (29), which lack the terminal carboxyl group. On the other hand, E. coli γGCS is only weakly inhibited by BSO (14, 36), whereas the sulfoximine 1 affects it potently (14, 15). Therefore, the structure of the Cys-binding site is important in determining the enzyme's sensitivity to inhibitors and specificity for its substrates. In this regard, the predicted Cys-binding site shown here provides hints for the rational design of novel γGCS inhibitors to control the cellular levels of glutathione in biological and medical contexts.
Acknowledgments
Synchrotron radiation experiments were conducted at SPring-8 with the approval of the Japan Synchrotron Radiation Research Institutes (JASRI) (Proposal No. 2001A0084-NL-np, 2002A0440-NL1-np, and 2002B0341-NL1-np). This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture and the National Project on Protein Structural and Functional Analysis. This work was also supported in part by the Protein 3000 Project from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan. J.O. and T.H. were supported by a grant-in-aid for scientific research from Fukui-ken University.
Author contributions: T.H., H.K., J.H., and J.O. designed research; T.H., H.N., and J.H. performed research; A.K. and J.H. contributed new reagents/analytical tools; T.H., H.N., T.N., and H.K. analyzed data; and T.H., T.N., H.K., J.H., and J.O. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: γGCS, γ-glutamylcysteine synthetase; BSO, l-buthionine sulfoximine; rmsd, rms distance.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1V4G (unliganded model) and 1VA6 (complexed model)].
References
- 1.Meister, A. (1974) in The Enzymes, ed. Boyer, P. D. (Academic, New York), pp. 671–697.
- 2.Anderson, M. E. (1998) Chem. Biol. Interact. 111–112, 1–14. [DOI] [PubMed] [Google Scholar]
- 3.Soltaninassab, S. R., Sekhar, K. R., Meredith, M. J. & Freeman, M. L. (2000) J. Cell. Physiol. 182, 163–170. [DOI] [PubMed] [Google Scholar]
- 4.Lu, S. C. (2000) in Current Topics in Cellular Regulation (Academic, New York), pp. 95–116. [DOI] [PubMed]
- 5.Griffith, O. W. (1999) Free Radical Biol. Med. 27, 922–935. [DOI] [PubMed] [Google Scholar]
- 6.Bailey, H. H. (1998) Chem. Biol. Interact. 111–112, 239–254. [DOI] [PubMed] [Google Scholar]
- 7.Arrick, B. A., Griffith, O. W. & Cerami, A. (1981) J. Exp. Med. 153, 720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lüersen, K., Walter, R. D. & Müller, S. (2000) Biochem. J. 346, 545–552. [PMC free article] [PubMed] [Google Scholar]
- 9.Rappa, G., Gamcsik, M. P., Mitina, R. L., Baum, C., Fodstad, O. & Lorico, A. (2003) Eur. J. Cancer 39, 120–128. [DOI] [PubMed] [Google Scholar]
- 10.Fojo, T. & Bates, S. (2003) Oncogene 22, 7512–7523. [DOI] [PubMed] [Google Scholar]
- 11.Griffith, O. W. (1999) Adv. Enzymol. Relat. Areas Mol. Biol. 73, 209–267. [DOI] [PubMed] [Google Scholar]
- 12.Abbott, J. J., Pei, J., Ford, J. L., Qi, Y., Grishin, V. N., Pitcher, L. A., Phillips, M. A. & Grishin, N. V. (2001) J. Biol. Chem. 276, 42099–42107. [DOI] [PubMed] [Google Scholar]
- 13.Abbott, J. J., Ford, J. L. & Phillips, M. A. (2002) Biochemistry 41, 2741–2750. [DOI] [PubMed] [Google Scholar]
- 14.Tokutake, N., Hiratake, J., Katoh, M., Irie, T., Kato, H. & Oda, J. (1998) Bioorg. Med. Chem. 6, 1935–1953. [DOI] [PubMed] [Google Scholar]
- 15.Hiratake, J., Irie, T., Tokutake, N. & Oda, J. (2002) Biosci. Biotechnol. Biochem. 66, 1500–1514. [DOI] [PubMed] [Google Scholar]
- 16.Hibi, T., Hisada, H., Nakatsu, T., Kato, H. & Oda, J. (2002) Acta Crystallogr. D 58, 316–318. [DOI] [PubMed] [Google Scholar]
- 17.Duyne, G. D. V., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993) J. Mol. Biol. 229, 105–124. [DOI] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project No. 4. (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 19.Terwilliger, T. C. (1994) Acta Crystallogr. D 50, 17–23. [DOI] [PubMed] [Google Scholar]
- 20.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, T. A., Bergdoll, M. & Kjeldgaard, M. (1990) in Crystallographic and Modeling Methods in Molecular Design (Springer, New York), pp. 189–195.
- 22.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 24.Winn, M., Isupov, M. & Murshudov, G. N. (2001) Acta Crystallogr. D 53, 122–133. [DOI] [PubMed] [Google Scholar]
- 25.Vagin, A. & Teplyakov, A. (1997) J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- 26.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. App. Crystallogr. 26, 283–291. [Google Scholar]
- 27.Vriend, G. (1990) J. Mol. Graphics 8, 52–56. [DOI] [PubMed] [Google Scholar]
- 28.Copley, S. D. & Dhillon, J. K. (April 29, 2002) Genome Biol. 3, 10.1186/gb-2002-3-5-research0025. [DOI] [PMC free article] [PubMed]
- 29.Griffith, O. W. (1982) J. Biol. Chem. 257, 13704–13712. [PubMed] [Google Scholar]
- 30.Baek, Y. H. & Nowak, T. (1982) Arch. Biochem. Biophys. 217, 491–497. [DOI] [PubMed] [Google Scholar]
- 31.Hara, T., Kato, H., Katsube, Y. & Oda, J. (1996) Biochemistry 35, 11967–11974. [DOI] [PubMed] [Google Scholar]
- 32.Fan, C., Moews, P. C., Walsh, C. T. & Knox, J. R. (1994) Science 266, 439–443. [DOI] [PubMed] [Google Scholar]
- 33.Kuzin, A. P., Sun, T., Jorczak-Baillass, J., Healy, V. L., Walsh, C. T. & Knox, J. R. (2000) Structure 8, 463–470. [DOI] [PubMed] [Google Scholar]
- 34.Jenks, W. P. (1987) in Catalysis in Chemistry and Enzymology (Dover, New York), pp. 111–115.
- 35.Matte, A., Tari, L. W. & Delbaere, L. T. J. (1998) Structure 6, 413–419. [DOI] [PubMed] [Google Scholar]
- 36.Kelly, B. S., Antholine, W. E. & Griffith, O. W. (2002) J. Biol. Chem. 277, 50–58. [DOI] [PubMed] [Google Scholar]
- 37.Hiratake, J., Kato, H. & Oda, J. (1994) J. Am. Chem. Soc. 116, 12059–12060. [Google Scholar]
- 38.Richman, P. & Meister, A. (1975) J. Biol. Chem. 250, 1422–1426. [PubMed] [Google Scholar]
- 39.Koshland, D. E., Jr. (1963) Science 142, 1533–1541. [DOI] [PubMed] [Google Scholar]
- 40.DeLano, W. L. (2002) pymol Molecular Graphics System (DeLano Scientific, San Carlos, CA).
- 41.Beutler, E., Gelbart, T., Kondo, T. & Matsunaga, A. T. (1999) Blood 94, 2890–2894. [PubMed] [Google Scholar]