Abstract
Schizophrenia is a complex polygenic disorder of unknown etiology. Over 3,000 candidate genes associated with schizophrenia have been reported, most of which being mentioned only once. Alterations in cognitive processing - working memory, metacognition and mentalization - represent a core feature of schizophrenia, which indicates the involvement of the prefrontal cortex in the pathophysiology of this disorder. Hence we compared the gene expression in postmortem tissue from the left and right dorsolateral prefrontal cortex (DLPFC, Brodmann's area 46), and the medial part of the orbitofrontal cortex (MOFC, Brodmann's area 11/12), in six patients with schizophrenia and six control brains. Although in the past decade several studies performed transcriptome profiling in schizophrenia, this is the first study to investigate both hemispheres, providing new knowledge about possible brain asymmetry at the level of gene expression and its relation to schizophrenia. We found that in the left hemisphere, twelve genes from the DLPFC and eight genes from the MOFC were differentially expressed in patients with schizophrenia compared to controls. In the right hemisphere there was only one gene differentially expressed in the MOFC. We reproduce the involvement of previously reported genes TARDBP and HNRNPC in the pathogenesis of schizophrenia, and report seven novel genes: SART1, KAT7, C1D, NPM1, EVI2A, XGY2, and TTTY15. As the differentially expressed genes only partially overlap with previous studies that analyzed other brain regions, our findings indicate the importance of considering prefrontal cortical regions, especially those in the left hemisphere, for obtaining disease-relevant insights.
Keywords: Brain asymmetry, Dorsolateral prefrontal cortex, Gene expression, Orbitofrontal cortex, Schizophrenia, Transcriptomics
1. Introduction
Schizophrenia is a common, severe mental disorder of unknown etiology. It usually appears during adolescence and early adulthood and is characterized by positive and negative symptoms that reflect disruption of thought, mood, perception, and volition. Neurobehavioral fMRI studies have shown that impaired attention, working memory and decision-making (as functions of the dorsolateral prefrontal cortex, DLPFC), and impaired sensory integration, affect regulation and emotion processing, social cognition and metacognition (as functions of the medial orbitofrontal cortex, MOFC), are core features of schizophrenia [1, 2]. Whereas the concordance for schizophrenia in identical twins is about 48%, the incidence decreases with decreased level of genetic similarity to about 6-17% in first-degree relatives, 2-6% in second-degree relatives, and 1-2% in third-degree relatives and the general population [3]. The earlier mentioned domains of cognitive function appear to be mildly impaired in the unaffected relatives of individuals with schizophrenia, which indicates that cognitive dysfunction likely reflects the genetic liability for the illness [4].
Some of the main candidate genes for genetic susceptibility for schizophrenia are DISCI (disrupted in schizophrenia 1), DTNBP1 (dystrobrevin binding protein 1),RGS4 (regulator of G-protein signaling 4), PRODH (proline dehydrogenase), NRG1 (neuroregulin 1), COMT (catechol-O-methyl transferase), but many others have been reported (e.g., TRAR4, GABAA, PIP5K2A, ZDHHC8, G72/G30, CAPON, SYN2, GRM3, CHI3L1, SPEC2/PDZ-GEF2/ACSL6, FXYD6, PPP3CC) [5,6]. In addition, a recent large genome-wide association study identified 108 schizophrenia-associated loci, out of which 83 were previously not reported [7].
The main goal of this study was to assess whether gene expression patterns differ between normal control and schizophrenic brain in regions of the prefrontal cortex implicated in schizophrenia etiology and symptomatology. Although psychosis (hallucinations, delusions, and disorganized behavior) is the most pronounced clinical feature of schizophrenia, impaired cognition has been suggested to be a core domain of dysfunction [8]. Therefore, we analyzed gene expression in the dorsolateral prefrontal cortex (DLPFC, Brodmann's area 46) and in the medial orbitofrontal cortex (MOFC, Brodmann's area 11/12) of subjects with schizophrenia and normal controls. DLPFC was selected primarily because impaired working memory is a hallmark endophenotype of schizophrenia [9], and because working memory deficits are among the key cognitive impairments of schizophrenia that remain relatively stable despite fluctuations in symptoms over the course of the disease [10]. MOFC has been proposed to be involved in sensory integration, in representing the affective value of reinforcers, in decision-making and expectation, as well as intuitive coherence judgments, subjective control of action and believing [11]. Imaging studies have confirmed that the activity in the left orbitofrontal region (area 11) is impaired in schizophrenic patients. Together with several other regions, the left area 11 is critically associated with directed efforts (i.e., realizing control of internal thoughts) such as the ability to represent, monitor and control the thoughts, feelings, and actions of self (a process called metacognition) [12], and of others across time (mentalization) [13]. As the aberrant activity and connectivity of the left MOFC may reflect impairments in the perception and initiation of action and be responsible for both the positive and negative symptoms of schizophrenia, we additionally tested whether the difference in gene expression observed between the left and the right hemispheres also differed between schizophrenic and control brains. The level of gene expression in these areas of the brain could offer valuable insight into the cause of symptoms seen in schizophrenia as well as its etiology.
2. Materials and methods
2.1. Study samples
Postmortem samples of DLPFC and MOFC were obtained from 6 brains from subjects with schizophrenia and 6 normal controls from the Zagreb Collection of human brains [14]. The normal control subjects were chosen to match the schizophrenic group closely for age and postmortem delay (Table 1). All schizophrenic patients met the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, American Psychiatric Association, 2000 (DSM- IV-TR) [15] diagnostic criteria for schizophrenia and were, until their death, under long-term treatment (with either clozapine, olanzapine, or risperidone), and had regular clinical follow-ups by at least one experienced psychiatrist. Both the schizophrenic subjects and controls were carefully chosen so that the brains selected for analysis were not taken from subjects who had a prior history of head trauma, alcohol or drug abuse, or other major neuropsychiatric disorders.
Table 1.
Demographic and tissue characteristics of the schizophrenia (SZ) and normal control (NC) brain samples analyzed. PMD, postmortem delay; RIN, RNA integrity number; SD, standard deviation. RIN was calculated using a proprietary algorithm of the Agilent Technologies, Santa Clara, CA, USA.
| Subject Code | Gender | Age (years) | PMD (h) | RIN | Cause of death |
|---|---|---|---|---|---|
| SZ group | |||||
| SZ1 | M | 59 | 26 | 8.65 ±0.17 | Suicide by hanging |
| SZ2 | F | 56 | 20 | 7.40±0.56 | Pneumonia |
| SZ3 | F | 57 | 6 | 4.93±0.50 | Sudden cardiac arrest |
| SZ4 | M | 50 | 14 | 8.13±0.22 | Heart failure |
| SZ5 | F | 47 | 24 | 8.03±0.26 | Sudden cardiac arrest |
| SZ6 | F | 42 | 24 | 4.98±0.43 | Drug poisoning |
| Mean±SD | 51.83±6.62 | 19±7.67 | 7.02±1.65 | ||
| NC group | |||||
| NC1 | M | 54 | 17 | 7.53±0.36 | Heart failure |
| NC2 | F | 60 | 6 | 6.90±0.71 | Sudden cardiac arrest |
| NC2 | F | 61 | 24 | 8.55±0.64 | Heart failure |
| NC4 | F | 40 | 30 | 7.65±0.37 | Heart failure |
| NC5 | M | 55 | 13 | 8.55±0.21 | Heart failure |
| NC6 | M | 42 | 24 | 8.75±0.07 | Heart failure |
| Mean±SD | 52.00±8.97 | 19±8.72 | 7.99±0.74 | ||
2.2. Dissection of tissue samples
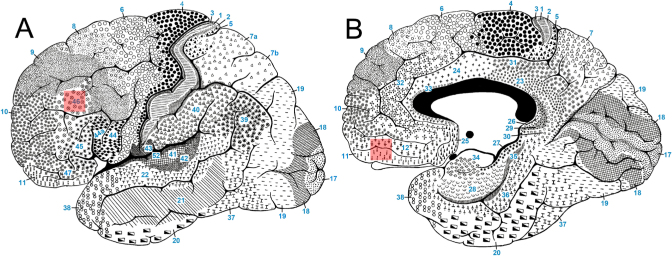
Gray matter tissue samples were carefully dissected from the mid frontal (area 46) and orbitofrontal (area 11/12) regions, which correspond to DLPFC and MOFC, respectively (Figure 1) [16-18]. Each sample had an approximate volume of about 0.5 cm3. The caudal part of each block was used for staining with cresyl-violet (Nissl stain). The Nissl stain served to check for the cytoarchitectural features of the Brodmann's areas analyzed. Area 46 was identified by the criteria described in Rajkowska et al. 1995 [19]. Area 46 is bounded dorsally by the granular frontal area 9, rostroventrally by the frontopolar area 10, and caudally it joins the triangular area 45 (16, 17, 19). Area 11 is bordered on the rostral and lateral aspects of the hemisphere by the frontopolar area 10, the orbital area 47, and the triangular area 45; on the caudal aspect it abuts subgenual area 25. On the medial surface it continues into rostral area 12.
Figure 1.
Locations of dissected samples. Both areas are labeled in transparent red color. A. Area 46, which corresponds to dorsolateral prefrontal cortex (DLPFC), B. Area 11/12 corresponds to medial orbitofrontal cortex (MOFC). See text for details.
2.3. RNA extraction, purification and quality control
After dissection, the samples were immediately placed in the RNAlater solution (Thermo Fischer Scientific, Waltham, MA, USA), in which RNA was stabilized, and then stored in the freezer at -80°C until further analysis. For each microarray analysis, we used 20-30 mg of brain tissue from either area 46 or area 11/12. Isolation of RNA from brain tissue samples was performed using RNeasy Plus Mini kit from Qiagen (Venlo, The Netherlands) according to the manufacturer's protocol. Concentrations and quality of isolated RNA were measured by using Bioanalyzer 2100 and RNA 6000 Nanochip Kit (both from Agilent, Palo Alto, CA, USA). RNA degradation was assessed by quantifying the resulting 28S/18S ribosomal band peak height ratios: all samples were in the acceptable range of values between 1.5 and 2.0. The quality of each sample was further assured by a RNA integrity number (RIN) greater than 5 (Table 1). Only 2 out of 48 samples had RIN value smaller than 5, and were discarded from the analysis.
2.4. Microarray procedure
Total RNA was reverse-transcribed and hybridized onto Affymetrix HG-U133 Plus 2.0 array, using the identical protocol and ID names for genes as GeneChip Human Exon 1.0ST Arrays (Affymetrix, Santa Clara, CA, USA). The Affymetrix Human Gene Chip Exon 1.0 ST Array interrogates over one million exons representing over 17,868 NCBI Reference Sequence (RefSeq) transcripts. Arrays were run using the manufacturer's technical protocol (Affymetrix, Santa Clara, CA). Briefly, 2 qg of total RNA was subjected to a ribosomal RNA removal procedure (RiboMinus Human/ Mouse Transcriptome Isolation Kit, Invitrogen - Thermo Fischer Scientific) to reduce the 28S and 18S rRNA population to minimize background and increase sensitivity of the assay. Reduced RNA was reverse-transcribed to cDNA using random hexamers tagged with a T7 promoter sequence followed by a second strand cDNA synthesis using DNA polymerase (GeneChip WT cDNA Synthesis Kit, Affymetrix). The resulting double-stranded cDNA was used for amplification of antisense cRNA and cleaned using the Gene Chip Sample Cleanup Module (Affymetrix). A second cycle cDNA synthesis was performed using random primers to reverse transcribe the cRNA into sense single stranded DNA, which was fragmented, labeled, and hybridized to arrays. Arrays were washed, stained, and scanned on the Affymetrix Fluidics Station and G7 Affymetrix high-resolution scanner using GCOS 1.3.
2.5. Microarray data preparation and analysis
The resulting gene expression values were derived from the microarray scan data (.cel files) and analyzed by using Partek Genomic Suite software ver. 6.5 (Partek Incorporated, St. Louis, MO, USA). Data preparation included corrections for background signal noise made by a robust multi-array average (RMA) procedure, as described in Irizarry et al. [20], and log2 transformation of yielded fluorescence intensities. After quantile normalization of the data and the averaging of probe signals by the arithmetic mean, the average expression values were calculated for each sample group and those showing a fold change of less than 1.5 were omitted, followed by removal of those with a p-value greater than 0.05. It is possible that these subjective filtering criteria resulted in some genuinely differentially expressed genes being omitted from the final analysis. As such we provide the raw data as a supplementary file to allow for reanalysing of the data with other approaches (Appendix A. Supplementary data). Schizophrenia and normal control groups were compared on mean expression levels of all transcripts using Student's t test, with a significance level of p< 0.05. To gain insight into the likely functions of the differentially expressed genes, gene ontology analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) [21,22]. Functional annotations that were assigned with a p-value higher than 0.05 were eliminated, followed by elimination of redundant terms.
3. Results
3.1. Differentially expressed genes
After applying the filtering criteria described in the Materials and Methods, five genes were found to be differentially expressed with statistical significance when the DLPFC from patients with schizophrenia (left and right hemispheres combined) was compared to the DLPFC from healthy controls (Table 2). Of these, one, TARDBP, had increased expression in the schizophrenic DLPFC compared to controls, while the remaining four genes had decreased expression. Comparison of the schizophrenic MOFC (left and right combined) to the MOFC from controls also identified five genes that were differentially expressed (Table 2), although only two of these (TARDBP and KRT18) were also detected in the DLPFC comparison.
Table 2.
Differences in gene expression for DLPFC (area 46) and MOFC (area 11/12) between schizophrenia (SZ) and control (NC) group.
| Gene name | Identifier | Average expression change in SZ | Significance (P values) | Name and function(s) |
|---|---|---|---|---|
| SZ vs NC: DLPFC and MOFC together from both hemispheres | ||||
| KRT18 | 4028716 | −1.67 | 0.000512*** | Keratin 18; intermediate filament involved in poly(A) RNA binding, scaffold protein binding and filament reorganization |
| XGY2 | 4028512 | −1.64 | 0.000028**** | XG pseudogene on Y chromosome; function unknown |
| DDX3Y | 4030162 | −1.45 | 0.000081**** | RNA helicase on Y chromosome; involved in RNA splicing, cell cycle, apoptosis and differentiation |
| TTTY15 | 4030063 | −1.43 | 0.000431*** | Testis-specific transcript on Y chromosome; non-coding RNA, function unknown |
| TARDBP | 2320116 | 1.52 | 0.000062**** | Transactive response DNA binding protein; bind both DNA and RNA and have multiple functions in transcriptional repression (e.g. represses HIV-1 transcription), pre-mRNA splicing and translational regulation; hyperphosphorylated, ubiquinated and cleaved form (TDP-43 protein) is the major disease protein in FTLD-TDP and ALS. |
|
| ||||
| SZ vs NC: DLPFC from both hemispheres | ||||
| KRT18 | 4028716 | −1.69 | 0.012059* | Keratin 18; intermediate filament involved in poly(A) RNA binding, scaffold protein binding and filament reorganization |
| XGY2 | 4028512 | −1.63 | 0.003009** | XG pseudogene on Y chromosome; function unknown |
| DDX3Y | 4030162 | −1.52 | 0.006221** | RNA helicase on Y chromosome; involved in RNA splicing, cell cycle, apoptosis and differentiation |
| TTTY15 | 4030063 | −1.47 | 0.011882* | Testis-specific transcript on Y chromosome; non-coding RNA, function unknown |
| TARDBP | 2320116 | 1.51 | 0.002267** | Transactive response DNA binding protein; bind both DNA and RNA and have multiple functions in transcriptional repression (e.g. represses HIV-1 transcription), pre-mRNA splicing and translational regulation; hyperphosphorylated, ubiquinated and cleaved form (TDP-43 protein) is the major disease protein in FTLD-TDP and ALS. |
|
| ||||
| SZ vs NC: MOFC from both hemispheres | ||||
| XGY2 | 4028512 | −1.64 | 0.005236** | XG pseudogene on Y chromosome; function unknown |
| KRT18 | 4028716 | −1.64 | 0.022923* | Keratin 18; intermediate filament involved in poly(A) RNA binding, scaffold protein binding and filament reorganization |
| unconfirmed | 3275188 | 1.51 | 0.001394** | Probably a non-protein coding RNA |
| TARDBP | 2320116 | 1.52 | 0.011170* | Transactive response DNA binding protein; bind both DNA and RNA and have multiple functions in transcriptional repression (e.g. represses HIV-1 transcription), pre-mRNA splicing and translational regulation; hyperphosphorylated, ubiquinated and cleaved form (TDP-43 protein) is the major disease protein in FTLD-TDP and ALS. |
| SNX3 | 2968446 | 1.55 | 0.006198** | Sorting nexin 3 protein; involved in endosome trafficking; interacts with PtdIns3P |
Table legend: ALS, amyotrophic lateral sclerosis; FTLD-TDP, frontotemporal lobar degeneration due to TDP-43-positive neuronal inclusions; HIV1, human immunodeficiency virus 1; PtdIns3P, phophatidylinositol 3-phosphate; TDP-43, transactive response DNA protein of 43 kDa; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
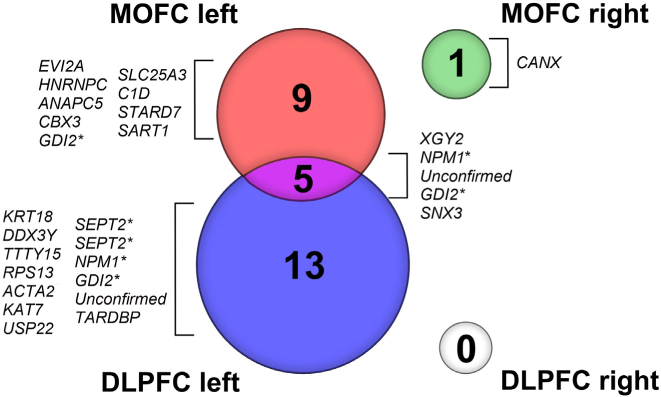
By separating the data and analyzing them according to which hemisphere the tissue was sampled from, we observed some differences compared to the analysis of the values from both hemispheres combined. Comparison of the left schizophrenic DLPFC to the left control DLPFC identified 18 genes that were differentially expressed with statistical significance, while comparison of the right DLPFC did not detect any genes that met the criteria for inclusion (Table 3 and Fig. 2). Similarly, 14 genes were differentially expressed when the left schizophrenic MOFC was compared to controls, and only one gene was differentially expressed in the right schizophrenic MOFC (Table 4 and Fig. 2). For the purpose of these comparisons, genes detected as repeated hits with separate identifiers (e.g.,SEPT2 and GDI2; Table 3) were each counted separately because they most probably account for different splice variants. It is possible, however, that duplicate hits arose due to the presence of a palindromic sequence.
Table 3.
Differences in gene expression in the dorsolateral prefrontal cortex (DLPFC, area 46) between schizophrenia (SZ) and control (NC) brains.
| Gene name | Identifier | Average expression change in SZ | Significance (P value) | Name and function(s) |
|---|---|---|---|---|
| SZ vs NC: DLPFC (area 46) left side only | ||||
| KRT18 | 4028716 | −1.81 | 0.040313* | Keratin 18; intermediate filament involved in poly(A) RNA binding, scaffold protein binding and filament reorganization; its promotor is responsive to EGFR activation |
| XGY2 | 4028512 | −1.75 | 0.007113** | XG pseudogene on Y chromosome; function unknown |
| DDX3Y | 4030162 | −1.68 | 0.010251* | RNA helicase on Y chromosome; involved in RNA splicing, cell cycle, apoptosis and differentiation |
| TTTY15 | 4030063 | −1.55 | 0.024267* | Testis-specific transcript on Y chromosome; non-coding RNA, function unknown |
| RPS13 | 3364736 | −1.51 | 0.006037** | Ribosomal protein S13; involved in poly(A) RNA binding and mRNA binding |
| ACTA2 | 3299504 | −1.50 | 0.012868* | Actin a2; recently described mutations support the role in development of frontal lobe, cingulate gyrus, corpus callosum and cerebellum |
| KAT7 | 3725964 | 1.49 | 0.005552** | Lysine acetyltransferase 7; forms histone acetyltransferase (HAT) complex |
| USP22 | 3749728 | 1.49 | 0.001972** | Ubiquitin-specific peptidase 22; a histone deubiquitinating component of the transcription regulatory HAT complexes, such as SAGA (Spt/Ada/Gcn5L acetyltransferase) |
| GDI2# | 3275374 | 1.50 | 0.011052* | GDP dissociation inhibitor 2; involved in vesicular trafficking of molecules between cellular organelles and G-protein coupled receptors (GPCR) signalling (such as dopamine receptors) |
| SEPT2# | 2536335 | 1.51 | 0.000687*** | Septin 2; involved in GTP binding and ERK signalling; may also play a role in the internalization of intracellular microbial pathogens (e.g. Listeria monocytogenes and Shigella flexneri) |
| SEPT2# | 2536333 | 1.51 | 0.002780** | Septin 2; involved in GTP binding and EGFR signalling; may also play a role in the internalization of intracellular microbial pathogens |
| NPM1# | 2840668 | 1.51 | 0.002012** | Nucleophosmin; involved in ribosome biogenesis, protein chaperoning, histone assembly and many other functions with multiple binding partners |
| SNX3 | 2968446 | 1.52 | 0.001712** | Sorting nexin 3 protein; involved in endosome trafficking and interacts with PtdIns3P |
| NPM1# | 2840726 | 1.54 | 0.011123* | Nucleophosmin; involved in ribosome biogenesis, protein chaperoning, histone assembly and many other functions with multiple binding partners |
| GDI2* | 3275252 | 1.54 | 0.017255* | GDP dissociation inhibitor 2; involved in vesicular trafficking of molecules between cellular organelles and G-protein coupled receptors (GPCR) signaling (such as dopamine receptors) |
| unconfirmed | 3275254 | 1.59 | 0.002173** | Probably a non-protein coding RNA |
| unconfirmed | 3275188 | 1.63 | 0.002547** | Probably a non-protein coding RNA |
| TARDBP | 2320116 | 1.73 | 0.006721** | Transactive response DNA binding protein; bind both DNA and RNA and have multiple functions in transcriptional repression (e.g. represses HIV-1 transcription), pre-mRNA splicing and translational regulation; hyperphosphorylated, ubiquinated and cleaved form (TDP-43 protein) is the major disease protein in FTLD-TDP and ALS. |
|
| ||||
| SZ vs NC: DLPFC (area 46) right side only | ||||
| No significant changes with a 1.5 fold expression level cut-off | ||||
Table legend: EGFR, epidermal growth factor receptor; FTLD-TDP, frontotemporal lobar degeneration due to TDP-43-positive neuronal inclusions; GDP, guanosine diphosphate; GTP, guanosine triphosphate; TDP-43, transactive response DNA protein of 43 kDa; *P . 0.05, **P . 0.01, ***P . 0.001, ****P . 0.0001. Genes marked with # are different splicing variants of the same gene.
Figure 2.
Representation of a number of differentially expressed genes between schizophrenia and normal control brains using Venn diagram. DLPFC, dorsolateral prefrontal cortex, MOFC, medial orbitofrontal cortex. Genes marked with asterisk are different splicing variants of the same gene.
Table 4.
Differences in gene expression in the MOFC (area 11/12) between schizophrenia (SZ) and control (NC) brains.
| Gene name | Identifier | Average expression change in SZ | Significance (P value) | Name and function(s) |
|---|---|---|---|---|
| SZ vs NC: MOFC (area 11/12) left side only | ||||
| GDI2* | 3275388 | 1.76 | 0.000033**** | GDP dissociation inhibitor 2; involved in vesicular trafficking of molecules between cellular organelles and G-protein coupled receptors (GPCR) signaling (such as dopamine receptors) |
| CBX3 | 2993696 | 1.52 | 0.000032**** | Chromobox protein homolog 3; a heterochromatin protein that can bind DNA and lamin; it is recruited to sites of DNA damage and binds to histone H3 tails (thus can be involved in transcriptional reprogramming) |
| SNX3 | 2968446 | 1.62 | 0.000893*** | Sorting nexin 3 protein; involved in endosome trafficking and interacts with PtdIns3P |
| SART1 | 3335846 | 1.64 | 0.001175** | Squamous cell carcinoma antigen recognized by T-cells 1; protein involved in RNA splicing and pre-mRNA processing |
| STARD7 | 2565180 | 1.63 | 0.009539** | StAR-related lipid transfer domain protein 7; lipid transporter that specifically bind and transports phosphatidylcholine between membranes |
| GDI2* | 3275252 | 1.58 | 0.002656** | GDP dissociation inhibitor 2; involved in vesicular trafficking of molecules between cellular organelles and G-protein coupled receptors (GPCR) signaling (such as dopamine receptors) |
| SLC25A3 | 3427855 | 1.53 | 0.003733** | Solute carrier family 25 member 3; catalyzes the transport of phosphate into the mitochondrial matrix |
| ANAPC5 | 3475146 | 1.51 | 0.003543** | Anaphase promoting complex subunit 5; a large E3 ubiquitin ligase required for the proper ubiquitination; also controls cell cycle progression and interacts with poly (A) binding protein and represses internal ribosome entry site-mediated translation |
| HNRNPC | 3556086 | 1.50 | 0.002927** | Heterogeneous nuclear ribonucleoprotein C; involved in pre-mRNA processing, mRNA transport and metabolism |
| C1D | 2557627 | 1.59 | 0.013190* | Nuclear nucleic acid-binding protein C1D; apoptosis-inducing and DNA binding protein |
| XGY2 | 4028512 | −1.58 | 0.031729* | XG pseudogene on Y chromosome; function unknown |
| unconfirmed | 3275188 | 1.57 | 0.012645* | Probably a non-protein coding RNA |
| EVI2A | 3752271 | −1.56 | 0.024747* | Ecotropic viral integration site 2A; protein involved in transmembrane signaling receptor receptor activity |
| NPM1 | 2840726 | 1.51 | 0.010982* | Nucleophosmin; involved in ribosome biogenesis, protein chaperoning, histone assembly and many other functions with multiple binding partners |
| SZ vs NC: mOFC (area 11/12) right side only | ||||
| CANX | 2844385 | 1.48 | 0.006169** | Calnexin; a chaperone protein involved in ensuring that only properly folded and assembled proteins proceed along the secretory pathway of endoplasmatic reticulum and Golgi apparatus |
Table legend: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. #Genes under identifiers 3275252 and 3275388 are different splicing variants of the same GDI2 gene.
3.2. Gene ontology analysis
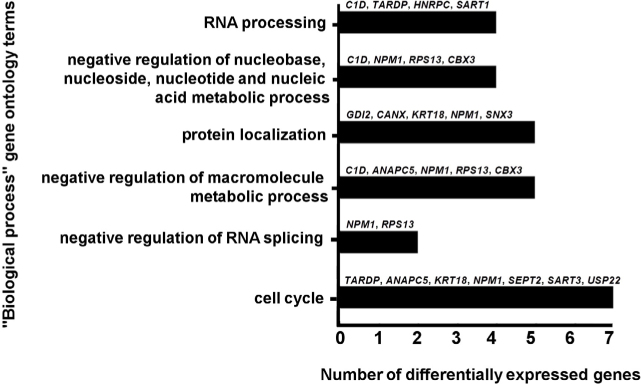
To gain insight into the possible functions of the differentially expressed genes in Tables 2 and 3, we performed gene ontology analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool [21, 22]. Functional annotations that were assigned with a p-value higher than 0.05 were eliminated, followed by elimination of redundant terms. Several biological process terms were enriched among the list of differentially expressed genes and these are summarized in Figure 3. In addition, the molecular function term "RNA binding" was assigned to six of the genes.
Figure 3.
Schematic representation of the number of differentially expressed genes with respect to gene ontology terms.
3.3. Interhemispheric transcriptome differences
Furthermore, to assess possible asymmetric gene expression between the left and right hemispheres, we performed an expression analysis between the right and left hemispheres of control brains, and between the two hemispheres within the schizophrenia samples. The only difference we found was in the DLPFC of normal subjects, that showed a reduced expression of KAT7 gene in the left hemisphere by 41% (p = 0.014), an increased expression of gene NONO in the left hemisphere by 52% (p = 0.005).
Other genes that were detected to have significantly different expression among the schizophrenia and control cases were catalogued for the left hemisphere DLPFC (Table 3) and left hemisphere MOFC (Table 4).
4. Discussion
Our transcriptomic study brings new knowledge about gene expression differences areas 46 and 11/12 from the left and right hemispheres of schizophrenic and control brains. Our results only partially overlap with the results of other gene expression studies, probably due to the analysis of RNA from different tissues and brain regions [23-33]. For the majority of differentially expressed genes between schizophrenia and control samples in the left hemisphere, an association with schizophrenia has already been reported. Particularly strong evidence exists for TARDBP and HNRNPC [34-36], followed by CANX areas 46 and 11/12 from the left and right hemispheres of schizophrenic and control brains. Our results only partially overlap with the results of other gene expression studies, probably due to the analysis of RNA from different tissues and brain regions [23-33]. For the majority of differentially expressed genes between schizophrenia and control samples in the left hemisphere, an association with schizophrenia has already been reported. Particularly strong evidence exists for TARDBP and HNRNPC [34-36], followed by CANX [37], SNX3 [38], GDI2 [39], and SLC25A3 [40], Association of CBX3 [41], DDX3Y, USP22 [42], RPS13 [43], STARD7 [44], ACTA2 [45], and KRT18 [46] with schizophrenia was reported in few studies, while SART1, KAT7, C1D, NPM1, EVI2A, XGY2, and TTTY15 genes were not previously related to schizophrenia. In the right hemisphere, only CANX showed a higher expression in the MOFC of subjects with schizophrenia, while no gene in the DLPFC had a difference in expression.
4.1. Genes associated with RNA processing
The subfamily of heterogeneous nuclear ribonucleoproteins (hnRNPs) forms complexes with heterogeneous nuclear RNA and influence pre-mRNA processing, and mRNA transport and metabolism. Here, we identified an increased expression of HNRNPC (heterogeneous nuclear ribonucleoprotein C (C1/C2)) in MOFC of schizophrenic brains, along with an increased expression of the gene encoding the hnRNPC-interacting protein, SART1 (squamous cell carcinoma antigen recognized by T-cells 1) [47]. Cohen et al. [25] also reported increased expression of HNRNPC, along with two other hnRNPs, HNRNPH1 and HNRNPH3, in area 10 of schizophrenic brains, although they did not precise whether their samples were derived from the left, right, or both hemispheres. Similarly, Focking et al. identified increased levels of HNRNPC protein in the postsynaptic density of supragenual anterior cingulate cortex (area 24) of schizophrenic brains, and implicated other members of the hnRNP subfamily such as HNRNPA3, HNRNPK, and HNRNPU in the pathogenesis of schizophrenia [34]. One of the functional consequences of HNRNPC dysregulation might involve myelin-related processes since HNRNPC regulates the expression of myelination-related genes MBP, QKI-5, QKI-6 and QKI-7 [48]. This finding is in line with numerous evidences about involvement of oligodendrocytes in the pathology of schizophrenia at the histological [27, 49-52] and gene expression level [52].
Further support for this concept arises from our gene ontology analysis, suggesting that five additional genes involved in RNA processing or splicing were dysregulated in the schizophrenia brains. Among them, the translational regulator and repressor, TARDBP (TAR DNA binding protein, also known as TDP-43), was increased in expression in the left hemisphere of DLPFC from schizophrenic brains. Abnormal nuclear expression of TARDBP in the hippocampus of patients with late-onset psychosis and familial history of psychiatric illness was previously reported [35, 36], and patients with young-onset FTD and TARDBP-positive pathology exhibit schizophrenia-like psychosis [49].
The decreased expression of RPS13 (encoding the ribosomal protein S13) in DLPFC of schizophrenic brains is of particular interest, considering that the RPS13 gene falls within the region around the novel single nucleotide polymorphism (SNP) associated with schizophrenia [43]. In contrast, an increased expression of RPS13 was recently reported in olfactory cells derived from schizophrenia patients [53]. It will be of interest to determine whether differences in gene and protein expression between cell types and brain regions may help to explain the region-specific pathophysiological symptoms in schizophrenia.
C1D and NPM1 were also differentially expressed genes with RNA-associated functions. NPM1, an ARF/p53 pathway regulator, showed increased expression in orbitofrontal cortex of schizophrenic brains and was identified as part of schizophrenia-associated protein network [35, 36]. C1D has apoptotic-inducing activity and was previously not correlated to schizophrenia in other studies. As there is a dysregulation of genes coding for similar functions in multiple studies, perturbation of RNA splicing proteins may play a causative role in the well-characterized decreased working memory function, attributed to the DLPFC and area 10, in the schizophrenic brain [54].
4.2. Genes associated with dendritic spine structure
It has been shown that genetic aberrations that affect the regulation and dynamics of actin filaments could be involved in the pathophysiology of schizophrenia by disrupting dendritic spine architecture and glutamatergic signaling (for review see [8]). Cohen et al. [25] found that the ENAH gene, which codes for actin-related cytoskeleton remodeling and the CPNE3 gene, which codes for actin-binding domains, have decreased expression in area 10 in subjects with schizophrenia. Our findings that ACTA2 gene, which codes for actin a2, has a decreased expression, whereas SEPT2, which assists in actin cytoskeleton formation, is overexpressed in the left DLPFC in schizophrenia (other members of septin family have also been associated with schizophrenia: SEPT4, SEPT5, SEPT8, and SEPT11 [39]), fit well to these reports. As both genes are associated with dendritic development and are ubiquitously expressed in the brain, their involvement could also partlyexplain the pathological findings in the frontal and cingulate cortex, corpus callosum, and cerebellum in schizophrenia (Table 3). At the systems level, multiple neurotransmitter systems, including dopaminergic, glutamatergic and GABAergic can be affected through the dysfunction of these genes. For example, if the ACTR2-ACTR3 system of actin-related proteins is disrupted, the result will be excessive pruning, with a loss of synaptic contact on pyramidal cells leading to an overactivation of long-range dopaminergic pathways [55, 56].
Following the trend of other cytoskeleton encoding genes such as ACTA2 and SEPT2, we show that the expression of KRT18 (keratin 18), as a member of the intermediate filament gene family, is significantly decreased in DLPFC and MOFC areas in subjects with schizophrenia. There has been one report about altered KRT17 expression in lymphocytes of patients with schizophrenia [57] and overexpression of KRT18 in rodent model of psychosis and patients with schizophrenia [46].
4.3. Genes associated with dopamine and glutamate signaling pathways
Calnexin is a chaperone molecule that facilitates protein folding and assembly, and may have a central role in sorting incorrectly folded proteins for degradation. Both differential splicing and de novo mutations [37] within the calnexin (CANX) gene have been associated with schizophrenia. Amongst others, it was shown that calnexin is involved in trafficking and expression of dopamine D1 and D2 [58], GABA-A [59] and AMPA glutamate receptors [60].
Our findings of differential expression of chromobox 3 (CBX3) and GDP dissociation inhibitor 2 (GDI2) provide additional evidence for the involvement of glutamate receptors in schizophrenia. CBX3 is involved in transcriptional silencing in heterochromatinlike complexes, whereas GDI2 regulates the GDP-GTP exchange reaction of rab proteins, thus influencing membrane trafficking and vesicle formation. Kim et al. [43] have reported somatic deletions of CBX3 gene in the PFC and cerebellum of patients with schizophrenia. Treatment of rodents with N-methyl-D- aspartate receptor (NMDAR) agonists has been an established animal model of schizophrenia (for review, see [61]) and these animals show aberrant expression of both CBX3 and GDI2. Furthermore, in vitro administration of MK-801, NMDAR antagonist, has been shown to increase GDI2 protein expression [39]. The increased expression of GDI2 observed in this study therefore could be the consequence of longterm usage of antipsychotics by schizophrenic patients.
4.4. Genes associated with mitochondrial dysfunction
Mitochondrial dysfunction is another component of the etiology of schizophrenia thought to be involved in the abnormal neuronal function, plasticity, and brain circuitry that characterize the cognitive and behavioral symptoms of the disease. Additionally, there is evidence from rat models that a hyperdopaminergic state may have inhibitory effects on the mitochondrial complex 1 and 3 [62].
Sorting nexin 3 (SNX3) is involved in protein transport between cellular compartments and interacts with phosphatidylinositol-3- phosphate [63]. Our finding of increased expression of SNX3 in the left hemisphere from DLPFC and MOFC of schizophrenic brains is in accordance with the study by Huang et al., who showed the same but in the superior temporal gyrus (area 22), and connecting this finding with mitochondria-associated genes and mitochondrial dysfunction in schizophrenia [38]. Importantly, other members of sorting nexin family have also been associatedwith schizophrenia, such as SNX2, SNX6, SNX8, SNX17, SNX19, SNX29, and SNX31 [64,65],
As for SNX3, we found an increased expression of solute carrier family member 25 member 3 gene (SLC25A3) in the MOFC of schizophrenia samples. The product of this gene catalyzes the transport of phosphate into the mitochondrial matrix. Other solute carrier family members have been also associated with the development of schizophrenia, such as SLC6A3 (dopamine transporter) [66] and SLC18A2 (vesicular monoamine transporter 2) [66]. Interestingly, SLC25A3 interacts with CANX that is also differentially expressed in schizophrenia (see above). It has been reported that rare variants of SLC25A3 contribute to a higher risk of schizophrenia [41] and that carriers of SLC39A13 polymorphisms or novel mutations have global cognitive deficits, severe negative symptoms, and significantly more suicide attempts [67].
STARD7 (StAR related lipid transfer domain containing 7) acts as a lipid transporter in the transport of phosphatidylcholine between membranes [68]. We observed an increase in STARD7 expression in MOFC in our samples. Kim and Webster already reported a negative correlation of illness duration and STARD7 expression in areas 8, 9, 10 and 46 [46]. A possible link of STARD7 and schizophrenia is through TCF4 (T cell-specific transcription factor 4), which activates its gene promotor (for a review, see [69]).
4.5. Genes associated with the cell-cycle
Gene ontology analysis indicated that seven of the differentially expressed genes are involved in promoting the cell cycle: TARDBP, KRT18, SEPT2, SART3, ANAPC5, USP22 and NPM1, Here we will discuss the significance of ANAPC5 and USP22, and KAT7, as the other genes have been already discussed above.
We show that ANAPC5 (anaphase promoting complex subunit 5), a component of anaphase promoting complex/cyclosome (APC/C) that is connected with centrosomes, was overexpressed in schizophrenia. Through its connection with the centrosome complex, ANAPC5 could interact with centrosome-associated protein DISC1, a widely accepted schizophreniarisk-gene [42, 70].
USP22 (ubiquitin specific peptidase 22) is a histone-deubiquitinating component of a transcription regulatory histone acetylation complex, required for cell-cycle progression. We found an increased expression of USP22 in DLPFC in schizophrenia. This gene has been identified among top ranking gene sets as significantly enriched in schizophrenia [44] (Szatkiewicz et al., 2014). Moreover, a single nucleotide polymorphism found near the USP22 gene was associated with negative symptoms in schizophrenia [71].
The expression of KAT7 (lysine acetyltransferase 7) was increased in DLPFC of schizophrenic brains. While the association of KAT7 with schizophrenia has not been described previously, bromodomain-containing protein 1 (BRD1) - protein with which KAT7 forms a histone acetyltransferase (HAT) complex - has been implicated in schizophrenia, as well as in brain development [72-74].
4.6. Limitations of the study
The present study took care to match brains as closely as possible for age, an exclusive medical history of schizophrenia, and comparable postmortem delay so that confounding biases could be minimized. Nevertheless, other biases that could have contributed to influencing the results such as the specific pharmacological treatment regimen of each patient or the severity of pathological changes could not be accounted for. We are aware that newer, increasingly recognized methods are now commonly used instead of RNA microarray, such as RNA sequencing. RNA sequencing has a higher signal-to-noise ratio than RNA microarray because it allows for the DNA to bind to specific regions of the genome, and does not have problems with cross-hybridization or non-ideal hybridization kinetics that are seen with RNA microarray [75, 76]. As described in details in the Materials and methods section, to accommodate for this disadvantage, the data were subjected to a procedure to minimize the background noise, as described by Irizarry et al. [20], and log2 transformation of yielded fluorescence intensities. Microarray is limited to detection of only transcripts of existing genomic sequencing, whereas RNA sequencing can also be used for discovering new transcripts. Moreover, RNA sequencing can quantify the transcripts with an absolute number value, rather than yielding a relative value as in microarray. Other research has suggested, even though relatively uncommon, that copy number variation (CNV) deletions or replications can lead to variable gene expression and cause schizophrenia with symptoms of differing severity [77, 78]. Thus, one of the limitations of our study is that, by using a RNA microarray, these CNV could not be detected. Additionally, the small sample size of six pathological brains in this study limits conclusions that can be made in relation to bias in the experimental design or the presence of CNV causing altered gene expression. Finally, in this study, we did not perform a statistical analysis to test for false positive results or determine the false discovery rate. Despite the small sample size, our P values, generally less than 0.01 confirm the high precision of the present data.
5. Conclusions
In this study we used Affymetrix microarray technology to perform a comprehensive transcriptome comparison of the cognitive regions of the frontal lobe, DLPFC and MOFC, from six clinically diagnosed schizophrenic brains and six control brains. Our analysis shows that the genes that exhibit differential expression have functions in RNA processing, nucleic acid metabolism, macromolecule metabolic processes, RNA splicing and cell cycle (Figure 3). Additionally, some of the other differentially expressed genes could be involved in abnormal brain development and activation of immunological mechanisms. Therefore, we compared our results with data from other studies regarding the expression of these genes. We gained valuable insights for further bioinformatics studies aiming at the detection of schizophrenia-associated molecular networks. Future studies should aim to validate these changes in larger cohorts of schizophrenic brains at gene and protein levels (including consideration of epigenetic and posttranslational changes). Also, as shown in the present study, additional comparisons of differential gene expression in the right and left hemispheres of the prefrontal cortex are required to help elucidate the mechanisms leading to decreased working memory, mentalization, and metacognition in schizophrenia. Bioinformatics and network pharmacology approaches could then be useful for identification of pharmacological compounds that may regulate these pathways.
Acknowledgements
Conflict of interest statement: The authors declare to have no conflicts of interest. Authors' contributions: GŠ conceived and designed the study, interpreted the results and prepared the article. MM, GS and DM were involved in acquisition and analysis of data and, together with FB, performed the microarray analysis. HRF, GS and AŠ performed the statistical analysis. MM, HRF, MBL, JK, and PRH helped with drafting the article and revising it critically for important intellectual content. All authors contributed edits and approved the contents of the manuscript. GŠ received funding from the Croatian Science Foundation (IP-2014-099730).
Appendix A. Supplementary data
Summary of gene expression data using the Affymetrix HG (human genome)-U133 Plus 2.0 array. The MS Excel (.xls) table is formatted according to Affymetrix hg18_ refseq_16_02_02_v2 protocol using the identical protocol and ID names for genes as GeneChip Human Exon 1.0ST Arrays (Affymetrix, Santa Clara, CA, USA).
References
- [1].Anticevic A., Schleifer C., Youngsun T.C.. Emotional and cognitive dysregulation in schizophrenia and depression: understanding common and distinct behavioral and neural mechanisms. Dialogues Clin. Neurosci. 2015;17:421–434. doi: 10.31887/DCNS.2015.17.4/aanticevic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gur R.E., Gur R.C.. Functional magnetic resonance imaging in schizophrenia. Dialogues Clin. Neurosci. 2010;12:333–43. doi: 10.31887/DCNS.2010.12.3/rgur. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gottesmann I.I., Erlenmeyer-Kimling L.. Family and twin strategies as a head start in defining prodromes and engophenotypes for hypothetical early-interventions in schizophrenia. Schizophr. Res. 2001;51:93–102. doi: 10.1016/s0920-9964(01)00245-6. [DOI] [PubMed] [Google Scholar]
- [4].Wisner K.M., Elvevag B., Gold J.M., Weinberger D.R., Dickinson D.. A closer look at siblings of patients with schizophrenia: the association of depression history and sex with cognitive phenotypes. Schizophr. Res. 2011;126:164–173. doi: 10.1016/j.schres.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Charney D.S., Buxbaum J.D., Sklar P., Nestler E.J. Neurobiology of Mental Illness. Oxford University Press; Oxford, UK: 2013. [Google Scholar]
- [6].O'Tuathaigh C.M.P., Babovic D., O'Meara G., Clifford J.J., Croke D.T., Waddington J.L.. Susceptibility genes for schizophrenia: characterization of mutant mouse models at the level of phenotypic behaviour. Neurosci. Biobehav. Rev. 2007;31:60–78. doi: 10.1016/j.neubiorev.2006.04.002. [DOI] [PubMed] [Google Scholar]
- [7].Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. Schizophrenia Working Group of the Psychiatric Genomics Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lewis D.A., Glausier J.R. Li M., Spaulding W.D. The Neuropsychopathology of Schizophrenia. Molecules, Brain Systems, Motivation, and Cognition, Springer International; Switzerland: 2016. Alterations in prefrontal cortical circuitry and cognitive dysfunction in schizophrenia; pp. 31–75. [DOI] [PubMed] [Google Scholar]
- [9].Gur R.E., Calkins M.E., Gur R.C., Horan W.P., Nuecheterlein K.H., Seidman L.J.. et al. The Consortium on the Genetics of Schizophrenia: neurocognitive endophenotypes. Schizophr. Bull. 2007;33:49–68. doi: 10.1093/schbul/sbl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weinberger D.R., Gallhofer B.. Cognitive function in schizophrenia. Int. Clin. Psychopharmacol. 1997;12(Suppl. 4):S29–36. doi: 10.1097/00004850-199709004-00006. [DOI] [PubMed] [Google Scholar]
- [11].Seitz R.J., Angel H.-F.. Processes of believing - a review and conceptual account. Rev. Neurosci. 2012;23:303–309. doi: 10.1515/revneuro-2012-0034. [DOI] [PubMed] [Google Scholar]
- [12].Lysaker P., Bob P., Pec O., Hamm J., Kukula M., Vohs J.. et al. Synthetic metacognition as a link between brain and behaviour in schizophrenia. Transl. Neurosci. 2013;4:368–377. [Google Scholar]
- [13].Penner J., Ford K.A., Taylor R., Schaefer B., Theberge J., Neufeld R.WJ.. et al. Medial prefrontal and anterior insular connectivity in early schizophrenia and major depressive disorder: a resting functional MRI evaluation of large-scale brain network models. Front. Hum. Neurosci. 2016;10:132. doi: 10.3389/fnhum.2016.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Judas M., Simic G., Petanjek Z., Jovanov-Milosevic N., Pletikos M., Vasung L.. et al. The Zagreb Collection of human brains: a unique, versatile, but underexploited resource for the neuroscience community. Ann. NY Acad. Sci. 2011;1225(Suppl. 1):E105–130. doi: 10.1111/j.1749-6632.2011.05993.x. [DOI] [PubMed] [Google Scholar]
- [15].4th ed. American Psychiatric Press; Washington, DC, USA: 1994. American Psychiatric Association, Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- [16].Brodmann K. in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Johann Ambrosius Barth, Leipzig; Germany: 1909. Vergleichende Lokalisationslehre der GroBhirnrinde. [Google Scholar]
- [17].Hof P.R., Mufson E.J., Morrison J.H.. Human orbitofrontal cortex: cytoarchitecture and quantitative immunohistochemical parcellation. J. Comp. Neurol. 1995;359:48–68. doi: 10.1002/cne.903590105. [DOI] [PubMed] [Google Scholar]
- [18].Simic G., Hof P.R.. In search of the definitive Brodmann's map of cortical areas in human. J. Comp. Neurol. 2015;523:5–14. doi: 10.1002/cne.23636. [DOI] [PubMed] [Google Scholar]
- [19].Rajkowska G., Goldman-Rakic P.S.. Cytoarchitectonic definition of prefrontal areas in the normal human cortex: II. Variability in locations of areas 9 and 46 and relationship to the Talairach Coordinate System. Cereb. Cortex. 1995;5:323–337. doi: 10.1093/cercor/5.4.323. [DOI] [PubMed] [Google Scholar]
- [20].Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis KJ., Scherf U.. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- [21].Huang D.W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- [22].Huang D.W., Sherman B.T., Lempicki R.A.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Altar C.A., Jurata L.W., Charles V., Lemire A., Liu P., Bukhman Y.. et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenic cohorts. Biol. Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- [24].Arion D., Unger T., Lewis D.A., Levitt P., Mirnics K.. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol. Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cohen O.S., Mccoy S.Y., Middleton F.A., Bialosuknia S., Zhang-James Y., Liu L.. et al. Transcriptomic analysis of postmortem brain identifies dysregulated splicing events in novel candidate genes for schizophrenia. Schizophr. Res. 2012;142:188–199. doi: 10.1016/j.schres.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hagihara H., Ohira K., Takao K., Miyakawa T.. Transcriptomi evidence for immaturity of the prefrontal cortex in patients with schizophrenia. Mol. Brain. 2014;7:41. doi: 10.1186/1756-6606-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hakak Y., Walker J.R., Li C., Wong W.H., Davis K.L., Buxbaum J.D.. et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc. Natl. Acad. Sci. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hemby S.E., Ginsberg S.D., Brunk B., Arnold S.E., Trojanowski J.Q., Eberwine J.H.. Gene expression profile in schizophrenia: discrete neuron transcription patterns in the entorhinal cortex. Arch. Gen. Psychiatry. 2002;59:631–640. doi: 10.1001/archpsyc.59.7.631. [DOI] [PubMed] [Google Scholar]
- [29].Iwamoto K., Bundo M., Kato T.. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder and schizophrenia, as revealed by large-scale DNA microarray analysis. Hum. Mol. Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- [30].Mexal S., Frank M., Berger R., Adams C.E., Ross R.G., Freedman R.. et al. Differential modulation of gene expression in the NMDA postsynaptic density of schizophrenic and control smokers. Mol. Brain Res. 2005;139:317–332. doi: 10.1016/j.molbrainres.2005.06.006. [DOI] [PubMed] [Google Scholar]
- [31].Middleton F.A., Mirnics K., Pierri J.N., Lewis D.A., Levitt P.. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J. Neurosci. 2002;22:2718–2719. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mirnics K., Middleton F.A., Marquez A., Lewis D.A., Levitt P.. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- [33].Vawter M.P., Crook J.M., Hyde T.M., Kleinman J.E., Weinberger D.R., Becker K.G.. et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr. Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- [34].Focking M., Lopez L.M., English J.A., Dicker P., Wolff A., Brindley E.. et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol. Psychiatry. 2015;20:424–432. doi: 10.1038/mp.2014.63. [DOI] [PubMed] [Google Scholar]
- [35].Velakoulis D., Walterfang M., Mocellin R., Pantelis C., Dean B., McLean C.. Abnormal hippocampal distribution of TDP-43 in patients with late-onset psychosis. Aust. NZ J. Psychiatry. 2009;43:739–745. doi: 10.1080/00048670903001984. [DOI] [PubMed] [Google Scholar]
- [36].Velakoulis D., Walterfang M., Mocellin R., Pantelis C., McLean C.. Frontotemporal dementia presenting as schizophrenia-like psychosis in young people: clinicopathological series and review of cases. Br. J. Psychiatry. 2009;194:298–305. doi: 10.1192/bjp.bp.108.057034. [DOI] [PubMed] [Google Scholar]
- [37].Xu B., Ionita-Laza I., Roos J.L., Boone B., Woodrick S., Sun Y.. et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia, Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Huang K.-C., Yang K.-C., Lin H., Tsao T.T., Lee S.-A.. Transcriptome alterations of mitochondrial and coagulation function in schizophrenia by cortical sequencing analysis. BMC Genomics. 2014;15(Suppl. 9):S6. doi: 10.1186/1471-2164-15-S9-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pennington K., Beasley C.L., Dicker P., Fagan A., English J., Pariante C.M.. et al. Prominent synaptic and metabolic abnormalities revealed by proteomic analysis of the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder. Mol. Psychiatry. 2008;13:1102–1117. doi: 10.1038/sj.mp.4002098. [DOI] [PubMed] [Google Scholar]
- [40].Cassoli J.S., Iwata K., Steiner J., Guest P.C., Turck C.W., Nascimento J.M.. et al. Effect of MK-801 and clozapine on the proteome of cultured human oligodendrocytes. Front. Cell. Neurosci. 2016;10:52. doi: 10.3389/fncel.2016.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].International Schizophrenia Consortium, Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Morris J.A., Kandpal G., Ma L., Austin C.P.. DISC1 (Disrupted-inSchizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum. Mol. Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- [43].Kim J., Shin J.-Y., Kim J.-I., Seo J.-S., Webster M.J., Lee D.. et al. Somatic deletions implicated in functional diversity of brain cells of individuals with schizophrenia and unaffected controls. Sci. Rep. 2014;4:3807. doi: 10.1038/srep03807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Szatkiewicz J.P., O'Dushlaine C., Chen G., Chambert K., Moran J.L., Neale B.M.. et al. Copy number variation in schizophrenia in Sweden. Mol. Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ruderfer D.M., Fanous A.H., Ripke S., McQuillin A., Amdur R.L.. et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol. Psychiatry. 2014;19:1017–10124. doi: 10.1038/mp.2013.138. Schizophrenia Working Group of Psychiatric Genomics Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kim S., Webster M.J.. Correlation analysis between genome-wide expression profiles and cytoarchitectural abnormalities in the prefrontal cortex of psychiatric disorders. Mol. Psychiatry. 2010;15:326–336. doi: 10.1038/mp.2008.99. [DOI] [PubMed] [Google Scholar]
- [47].Miklos G.L., Maleszka R.. Microarray reality checks in the context of a complex disease. Nat. Biotechnol. 2004;22:615–621. doi: 10.1038/nbt965. [DOI] [PubMed] [Google Scholar]
- [48].Ingason A., Giegling I., Hartmann A.M., Genius J., Konte B., Friedl M.. et al. Expression analysis in a rat psychosis model identifies novel candidate genes validated in a large case-control sample of schizophrenia. Transl. Psychiatry. 2015;5:e656. doi: 10.1038/tp.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dracheva S., Davis K.L., Chin B., Woo D.A., Schmeidler J., Haroutunian V.. Myelin-associated mRNA and protein expression deficits in the anterior cingulate cortex and hippocampus in elderly schizophrenia patients. Neurobiol. Dis. 2006;21:531–540. doi: 10.1016/j.nbd.2005.08.012. [DOI] [PubMed] [Google Scholar]
- [50].Havugimana P.C., Hart G.T., Nepusz T., Yang H., Turingsky A.L., Li Z.. et al. A census of human soluble protein complexes, Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Iwata K., Matsuzaki H., Manabe T., Mori N.. Altering the expression balance of hnRNP C1 and C2 changes the expression of myelination-related genes. Psychiatry Res. 2011;190:364–366. doi: 10.1016/j.psychres.2011.05.043. [DOI] [PubMed] [Google Scholar]
- [52].Tkachev D., Mimmack M.L., Ryan M.M, Wayland M., Freeman T., Jones P.B.. et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder, Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- [53].Martins-de-Souza D.. Proteome and transcriptome analysis suggests oligodendrocyte dysfunction in schizophrenia. J. Psychiatr. Res. 2010;44:149–156. doi: 10.1016/j.jpsychires.2009.07.007. [DOI] [PubMed] [Google Scholar]
- [54].English J.A., Fan Y., Focking M., Lopez L.M., Hryniewiecka M., Wynne K.. et al. Cotter, Reduced protein synthesis in schizophrenia patient-derived olfactory cells. Transl. Psychiatry. 2015;5:e663. doi: 10.1038/tp.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huang K.-C., Yang K.-C., Lin H., Tsao T.T., Lee W.-K., Lee S.-A.. et al. Analysis of schizophrenia and hepatocellular carcinoma genetic network with corresponding modularity and pathways: novel insights to the immune system. BMC Genomics. 2013;14(Suppl. 5):S10. doi: 10.1186/1471-2164-14-S5-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Millan MJ., Andrieux A., Bartzokis G., Cagenhead K., Dazzan P., Fusar-Poli P.. et al. Altering the course of schizophrenia: progress and perspectives. Nat. Rev. Drug Discov. 2016;15:485–515. doi: 10.1038/nrd.2016.28. [DOI] [PubMed] [Google Scholar]
- [57].Christoff K., Gabrieli J.D.E.. The frontopolar cortex and human cognition: evidence for a rostrocaudal hierarchical organization within the human prefrontal cortex. Psychobiology. 2000;28:168–186. [Google Scholar]
- [58].Maschietto M., Silva A.R., Puga R.D., Lima L., Pereira C.B., Nakano E.Y.. et al. Gene expression of peripheral blood lymphocytes may discriminate patients with schizophrenia from controls. Psychiatry Res. 2012;200:1018–1021. doi: 10.1016/j.psychres.2012.04.030. [DOI] [PubMed] [Google Scholar]
- [59].Wang P., Eshaq R.S., Meshul C.K., Moore C., Hood R.L., Leidenheimer N.J.. Neuronal gamma-aminobutyric acid (GABA) type A receptors undergo cognate ligand chaperoning in the endoplasmic reticulum by endogenous GABA. Front. Cell. Neurosci. 2015;9:188. doi: 10.3389/fncel.2015.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rubio M.E., Wanthold R.J.. Calnexin and the immunoglobulin binding protein (BiP) coimmunoprecipitate with AMPA receptors. J. Neurochem. 1999;73(1999):942–948. doi: 10.1046/j.1471-4159.1999.0730942.x. [DOI] [PubMed] [Google Scholar]
- [61].Moghaddam B., Javitt D.. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology. 2012;37:4–15. doi: 10.1038/npp.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Free R.B., Hazelwood L.A., Cabrera D.M., Spalding H.N., Namkung Y., Rankin M.L.. et al. D1 and D2 dopamine receptor expression is regulated by direct interaction with the chaperon protein calnexin. J. Biol. Chem. 2007;(282):21285–21300. doi: 10.1074/jbc.M701555200. [DOI] [PubMed] [Google Scholar]
- [63].Oldmeadow C., Mossman D., Evas T.-J., Holliday E.G., Tooney P.A., Cairns M.J.. et al. Combined analysis of exon splicing and genome wide polymorphism data predict schizophrenia risk loci. J. Psychiatr. Res. 2014;52:44–49. doi: 10.1016/j.jpsychires.2014.01.011. [DOI] [PubMed] [Google Scholar]
- [64].Ben-Shachar D.. Mitochondrial dysfunction in schizophrenia: a possible linkage to dopamine. J. Neurochem. 2002;83:1241–1251. doi: 10.1046/j.1471-4159.2002.01263.x. [DOI] [PubMed] [Google Scholar]
- [65].Xu Y., Hortsman H., Seet L., Wong S.H., Hong W.. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- [66].Talkowski M.E., Kirov G., Bamne M., Georgieva L., Torres G., Mansour H.. et al. A network of dopaminergic gene variations implicated as risk factors for schizophrenia. Hum. Mol. Genet. 2008;17:747–758. doi: 10.1093/hmg/ddm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kranz T.M., Berns A., Shields J., Rothman K., Walsh-Messinger J., Goetz R.R.. et al. Phenotypically distinct subtypes of psychosis accompany novel or rare variants in four different signaling genes. EBioMedicine. 2016;6:206–214. doi: 10.1016/j.ebiom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Horibata Y., Sugimoto H.. StarD7 mediates the intracellular trafficking of phosphatidylcholine to mitochondria. J. Biol. Chem. 2010;285:7358–7365. doi: 10.1074/jbc.M109.056960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Quednow B.B., Brzozka M.M., Rossner M.J.. Transcription factor 4 (TCF4) and schizophrenia: integrating the animal and the human perspective. Cell. Mol. Life Sci. 2014;71:2815–2835. doi: 10.1007/s00018-013-1553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Watanabe Y., Khodosevich K., Monyer H.. Dendrite development regulated by the schizophrenia-associated gene FEZ1 involves the ubiquitin proteasome system. Cell Rep. 2014;7:552–564. doi: 10.1016/j.celrep.2014.03.022. [DOI] [PubMed] [Google Scholar]
- [71].Xu C., Aragam N., Li X., Villa E.C., Wang L., Briones D.. et al. BCL9 and C9orf5 are associated with negative symptoms in schizophrenia: meta-analysis of two genome-wide association studies. PLoS One. 2011;8:e51–674. doi: 10.1371/journal.pone.0051674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Severinsen J.E., Bjarkam C.R., Kiaer-Larsen S., Olsen I.M., Nielsen M.M., Blechingberg J.. et al. Evidence implicating BRD1 with brain development and susceptibility to both schizophrenia and bipolar affective disorder. Mol. Psychiatry. 2006;11:1126–1138. doi: 10.1038/sj.mp.4001885. [DOI] [PubMed] [Google Scholar]
- [73].Bjarkam C.R., Corydon T.J., Olsen I.M., Pallesen J., Nyegaard M., Fryland T.. et al. Further immunohistochemical characterization of BRD1: a new susceptibility gene for schizophrenia and bipolar affective disorder. Brain Struct. Funct. 2009;214:37–47. doi: 10.1007/s00429-009-0219-3. [DOI] [PubMed] [Google Scholar]
- [74].Kushima I., Aleksic B., Ikeda M., Yamanouchi Y., Kinoshita Y., Ito Y.. et al. Association study of bromodomain-containing 1 gene with schizophrenia in Japanese population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B:786–791. doi: 10.1002/ajmg.b.31048. [DOI] [PubMed] [Google Scholar]
- [75].Ozsolak F., Milos P.M.. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mantione KJ., Kream R.M., Kuzelova H., Ptacek R., Raboch J., Samuel J.M.. et al. Comparing bioinformatics gene expression profiling methods: microarray and RNA-seq. Med. Sci. Monit. Basic Res. 201420:138–141. doi: 10.12659/MSMBR.892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lee C.H., Liu C.M., Wen C.C., Chang S.M., Hwu H.G.. Genetic copy number variants in sib pairs both affected with schizophrenia. J. Biomed. Sci. 2010;17(2) doi: 10.1186/1423-0127-17-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Searles Quick V.B., Davis J.M., Olincy A., Sikela J.M.. DUF1220 copy number is associated with schizophrenia risk and severity: implications for understanding autism and schizophrenia as related diseases. Transl. Psychiatry. 2015;5:e697. doi: 10.1038/tp.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.