Abstract
We propose to estimate the proportion of variance explained by regression on genome-wide markers (or genomic heritability) when wild/domestic status is considered the phenotype of interest. This approach differs from the standard Fst in that it can accommodate genetic similarity between individuals in a general form. We apply this strategy to complete genome data from 47 wild and domestic pigs from Asia and Europe. When we partitioned the total genomic variance into components associated to subsets of single nucleotide polymorphisms (SNPs) defined in terms of their annotation, we found that potentially deleterious non-synonymous mutations (9566 SNPs) explained as much genetic variance as the whole set of 25 million SNPs. This suggests that domestication may have affected protein sequence to a larger extent than regulatory or other kinds of mutations. A pathway-guided analysis revealed ovarian steroidogenesis and leptin signaling as highly relevant in domestication. The genomic regression approach proposed in this study revealed molecular processes not apparent through typical differentiation statistics. We propose that at least some of these processes are likely new discoveries because domestication is a dynamic process of genetic selection, which may not be completely characterized by a static metric like Fst. Nevertheless, and despite some particularly influential mutation types or pathways, our analyses tend to rule out a simplistic genetic basis for the domestication process: neither a single pathway nor a unique set of SNPs can explain the process as a whole.
Introduction
It is now widely accepted that animal and plant domestication, arguably the most important event in the last 13 000 years of humanity (Diamond, 2002), was no simple process. It involved complex humanistic, environmental and ecological factors (Larson et al., 2014; Zeder et al., 2006). The process was likely quite different dependent on species, with varied contributions of mutualism versus commensalism and humanistic intention. A common component has been selected, for both adaptations to human agricultural environments as well as to preferred phenotypic characteristics that meet human preferences. In several species spanning the Plant and Animal Kingdoms, for example, cattle, pig, rice, domestication has occurred at multiple locations. Further, rather than being an abrupt change, domestication plausibly involved gradual discontinuities in gene flow between the domestic and the wild populations (Frantz et al., 2015). This gene interchange, even if small, has continued to the present day in some cases like Mediterranean pigs (Ramírez et al., 2014).
Nevertheless, domestication has resulted in a number of commonly shared phenotypes across animal species. Unsurprisingly, some of the most salient features modified by human intervention in several animal species include coat color and behavior, for example, increased tameness and lack of fear towards humans. Domestic animals are also characterized by modified feeding habits, growth or reproductive features, such as the loss of estrus seasonality in ruminants (sheep, cattle and goat) as well as in pigs or rabbits (Chemineau et al., 2008; Carneiro et al., 2014). For instance, in terms of feeding, it was recently found that increased starch digestion capacity has occurred in dogs as compared with wolves, and that although one gene was convincingly identified (AMY2B), other unknown genes are certainly involved (Axelsson et al., 2013). In general, domestication has modified numerous phenotypes that are polygenic. Even clear domestication-driven phenotypes may vary within a species. For instance, Freedman et al. (2014) found that not all domestic dogs exhibited the genetic mark for increased starch digestion ability. As a result, identifying the specific causal mutations involved in domestication will remain, for the most part, elusive (Carneiro et al., 2014). Further, the impact exerted by recent human-mediated positive selection may act to mask the earlier genomic signatures of the domestication process. This may be particularly true for livestock species that have experienced increasingly strong positive selection over the past couple of hundred years following the assortment of animals into breeds and the introduction of sophisticated breeding practices.
Animal and plant breeders have recognized the polygenic nature of trait variation for a long time and applied the infinitesimal model for analysis and prediction of complex trait breeding values. This is equivalent to fitness in a population genetics context. This model circumvents the need to identify the causal mutations, and can be used to estimate the proportion of variance explained by genetic factors without explicit knowledge about what these factors are. The estimation of genetic variance and of heritability exploits the degree of resemblance between relatives. Traditionally, these models have been applied to family data with known pedigrees. However, nowadays, it has become more common to assess genetic relationships using molecular markers. Genomic relationships (G) can then be used to estimate the fraction of phenotypic variance explained by regression on the markers used to compute G, the so-called ‘genomic heritability', hG2, for example, de los Campos et al. (2015).
The use of markers to compute relatedness between individuals has a long history (Thompson, 1975). However, it was only with the recent advent of large-scale genotyping tools that they have been applied to replace pedigree-based relationships. As a result, whole-genome regression was adopted for analysis and prediction of complex traits in breeding programs as genomic selection (Meuwissen et al., 2001). So far, these techniques have been applied to classical quantitative traits such as height in humans (Yang et al., 2010), or performance in livestock (Hayes et al., 2009) and plant breeding (Crossa et al., 2010). However, the same principles can be applied to less conventional traits, such as domestication status. The proposed approach uses the Genomic Best Linear Predictor G-BLUP (VanRaden, 2008), extended with addition of a probit link, to regress domestic status on a set of single nucleotide polymorphisms (SNPs). The model renders estimates, such as genomic heritability, that can be used to assess the relative proportion of variance in domestication explained by regression on the marker set.
In this article we demonstrate how the G-BLUP model can be used to estimate the proportion of variance in domestication that can be explained by regression on SNPs, or genomic heritability. Perhaps more importantly, we discuss how mixed models can be used to partition the genetic variance for domestication into components attributable to sets of variants defined based on annotation (for example, functional, regulatory, deleterious, and so on). An appeal of mixed model theory, on which heritability estimation is founded, lies in its flexibility to accommodate covariance structures in a rather general form. For instance, hG2 can be estimated using different genome-wide markers (for example, SNP) that can be used to infer their relevance in discriminating wild from domestic specimens. We shall exploit these features in this work to partition the amount of genetic similarity in terms of functional classifications of marker sets.
We present the application of these concepts to whole-genome data from 47 wild and domestic pigs from Europe and Asia. The pig is a species with a complex demographic history: their native distribution ranges through most of Eurasia, and it was independently domesticated in Europe and in Asia from local wild boars, Asian and European clades diverged ca. 1.2 MYA (million years ago; Larson et al., 2005; Groenen et al., 2012). To further complicate matters, most European domestic breeds were crossed with Chinese pigs starting in the seventeenth century. As a result, as much as 20% of its genome has been inferred to be of Asian origin (Bosse et al., 2014; Bosse et al., 2015; Bianco et al., 2015). A central objective of our study was to investigate whether similar biological conditions were targeted in these two domestication events and some sort of ‘convergent evolution' did occur. We propose to quantify, via the hG2 parameter, how much two conceptual replicates of the same ‘experiment' (domestication) can be explained by similar genetic processes.
Materials and methods
Samples
We analyzed complete genome sequence from 47 domestic and wild pigs from Asia and Europe (Supplementary Table 1). Our aim was to capture signals that were shared between the different domestic pig lineages. Therefore, the sampling scheme was designed to be as balanced as possible while simultaneously aiming at capturing variability within continents and within domestic status. Among all sequences available, we chose eight wild boars (WB) from Asia (WA) and eight from Europe (WE), and four samples from four domestic Chinese (DA) and four European (DE) breeds. In China, we chose breeds from four distant geographic regions: Tibetan pigs from Tibet (West), Meishan (East), Hetao (North) and Bamaxiang (South). As for Europe, two breeds considered as ‘local' breeds (Spanish Iberian and Hungarian Mangalica) and two worldwide used breeds (Large White and Duroc) were analyzed. Four samples from each of the domestic breeds were chosen, except for Mangalica, for which only three complete sequences were available. Most of the sample sequences were downloaded from the short read archive (SRA, http://www.ncbi.nlm.nih.gov/sra; Groenen et al., 2012; Rubin et al., 2012; Esteve-Codina et al., 2013; Molnár et al., 2014; Ai et al., 2015; Bianco et al., 2015), but five were specifically sequenced for this study: a wild boar from East Russia (Primorsky Krai Peninsula, WA6), a Tunisian wild boar (WE8), a Spanish wild boar from North Spain (Asturias region, WE6) and 2 Iberian pigs (IB2 and IB3). Supplementary Table 1 presents details from all samples analyzed. The new sequences were obtained in the Centro Nacional de Análisis Genómico (CNAG, www.cnag.cat, Barcelona, Spain) using the HiSeq2000 Illumina platform. Library preparation was done according to the Illumina paired-end sequencing protocol, with minor modifications. All samples analyzed in this work had been shotgun sequenced with Illumina technology, although employing different versions of HiSeq.
Bioinformatic analyses
Samples were sequenced to an average depth of 11 × approximately (Supplementary Table 1). One lane per pig was analyzed; when there was more than one lane available per sample, the one containing the largest number of reads was chosen. Raw reads or aligned bam files were downloaded from SRA (www.ncbi.nlm.nih.gov/sra). For raw reads, alignment was carried out with bwa —mem option (Li and Durbin, 2009). Alignment was against the latest pig reference genome v. 10.2. For all downloaded and in-house obtained bam files, PCR duplicates were removed with samtools v. 1.19 (Li et al., 2009) with rmdup option. The bam files were then realigned around indels with GATK IndelRealigner tool (McKenna et al., 2010).
SNP calling was performed with samtools/bcftools suite v. 1.2.1 (Li et al., 2009) for each individual separately. SNPs were called in positions with depth bounds between 5x and twice the average depth plus one; further, minimum mapping (RMS) quality of 20 and base quality of 20 were required. Finally, SNPs with a minimum quality of 10 were retained. To estimate nucleotide diversity, in addition to heterozygous positions, the number of bases sequenced is also required. The new version of samtools provides the ‘homozygous blocks', that is, the segments where the sample is equal to the reference. In principle, this should suffice but, unfortunately, there is no tool to filter homozygous blocks when both lower and upper depth limits are set. To remedy this, we extracted the regions with minimum and maximum depth using samtools depth, further filtering by minimum map and base qualities and then, using bedtools (Quinlan, 2014), we intersected these regions with the homozygous blocks provided in the individual gvcf file. This resulted in a modified gvcf file where both SNPs and homozygous blocks had been filtered by the same criteria.
Merging individual SNP files can be done with multiple tools (Danecek et al., 2011) but they do not consider homozygous blocks properly, as they do not distinguish between a missing SNP or a homozygous reference genotype. Here we followed a two-step approach. First, we extracted the consensus fasta from the gvcf file for each individual and second, we merged all fasta files identifying the polymorphic positions. This strategy allows us to extract, with modest memory and CPU requirements, the complete joint SNP file where individual missing and called positions can be easily tracked. For further analyses, singletons, SNPs missing in >30% of the samples and sex-linked markers were discarded. To build the marker relationship matrices, missing genotypes were imputed with Beagle 4 (Browning and Browning, 2013).
SNP-based relationship matrices
SNP-based relationship matrices (G) were computed from the multiple-individual vcf file using custom perl and fortran programs, according to the following equation (VanRaden, 2008):
where gij is the ij-th individual element of G, xjk is the genotype of j-th individual at k-th SNP coded for example, as 0,1,2 and pk is the k-th SNP minor allele frequency. To compute p, we employed the 47 sample data. The SNP list used in building G permits a fine decomposition of genetic components along the genome. Here we used the following SNP sets:
1. All autosomal SNPs.
2. Functional decomposition: all SNPs within genes of a given pathway (see below).
3. Annotation feature: we computed distinct G-matrices using synonymous, non-synonymous, enhancer or transcription factor SNPs (see below).
For the different SNP filtering steps, we used bcftools (Li et al., 2009) and vcftools (Danecek et al., 2011), embedded in shell scripts.
SNP annotation
SNPs were annotated with variant effect predictor tool from ensembl (database v. 79). Pig transcription factors, including chromatin remodeling factors, were downloaded from animal transcription factor database v. 2.0 (http://www.bioguo.org/AnimalTFDB/, Zhang et al. 2012). They comprised 1067 genes and 289 641 SNPs were found in our data within these genes. Pig enhancer regions were downloaded from Villar et al. (2015). These consisted of regions enriched in H3K27 acetylation (32 979 regions, 1 290 312 SNPs) and H3K4 trimethylation (10 756 regions, 122 311 SNPs) along the pig genome.
Functional partitioning
The complete data set from NCBI biosystems v. 20150108 (www.ncbi.nlm.nih.gov/biosystems/) was downloaded, and the human data set was selected. The downloaded file contained 135 652 records from 2837 pathways and 10 933 genes. The average number of genes per pathway was 47.8 (s.d.=101.3 genes) and ranged from 1 to 2141. This database nevertheless contains a variety of pathway sources that are often redundant, for example, KEGG, wikipathways, interactome. Subsequently, we pruned the pathways according to size and redundancy via the following two-step protocol:
1. Very small and very large pathways were deemed to be uninformative and too generic (for example, such as metabolism and disease resistance), respectively. Hence, pathways with <10 and >150 genes were removed.
2. A pairwise comparison between pathways was computed removing pathways with an overlap in the number of genes >50%, and the smallest pathway in terms of genes was removed if the overlap exceeded 50%.
Following this protocol we retained a final number of 605 pathways and 8972 genes. Next, pig ortholog positions were determined using biomart (www.ensembl.org/biomart). To compute the G matrix associated to each pathway, we included all SNPs within the ortholog pig genes in that pathway, using the gene bounds defined in ensembl database v. 79. We considered genes mapped only in one of the 18 pig autosomes, that is, we excluded genes in unassembled contigs, in the mitochondria or in sex chromosomes.
To analyze in detail the ovarian steroidogenesis pathway, we downloaded the KEGGscape plugin version 0.7.0 (Nishida et al., 2014) using Cytoscape version 3.2.1 (Shannon et al., 2003), which can read and visualise KEGG Markup Language (KGML) files. We downloaded the KGML file for Sus scrofa ‘ovarian steroidogenesis' (SSC04913) and imported it into Cytoscape using the plugin. This particular pathway annotation shows the signaling and transcriptional cascade of ovarian steroidogenesis including relevant substrates and metabolites and the cellular compartments where the various events occur. The heritability scores for the genes in the ovarian steroidogenesis pathway were continuously mapped to node color. In circumstances where we had heritability values for multiple isoforms of the same protein we always selected the most extreme by heritability score. The mapped figure produced by Cytoscape was pasted into Powerpoint. Any pathway information lost during Cytoscape import (some of the flux arrows, the position of the cell membrane) was recreated manually using the original KEGG pathway as a guide.
Gene ontology enrichment analyses
We employed the GOrilla tool (cbl-gorilla.cs.technion.ac.il, Eden et al., 2009) to identify enriched gene ontology (GO) terms using two alternative approaches as follows:
1. Searching for enriched GO terms that appear densely at the top of a ranked list of genes. In this ‘single list' approach, genes were ranked according to the average heritability of the pathway(s) in which they were annotated.
2. Searching for enriched GO terms in a target list of genes compared with a background list of genes. In this ‘double list' approach, genes in the background list comprised all the 7502 genes under scrutiny, whereas genes in the target genes comprised 1084 genes from the top 30 pathways (or 5% of 605 total pathways) according to their heritability in the domestic status (wild versus domestic) analysis.
After running GOrilla, we selected molecular function GO terms with FDR-corrected P-value <0.001. These resulted in 52 and 73 enriched GO terms from ‘single list' and ‘double list' approach, respectively.
Quantifying genetic variance
We considered the domestic status (wild versus domestic) as the ‘phenotype' of interest. For the analysis, the Bayesian approach implemented in BGLR package (Pérez and de Los Campos, 2014) was employed because of its flexibility. BGLR implements various methods for genomic regressions, including the so-called G-BLUP model (VanRaden, 2008) and supports quantitative, binary and ordinal traits, these two are implemented using the probit link. Given the low amount of samples, a Bayesian approach is interesting because it allows a thorough quantification of incertitude via the posterior distribution of variance components. A principal component analysis decomposition of G was employed, given its good convergence properties (De los Campos et al., 2010). Default prior parameters and 150 000 iterations plus 2000 burn-in cycles were employed.
Population genetic analyses
R (R Core Team, 2014) was used to obtain principal components on matrix G, as well as its function ‘heatmap' to represent distances between samples. Fst Weir—Cockerham estimate between wild and domestic pigs was obtained with vcftools (Danecek et al., 2011).
Results and discussion
Genomic relationship matrix
After quality filtering and discarding singletons, we retrieved a total of 25 109 267 autosomal SNPs that covered 20 033 genes out of all annotated genes (25 322) by ensembl v. 79. A summary of predicted annotations is in Supplementary Table 2; among the non-synonymous SNPs, 10 007 (24%) were predicted to be deleterious by the SIFT algorithm (Sim et al., 2012).
To gain insight into the fine-grained structure of Sus scrofa (and as a prior step to estimating heritabilities), we computed the genomic relationship matrix between samples (G) using all available autosomal SNPs, after excluding singletons. A heatmap allows us to visualize the data but also to perform a quality control (Supplementary Figure 1). Reassuringly, samples from the same origin clustered together despite the fact that some were sequenced in different facilities and reported in independent publications. The only exception is a Tibetan pig (TT3), but this may be due to within-breed heterogeneity and not to sequencing bias because this sample was sequenced together with the rest of Tibetan samples (Supplementary Table 1). As expected, samples clustered by continent. Within Europe, wild boar (WE), Mangalica (MG) and Iberian (IB) pigs were the closest populations, whereas international pig breeds Duroc (DU) and Large White (LW) clustered apart. In Asia, the portrayed structure is more complex, especially within wild boars (WA).
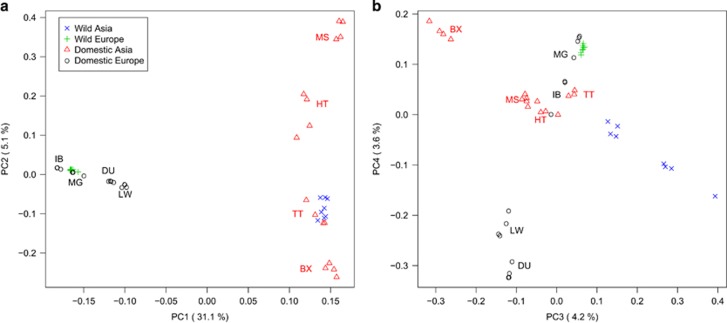
Logically, the well-known large geographic divide between the European and Asian subspecies, estimated in ~1.2 MYA, is also clearly visible in the principal component analysis (Figure 1a). In agreement with other studies, for example, Frantz et al. (2015), the first principal component neatly represents either the Chinese or European origin, proving that continent is the single most influential factor in explaining genetic structure of S. scrofa. Note that the international breeds, Duroc and Large White, are shifted towards Asia: a result of the well-known admixing process with Chinese pigs. Following on McVean (2009) the percentage of admixing can be estimated by the relative position of the admixed population in the first axis, compared with those of the founder populations. This results in an Asian component estimate of ~20% for Large White, which is quite similar to the figure obtained by Bianco et al. (2015) using high-density genotyping array data or by Ai et al. (2015) and Bosse et al. (2015) with sequence. Moreover, Figure 1a indicates that neither Spanish Iberian nor Hungarian Mangalica have been crossed with Chinese pigs, as they cluster tightly with European wild boars. Although the absence of any Asian signature had already been conclusively shown in Iberian pigs (Ramírez et al., 2014), the evidence in Mangalica was much more limited (Molnár et al., 2013), but is conclusively shown in these data.
Figure 1.
Principal component analyses of the molecular-based relationship matrix. (a) PC1 versus PC2. (b) PC3 versus PC4. IB, Iberian; MG, Mangalica; DU, Duroc; LW, Large White; MS, Meishan; HT, Hetao; TT, Tibetan pig (Tibet); BX, Bamaxiang.
The main goal of this work was to determine whether there exists a genetic basis, shared across continents, for the domestication process and if so, to functionally characterize it at the gene and biological pathway level. Such a signal, if it exists, is manifestly of lower intensity than the geography-driven signal. Although the first principal component, explaining ~31% of variance, discriminates between continents, the third PC separates domestic from wild pigs, predominantly in Asia, but explains a much lower fraction of variance, ~4% (Figures 1a and b). Nevertheless, it can be suspected that domestication-affected loci will be concentrated in genomic regions, assuming that the shared domestication signal is primarily selection driven, whereas drift affects the whole genome and likely dominate the changes that have occurred as a result of the European bottleneck and geographic split.
Heritability and the contribution of potentially non-neutral SNPs
We considered domestication status as a binary phenotype (wild or domestic) for which the genetic basis can be inferred as for any other complex trait using the genomic relationship matrix G, as described in methods. There are numerous approaches for quantifying the genetic variance using high-density genotyping data. In this case, we applied a Bayesian method because it allows us to easily compute the uncertainty associated with the estimates; this is important given the small sample size.
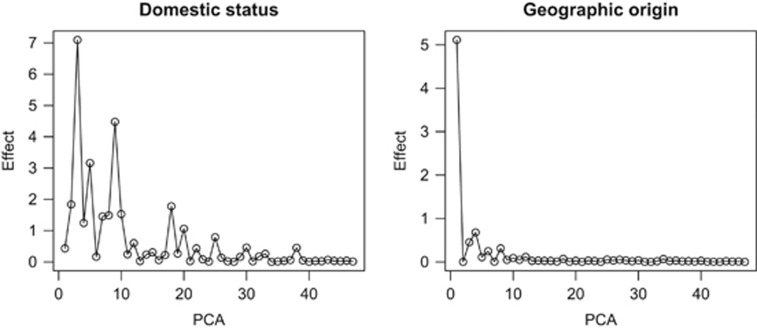
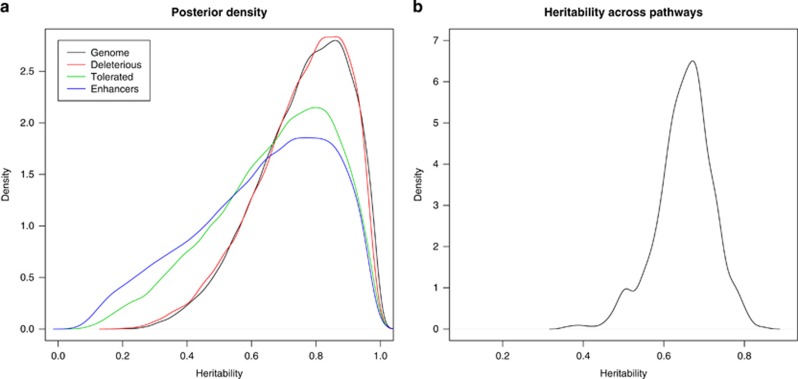
The Eigen-value decomposition of G employed allowed us to quantify the contribution of each principal component to variance in the liability scale (Janss et al., 2012). Figure 2 shows the absolute value of the regression coefficient on each PC. Compared with geographic origin, which can be basically explained from its first PC, domestication is a much more subtle and complex process. It needs a large number of components to be explained. As could be guessed from Figure 1b, PC3 is the most important to infer domestic status, but there are also other PCs of interest. The posterior distribution of heritability using all autosomal SNPs is plotted in Figure 3a. Given that the prior for hG2 was 0.5, but only 4% of the posterior distribution mass is below that value, the analysis reveals a clear genetic component for the domestication status (Figure 3a).
Figure 2.
Posterior means of the regression coefficients (absolute values) on each principal component when the samples are classified by domestic or wild status or continent (Asia versus Europe). All autosomal SNPs are used for the analysis.
Figure 3.
(a) Posterior density of heritability when computed using either all autosomal markers (black line), non-synonymous predicted deleterious SNPs (red line), non-synonymous predicted tolerated SNPs (green line) or SNPs within enhancers (blue line). (b) Distribution of mean posterior heritabilities of domestic status across pathways. The values are the means of the posterior distribution per path. A full color version of this figure is available at the Heredity journal online.
Next, we investigated the effect of different SNP sets based on SNP annotations obtained using variant effect prediction. Various SNP subsets were used for computation of separate G-matrices (Table 1) and estimating hG2 using each G matrix in turn, that is, only one matrix was fitted at a time. There were few differences across SNP partitions (Table 1), suggesting that there is not enough power in the experiment to discriminate between SNPs and/or that the differences among the SNP effects are small. Furthermore, linkage disequilibrium will act to smooth any effect across nearby SNPs, meaning that functionally distinct SNP sets may explain similar proportions of variance. For instance, synonymous and non-synonymous SNPs will be in tight disequilibrium within the same gene, and lead to similar variance partitioning, even if they are functionally different.
Table 1. Heritability (median posterior distribution and s.d.) using different SNP subsets.
| SNP subset | No. SNPs | Median hG2 | S.d. hG2 |
|---|---|---|---|
| All autosomal | 25 109 276 | 0.78 | 0.14 |
| Synonymous | 6 175 640 | 0.68 | 0.20 |
| Non-synonymous | 43 254 | 0.70 | 0.19 |
| Tolerated non-synonymousa | 31 145 | 0.70 | 0.19 |
| Deleterious non-synonymousa | 9566 | 0.78 | 0.14 |
| Enhancersb | 1 294 669 | 0.67 | 0.21 |
| Transcription factorsc | 1 290 312 | 0.67 | 0.21 |
| Potentially regulatoryd | 1 707 865 | 0.72 | 0.18 |
According to SIFT algorithm implemented in ensembl's variant effect predictor (database v. 79).
Obtained from Villar et al. (2015).
Using SNPs in TFs obtained from animal transcription factor database (www.bioguo.org/AnimalTFDB/, Zhang et al., 2012).
Comprises miRNAs, UTR, upstream, downstream and non-mediated decay transcript variants.
As expected, the best fit was with the complete data set (all SNPs). However, the most noticeable aspect was the finding of similar hG2 estimates using the 9566 potentially deleterious SNPs (according to the SIFT algorithm) or using the whole 25 million SNP data set. In contrast, hG2 estimates were somewhat lower with other functionally different subsets, and variances 35% larger than when all SNPs were used (Table 1; Figure 3a). In particular, it is noteworthy that we did not find an increased heritability using regulatory variants such as enhancer SNPs or transcription factors. Therefore, contrary to what has been often advocated (Pai et al., 2015), recent evolution may have been mediated to a larger extent by changes in potentially deleterious changes in amino acid sequence than by regulatory motifs. Certainly, this does not preclude that regulatory changes are important in domestication (Carneiro et al., 2014), but rather hints that its global effect, weighed by frequency, could be no higher than those of deleterious protein sequence changes. Certainly, this is a hypothesis that needs to be verified with larger data sets and more detailed analyses.
Pathway-guided partitioning of heritability
A convenient property of the theory applied here is its flexibility, as the same procedure can be applied to any SNP set, which simply results in different G-matrices. Further to quantifying the importance of each mutation type, we were also interested in assessing the contribution of each pathway to ‘domesticability'. Figure 3b shows the hG2 associated with each of the 605 pathways that fulfilled our conditions, namely minimum number of genes of 10, maximum number 150 and limited overlap (see Materials and methods for details). All 605 pathways together involved 8972 or about 35% of all annotated genes in the pig genome. Clearly, not all pathways were equally relevant and this was not a consequence of different number of SNPs per pathway, as we did not observe a large correlation between hG2 and number of markers (Supplementary Figure 2). We interpret this result as a consequence of domestication having affected non-random regions of the genome, particularly coding regions and where pathway is a meaningful analysis unit.
The top 5% pathways in terms of their genomic heritability are listed in Supplementary Table 3. Some pathways are associated with disease resistance. This presumably reflects annotation bias and prior investment in human biomedical studies and is probably irrelevant in our context. However, we found a number of pathways consistent with expectation based on earlier studies into the consequences of animal domestication. These relate to reproductive performance, metabolism, body conformation and coat color. For instance, the second most relevant pathway is ‘Ovarian steroidogenesis', comprising genes critical to reproductive performance. Prolactin signaling pathway is another significant pathway related to reproduction. Additional pathways worth mentioning are related to appetite, feed efficiency and glucose metabolism (signaling by leptin, type II diabetes), coat color (melanogenesis) or muscle differentiation (YAP1 stimulated gene expression, SRF and miRs in smooth muscle differentiation and proliferation, Syndecan-2-mediated signaling events). The presence of the YAP1 pathway in Supplementary Table 3 is of particular interest; YAP1 is a member of the HIPPO pathway that governs mammalian organ size, and the process of livestock domestication has led to systematic increase in muscle mass and modifications in size to a range of other metabolic tissues and organs. The original discovery of YAP1's role in organ size was made in liver and gut (Camargo et al., 2007), but this finding has subsequently been expanded to heart and skeletal muscle (Wackerhage et al., 2014). In summary, given that domestication has influenced many aspects of reproduction, growth and feeding, it is therefore reassuring that some of the pathways with highest heritability are related to these phenotypes.
Fst and heritability are distinct metrics
Population structure is generally measured with statistics such as Wright's fixation index or Fst. It can be thought that Fst and hG2 are equivalent metrics reflecting the same phenomena. Yet, importantly, Fst between domestic and wild populations and hG2 for domestic status captured different signals (Figure 4); the correlation coefficients (ρ) between the two were positive, but moderate across either windows (ρ=0.38) or pathways (ρ=0.29). The reason is that only the first principal component of the matrix G can be interpreted in terms of Fst in a straightforward manner (McVean, 2009). Furthermore, Fst can be wholly explained by frequency differences between populations, whereas no explicit interpretation based on frequencies is known for hG2. Estimating hG2 involves weighing all principal components and treating each one as a random regressor. Although hG2 depends on variability, there was no clear relationship between hG2 estimates and other known parameters. We employed a multiple regression to investigate the main factors influencing hG2 across windows. As can be seen in Supplementary Table 4, Fst was the single most influential factor, but not the only one. In particular, diversity and Tajima's D in the wild population were also significantly associated with heritability estimate.
Figure 4.
Heritability in domestic status versus Fst between domestic and wild populations across pathways.
Nevertheless, Fst is a popular measure of differentiation widely used to infer selection, although see Vilas et al. (2012) for some caveats on its use, and so we compared the results obtained with either Fst or hG2. Following the same logic as for hG2, we ranked pathways according to their Fst, averaged across SNPs in that pathway (Supplementary Table 5). Not unexpectedly, given their modest correlation (Supplementary Figure 2), there was limited agreement between the two metrics. Only five pathways were shared (marked with * in the Supplementary Tables 3 and 5). The single most consistent pathway both in terms of Fst and hG2 was glycosphingolipid biosynthesis. Glycosphingolipid molecules are predominantly found in nervous tissue and they are involved in protection against infectious diseases. Remarkably, this pathway was also among the 14 most differentiated ones in human populations, as reported by Daub et al. (2013).
A closer look at reproductive pathways
We analyzed in more detail the ovarian steroidogenesis pathway due to its importance in all domestic animals. Although this pathway was identified using the heritability approach, note that ‘Steroid hormone biosynthesis', identified by Fst criterion, comprises ovarian steroidogenesis (Supplementary Table 5). Ovarian steroidogenesis pathway consisted of 34 pig genes with coordinates in any of the autosomes (sex and unassembled contig locations were discarded) and containing more than five non singleton SNPs. It involved 37 733 SNPs. In this pathway (Supplementary Table 6), we find highly relevant genes such as follicle-stimulating hormone beta subunit (FSHB), Insulin-like growth factor 1 (IGF1), IGF1 receptor, follicle-stimulating hormone receptor (FSHR) among others (Supplementary Table 6). Importantly, these four genes are also much more differentiated between domestic and wild pigs (Fst=0.08) than genome-wide average (Fst=0.02). FSHB and FSHR are genes well-known to be critical for reproductive performance (Laan et al., 2012); FSHB encodes the beta polypeptide of FSH and FSHR is its cognate receptor, together they drive egg and sperm production in a hormonal pathway that also incorporates signaling from luteinising hormone. It has previously been documented in sheep that the domestic breeds have an extended reproductive cycle compared with progenitor mouflon and that this is reflected in altered patterns of circulating FSH (Lincoln et al., 1990).
These genes were also among those with largest gene heritability in the pathway, that is, the heritability obtained when the G was computed using the SNPs within that gene (Supplementary Table 6). In agreement with genome-wide analyses (Figure 4), there was a positive correlation (0.47) between Fst and hG2. Nevertheless, there were some outliers like SCARB1 gene, which showed moderate hG2 and low Fst. Further analysis showed that SCARB1 contained highly differentiated SNPs (24 had Fst>0.10) but, combined with slightly negative Fsts, made it a global Fst near zero, whereas heritability depends as well on other parameters (Supplementary Table 4). Next, to visualize whether there was a relation between heritability and biological function, we downloaded the S. scrofa ‘ovarian steroidogenesis' pathway (SSC04913) from KEGG, imported it into Cytoscape, and we colored each gene according to its associated heritability (see Materials and methods). The resulting graph (Supplementary Figure 3) makes it clear that both ovarian cell types (interstitial theca and granulosa) appear modified by domestication. However, four serial components (FSHB, FSHR, GNAS and ADCY6) of the granulosa cell-specific pathway suggest modification of signaling in these cells is particularly convincing (bottom panel). Therefore, this analysis suggests a differential effect of domestication along the ovarian steroidogenesis pathway.
A global functional outlook
The pathway-guided approach identified pathways involved in some of the biological changes known to be caused by domestication (Supplementary Table 3). Yet, it also follows from these analyses that domestication is complex and mediated through numerous signals. Our analyses tend to rule out a simplistic genetic basis of the domestication process: neither a single pathway nor a unique set of SNPs explains the process as a whole. To get a more holistic view, we carried out a gene ontology enrichment analysis using GOrilla according to the ordered list of genes, ranked according to the average hG2 of the pathways in which they were annotated, and following a single or double list approach (see Materials and methods). Supplementary Table 7 shows the list of 30 GO terms found to be in either the top 10 enriched from each single or double list approach or in the intersection of the two approaches at P<0.001. We found nuclear hormone receptors were prominently enriched by this approach. Nuclear hormone receptors are considered key hubs in gene regulatory networks. They are ligand-activated transcription factors that sense an environmental or metabolic signal through binding to the ligand of interest in the cytoplasm, then enacting an appropriate gene expression response after translocation to the nucleus. Identified ligands include thyroid and steroid hormones, fatty acids and other key molecules that allow the fine regulation of development, homeostasis and metabolism.
General discussion
Using genome-wide sequence, we have addressed the process of pig domestication in the two continents simultaneously. The study was designed to capture the shared signals between disparate breeds that have few phenotypic characteristics in common, except for the fact of being domestic. To characterize the genetic basis of domestication, we have capitalized on well-known robust approaches in quantitative genetics but that have not been applied in this context. We have shown that treating the status of domestics versus wild as a binary trait and assuming an infinitesimal genetic basis reveals genetic phenomena that are not detectably by the usual differentiation statistics. In this sense, a high heritability in jointly analyzing two separate events, such as domestication in Asia and in Europe, can be interpreted as the method being able to identify subjacent shared biological processes (for example, reproduction) that are however modified by different SNPs or genes of the same pathway. We have employed a threshold model, which is suited for binary traits where the probability of outcome (that is, phenotype of either class) depends on an underlying normally distributed genetic merit. In this scenario, we justify the threshold model by assuming the presence of a large number of variants that have been collectively selected during independent domestication processes. Although we have applied this strategy to domestication treated as a binary status, the same principle could be applied to other questions such as, for example, polledness in ruminants or average quantitative phenotypes by breed.
Importantly, Fst and genomic heritability are distinct metrics. Fst measures the proportion of genetic variance that can be explained as between-population variance. This statistic treats all individuals within a group (for example, domestic) as alike and individuals of different groups as different. On the other hand, the genomic heritability approach incorporates, via genomic similarity, variations and differences in genotypes between and within groups in a more continuous fashion. Therefore, we argue, the genomic heritability approach seems specifically suited for low differentiation and complex scenarios such as the one studied here, where structuring is caused primarily by geography rather than by the domestication process. Furthermore, as with any mixed model approach, any number of environmental or genetic effects could be included into the model. For instance, different G-matrices corresponding to any number of pathways or SNP sets could be jointly estimated, provided larger sample sizes than the one studied here are available.
The interpretation of heritability, although debatable (Tenesa and Haley, 2013), has a straightforward interpretation as a proportion of variance when applied to standard quantitative phenotypes. The parameter is related to response to selection and to the correlation of phenotypes between relatives. In the context of this work, heritability can be considered as a measure of differentiation, complementary to that of Fst. However, this relation is not so straightforward (Figure 4). McVean (2009) did show that the first principal component of G is proportional to Fst, but the computation hG2 of involves weighing all principal components and, as shown (Figure 1), domestication in pigs is not explained by only the first PC, due to the deep genetic divide between Asian and European S. scrofa. This likely explains that the correlation between Fst and hG2 was only 0.38 in 100 kb windows genome wide. We propose that hG2 in the context of this research is best understood as a measure of how much a given SNP set has been collectively influenced (that is, change in allele frequency) by domestication. We do not claim though that the most relevant SNP sets are causative, as this could be due to linkage or other factors, for example, intermediate allele frequency SNPs are more informative and hence more prone to have an increased weight in G. In turn, we do claim that the hG2 metrics can shed light on the underlying genetic phenomena. In this respect, a relevant result from our analyses is that non-synonymous potentially deleterious variants may have had a larger role in the domestication process than previously thought (Figure 3a; Table 1), although we do not disregard the relevance of regulatory mutations. Importantly, this was also suggested for domestication in dogs (Cruz et al., 2008) and, more recently, by Renaut and Rieseberg (2015) in crops. Note, however, that other evidence supports that regulatory mutations are relevant in other species, such as in rabbit domestication (Carneiro et al., 2014), so quantifying the relevance of each mutation type is an open question.
Although the genetics of domestication has been a hot topic for a while, studies in livestock using complete genome sequence are still scarce. In pigs, Rubin et al. (2012) used complete genome sequence with the purpose of studying domestication. These authors combined both pool and individual sequencing and used an excess of homozygosity in domestic breeds as a proxy for selection targets. They reported a list of 228 highly differentiated SNPs between wild and domestic pigs that were in coding sequence (Supplementary Table 3). We retrieved 146 of these SNPs that fulfilled the required conditions of missing rate and quality in our data (see Materials and methods). We clearly found a higher Fst for this subset of SNPs (average Fst=0.17, s.d.=0.13) than for genome-wide SNPs (Fst=0.03, Supplementary Figure 4). Therefore, most of these SNPs are promising diagnostic markers for the wild/domestic status. In contrast, the 157 916 SNPs within the selective regions also reported by Rubin et al. and found in our data, did not show any increased differentiation between wild and domestics (mean Fst=0.029). Certainly, hard selective sweeps causing a decrease in nucleotide diversity may not necessarily result in an increased differentiation. An explanation can be that Rubin et al. focused on international European breeds, whereas here we combine both Asian and European breeds. In our opinion, it is likely also that low diversity regions are shared across populations, as found in Bianco et al. (2015).
More recently, Frantz et al. (2015) reported an analysis of 100 Asian and European complete pig genomes. They looked for potential selective signals that were shared between Asian and European populations. The most significant one was on SSC4, 82.37–82.39 Mb. This region does not contain any known gene but is close to gene PLAG1 (SSC4: 82, 606, 862–82, 608, 850 bp), which is under strong selection in European domestics (Rubin et al., 2012; Frantz et al., 2015). To contrast these results, we carried a 100 kb genome-wide analysis with our approach and we found that the window containing PLAG1, made up of 891 SNPs, has a hG2=0.69; this is the 493 largest heritability out of the 23 395 windows analyzed (2% upper distribution of hG2). The mean window heritability was 0.41 (s.d.=0.12). Therefore, our analysis also supports an important, but not exclusive, role for PLAG1 region in domestication. Other genes that have been consistently affected by domestication are coat color genes such as MC1R or KIT (Giuffra et al., 2002; Fang et al., 2009). We did not find any SNP on MC1R in our data, but 782 SNPs in the KIT gene. The associated heritability with KIT SNPs was moderate (hG2=0.51). Interestingly, despite the average domestic/wild Fst was similar to the genome-wide value (Fst=0.02), its distribution was highly skewed (Supplementary Figure 5): there were 27 SNPs with Fst>0.20, and 79 with Fst>0.10. Therefore, an average low heritability does not preclude that some smaller regions or SNPs may be under selection.
A major drawback of the window-based approach, that is, a typical whole-genome scan, is difficulty in integrating functional information. In particular, it can be challenging to deal with the fact that genes act in a concerted manner. This is well recognized and previous works, for example, Daub et al. (2013) and Ha et al. (2015) have employed a pathway-centered approach, but it is not clear though how to incorporate this fact into a coherent statistical inference framework. Daub et al. chose a function of the most differentiated SNP per gene, whereas Ha et al. (2015) used a gene-based score test; in either case the authors combined all genes in a pathway to obtain a pathway score. Certainly, additional limitations in this kind of studies are incorrect annotation and incomplete assemblies, which are normally more serious in complex repetitive gene families such as olfactory receptors or in the SLA region. In particular, about 14% of current porcine assembly is of low quality, according to (Warr et al., 2015). Ongoing annotation initiatives in livestock genomes such as FAANG (Andersson et al., 2015) and new assemblies should alleviate this problem.
In agreement with Edwards et al. (2015), we also believe mixed model and related theory is a most appropriate tool to accomplish a multiple gene/region analysis. Here, we computed genomic relationships using the SNPs located within all genes in a given pathway, and computed a genomic heritability associated to each pathway. As genetic response is a function of heritability, the framework used here seems a logical and justified choice. By using this method, we were able to retrieve pathways (ovarian hormones, leptin receptor, organ size among others), which play key roles in phenotypes well-known to have been modified by domestication (Supplementary Table 3).
To conclude, we present a novel application of standard statistical tools in animal breeding to study population genetic events from a novel perspective. Here we argue that shared genetic signals underlying complex, disparate and independent domestication events exist and that its relevance can be quantified. We hypothesize that some sort of convergent adaptation has occurred. Nevertheless, although genetic markers on the whole are powerful to detect whether a specimen is wild or domestic, the genetic contribution of any single marker is weak and is difficult to reach definitive answers on the effect of few genes or pathways. For the future, as the number of sequences increase, more sophisticated analyses could be performed, such as a multi-trait analysis where domestication in different continents is regarded as correlated traits. Such an analysis would yield continent-specific heritabilities and genetic correlations that may shed light on the extent of convergence between continents.
In all, despite the small sample utilized, our analyses do hint at some interesting results that merit further exploration with larger data sets and in additional species. In particular, it seems that deleterious mutations are at least as relevant as regulatory positions to explain domestication in the pig. Further, when genes are investigated in pathway units, some reproductive (ovarian steroidogenesis) and feeding (leptin regulation) pathways seemed especially relevant to explain the domestication process.
Data archiving
New sequence raw data is available at SRA (SRP074357). A vcf format file with all genotypes used for the analyses has been deposited in dryad (doi:10.5061/dryad.r6t26).
Acknowledgments
We thank S Ramos-Onsins for sharing software and for comments and help to use it, as well as personnel at CSIRO's high performance computing facilities. This work was developed whereas the senior author was visiting CSIRO Agriculture in St Lucia campus (Brisbane, Australia) funded by a CSIRO McMaster Visiting Fellowship. Work also funded by AGL2013-41834-R grants (Ministry of Economy and Science, Spain) to MPE and NIH grant R01GM101219 (USA) to GDLC.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Ai H, Fang X, Yang B, Huang Z, Chen H, Mao L et al. (2015). Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet 47: 217–225. [DOI] [PubMed] [Google Scholar]
- Andersson L, Archibald AL, Bottema CD, Brauning R, Burgess SC, Burt DW et al. (2015). Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol 16: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, Ratnakumar A, Arendt M-L, Maqbool K, Webster MT, Perloski M et al. (2013). The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495: 360–364. [DOI] [PubMed] [Google Scholar]
- Bianco E, Soto HW, Vargas L, Pérez-Enciso M. (2015). The chimerical genome of Isla del Coco feral pigs (Costa Rica), an isolated population since 1793 but with remarkable levels of diversity. Mol Ecol 24: 2364–2378. [DOI] [PubMed] [Google Scholar]
- Bosse M, Madsen O, Megens HJ, Frantz LA, Paudel Y, Crooijmans RP et al. (2015). Hybrid origin of European commercial pigs examined by an in-depth haplotype analysis on chromosome 1. Front Genet 5: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse M, Megens HJ, Madsen O, Frantz LA, Paudel Y, Crooijmans RP et al. (2014). Untangling the hybrid nature of modern pig genomes: a mosaic derived from biogeographically distinct and highly divergent Sus scrofa populations. Mol Ecol 23: 4089–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. (2013). Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R et al. (2007). YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 17: 2054–2060. [DOI] [PubMed] [Google Scholar]
- Carneiro M, Rubin C-J, Di Palma F, Albert FW, Alfoldi J, Barrio AM et al. (2014). Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 345: 1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemineau P, Guillaume D, Migaud M, Thiéry JC, Pellicer-Rubio MT, Malpaux B. (2008). Seasonality of reproduction in mammals: intimate regulatory mechanisms and practical implications. Reprod Domest Anim 43: 40–47. [DOI] [PubMed] [Google Scholar]
- Crossa J, De Los Campos G, Pérez P, Gianola D, Burgueño J, Araus JL et al. (2010). Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186: 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F, Vilà C, Webster MT. (2008). The legacy of domestication: accumulation of deleterious mutations in the dog genome. Mol Biol Evol 25: 2331–2336. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011). The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub JT, Hofer T, Cutivet E, Dupanloup I, Quintana-Murci L, Robinson-Rechavi M et al. (2013). Evidence for polygenic adaptation to pathogens in the human genome. Mol Biol Evol 30: 1544–1558. [DOI] [PubMed] [Google Scholar]
- Diamond J. (2002). Evolution, consequences and future of plant and animal domestication. Nature 418: 700–707. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SM, Thomsen B, Madsen P, Sørensen P. (2015). Partitioning of genomic variance reveals biological pathways associated with udder health and milk production traits in dairy cattle. Genet Sel Evol 47: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Codina A, Paudel Y, Ferretti L, Raineri E, Megens H-J, Silió L et al. (2013). Dissecting structural and nucleotide genome-wide variation in inbred Iberian pigs. BMC Genomics 14: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Larson G, Ribeiro HS, Li N, Andersson L. (2009). Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet 5: e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz LAF, Schraiber JG, Madsen O, Megens H-J, Cagan A, Bosse M et al. (2015). Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet 47: 1141–1148. [DOI] [PubMed] [Google Scholar]
- Freedman AH, Gronau I, Schweizer RM, Ortega-Del Vecchyo D, Han E, Silva PM et al. (2014). Genome sequencing highlights the dynamic early history of dogs. PLoS Genet 10: e1004631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffra E, Törnsten A, Marklund S, Bongcam-Rudloff E, Chardon P, Kijas JMH et al. (2002). A large duplication associated with dominant white color in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm Genome 13: 569–577. [DOI] [PubMed] [Google Scholar]
- Groenen MAM, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF et al. (2012). Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491: 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha N-T, Gross JJ, van Dorland A, Tetens J, Thaller G, Schlather M et al. (2015). Gene-based mapping and pathway analysis of metabolic traits in dairy cows. PLoS One 10: e0122325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BJ, Bowman PJ, Chamberlain AJ, Goddard ME. (2009). Invited review: genomic selection in dairy cattle: progress and challenges. J Dairy Sci 92: 433–443. [DOI] [PubMed] [Google Scholar]
- Janss L, de los Campos G, Sheehan N, Sorensen D. (2012). Inferences from genomic models in stratified populations. Genetics 192: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan M, Grigorova M, Huhtaniemi IT. (2012). Pharmacogenetics of follicle-stimulating hormone action. Curr Opin Endocrinol Diabetes Obes 19: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, Robins J et al. (2005). Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307: 1618–1621. [DOI] [PubMed] [Google Scholar]
- Larson G, Piperno DR, Allaby RG, Purugganan MD, Andersson L, Arroyo-Kalin M et al. (2014). Current perspectives and the future of domestication studies. Proc Natl Acad Sci USA 111: 6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln GA, Lincoln CE, McNeilly AS. (1990). Seasonal cycles in the blood plasma concentration of FSH, inhibin and testosterone, and testicular size in rams of wild, feral and domesticated breeds of sheep. J Reprod Fertil 88: 623–633. [DOI] [PubMed] [Google Scholar]
- De los Campos G, Gianola D, Rosa GJM, Weigel Ka, Crossa J. (2010). Semi-parametric genomic-enabled prediction of genetic values using reproducing kernel Hilbert spaces methods. Genet Res (Camb) 92: 295–308. [DOI] [PubMed] [Google Scholar]
- De los Campos G, Sorensen D, Gianola D. (2015). Genomic heritability: what is it? PLOS Genet 11: e1005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. (2009). A genealogical interpretation of principal components analysis. PLoS Genet 5: e1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen TH, Hayes BJ, Goddard ME. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár J, Nagy T, Stéger V, Tóth G, Marincs F, Barta E. (2014). Genome sequencing and analysis of Mangalica, a fatty local pig of Hungary. BMC Genomics 15: 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár J, Tóth G, Stéger V, Zsolnai A, Jánosi A, Mohr A et al. (2013). Mitochondrial D-loop analysis reveals low diversity in Mangalica pigs and their relationship to historical specimens. J Anim Breed Genet 130: 312–320. [DOI] [PubMed] [Google Scholar]
- Nishida K, Ono K, Kanaya S, Takahashi K. (2014). KEGGscape: a Cytoscape app for pathway data integration. F1000Res 3: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Pritchard JK, Gilad Y. (2015). The genetic and mechanistic basis for variation in gene regulation. PLoS Genet 11: e1004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez P, de Los Campos G. (2014). Genome-wide regression & prediction with the BGLR Statistical Package. Genetics 198: 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. (2014). BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2014) R: A Language and Environment for Statistical Computing. Veinna: R Foundation for Statistical Computing. [Google Scholar]
- Ramírez O, Burgos-Paz W, Casas E, Ballester M, Bianco E, Olalde I et al. (2014). Genome data from a sixteenth century pig illuminate modern breed relationships. Heredity (Edinb) 114: 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaut S, Rieseberg LH. (2015). The accumulation of deleterious mutations as a consequence of domestication and improvement in sunflowers and other compositae crops. Mol Biol Evol 32: 2273–2283. [DOI] [PubMed] [Google Scholar]
- Rubin C-J, Megens H-J, Martinez Barrio A, Maqbool K, Sayyab S, Schwochow D et al. (2012). Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci USA 109: 19529–19536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. (2012). SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40: W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A, Haley CS. (2013). The heritability of human disease: estimation, uses and abuses. Nat Rev Genet 14: 139–149. [DOI] [PubMed] [Google Scholar]
- Thompson EA. (1975). The estimation of pairwise relationships. Ann Hum Genet 39: 173–188. [DOI] [PubMed] [Google Scholar]
- VanRaden PM. (2008). Efficient methods to compute genomic predictions. J Dairy Sci 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Vilas A, Pérez-Figueroa A, Caballero A. (2012). A simulation study on the performance of differentiation-based methods to detect selected loci using linked neutral markers. J Evol Biol 25: 1364–1376. [DOI] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M et al. (2015). Enhancer Evolution across 20 Mammalian Species. Cell 160: 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackerhage H, Del Re DP, Judson RN, Sudol M, Sadoshima J. (2014). The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal 7: re4. [DOI] [PubMed] [Google Scholar]
- Warr A, Robert C, Hume D, Archibald AL, Deeb N, Watson M. (2015). Identification of low-confidence regions in the pig reference genome (Sscrofa10.2). Front Genet 6: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR et al. (2010). Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder M, Bradley D, Emshwiller E, Smith B. (2006) Documenting Domestication In: New Genetic and Archaeological Paradigms. University of California Press: Berkeley, CA, USA. [Google Scholar]
- Zhang H-M, Chen H, Liu W, Liu H, Gong J, Wang H et al. (2012). AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res 40: D144–D149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.