Abstract
Risk stratification using number, size, and histology of colorectal adenomas is currently suboptimal for identifying patients at increased risk for future colorectal cancer (CRC). We hypothesized that molecular markers of carcinogenesis in adenomas, measured via immunohistochemistry, may help identify high-risk patients. To test this hypothesis, we conducted a retrospective, 1:1 matched case-control study (n=216; 46% female) in which cases were patients with CRC and synchronous adenoma and controls were patients with adenoma but no CRC at baseline or within 5 years of follow-up. In phase I of analyses, we compared expression of molecular markers of carcinogenesis in case and control adenomas, blind to case status. In phase II of analyses, patients were randomly divided into independent training and validation groups to develop a model for predicting case status.
We found that seven markers (p53, p21, Cox-2, beta-catenin, DNA dependent protein kinase (DNApkcs), survivin, and 0(6)-methylguanine-DNA methyltransferase (MGMT)) were significantly associated with case status on unadjusted analyses, as well as analyses adjusted for age and advanced adenoma status (p<0.01 for at least one marker component). When applied to the validation set, a predictive model using these 7 markers showed substantial accuracy for identifying cases (AUC=0.83; 95% CI:0.74–0.92). A parsimonious model employing 3 markers performed similarly to the 7 marker model (AUC=0.84). In summary, we found that molecular markers of carcinogenesis distinguished adenomas from patients with and without CRC. Further, we speculate that prospective studies utilizing molecular markers to identify individuals with polyps at risk for future neoplasia are warranted.
Keywords: colorectal cancer, colonic polyps, colorectal adenomas, epidemiology, molecular epidemiology, immunohistochemistry
INTRODUCTION
Risk stratification of individuals with colorectal polyps using current surveillance guidelines is suboptimal(1). Current guidelines stratify patients into high and low risk categories based on number, size, and basic histologic analysis of polyps detected at colonoscopy(2–4). A number of studies have confirmed that the incidence of advanced neoplasia on follow up among high versus low risk patient groups based on these criteria are clearly different –approximately 16% for patients with high risk adenomas at baseline versus 7% for patients with low risk adenomas at baseline (5, 6). These findings suggest that genetic and environmental factors that cause high risk adenomas continue to promote development of incident high risk adenomas in the future. However, risk stratification based on current high and low risk criteria are neither sensitive nor specific for detecting all patients who will have incident advanced neoplasia(5, 6). Prior studies suggest that the current approach to risk stratification is at most 68% sensitive, and 53% specific for predicting which patients will develop high risk polyps on follow up(5, 6).
Thus, despite guideline-recommended risk stratification based on number, size, and histology of polyps, many patients who go on to develop high risk neoplasia on follow-up are incorrectly characterized as being low risk at baseline. Similarly, use of current guidelines also results in characterizing many patients who only develop low risk neoplasia on follow up as high risk at index evaluation. The consequences of imprecise risk stratification are substantial, including under-surveillance of patients who will develop high risk neoplasia, and over-surveillance of patients who will develop only low risk or no neoplasia. New approaches for risk stratification are required.
One approach would be to apply the long-standing concept of “field carcinogenesis,” (also known as “field effect,” “field defect,” and “field cancerization”), and the newly emerging concept of “etiologic field effect” to risk stratification of individuals with colorectal polyps(7–10). The concept of “field carcinogensis” includes the principle that cancerous and non-cancerous tissue within the same organ may share molecular characteristics that promote carcinogenesis. The newly emerging concept of “etiologic field effect” expands on the theory of “field carcinogenesis” by postulating that an environmental milieu (which may be represented by the microbiome, diet factors, and/or other exposures, such as smoking), exerts pressure (through interactions with host factors such as constitutive genetic makeup) towards development of cancers with a specific molecular phenotype. While in our current approach to risk stratification of individuals with polyps we use baseline number, size, and histology of adenomas as markers of the sum effect of genetic and environmental factors, more precise markers that are a result of these sum effects might improve risk stratification(11).
Use of molecular markers of carcinogenesis may offer such an opportunity for developing a more precise approach to risk stratification of patients with polyps(12, 13). In this study, we have postulated that molecular changes within polyps may reflect both “field carcinogenesis” (changes that may be more likely to be present in adjacent normal tissue) as well as “etiologic field effects” (environmental pressures such as diet and intestinal microbiome) that will continue to be exerted on adjacent normal tissue, and thus be valuable markers for prediction of future neoplasia risk. This postulate is supported by work that has shown that synchronous neoplasia and normal tissue may share molecular changes such as O6-methylguanine-DNA methyltransferase (MGMT) promotor methylation(14), and that environmental exposures (such as smoking) may predispose to development of neoplasia with specific molecular features (such as CpG Island methylation and/or BRAF mutated neoplasia in the case of smoking exposure)(9). Accordingly, we further hypothesized that immunohistochemical expression of molecular markers of carcinogenesis may differ between polyps from patients at high and low risk for colorectal cancer (CRC). To test this hypothesis, we conducted a case control study, in which cases were patients with CRC and a synchronous adenoma (high risk patients) and controls were patients with an adenoma but no synchronous CRC and at least 5 years of cancer-free follow up (low risk patients). We compared expression of a defined panel of molecular markers of carcinogenesis among adenomas from case and control patients, and found several markers that may improve ability to distinguish patients at high versus low risk for CRC. Our findings provide a proof of concept that use of molecular markers can potentially improve risk stratification of patients with colorectal polyps.
MATERIALS and METHODS
Study Overview
We conducted a 1:1 matched retrospective case-control study, 2007–2012. Cases were patients with CRC and a synchronous adenoma at time of CRC diagnosis. Controls were patients with adenoma, but no CRC at time of index polyp diagnosis, and at least 5 years of cancer free clinical follow-up, determined by medical record review. We considered adenomas from cases as indicative of high risk patients because all of these patients had already had the outcome of greatest interest for risk prediction: CRC. We considered adenomas from controls to be indicative of a low risk patient because none of these patients had CRC at baseline or on follow up. These case and control groups were chosen based on our reasoning that if polyp biomarkers could not distinguish adenomas from these groups, future prospective studies to evaluate marker utility among adenoma patients identified by routine colonoscopy would not be warranted. Archived, paraffin embedded adenoma samples were subjected to IHC as described below, blind to case/control status. The analysis consisted of two phases. In phase I, we compared ability of each molecular marker to distinguish cases from controls. In phase II, patients were randomly divided into independent training and validation groups to develop a model for predicting case status using markers from phase I that distinguished cases from controls. All cases and controls were identified from Parkland Health and Hospital System (Parkland), the primary safety-net health system for Dallas, TX. The UT Southwestern Institutional Review Board approved the study. A waiver of written informed consent was approved by the Institutional Review Board for the study, including for data collection and analyses.
Case and Control Selection
Case and Control Adenoma Inclusion/Exclusion Criteria
We included case and control patients age 40 or older who had CRC and one or more synchronous adenomas 0.5 cm or larger in size. For cases, we required that synchronous adenomas had been diagnosed within 6 months of CRC diagnosis, either at time of colonoscopy or surgical resection. When more than one adenoma was present for the same candidate case or control patient, we selected the adenoma closest to 1 cm in size. If multiple adenomas were present and varied in dysplasia or presence of villous features, we selected the adenoma with the least advanced features for study inclusion.
We excluded candidate cases and controls when there was insufficient tissue for analysis, Familial Adenomatous Polyposis, Lynch syndrome, or when there was a history of inflammatory bowel disease or colorectal surgery prior to the index adenoma diagnosis. We also excluded candidate cases if we were unable to find a sex and 5-year age matched control patient, or if pathology records did not confirm CRC diagnosis.
Candidate Case and Control Selection
To identify cases, we obtained a random sample of CRC patients meeting inclusion and exclusion criteria diagnosed 1997 through 2007 using our American College of Surgeons Committee on Cancer-certified cancer registry.
To identify candidate controls, we used a stepwise approach to minimize the chances of including patients with CRC on follow up. First, we queried our clinical pathology database to identify all patients diagnosed with an adenoma 1999 through 2002. This time period allowed us to identify patients with at least 5 years of cancer-free clinical follow up after index adenoma diagnosis for exclusion. Next, the list of candidate controls was stratified by sex, 5-year age categories, as well as by whether patients had any follow up colonoscopy <3 or 3 or more years after index adenoma diagnosis. Once a case was identified, we searched the list of candidate controls to randomly select patients with an adenoma meeting inclusion/exclusion criteria below. To optimize inclusion of controls from patients with at least one follow up colonoscopy after index adenoma diagnosis, we first matched cases to controls who had colonoscopy 3 or more years after index adenoma diagnosis. If no match was identified, we then matched to controls with colonoscopy within 3 years of index adenoma diagnosis. If still no match was found, we matched the case to a control with at least 5 years of clinical follow up documenting CRC free status.
Measurements
Clinical and Demographic Data
Clinical and demographic data for cases and controls were abstracted from paper and electronic medical records. Abstracted data included age, race/ethnicity, sex, height, weight, date of last follow up, vital status, timing and findings of any follow up colonoscopies, and number, histology, location, and size of any polyps diagnosed. For cases, summary Surveillance Epidemiology and End Results (SEER) cancer stage and CRC location were recorded.
Molecular Markers
After identifying archived, paraffin embedded case and control adenomas, we obtained up to 13, 5 micron in thickness slides of each specimen for IHC analysis. We selected the following protein markers as potential predictors of case versus control status for analysis: nuclear p53, nuclear p21, nuclear p27, nuclear beta-catenin (BCAT), cytoplasmic Cox-2, nuclear DNA dependent protein kinase (DNApkcs), cytoplasmic survivin, cytoplasmic BRAF, cytoplasmic p27, nuclear 0(6)-methylguanine-DNA methyltransferase (MGMT), nuclear mutS homolog 2 (MSH2), nuclear mutL homolog 1 (MLH1), and human telomerase reverse transcriptase (hTERT) (Supplementary Table 1). These markers were selected because of their hypothesized roles in colorectal carcinogenesis or as markers of carcinogenesis pathways(15–19), or because of prior studies suggesting a role for risk prediction among patients with colorectal polyps(12, 13). We hypothesized that some might be associated with rare but more advanced changes within an adenoma, while others might be more common across the spectrum of adenomas available. Brief rationale for inclusion of each marker studied, as well as details regarding antibodies used are provided in Supplementary Table 1 (20–40).
IHC technique and interpretation
IHC was performed using either Ventana or the DAKO automated staining systems (Ventana Ultraview or the DAKO Biocare Polymer Detection System, respectively) using standardized protocols. (Details on IHC technique can be found in Supplementary Table 1). Interpretation and scoring of IHC results was done by a single pathologist (RA) in a blinded fashion. Visual semi quantitative staining was performed for markers with cytoplasmic localization (BCAT, survivin, COX-2, BRAF). Visual scores were performed using an intensity scale of 0 (negative) to 3 + and percentage of cells staining (0–100%). Quantitative automated cellular imaging system (ACIS, DAKO, Carpenteria, CA) scores for staining intensity and percentage of cells staining were generated for markers with predominant nuclear expression (p53, MGMT, p21, DNA Pkcs, p27, MSH2).
Outcome Assessment
The primary outcome of interest was the probability of being a case (i.e., having CRC, case status). The primary predictors of interest were levels of expression of the panel of molecular markers of carcinogenesis within case and control adenomas. After conducting all molecular analyses, we first eliminated markers that were non-informative for the majority of case and/or control specimens due to either insufficient tissue, or technical failure of IHC staining. This occurred for the p27, BRAF, telomerase, and MSH2, and MLH1 antibodies. Next, we examined the distribution of marker results blind to case/control status. Because results were not normally distributed, we used Box-Cox transformation(41) and used these transformed data for our subsequent analyses. We conducted analyses in two phases. Phase I was our primary case-control analysis, in which we conducted univariate and multivariate logistic regression analyses to evaluate the association of individual marker results with case status. In these analyses, we included all case/control adenomas, regardless of whether a given adenoma had missing data for one or more markers. We constructed multivariate logistic regression models for each marker result, after adjusting for age and advanced adenoma status, as well as for results of other markers. Even though cases and controls were matched on age, we further adjusted for age in regression models because of observed inconsistencies in the protocol for age-matching in our study. P values of <0.05 were considered statistically significant for all comparisons.
In phase II, we created and validated a prediction model for case status using marker results. We randomly assigned half of all case and control adenomas with informative results for all IHC markers into a training set, and the other half into a test validation set. The training set prediction model was developed using a Random Forest algorithm, and then tested in the validation set. Random Forest is an ensemble learning method using classification tree as the base classifier, and the prediction results are obtained by majority votes of the classification trees. The Random Forest algorithm has been shown to perform well in many classification and prediction problems, especially with high dimensional data(42, 43). To evaluate the prediction performance in the validation set, we plotted receiver operation characteristic (ROC) curves for case status prediction. Predictive accuracy in the validation set for the entire model was summarized using sensitivity and specificity for case status, as well as area under the ROC curve (AUC), with associated 95% confidence intervals (95%CI).
Secondary analyses
Using the test validation set created for our phase II analysis, we also used data on the three most predictive markers in the training Random Forest model to determine the predictive accuracy of these three markers alone in predicting case status. All analyses were conducted R Linux 64 bit 2.14.
RESULTS
Demographic and Clinical Characteristics
We reviewed the medical records for 963 patients with CRC age 40 and older from the Parkland cancer registry 1997 through 2007; 108 patients meeting eligibility criteria with CRC and at least one synchronous adenoma were identified and matched to 108 controls with adenoma but no CRC. The most common reason for case exclusion was lack of a synchronous adenoma (n=445). Less frequent reasons for exclusion included insufficient archived tissue for analysis, incorrect record of age in the tumor registry, presence of a tumor other than colorectal adenocarcinoma, or inability to find an age/sex matched control. The process of case selection is depicted in the Supplementary Figure 1.
The median age for case subjects was slightly higher than for controls (60 vs. 58 years, Table 1). Women accounted for 46% of case/control pairs. A similar proportion of advanced adenomas was present among cases and controls. Controls had longer follow up after time of adenoma diagnosis, and as expected, were more likely to be alive at last follow up. All controls had at least 5 years of cancer free follow up after index adenoma diagnosis, and 81 had at least one colonoscopy in the follow up period (n=74 three or more years after adenoma diagnosis, and n=7 within 3 years of adenoma diagnosis).
Table 1.
Demographic and Clinical Characteristics
| Case (n=108) | Control (n=108) | p value | |
|---|---|---|---|
| Age, median years (IQR) | 60.3 (55.6–65.5) | 57.9 (53.6–63.1) | 0.04 |
| Female, n (%) | 50 (46.3) | 50 (46.3) | 1 |
| Race/ethnicity, n (%) | 0.46 | ||
| White | 28 (25.9) | 24 (22.2) | |
| Black | 62 (57.4) | 57 (52.8) | |
| Hispanic | 16 (14.8) | 25 (23.1) | |
| Other | 2 (1.9) | 2 (1.9) | |
| Follow Up | |||
| Median follow time, months (IQR) | 32.2 (13.0–50.6) | 102.4 (91.3–121.3) | <0.0001 |
| Dead at last follow up, n (%) | 32 (29.6) | 5 (4.6) | <0.0001 |
| Follow up colonoscopy after index adenoma (controls) | --- | 81 | --- |
| Time to follow up colonoscopy (controls), median yrs | --- | 5.5 | --- |
| Follow up colonoscopy ≥3 years after index adenoma (controls), n (%) | --- | 74 (91.2) | --- |
| CRC Location (cases)* | |||
| Right Colon | 54 (50) | --- | --- |
| Left Colon/Rectum | 54 (50) | --- | --- |
| Summary SEER Stage (cases), n (%) | |||
| Local | 41 (37.96) | --- | --- |
| Regional | 33 (30.56) | --- | --- |
| Distant | 27 (25.00) | --- | |
| Unknown/unstaged | 7 (6.48) | --- | --- |
| Features of Adenoma Included for Analysis | |||
| Mean Size, cm (SD) | 1.18 (0.7) | 1.06 (0.5) | 0.16 |
| Advanced Adenoma§, n (%) | 64 (59.3) | 59 (54.6) | 0.5826 |
Right colon=proximal to descending colon;
adenoma ≥10mm in size, and/or containing high grade dysplasia, tubulovillous, or villous features; IQR, interquartile range; SEER, Surveillance Epidemiology and End Results
Molecular Marker Results: Phase I Case/Control Analyses
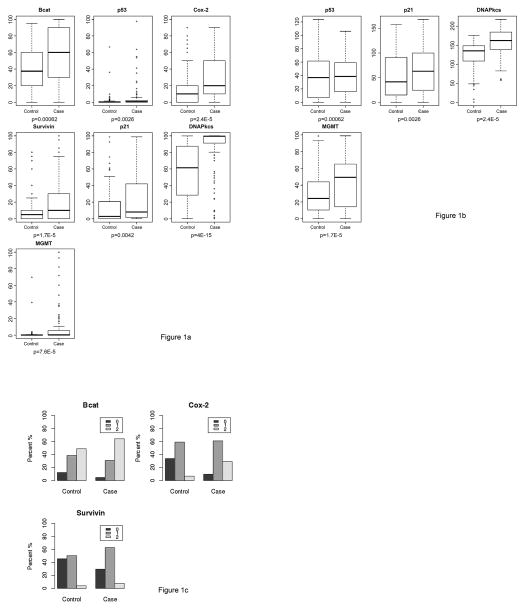
Informative marker results were available for the majority of cases and controls for testing with p53, p21, Cox-2, BCAT, DNAPkcs, survivin, and MGMT. Thus the final marker set consisted of 7 markers (Table 2). Both percentage and intensity score for BCAT, Cox 2, Survivin, p21, DNAPkcs, and MGMT were significantly different between cases and controls, while p53 differed between cases and controls only on percentage score (all p<0.05, Table 2). Univariate analyses showed results of nearly all molecular markers were associated with case adenoma status (Table 3, Figures 1a, 1b, and 1c; On multivariable adjusted analyses, p53, p21, Cox-2, BCAT, DNAPkcs, survivin, and MGMT were all found to have a statistically significant association with case status (p<0.05 for all markers, Table 3). Marker distribution results for cases versus controls stratified by adenoma type (non-advanced or advanced adenoma) were qualitatively similar (data not shown).
Table 2.
Distribution of molecular marker results for case and control adenomas
| All Adenomas (n=216) | Case Adenomas (n=108) | Control Adenomas (n=108) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Marker | n | Median Score | (IQR) | n | Median Score | (IQR) | N | Median Score | (IQR) | p value** |
| BCAT percentage score* | 213 | 47.08 | (20–70) | 107 | 54.6 | (30–90) | 106 | 39.49 | (20–60) | <0.001 |
| BCAT intensity score* | 213 | 1.48 | (1–2) | 107 | 1.6 | (1–2) | 106 | 1.37 | (1–2) | 0.013 |
| p53 percentage score | 214 | 3.42 | (0.00–1.37) | 107 | 5.18 | (0.02–2.34) | 107 | 1.66 | (0.00–0.74) | 0.003 |
| p53 intensity score | 204 | 40.12 | (14.12–61.52) | 103 | 39.4 | (15.9–59.07) | 101 | 40.86 | (6.80–61.60) | 0.95 |
| Cox 2 percentage score* | 206 | 22.22 | (5–30) | 103 | 27.79 | (10–50) | 103 | 16.65 | (0–20) | <0.0001 |
| Cox 2 intensity score* | 206 | 0.96 | (1–1) | 103 | 1.19 | (1–2) | 103 | 0.73 | (0–1) | <0.0001 |
| Survivin percentage score* | 210 | 14.86 | (0–20) | 105 | 21.1 | (0–30) | 105 | 8.62 | (0–10) | <0.0001 |
| Survivin intensity score* | 210 | 0.68 | (0–1) | 105 | 0.78 | (0–1) | 105 | 0.58 | (0–1) | 0.012 |
| p21 percentage score | 208 | 19.01 | (1.11–30.32) | 104 | 24.23 | (1.67–41.49) | 104 | 13.8 | (0.10–20.43) | 0.004 |
| p21 intensity score | 208 | 57.75 | (18.67–97.3) | 104 | 63.74 | (24.65–99.42) | 104 | 51.76 | (14.78–90.77) | 0.028 |
| DNAPkcs percentage score | 208 | 71.35 | (42.98–99.73) | 105 | 85.75 | (91.08–99.98) | 103 | 56.68 | (28.39–87.38) | <0.0001 |
| DNAPkcs intensity score | 208 | 141.86 | (120.60–166.64) | 105 | 158.61 | (139.30–184.80) | 103 | 124.79 | (108.50–148.75) | <0.0001 |
| MGMT percentage score | 211 | 5.35 | (0.02–1.1) | 108 | 9.16 | (0.04–4.84) | 103 | 1.36 | (0.00–0.36) | <0.0001 |
| MGMT intensity score | 211 | 36.21 | (12.00–58.05) | 108 | 43.22 | (14.38–65.02) | 103 | 28.85 | (10.45–43.9) | <0.001 |
subjectively measured, no decimals used;
comparison case vs. control adenoma; IQR, interquartile range; BCAT, beta catenin; DNAPkcs, DNA dependent protein kinase; MGMT, 0(6)-methylguanine-DNA methyltransferase.
Table 3.
Unadjusted and adjusted analysis of clinical and molecular factors associated with case status*
| OR | 95% CI | AjOR | 95%CI | |
|---|---|---|---|---|
| Age | 1.04 | 1.00--1.075 | -- | -- |
| Advanced Adenoma | 1.21 | 0.70--2.08 | -- | -- |
| BCAT percentage score | 1.10 | 1.03--1.17 | 1.10 | 1.03--1.17 |
| BCAT subjective intensity score | 1.76 | 1.14--2.71 | 1.57 | 1.00--2.46 |
| P53 percentage score | 1.78 | 1.25--2.54 | 1.78 | 1.25--2.54 |
| P53 intensity score | 1.00 | 0.92--1.10 | 1.00 | 0.92--1.10 |
| Cox-2 percentage score | 4.05 | 2.10--7.83 | 4.05 | 2.10--7.83 |
| Cox-2 subjective intensity score | 3.85 | 2.25--6.57 | 3.59 | 2.09--6.18 |
| Survivin percentage score | 1.46 | 1.21--1.77 | 1.46 | 1.21--1.77 |
| Survivin subjective intensity score | 1.86 | 1.14--3.02 | 1.84 | 1.12--3.04 |
| P21 percentage score | 1.32 | 1.09--1.58 | 1.32 | 1.09--1.58 |
| P21 intensity score | 1.10 | 1.01--1.21 | 1.10 | 1.01--1.21 |
| DNApkcs percentage score | 1.01 | 1.01--1.02 | 1.01 | 1.01--1.02 |
| DNApkcs intensity score | 1.00 | 1.00--1.00 | 1.00 | 1.00--1.00 |
| MGMT percentage score | 2.40 | 1.60--3.55 | 2.38 | 1.60--3.55 |
| MGMT intensity score | 1.22 | 1.09--1.36 | 1.22 | 1.09--1.36 |
rounded to hundredth; OR, odds ratio; CI, confidence interval; BCAT, beta catenin; DNApkcs, DNA dependent protein kinase; MGMT, 0(6)-methylguanine-DNA methyltransferase; Aj, adjusted for age, advanced adenoma status, and mutually adjusted for other marker results.
Figure 1. Distribution of molecular markers (percentage and intensity scores).
In (a–c), IHC was performed on control and case subject polyps as in methods. In (a), distribution of percent of cells staining for each marker is depicted for cases and controls. The upper and lower box boundaries represent the 25th and 75th percentiles, and the middle bar represents the median score. In (b), distribution of staining intensity for markers with objective measurement of intensity is depicted for cases and controls. The upper and lower box boundaries represent the 25th and 75th percentiles, and the middle bar represents the median score. In (c), distribution of intensity scores for markers with subjective assessment of intensity are depicted for cases and controls as 0 (lowest intensity), 1 (moderate intensity), or 2 (highest intensity). BCAT, beta catenin; DNApkcs, DNA dependent protein kinase; MGMT, 0(6)-methylguanine-DNA methyltransferase.
Molecular Marker Results: Phase II Case Status Prediction
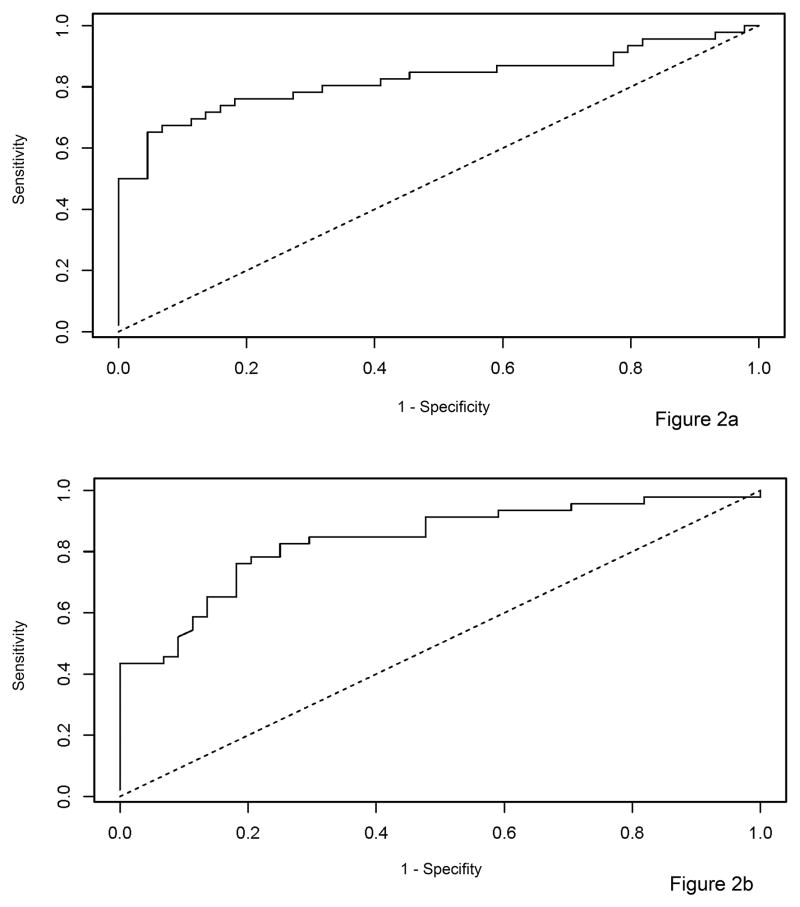
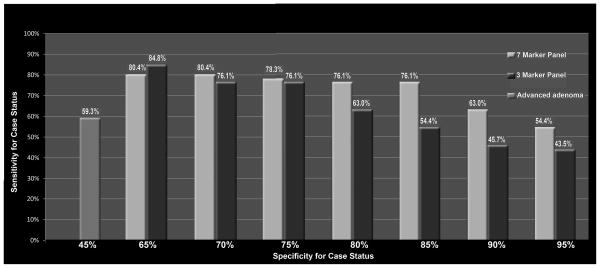
One-hundred seventy-nine patients (85% of cases, 82% of controls) had informative marker results for all 7 markers; data from these patients were used to predict the ability of molecular markers to identify cases. Characteristics of patients randomly assigned to the training and validation sets were similar (Supplementary Table 2). The Random Forest Prediction model for case adenoma prediction developed in the training set of 90 patients (46 cases, 44 controls) was applied to the independent validation set of 89 patients (46 cases, 43 controls). In the test validation set, the model demonstrated high accuracy for predicting case status, with AUC=0.83, 95% CI: (0.74 – 0.92), Figure 2a. To determine if a more focused molecular panel could maintain high accuracy for identifying case adenomas, we identified the three individual markers in the training set with best predictive value (DNApkcs, MGMT, and p21), and determined their predictive value in the test validation set. The 3-panel molecular marker set maintained a high level of accuracy, with AUC=0.84, (95% CI: 0.74–0.92), Figure 2b. When compared to use of advanced adenoma status as a predictor, both our 7 and 3-marker panels appeared to be more sensitive for identifying case status at various specificity cut points (Figure 3).
Figure 2. Predictive value of molecular markers for distinguishing case from control adenomas.
In (a), is shown a receiver operating curve (ROC) for ability of a panel of molecular markers of carcinogenesis including p53, BCAT, MGMT, DNApcks, p21, Cox-2, and survivin to distinguish adenomas from high risk case patients with CRC from low risk control patients without CRC. The ROC demonstrates substantial predictive ability, AUC=0.83, 95% CI (0.74 -0.92). In (b), is shown a ROC for a parsimonious, 3 marker panel, with DNApkcs, MGMT, and p21 for prediction of case adenoma status. The ROC demonstrates substantial predictive ability, AUC=0.84, 95% CI: (0.74 – 0.92). BCAT, beta catenin; DNApkcs, DNA dependent protein kinase; MGMT, 0(6)-methylguanine-DNA methyltransferase.
Figure 3. Sensitivity of the 7-marker panel, 3-marker panel, and presence of advanced adenoma for case status.
Across a range of specificity cut points (shown on the X axis), both the 7- and 3-marker panels had superior sensitivity (Y axis) for identifying high risk case patients compared to sensitivity conferred by presence/absence of advanced adenoma (shown on the far left).
DISCUSSION
Our primary aim was to explore the hypothesis that use of molecular markers for risk stratification could distinguish adenomas from high risk case patients with CRC from low risk control patients without CRC, and thereby support the proof of concept for using this approach to improve surveillance for patients with adenomas. Current polyp surveillance guidelines recommend risk stratification of patients with colorectal polyps into high and low risk categories based the number, size, and histology of polyps identified(2–4). While prior data support that patients with high risk findings based on these criteria have a higher incidence of advanced neoplasia at follow up colonoscopy, the criteria are neither sensitive nor specific for identifying a patient who will develop incident advanced neoplasia(1, 2, 5, 6, 44, 45).
We found 7 molecular markers that were statistically significantly associated with case status: BCAT, p53, Cox-2, survivin, p21, DNAPkcs, and MGMT. Using independent model training and model validation sets, we found that a predictive model including results from our 7 marker panel had substantial accuracy for predicting case status. A more parsimonious model using 3 markers (DNAPkcs, p21, and MGMT) maintained similar accuracy. In contrast, advanced adenoma status--the main current guideline recommended criteria for risk stratification of patients with polyps--was not associated with case status. Moreover, advanced adenoma status had lower sensitivity and specificity for identification of high risk case patients than our predictive model employing 7 molecular markers. Taken together, our findings suggest that there are inherent differences in the cancer biology of polyps in those with and without colon cancer and support the proof of concept that use of molecular markers in clinical practice has great potential to improve risk stratification of patients with colorectal polyps.
Few studies have examined the use of molecular markers for risk stratification of individuals with colorectal polyps. Two retrospective cohort studies of patients with adenomas at baseline examined whether IHC for several markers of carcinogenesis could predict incident colorectal cancer(12, 13). The first report included 147 patients with adenomas at baseline, 10 of whom developed CRC on median follow up over 144 months(12). In this report, Soreide K et al. found that IHC expression of BCAT, p21, p16, survivin, and human telomerase reverse transcriptase (hTERT) among baseline adenomas was significantly different for patients who developed incident CRC compared to controls. The most predictive molecular markers were survivin, hTERT, and nuclear BCAT. No association was found for several other tested markers including Cox-2 and p53. In contrast, we observed an association between Cox-2 and p53 expression and case status. Unlike our study, DNApkcs, p27, and MGMT were not studied. In a follow up report by Soreide K et al., the association of hTERT and survivin among a group of 274 patients with baseline adenomas was confirmed, 16 of whom developed incident CRC(13). Similar to these studies, we found that p21, and in particular survivin and BCAT, were associated with adenomas from high risk patients, validating the observations for these markers. Key distinguishing features of our report compared to these 2 previous studies include our examination of 108 high risk CRC case patients matched to controls to allow for greater discrimination of marker predictive ability, and validation of a broader panel of candidate predictive markers. Additionally, we identified novel associations between DNApkcs, p27, MGMT, and high risk case status. Finally, we focused on using cytoplasmic, rather than nuclear over-expression of BCAT as a biomarker, because of prior work suggesting that cytoplasmic over-expression may precede nuclear over-expression in carcinogenesis(46). Nonwithstanding, the findings previously reported by Soreide K et al., and now in this current novel data set provide strong proof of concept that molecular markers can be used to effectively risk stratify patients with adenomas.
We recognize limitations of our study. First, we employed a retrospective, case-control design. Case-control studies of diagnostic and predictive tests may overestimate the association of predictors with outcomes(47). To establish proof of concept that molecular markers within polyps can be used for post-polypectomy risk stratification, we employed a study design in which cases were adenomas from CRC patients, and controls were adenomas from non-CRC patients. In practice, post-polypectomy surveillance focuses on assessing risk for both CRC, as well as advanced adenomas, which are thought to be potential biologic precursors of CRC. Thus, an ideal biomarker strategy would address risk for both CRC and advanced adenomas; this should be addressed in future research. Among patients with adenomas detected by average risk screening, approximately 25% have advanced adenomas(48). Thus, the spectrum of adenomas included in our study differs from usual practice, and could have resulted in spectrum bias that may overestimate the association of our predictors with case status. Similar to most studies examining prevalence of molecular changes in colorectal neoplasia, we were unable to determine whether differential patterns or marker expression between case and control adenomas represent critical carcinogenic steps, or biomarkers of other carcinogenic processes(19). Plausibility of our findings hinges on the concept that polyp biomarker expression patterns found in high versus low risk patients reflect genetic factors and environmental exposures that will promote recurrence of similar high risk polyps on follow up. This concept is supported by the observation (employed in current polyp surveillance guidelines) that even gross polyp characteristics such as number, size, and histology are associated with future risk of high-risk neoplasia. It should also be noted that this was a single institution study. Thus, it is possible that characteristics of case and control patients, including factors such as polyp distribution, number, size, and histology may differ from all individuals with adenoma, requiring future studies of other populations to establish generalizability. Of the 963 candidates considered for inclusion of cases, 108 were ultimately selected for our study. Though most exclusions were due to absence of synchronous adenoma, presence of cancer other than colorectal cancer, or absence of sufficient adenoma tissue for analysis (see Supplementary Figure 1), we cannot exclude the possibility that our CRC sample may not be representative of the general population of individuals with CRC, perhaps limiting generalizability of study findings. For 25% of control patients (n=27), data on follow up colonoscopy were not available. Additionally, among the 81 control patients who did have follow up colonoscopy, 7 patients had the colonoscopy performed within 3 years of baseline. Thus it is possible that a control patient might have had an undiagnosed CRC because follow up colonoscopy was not performed, or performed too soon for neoplasia to develop. However, the chances of such misclassification are low, given that median clinical follow up among controls without follow up colonoscopy was 102.6 months (8.5 years), and that cancer after colonoscopic polypectomy is rare, estimated at occurring among 0.4 to 2.2 individuals per 1000 person years of follow up(49). Further, among the 81 controls who had follow up colonoscopy, median time between baseline and repeat colonoscopy was 5.5 years, a time period long enough for some CRCs to develop. Of note, even if misclassification of a control as being CRC free did indeed occur, it would have biased towards finding no difference in marker expression between case and control polyps. Finally, a single study pathologist was responsible for review of subjective markers and confirmation of objective marker expression measurements. Inter-and intra-observer variability in IHC assessment were not assessed, potentially impacting generalizability(50–52). However, since the review was blind to case/control status of polyps, this should not have biased our results showing differential expression patterns between cases and controls. Limitations of the current study may be addressed by conduct of prospective validation of the predictive markers identified here, as well as study of other molecular markers for risk stratification of patients with polyps. Standardization and reproducibility of IHC reads among different pathologists will also have to be established. Importance of novel associations, such as that observed for DNAPkcs, can be explored through future studies of the importance of this protein in mouse and cell line models.
Several strengths of our study may balance potential limitations. The strengths include a) use of a large random sample of candidate cases and controls, b) blinded evaluation of marker expression among cases and controls, and c) and use of independent model development and model validation sets.
In conclusion, we have shown that molecular markers of carcinogenesis, measured by IHC, can distinguish adenomas from high risk case patients with CRC from low risk control patients who do not develop CRC. Our findings support the proof of concept that molecular markers can improve risk stratification of patients with polyps. Since the current approach to risk stratification of patients with polyps is insufficiently sensitive and specific for identifying patients who will develop incident advanced neoplasia, prospective studies of molecular markers for polyp risk stratification are warranted.
Supplementary Material
Acknowledgments
Grant Support: Funding Support was provided by: 1) American Society of Gastrointestinal Endoscopy 2006 Endoscopic Research Award (Gupta, PI); 2) National Institutes of Health grant number 1 KL2 RR024983-01, titled, “North and Central Texas Clinical and Translational Science Initiative” (Milton Packer, PI, Gupta, KL2 Scholar) from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research, and its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NCRR or NIH. Information on NCRR is available at http://www.ncrr.nih.gov/. Information on Re-engineering the Clinical Research Enterprise can be obtained from http://nihroadmap.nih.gov/clinicalresearch/overview-translational.asp; 3) Cancer Prevention and Research Institute of Texas Grant PP100039 (Gupta, PI); 4) National Institutes of Health/National Cancer Institute grant number 1U54CA163308-01 titled, “Parkland-UT Southwestern PROSPR Center: Colon cancer screening in a safety net.” (Celette Sugg Skinner, PI, Gupta Co-I). None of the fundings sources had a role in the design, analysis, interpretation, or manuscript preparation phases of the study.
The authors would like to thank HaoChi Zhang, Rodrigo Soto, Gretchen Troxler for assistance with data collection, Zhuo Gheng and Malaika Tobias for assistance with manuscript preparation, Dr. Andres Roig for assistance in project planning, and Jennifer Tinkler for assistance with conducting immunohistochemistry studies.
Footnotes
Financial Disclosure: Authors have no financial disclosures.
References
- 1.Laiyemo AO, Murphy G, Albert PS, Sansbury LB, Wang Z, Cross AJ, et al. Postpolypectomy colonoscopy surveillance guidelines: predictive accuracy for advanced adenoma at 4 years. Annals of internal medicine. 2008;148(6):419–26. doi: 10.7326/0003-4819-148-6-200803180-00004. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR. Guidelines for Colonoscopy Surveillance After Screening and Polypectomy: A Consensus Update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2012;143:844–57. doi: 10.1053/j.gastro.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Atkin WS, Saunders BP British Society for G, Association of Coloproctology for Great B Ireland. Surveillance guidelines after removal of colorectal adenomatous polyps. Gut. 2002;51 (Suppl 5):V6–9. doi: 10.1136/gut.51.suppl_5.v6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns SR, Scholefield JH, Steele RJ, Dunlop MG, Thomas HJ, Evans GD, et al. Guidelines for colorectal cancer screening and surveillance in moderate and high risk groups. Gut. 2010;59(5):666–89. doi: 10.1136/gut.2009.179804. [DOI] [PubMed] [Google Scholar]
- 5.Martinez ME, Baron JA, Lieberman DA, Schatzkin A, Lanza E, Winawer SJ, et al. A pooled analysis of advanced colorectal neoplasia diagnoses after colonoscopic polypectomy. Gastroenterology. 2009;136(3):832–41. doi: 10.1053/j.gastro.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta S. Colorectal polyps: the scope and management of the problem. The American journal of the medical sciences. 2008;336(5):407–17. doi: 10.1097/MAJ.0b013e31817d2402. [DOI] [PubMed] [Google Scholar]
- 7.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6(5):963–8. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 8.Lippman SM, Hawk ET. Cancer prevention: from 1727 to milestones of the past 100 years. Cancer research. 2009;69(13):5269–84. doi: 10.1158/0008-5472.CAN-09-1750. [DOI] [PubMed] [Google Scholar]
- 9.Lochhead P, Chan AT, Nishihara R, Fuchs CS, Beck AH, Giovannucci E, et al. Etiologic field effect: reappraisal of the field effect concept in cancer predisposition and progression. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2014 doi: 10.1038/modpathol.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy HK, Damania DP, DelaCruz M, Kunte DP, Subramanian H, Crawford SE, et al. Nano-architectural alterations in mucus layer fecal colonocytes in field carcinogenesis: potential for screening. Cancer prevention research (Philadelphia, Pa) 2013;6(10):1111–9. doi: 10.1158/1940-6207.CAPR-13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backman V, Roy HK. Advances in biophotonics detection of field carcinogenesis for colon cancer risk stratification. Journal of Cancer. 2013;4(3):251–61. doi: 10.7150/jca.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soreide K, Buter TC, Janssen EA, Gudlaugsson E, Skaland I, Korner H, et al. Cell-cycle and apoptosis regulators (p16INK4A, p21CIP1, beta-catenin, survivin, and hTERT) and morphometry-defined MPECs predict metachronous cancer development in colorectal adenoma patients. Cellular oncology : the official journal of the International Society for Cellular Oncology. 2007;29(4):301–13. doi: 10.1155/2007/457427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soreide K, Gudlaugsson E, Skaland I, Janssen EA, Van Diermen B, Korner H, et al. Metachronous cancer development in patients with sporadic colorectal adenomas-multivariate risk model with independent and combined value of hTERT and survivin. International journal of colorectal disease. 2008;23(4):389–400. doi: 10.1007/s00384-007-0424-6. [DOI] [PubMed] [Google Scholar]
- 14.Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. Journal of the National Cancer Institute. 2005;97(18):1330–8. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 15.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. The Journal of molecular diagnostics : JMD. 2008;10(1):13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50(1):113–30. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Fearon ER. Molecular genetics of colorectal cancer. Annual review of pathology. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 20.Guilera M, Connelly-Frost A, Keku TO, Martin CF, Galanko J, Sandler RS. Does physical activity modify the association between body mass index and colorectal adenomas? Nutrition and cancer. 2005;51(2):140–5. doi: 10.1207/s15327914nc5102_3. [DOI] [PubMed] [Google Scholar]
- 21.Martin C, Connelly A, Keku TO, Mountcastle SB, Galanko J, Woosley JT, et al. Nonsteroidal anti-inflammatory drugs, apoptosis, and colorectal adenomas. Gastroenterology. 2002;123(6):1770–7. doi: 10.1053/gast.2002.37053. [DOI] [PubMed] [Google Scholar]
- 22.Connelly-Frost A, Poole C, Satia JA, Kupper LL, Millikan RC, Sandler RS. Selenium, apoptosis, and colorectal adenomas. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2006;15(3):486–93. doi: 10.1158/1055-9965.EPI-05-0759. [DOI] [PubMed] [Google Scholar]
- 23.Connelly AE, Satia-Abouta J, Martin CF, Keku TO, Woosley JT, Lund PK, et al. Vitamin C intake and apoptosis in normal rectal epithelium. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2003;12(6):559–65. [PubMed] [Google Scholar]
- 24.Keku TO, Lund PK, Galanko J, Simmons JG, Woosley JT, Sandler RS. Insulin resistance, apoptosis, and colorectal adenoma risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14(9):2076–81. doi: 10.1158/1055-9965.EPI-05-0239. [DOI] [PubMed] [Google Scholar]
- 25.Il’yasova D, Hodgson ME, Martin C, Galanko J, Sandler RS. Tea consumption, apoptosis, and colorectal adenomas. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation. 2003;12(5):439–43. doi: 10.1097/00008469-200310000-00016. [DOI] [PubMed] [Google Scholar]
- 26.Miller EA, Keku TO, Satia JA, Martin CF, Galanko JA, Sandler RS. Calcium, vitamin D, and apoptosis in the rectal epithelium. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14(2):525–8. doi: 10.1158/1055-9965.EPI-04-0466. [DOI] [PubMed] [Google Scholar]
- 27.Markowitz SD. Aspirin and colon cancer--targeting prevention? The New England journal of medicine. 2007;356(21):2195–8. doi: 10.1056/NEJMe078044. [DOI] [PubMed] [Google Scholar]
- 28.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132(1):127–38. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Jin YM, Li BJ, Qu B, Du YJ. BRAF, K-ras and BAT26 mutations in colorectal polyps and stool. World journal of gastroenterology : WJG. 2006;12(32):5148–52. doi: 10.3748/wjg.v12.i32.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohue M, Tomita N, Monden T, Fujita M, Fukunaga M, Takami K, et al. A frequent alteration of p53 gene in carcinoma in adenoma of colon. Cancer research. 1994;54(17):4798–804. [PubMed] [Google Scholar]
- 31.Einspahr JG, Martinez ME, Jiang R, Hsu CH, Rashid A, Bhattacharrya AK, et al. Associations of Ki-ras proto-oncogene mutation and p53 gene overexpression in sporadic colorectal adenomas with demographic and clinicopathologic characteristics. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2006;15(8):1443–50. doi: 10.1158/1055-9965.EPI-06-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curtin K, Slattery ML, Holubkov R, Edwards S, Holden JA, Samowitz WS. p53 alterations in colon tumors: a comparison of SSCP/sequencing and immunohistochemistry. Applied immunohistochemistry & molecular morphology : AIMM / official publication of the Society for Applied Immunohistochemistry. 2004;12(4):380–6. doi: 10.1097/00129039-200412000-00017. [DOI] [PubMed] [Google Scholar]
- 33.Balbinotti RA, Ribeiro U, Jr, Sakai P, Safatle-Ribeiro AV, Balbinotti SS, Scapulatempo C, et al. hMLH1, hMSH2 and cyclooxygenase-2 (cox-2) in sporadic colorectal polyps. Anticancer research. 2007;27(6C):4465–71. [PubMed] [Google Scholar]
- 34.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(12):6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell research. 2008;18(1):134–47. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 36.Rigas B, Borgo S, Elhosseiny A, Balatsos V, Manika Z, Shinya H, et al. Decreased expression of DNA-dependent protein kinase, a DNA repair protein, during human colon carcinogenesis. Cancer research. 2001;61(23):8381–4. [PubMed] [Google Scholar]
- 37.Zhivotovsky B, Kroemer G. Apoptosis and genomic instability. Nature reviews Molecular cell biology. 2004;5(9):752–62. doi: 10.1038/nrm1443. [DOI] [PubMed] [Google Scholar]
- 38.Petko Z, Ghiassi M, Shuber A, Gorham J, Smalley W, Washington MK, et al. Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(3):1203–9. [PubMed] [Google Scholar]
- 39.Ogino S, Hazra A, Tranah GJ, Kirkner GJ, Kawasaki T, Nosho K, et al. MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis. 2007;28(9):1985–90. doi: 10.1093/carcin/bgm160. [DOI] [PubMed] [Google Scholar]
- 40.McKay JA, Douglas JJ, Ross VG, Curran S, Loane JF, Ahmed FY, et al. Analysis of key cell-cycle checkpoint proteins in colorectal tumours. The Journal of pathology. 2002;196(4):386–93. doi: 10.1002/path.1053. [DOI] [PubMed] [Google Scholar]
- 41.Kutner M, Nachtsheim C, Neter J, Li W. Applied Linear Statistical Models. Homewood, IL: McGraw-Hill/Irwin; 2004. [Google Scholar]
- 42.Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu B, Abbott T, Fishman D, McMurray W, Mor G, Stone K, et al. Comparison of statistical methods for classification of ovarian cancer using mass spectrometry data. Bioinformatics. 2003;19(13):1636–43. doi: 10.1093/bioinformatics/btg210. [DOI] [PubMed] [Google Scholar]
- 44.Winawer SJ, Zauber AG, Fletcher RH, Stillman JS, O’Brien MJ, Levin B, et al. Guidelines for colonoscopy surveillance after polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer and the American Cancer Society. CA: a cancer journal for clinicians. 2006;56(3):143–59. doi: 10.3322/canjclin.56.3.143. [DOI] [PubMed] [Google Scholar]
- 45.Saini SD, Kim HM, Schoenfeld P. Incidence of advanced adenomas at surveillance colonoscopy in patients with a personal history of colon adenomas: a meta-analysis and systematic review. Gastrointest Endosc. 2006;64(4):614–26. doi: 10.1016/j.gie.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi M, Honma T, Matsuda Y, Suzuki Y, Narisawa R, Ajioka Y, et al. Nuclear translocation of beta-catenin in colorectal cancer. British journal of cancer. 2000;82(10):1689–93. doi: 10.1054/bjoc.1999.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pepe M. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford: Oxford University Press; 2003. [Google Scholar]
- 48.Heitman SJ, Ronksley PE, Hilsden RJ, Manns BJ, Rostom A, Hemmelgarn BR. Prevalence of adenomas and colorectal cancer in average risk individuals: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2009;7(12):1272–8. doi: 10.1016/j.cgh.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 49.Loeve F, van Ballegooijen M, Snel P, Habbema JD. Colorectal cancer risk after colonoscopic polypectomy: a population-based study and literature search. Eur J Cancer. 2005;41(3):416–22. doi: 10.1016/j.ejca.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Walker RA. Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment I. Histopathology. 2006;49(4):406–10. doi: 10.1111/j.1365-2559.2006.02514.x. [DOI] [PubMed] [Google Scholar]
- 51.Taylor CR, Levenson RM. Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49(4):411–24. doi: 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- 52.Seidal T, Balaton AJ, Battifora H. Interpretation and quantification of immunostains. The American journal of surgical pathology. 2001;25(9):1204–7. doi: 10.1097/00000478-200109000-00013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.