Abstract
The process parameters governing the production of fibrinolytic enzyme in solid state fermentation employing Bacillus cereus IND5 and using cuttle fish waste and cow dung substrate were optimized. The pH value of the medium, moisture content, sucrose, casein and magnesium sulfate were considered for two-level full factorial design and pH, casein and magnesium sulfate were identified as the important factors for fibrinolytic enzyme production. Central composite design was applied to investigate the interactive effect among variables (pH, casein and magnesium sulfate) and response surface plots were created to find the pinnacle of process response. The optimized levels of factors were pH 7.8, 1.1% casein and 0.1% magnesium sulfate. Enzyme production was increased 2.5-fold after statistical approach. The enzyme was purified up to a specific activity of 364.5 U/g proteins and its molecular weight was 47 kDa. It was stable at pH 8.0 and was highly active at 50 °C. The mixture of cuttle fish waste and cow dung could find great application as solid substrate for the production of fibrinolytic enzyme.
Keywords: Cuttle fish waste, Cow dung, Solid state fermentation, Fibrinolytic enzyme, Bacillus cereus IND5, Thrombolytic therapy
Introduction
Cardiovascular diseases (CVDs) namely, coronary heart disease, acute myocardial infarction and atherosclerosis are the leading cause of death. Among these CVDs, thrombosis is one of the most important diseases (Wang et al. 2006). CVDs are treated by the extensive use of tissue-type plasminogen activator (t-PA), urokinase and streptokinase among other fibrinolytic agents (Holden 1990). However, these fibrinolytic agents cause allergic reactions, bleeding complications and short half-lives (Blann et al. 2002; Bode et al. 1996; Turpie 2002). Hence, the search is still vibrant for new and safer fibrinolytic enzymes throughout the world. In recent years, microbial fibrinolytic enzymes have been considered cost effective among the other fibrinolytic enzymes. These enzymes have been extensively studied from insects (Ahn et al. 2003), snake venom (He et al. 2007), and marine organisms (Sumi et al. 1992). Fermented foods namely, natto (Sumi et al. 1987, 1990; Fujita et al. 1993, 1995), Chungkook-Jang (Kim et al. 1996) and Tempeh (Sugimoto et al. 2007) have also been the source of fibrinolytic enzyme isolation. Apart from therapeutic applications, fibrinolytic proteases continue to be attractive as their potent activity on blood clot (He et al. 2007), Keratin (Bressollier et al. 1999) and collagen (Itoi et al. 2006) could have important industrial waste management and medical applications.
Solid state fermentation (SSF) is emerging as an efficient technique for production of enzymes and bioconversion of metabolites. Agro-industrial wastes, waste water and fishery wastes were utilized as cost-effective substrates in processes of biomolecule production. The agro-wastes namely, green gram husk (Prakasham et al. 2006), cake of Jatropha curcas seed (Mahanta et al. 2008), ground nut husk (Salihu et al. 2014), copra waste (Dilipkumar et al. 2013) and wheat bran and deproteinized acid cheese whey (Raol et al. 2015) were used successfully as the substrate for the enzyme production. The ideal substrate should be cheap so as to reduce the production cost of biomolecules (Pandey et al. 2000). In SSF process, the solid substrate provides required nutrient to the growth of microbes and enzyme production. Hence, the solid substrate employed for any bioprocess should be rich in nutrients for the production of biomolecules. Recently, there is an increase in the exploitation of resources from marine wastes. About 105.6 million tons of fish resources were utilized for human consumption and 34.8 million tons (25% of the fish resource) are treated as waste. The fishery waste includes whole fish waste, viscera, bones, skin, fish head, gonads, frame liver, muscle tissue and other parts (Awarenet 2004). However, these wastes were left unused or buried along sea shore which causes pollution to the environment (Bozzano and Sardà 2002). Several studies were carried out to utilize these wastes as substrate for enzyme production. The fish wastes included fish meat wastes (Vázquez et al. 2006), head and viscera powder (Triki-Ellouz et al. 2003) and waste water (Haddar et al. 2010). These fish wastes were rich of proteins, amino acids and oils (Ghaly et al. 2013). On the other hand, cow dung is one of the cheapest biomass which proved to be a useful substrate in protease (Vijayaraghavan and Vincent 2012), cellulase (Vijayaraghavan et al. 2016a) and fibrinolytic enzyme (Vijayaraghavan et al. 2016b) production in SSF. It contains cellulose, carbon, hemicelluloses, ions and trace elements (Fulhage 2000) and it could be a novel feedstock for better growth of microorganisms (Adegunloye et al. 2007). Considering the nutritive value of fish waste and cow dung, this combination of feedstock was used as the solid substrate for fibrinolytic enzyme production.
Statistical designs of experiments help to find the important variable and concentration of variables for biomolecules production. These were traditionally evaluated by one–factor–at–a–time strategy. However, it fails most often to nail down the correct response in an enzyme bioprocess and this can be overcome by statistical experimental design. The statistical designs such as fractional factorial design (Liu et al. 2005), Plackett–Burman (Dilipkumar et al. 2011) and L18-orthogonal array (Mahajan et al. 2012) were used to screen the significant variables affecting biomolecule production. Response surface methodology (RSM) was used to find the optimum concentration of these important variables, and thereby used to enhance the production of acid proteases (Siala et al. 2012), fibrinolytic enzymes (Vijayaraghavan and Vincent 2014) and β-galactosidase (Raol et al. 2015). In this study, optimization of medium components involved in fibrinolytic enzyme production by Bacillus cereus IND5 using fish waste and cow dung substrate was carried out using RSM. Based on our knowledge, no work is available regarding fibrinolytic enzyme production using the mixture of these feedstock (fish waste and cow dung) in solid state fermentation. Hence, this substrate was selected for the production of fibrinolytic enzyme.
Materials and methods
Screening of organism producing fibrinolytic enzyme
About 1 g fermented rice was mixed with 100 ml distilled water. Sample was taken from it and screened for organisms showing proteolytic activity using skimmed milk agar plates. Ten bacterial cultures, which showed clear zone in the casein agar plates, were further screened for fibrinolytic enzyme producing ability. The protease positive isolates were cultured in a medium containing peptone (3.0%), glucose (1.0%), CaCl2 (0.50%), and MgSO4 (0.20%). The pH of culture medium was brought to 7.0, and incubated at 37 °C for 72 h at 150 rpm (Gad et al. 2014). After 72 h of incubation, the bacterial biomass was separated by centrifugation (8000 rpm, 10 min, and 4 °C) and the extract devoid of cells was used to screen fibrinolytic enzyme activity. Fibrin plate assay was used to assess the fibrinolytic activity of the extracts. The fibrinolytic protease activity appeared as a halo zone around the fibrin clot after incubation at 37 °C for a period of 5 h.
Identification of the bacterial isolate
A bacterial isolate (IND5), which showed good activity based on the above screening procedures, was further identified using biochemical and morphological tests (Bednarski 2006). The 16S rRNA sequencing was carried out using the forward (5′AGAGTTTGATCMTGGCTAG3′) and reverse primer (5′ACGGGCGGTGTGTRC3′) (Rejiniemon et al. 2015). Amplification of 16S rRNA gene was carried out using a gradient PCR machine (Peng et al. 2004). The available sequence was compared using NCBI BLAST and the organism was identified as Bacillus cereus IND5.
Solid state fermentation (SSF)
Cuttle fish was collected from Kanniyakumari coast and its by-product was prepared (Souissi et al. 2008). Briefly, gut, stomach and head were removed, rinsed with double distilled water, heated, minced and dried well at 80 ± 2 °C for 48 h. Cow dung substrate was processed as described previously (Vijayaraghavan et al. 2012). Equal amount of cuttle fish and cow dung waste were mixed and used as the substrate. 5 g of substrate (2.5 g cuttle fish waste +2.5 g cow dung) was taken in 100 ml Erlenmeyer flasks and the pH of the solid substrate medium was adjusted to 8.0 by the addition of 0.1 M tris–HCl buffer. The moisture content of SSF medium was adjusted to 70% (v/w). During the preliminary experiments, the Erlenmeyer flasks were incubated at 37 °C for 72 h, and at 96 h, the maximum production of fibrinolytic enzyme was registered. Hence, all fermentations were run for a period of 72 h.
Submerged fermentation
In the present study, B. cereus IND5 was cultured in submerged fermentation to compare the yield with that of SSF. For this, 100 ml nutrient broth medium (beef extract, 5 g/l; peptone, 5 g/l; yeast extract, 1.5 g/l; and sodium chloride) was prepared, sterilized and inoculated with 0.1 ml of 18 h grown B. cereus IND5. The Erlenmeyer flask was incubated at 37 °C for 48 h. The culture was centrifuged at 10,000 rpm for 10 min and the cell free supernatant was used as the crude enzyme.
Fibrinolytic enzyme assay
The enzyme (0.1 ml) was mixed with 2.5 ml of tris–HCl buffer (pH 7.8) containing calcium chloride (0.01 M). Fibrinolytic activity was assayed on fibrin substrate. The absorbance was read at 275 nm (Anson 1938; Chang et al. 2000). Enzyme activity was calculated based on the calibration curve drawn for standard solution of l-Tyrosine. One unit of fibrinolytic enzyme activity (U) was defined as the amount of enzyme which liberates 1 µg of tyrosine per min under the standard assay condition. The results of the determination of fibrinolytic activity were described in units of activity/gram of substrate (U/g).
Selection of important medium components for fibrinolytic enzyme production by one variable at a time approach
In the present study, the mixture of cuttle fish waste and cow dung was used as the substrate for the optimization of enzyme production. The effect of six different carbon sources namely, glucose, starch, trehalose, xylose, sucrose, and maltose on the production of fibrinolytic enzyme was studied. To evaluate the influence of nitrogen sources, beef extract, casein, gelatin, urea, and yeast extract were employed. The solid medium was supplemented with ammonium chloride, ammonium sulfate, calcium chloride, di-sodium hydrogen phosphate (Na2HPO4), ferrous sulfate, sodium di-hydrogen phosphate (NaH2PO4), and sodium nitrate, to evaluate the influence of ions on production of enzyme. These nutrients were supplemented with the substrate individually. The substrate was mixed carefully with tris HCl buffer (pH 8.0) to adjust the initial moisture to 70%. Then, 500 µl inoculum (10%, v/w) was introduced into the medium and incubated as described previously. Fifty milliliter of ice cold double distilled water was poured with the fermented medium and shaken at 150 rpm for 30 min. It was further centrifuged at a speed of 10,000 rpm for 20 min in a centrifuge maintained at 4 °C. The cell free extract was stored and utilized for fibrinolytic enzyme assay.
Screening of vital medium components using two-level full factorial design
A two-level full factorial statistical design, a preliminary medium optimization strategy to find the important medium components in fibrinolytic enzyme production, was employed in this study. For screening of vital medium components, five variables, pH, moisture, sucrose, casein and MgSO4 were selected. In two-level full factorial design, each variable is represented at the low (−) level and high (+) level. The design model is based on the first order polynomial equation:
| 1 |
where α 0, α i, α ij and α ijk represent the intercept, ith linear coefficient, ijth interaction coefficient and the ijkth interaction coefficient, respectively. SSF process was initiated in 100 ml Erlenmeyer flasks and the same were maintained at 37 °C for 72 h using an incubator. Each experiment was carried out in duplicate and average value was presented. After statistical analysis using analysis of variance (ANOVA), the important factors were identified. Design-Expert 9.0 (StatEase Inc, Minneapolis, USA) was sought for the design of experiments and statistical analysis of data.
Response surface methodology
The significant medium ingredients (pH, casein, and MgSO4) affecting fibrinolytic enzyme production, as observed by the two-level full factorial design, were tested for interactive effects using a central composite design (CCD) of response surface methodology. The selected variables were coded as A (pH), B (casein), and C (MgSO4) and the following second order model equation was used to predict the response (Eq. 2).
| 2 |
The experimental runs were performed in 100 ml Erlenmeyer flasks as per the central composite design in randomized manner. The substrate was sterilized at 121 °C for 20 min and cooled. After which, 10% inoculum (B. cereus IND5) were carefully inoculated under aseptic conditions. SSF was carried out in an incubator maintained at 37 °C for a period of 72 h. After fermentation, enzyme in the SSF medium was extracted and enzyme assay was carried out. The ANOVA was used to evaluate the significant model terms. The optimum medium composition was obtained using RSM and these optimum conditions were validated. Design-Expert 9.0 (StatEase Inc, Minneapolis, USA) was the software program employed to design the experiment and to analyze the data.
Purification of fibrinolytic enzyme from B. cereus IND5
The fibrinolytic enzyme was produced through SSF using B. cereus IND5 using the optimized culture medium. The crude enzyme was purified by performing ammonium sulfate precipitation, diethyl aminoethyl cellulose (DEAE cellulose) and casein-agarose affinity chromatography. 50 ml of crude extract was precipitated and 70% saturation was attained by the addition of ammonium sulfate salt. The precipitate was separated out by centrifugation (10,000 rpm, 4 °C) and was dissolved in 0.1 M tris–HCl buffer (pH 8.0). It was further dialyzed against water (two changes) and buffer (third change). The sample was further loaded on a DEAE-cellulose column which was pre equilibrated with 0.05 M tris–HCl buffer (pH 8.0). The bounded proteins were eluted with 0.05 M tris buffer containing 0–1.0 M NaCl. NaCl gradient was made using a gradient maker and all fractions were collected manually. The fractions (2.0 ml) were analyzed for their protein concentration (Absorbance 280 nm) and enzyme activity. The active fractions were loaded on casein-agarose affinity chromatography (Sigma, USA) which was pre equilibrated with 0.02 M tris–HCl buffer. Elution of the proteins was done with a gradient of buffer with strength of 0–0.8 M NaCl. The fractions resulting from elution were assayed for fibrinolytic activity. In each step of enzyme purification, enzyme assay and also total protein estimation were carried out. Further, specific activity of enzyme, yield, and purification fold were also measured.
SDS-PAGE analysis and zymography
The highly active fraction from DEAE cellulose chromatography and affinity chromatography was subjected to analysis for homogeneity. The enzyme sample was added with SDS sample buffer and boiled for 1 min before loading on to the SDS-PAGE gel (12%). The molecular markers used were soybean trypsin inhibitor of 20.1 kDa, carbonic anhydrase of 29 kDa, ovalbumin of 43 kDa, bovine serum albumin of 66 kDa and phosphorylase b of 97.4 kDa. The protein bands were visualized by staining using coomassie brilliant blue (R-250). For zymography analysis, a fibrin substrate containing polyacrylamide gel (12%) was prepared by adding 0.12% (w/v) fibrinogen and thrombin (100 NIH U/ml). The affinity chromatography purified-fraction was loaded on this gel. At the end of electrophoresis, the gel was taken out and incubated with buffer A containing 2.5% (v/v) Triton X-100 for 30 min at room temperature (30 ± 1 °C). The residual Triton X-100 was removed by washing the gel with double distilled water for 30 min. The gel was incubated with buffer A for 4 h. Then the gel was subjected to coomassie brilliant blue (R-250) staining for 2 h and then destained (overnight). The fibrinolytic activity of the enzyme was visualized as nonstained region on blue background.
Evaluation of characteristics of the purified enzyme
The optimal pH needed for the maximized fibrinolytic activity of enzyme, was estimated using buffers (0.1 M) of different pH namely, pH 3.0 and 4.0 (citrate buffer), pH 5.0 (succinate buffer), pH 6.0 and 7.0 (sodium phosphate buffer), pH 8.0 (tris buffer) and pH 9.0 and 10.0 (glycine-NaOH buffer). To evaluate the stability of enzyme activity with respect to pH, the fibrinolytic enzyme was incubated with the aforementioned buffers, separately and incubated at 37 °C for 1 h before adding the substrate. The influence of temperature on the activity of enzyme was estimated by performing the reactions at different temperatures of 30, 40, 50, 60, and 70 °C. The stability of the fibrinolytic enzyme with respect to temperature was estimated by incubating it in the absence of substrate at different temperatures, 30–70 °C for 1 h. Fibrinolytic activity of the enzyme was assayed as described earlier. To elucidate the influence of divalent ions on enzyme activity, the enzyme was incubated for 30 min along with different divalent ions, namely Ca2+, Cu2+, Co2+, Mg2+, Mn2+, Hg2+, Fe2+, and Zn2+ at 0.01 M concentrations (Lomate and Hivrale 2011).
Results and discussion
Screening and identification of the fibrinolytic enzyme producing bacterium
Ten protease producing bacterial isolates were selected after casein hydrolysis. These protease positive isolates were further evaluated for production of fibrinolytic enzyme using fibrin plate. Among the ten bacterial cultures, the cell free extract of an isolate named IND5 produced the highest halo zone than other isolates (Fig. 1). The halo zone represents the clearance of fibrin which in turn indicates the fibrinolytic activity of the strains. The isolate IND5 also produced more fibrinolytic enzyme in submerged fermentation compared to the other isolates. The 16S rDNA sequence of the isolate IND5 was subjected to BLAST and the sequence showed a high similarity with Bacillus cereus. The strain was identified as B. cereus IND5 and accession number was assigned (KF250421).
Fig. 1.
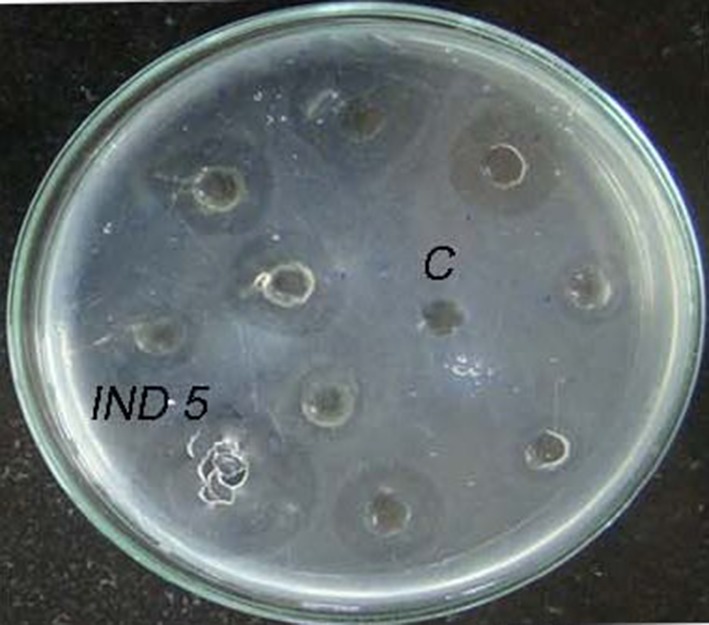
Fibrinolytic enzyme activity of the bacterial isolates on fibrin plate (C-control)
Production of fibrinolytic enzyme in submerged fermentation
B. cereus IND5 was cultured in nutrient broth medium and enzyme production was found to be 73 U/ml after 48 h of incubation at 37 °C. Submerged fermentation has been widely used for the production of enzymes. Recently, Bajaj et al. (2014) used submerged fermentation for the production of fibrinolytic enzyme from Bacillus sp.
Cuttle fish waste and cow dung mixture for fibrinolytic enzyme production in SSF
In SSF, enzyme production was found to be maximum after 72 h incubation at 37 °C (1205 ± 48 U/g). SSF technique has been widely used for the production of enzymes, antibiotics, secondary metabolites, flavoring compounds and also animal feeds. SSF process is defined as the one in which microbes get attached to solid materials and grow, without any free water. In SSF process, the solid substrate provides anchorage for the cells and supply nutrients. Hence, the native-like environment is created for the organism and maximum product production is expected. In recent years, many substrates were mixed and used as the fermentation medium to confirm its balanced nutritive value. The mixture of substrates, such as, protein and chitin (Wang et al. 2008b), powdered crab and shrimp shell (Oh et al. 2000), cuttlefish by-products powder and wastewater from fish processing industry (Souissi et al. 2008), cuttlefish and shrimp by-products (Ben Rebah et al. 2008) were used for the production of various enzymes. The by-products from fish processing support growth of microbes in an excellent manner. Due to its low cost and easily available nature, these substrates play a significant role in the production of enzyme (Rebah and Miled 2013). Mukherjee et al. (2008) used a mixture of potato peel with Imperata cylindrica grass as the substrate and produced fibrinolytic enzyme. Cow dung was reported as a novel substrate for use in SSF aimed at the production of cellulases (Vijayaraghavan et al. 2016a) and fibrinolytic enzymes (Vijayaraghavan et al. 2016b). Ghorbel et al. (2005) stated the importance of balanced nutrients for enzyme production by microbes. Considering the importance of balanced diet, the combination of cuttlefish by-product and cow dung was used as a substrate for the production of fibrinolytic enzyme. This result establishes the use of cuttle fish waste and cow dung mixture for fibrinolytic enzyme production. In enzyme bioprocess, no defined culture medium has been proposed to enhance the production of enzymes from various microbial sources. Every organism is unique in the requirement of its own environmental and nutritive factors for more enzyme production. Hence, optimization of enzyme production by an individual organism is a key to enhance enzyme production. These kinds of studies would provide low cost substrates for enzyme production in industries.
Use of one variable at a time approach (OVAT) for screening medium components
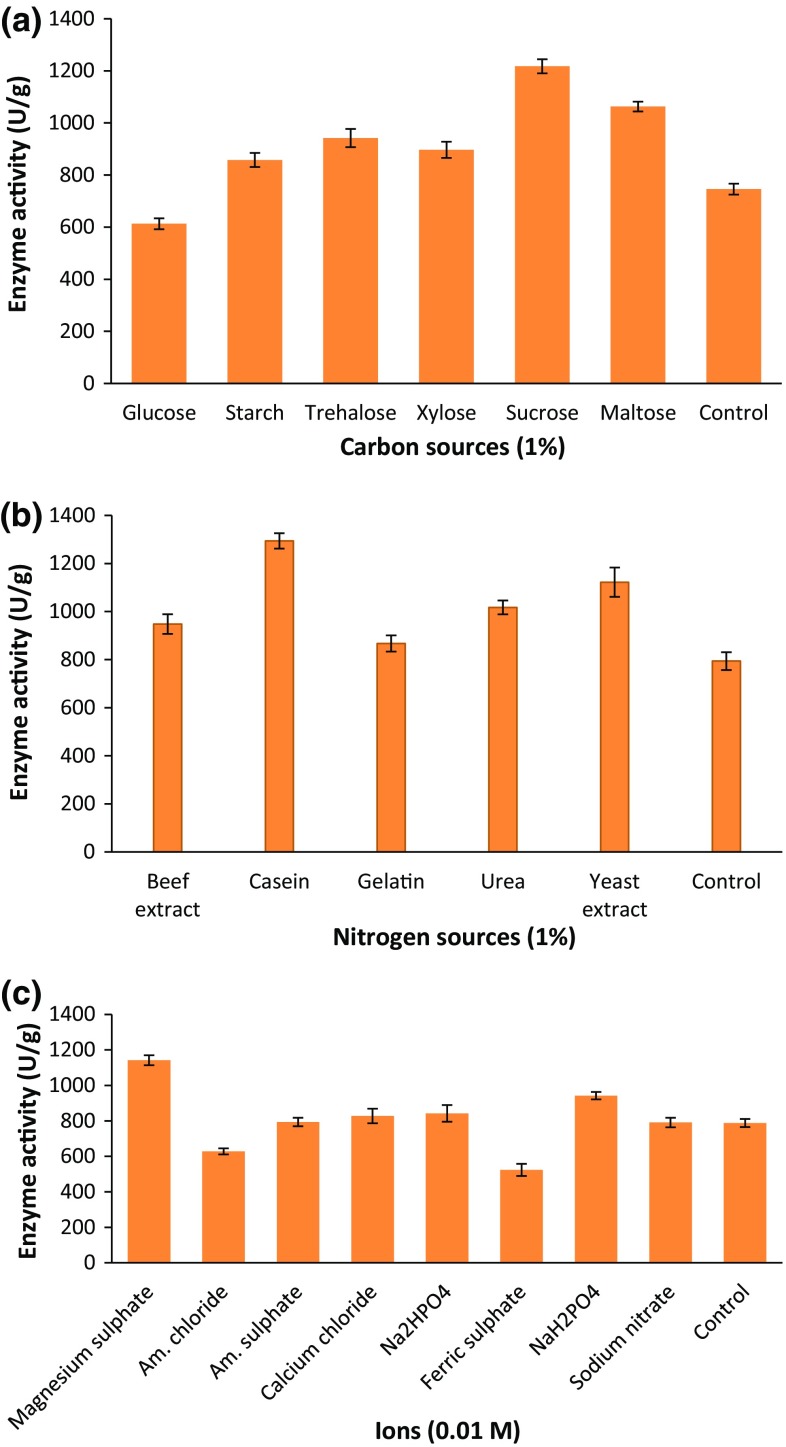
The traditional OVAT experiment helps to screen the variables without any complicated analysis. The mixture of cuttle fish waste and cow dung was used as the substrate for optimization of fibrinolytic enzyme production. In the present study, experiments were carried out with different carbon (glucose, starch, trehalose, xylose, sucrose, and maltose), nitrogen (beef extract, casein, gelatin, urea, and yeast extract) and ionic sources (ammonium chloride, magnesium sulfate, ammonium sulfate, calcium chloride, di-sodium hydrogen phosphate (Na2HPO4), sodium di-hydrogen phosphate (NaH2PO4), ferrous sulfate, and sodium nitrate) to evaluate the suitable nutrient source for the process of fibrinolytic enzyme production. All carbon sources, except glucose containing culture medium did not exhibit fibrinolytic activity than that of control and this indicated the inducible nature of B. cereus IND5 fibrinolytic enzyme. Among the carbon sources, sucrose stimulated more enzyme production (1218 ± 27 U/g) (Fig. 2a). Hence, sucrose was used as the suitable carbon source. Vijayaraghavan and Vincent (2014) concluded that sucrose was the best carbon source for fibrinolytic enzyme production for Paenibacillus sp. IND8. Among the nitrogen sources, casein considerably enhanced fibrinolytic enzyme production (1294 ± 32 U/g) (Fig. 2b). This result agreed with the observations made with Bacillus sp. strain AS-S20-I (Mukherjee and Rai 2011) and Proteus penneri SP-20 (Jhample et al. 2015). MgSO4 (0.1%, w/w) was found to be the best ionic source for production of fibrinolytic enzyme (1142 ± 28 U/g). However, addition of ammonium chloride (0.1%, w/w) negatively influenced the enzyme production (Fig. 2c). Based on OVAT approach, sucrose, casein and MgSO4 were chosen for further experiments.
Fig. 2.
Effect of carbon source (a) nitrogen source (b) and ion (c) on fibrinolytic enzyme production
Medium optimization using 25 full factorial design
In this study, cow dung and cuttle fish waste were (50:50) used for the production of enzyme. A two-level five factorial design is a best statistical tool for studying the production of fibrinolytic enzymes (Liu et al. 2005). This method analyzes the factors in two levels, i.e., high (+) and low (−), for the screening of important variables or factors (Table 1). From the results of preliminary screening carried out by OVAT approach, sucrose, casein and MgSO4 were selected as the suitable nutritional factors for evaluating fibrinolytic enzyme production. In this statistical design, along with these three variables, moisture and pH were also included as the critical factors for optimizing enzyme production in SSF. The variables and the results of the experiments were described in the Table 2. In this study, the production of fibrinolytic enzyme varied between 180 and 5044 U/g. The production of enzyme was observed to be maximum in the medium containing 80% moisture content, 0.1% sucrose, 0.1% casein, 0.1% MgSO4 and pH 7.0. ANOVA of the interactive effects were represented in the Table 3. The F value of this model was 128.47 and it was statistically significant. There is only a little chance (0.01%) that this much large “Model F-value” had occurred due to noise. Hence, the interactive tested variables were found to be statistically significant. The model terms A, B, D, E, AB, AC, AD, AE, BD, BE, CE, DE, ABC, ABE, ACD, ACE, ADE, BCD, BCE, BDE, ABCE, ACDE and BCDE were found to be significant. The predicted R 2 value (0.9263) was found to be close to the adjusted R 2 value (0.9911). The R 2 value of this model was 0.9988. The equation of the model can be written using coded levels of factors as:
| 3 |
Table 1.
The independent variables chosen for 25 factorial design and their coded levels
| Symbol | Variables | Units | Coded levels | |
|---|---|---|---|---|
| −1 | +1 | |||
| A | pH | 7 | 9 | |
| B | Moisture | (%) | 80 | 100 |
| C | Sucrose | (%) | 0.1 | 1.0 |
| D | Casein | (%) | 0.1 | 1.0 |
| E | MgSO4 | (%) | 0.01 | 0.10 |
Table 2.
Randomized runs of 25 factorial design and the measured response
| Run | A:pH | B:Moisture | C:Sucrose (%) | D:Casein (%) | E:MgSO4 (%) | Enzyme activity (U/g) |
|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | −1 | 1 | 1693 |
| 2 | −1 | −1 | −1 | 1 | 1 | 2730 |
| 3 | 1 | −1 | 1 | 1 | 1 | 1258 |
| 4 | −1 | 1 | 1 | 1 | −1 | 2019 |
| 5 | 1 | −1 | 1 | 1 | −1 | 2997 |
| 6 | −1 | −1 | 1 | −1 | 1 | 2320 |
| 7 | 1 | −1 | −1 | 1 | 1 | 821 |
| 8 | 1 | −1 | 1 | −1 | 1 | 1166 |
| 9 | 1 | 1 | 1 | −1 | −1 | 2233 |
| 10 | −1 | 1 | −1 | −1 | −1 | 2310 |
| 11 | 1 | −1 | −1 | −1 | −1 | 530 |
| 12 | 1 | 1 | −1 | −1 | −1 | 4251 |
| 13 | 1 | −1 | −1 | −1 | 1 | 1546 |
| 14 | −1 | 1 | −1 | −1 | 1 | 180 |
| 15 | 1 | −1 | −1 | 1 | −1 | 2031 |
| 16 | 1 | 1 | 1 | 1 | −1 | 2544 |
| 17 | −1 | −1 | 1 | −1 | −1 | 2754 |
| 18 | −1 | 1 | −1 | 1 | 1 | 1750 |
| 19 | 1 | 1 | −1 | −1 | 1 | 230 |
| 20 | −1 | −1 | −1 | −1 | −1 | 3587 |
| 21 | −1 | 1 | 1 | 1 | 1 | 3605 |
| 22 | 1 | −1 | 1 | −1 | −1 | 3198 |
| 23 | −1 | 1 | −1 | 1 | −1 | 910 |
| 24 | 1 | 1 | −1 | 1 | −1 | 4640 |
| 25 | −1 | −1 | −1 | −1 | 1 | 5044 |
| 26 | −1 | −1 | 1 | 1 | −1 | 2521 |
| 27 | −1 | 1 | 1 | −1 | 1 | 2818 |
| 28 | −1 | −1 | −1 | 1 | −1 | 3651 |
| 29 | 1 | 1 | −1 | 1 | 1 | 3840 |
| 30 | −1 | −1 | 1 | 1 | 1 | 3340 |
| 31 | −1 | 1 | 1 | −1 | −1 | 2251 |
| 32 | 1 | 1 | 1 | 1 | 1 | 1510 |
Table 3.
ANOVA results for 25 factorial design
| Source | Sum of squares | df | Mean square | F value | p value | |
|---|---|---|---|---|---|---|
| Model | 4.876+007 | 27 | 1.806E+006 | 128.47 | 0.0001 | Significant |
| A-pH | 1.759E+006 | 1 | 1.759E+006 | 125.13 | 0.0004 | |
| B-Moisture | 2.646E+005 | 1 | 2.646E+005 | 18.83 | 0.0123 | |
| C-Sucrose | 18.00 | 1 | 18.00 | 1.281E−003 | 0.9732 | |
| D-Casein | 4.646E+005 | 1 | 4.646E+005 | 33.06 | 0.0045 | |
| E-MgSO4 | 2.192E+006 | 1 | 2.192E+006 | 155.98 | 0.0002 | |
| AB | 9.788E+006 | 1 | 9.788E+006 | 696.37 | <0.0001 | |
| AC | 2.042E+005 | 1 | 2.042E+005 | 14.52 | 0.0189 | |
| AD | 1.027E+006 | 1 | 1.027E+006 | 73.05 | 0.0010 | |
| AE | 4.762E+006 | 1 | 4.762E+006 | 338.77 | <0.0001 | |
| BC | 41184.50 | 1 | 41184.50 | 2.93 | 0.1621 | |
| BD | 1.069E+006 | 1 | 1.069E+006 | 76.03 | 0.0010 | |
| BE | 2.258E+005 | 1 | 2.258E+005 | 16.06 | 0.0160 | |
| CD | 40186.13 | 1 | 40186.13 | 2.86 | 0.1661 | |
| CE | 2.384E+005 | 1 | 2.384E+005 | 16.96 | 0.0146 | |
| DE | 3.737E+005 | 1 | 3.737E+005 | 26.59 | 0.0067 | |
| ABC | 1.069E+007 | 1 | 1.069E+007 | 760.25 | <0.0001 | |
| ABE | 1.474E+005 | 1 | 1.474E+005 | 10.49 | 0.0317 | |
| ACD | 2.193E+006 | 1 | 2.193E+006 | 156.05 | 0.0002 | |
| ACE | 3.659E+005 | 1 | 3.659E+005 | 26.03 | 0.0070 | |
| ADE | 1.093E+005 | 1 | 1.093E+005 | 7.77 | 0.0494 | |
| BCD | 1.065E+006 | 1 | 1.065E+006 | 75.77 | 0.0010 | |
| BCE | 3.523E+006 | 1 | 3.523E+006 | 250.65 | <0.0001 | |
| BDE | 3.109E+006 | 1 | 3.109E+006 | 221.17 | 0.0001 | |
| CDE | 14620.50 | 1 | 14620.50 | 1.04 | 0.3654 | |
| ABCE | 2.869E+005 | 1 | 2.869E+005 | 20.41 | 0.0107 | |
| ACDE | 2.957E+005 | 1 | 2.957E+005 | 21.04 | 0.0101 | |
| BCDE | 4.512E+006 | 1 | 4.512E+006 | 321.00 | <0.0001 | |
| Residual | 56223.75 | 4 | 14055.94 | |||
| Cor total | 4.881E+007 | 31 |
The negative coefficients for medium components A (pH), B (moisture), C (sucrose) and E (MgSO4) indicated that the enzyme production can be increased by decreasing their concentrations in the fermentation medium. The positive coefficient for the model term D (casein) indicated that enzyme production could be increased by increasing the amount of casein in the SSF medium. Based on the F value from Table 3, the medium components casein, MgSO4 and pH were considered as the vital components for fibrinolytic enzyme production by B. cereus IND5 using cuttle fish waste and cow dung mixture.
Optimized enzyme production SSF process using RSM
RSM is an ideal statistical tool for optimizing enzyme production. Recently, RSM was applied for optimized production of cellulases (Singh et al. 2014; Premalatha et al. 2015), fibrinolytic enzyme (Majumdar et al. 2015) and xylanase (Khusro et al. 2016). The optimum concentration of individual process parameters considering the interactive effect were elucidated using RSM. Hence, the vital parameters were taken at five levels and SSF was done according to CCD. The factors and their levels were described in Table 4. The enzyme activity was found to vary from 1488 to 5364 U/g (Table 5). ANOVA was used to evaluate the results and the model was statistically significant with F value of 1276.39 (Table 6). The chance of interplay of noise in the appearance of this much large F value is as little as 0.01%. The model terms A, B, C, AC, AB, BC, A2, B2 and C2 were significant. The “Lack of fit F value” of 1.02 implies the insignificance of lack of fit as compared to the pure error. There was an estimated chance of 48.97% that a “Lack of Fit-value” of this magnitude could occur owing to noise. “The Pred R-squared” of 0.9960 was close to the “Adj R-Squared” of 0.9983. The second order polynomial equation is given below.
| 4 |
Table 4.
The independent variables selected for CCD and their coded values
| Variables | Symbol | Coded values | ||||
|---|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | ||
| pH | A | 6.32 | 7 | 8 | 9 | 9.68 |
| Casein | B | −0.21 | 0.10 | 0.55 | 1.00 | 1.31 |
| MgSO4 | C | 0.01 | 0.03 | 0.05 | 0.07 | 0.09 |
Table 5.
Central composite design runs and results
| Run | A:pH | B:Casein | C:MgSO4 | Enzyme activity (U/g) |
|---|---|---|---|---|
| 1 | −1.000 | 1.000 | −1.000 | 5201 |
| 2 | 0.000 | 0.000 | −1.682 | 4457 |
| 3 | 0.000 | 0.000 | 0.000 | 3421 |
| 4 | 1.000 | −1.000 | 1.000 | 5364 |
| 5 | 0.000 | −1.682 | 0.000 | 3682 |
| 6 | 0.000 | 1.682 | 0.000 | 3358 |
| 7 | 0.000 | 0.000 | 0.000 | 3425 |
| 8 | −1.682 | 0.000 | 0.000 | 2126 |
| 9 | 0.000 | 0.000 | 0.000 | 3410 |
| 10 | 0.000 | 0.000 | 0.000 | 3512 |
| 11 | 1.000 | 1.000 | −1.000 | 3528 |
| 12 | 0.000 | 0.000 | 0.000 | 3430 |
| 13 | −1.000 | −1.000 | 1.000 | 1488 |
| 14 | 0.000 | 0.000 | 0.000 | 3398 |
| 15 | 0.000 | 0.000 | 1.682 | 2569 |
| 16 | 1.000 | −1.000 | −1.000 | 4107 |
| 17 | 1.682 | 0.000 | 0.000 | 3852 |
| 18 | 1.000 | 1.000 | 1.000 | 2609 |
| 19 | −1.000 | 1.000 | 1.000 | 1617 |
| 20 | −1.000 | −1.000 | −1.000 | 2830 |
Table 6.
ANOVA for CCD design results
| Source | Sum of squares | df | Mean square | F value | p value | |
|---|---|---|---|---|---|---|
| Model | 1.910E+007 | 9 | 2.122E+006 | 1276.39 | <0.0001 | Significant |
| A-pH | 3.982E+006 | 1 | 3.982E+006 | 2395.44 | <0.0001 | |
| B-Casein | 1.392E+005 | 1 | 1.392E+005 | 83.74 | <0.0001 | |
| C-MgSO4 | 4.413E+006 | 1 | 4.413E+006 | 2654.43 | <0.0001 | |
| AB | 4.254E+006 | 1 | 4.254E+006 | 2559.06 | <0.0001 | |
| AC | 3.464E+006 | 1 | 3.464E+006 | 2083.43 | <0.0001 | |
| BC | 2.440E+006 | 1 | 2.440E+006 | 1467.57 | <0.0001 | |
| A2 | 3.471E+005 | 1 | 3.471E+005 | 208.77 | <0.0001 | |
| B2 | 15266.04 | 1 | 15266.04 | 9.18 | 0.0127 | |
| C2 | 13032.65 | 1 | 13032.65 | 7.84 | 0.0188 | |
| Residual | 16625.01 | 10 | 1662.50 | |||
| Lack of fit | 8413.68 | 5 | 1682.74 | 1.02 | 0.4897 | Not significant |
| Pure error | 8211.33 | 5 | 1642.27 | |||
| Cor total | 1.911E+007 | 19 |
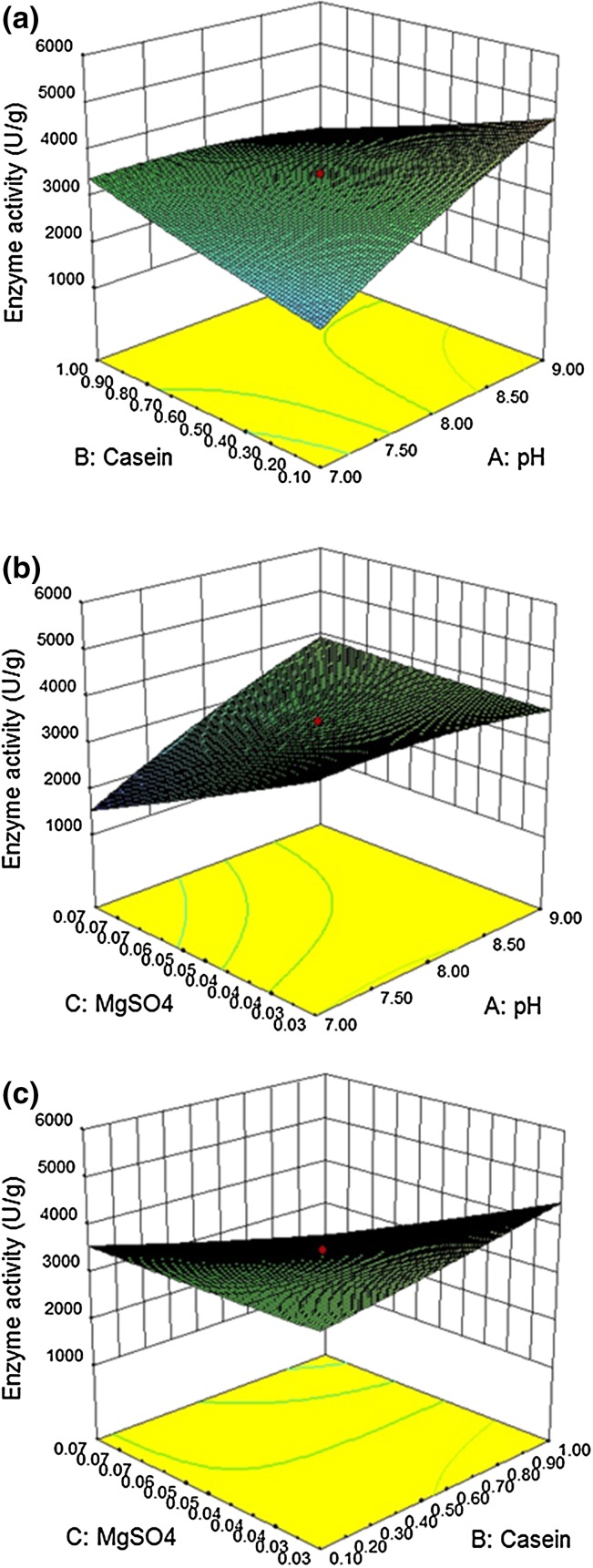
To deduce the optimum concentration of each variable for the enhancement of enzyme production 3D surface plots were constructed using RSM (Fig. 3). The optimized values of pH, casein and MgSO4 were 7.8, 1.1 and 0.08%, respectively. To check the predicted response, a validation experiment was conducted. Under the optimum levels of medium components, the fibrinolytic enzyme production was observed to be 5247 ± 37 U/g. The predicted response 5201 U/g was close to the observed results and this validated the selected model. This finding shows the aptness of the selected model, indicating that the optimized levels of medium components favor the fibrinolytic enzyme production in SSF.
Fig. 3.
Three dimensional response surface plots showing the effect of (a) pH and casein, (b) pH and MgSO4 and (c) casein and MgSO4
The culture medium cost is approximately 30–40% of total production cost of enzymes (Joo et al. 2003), hence the application of low cost substrate could reduce the overall enzyme cost. In the present observation, enzyme yield was found to be high in SSF than submerged fermentation. The processing cost of cow dung and cuttle fish waste was approximately 10 INR/kg material. The cost of one of the commercially available culture medium (Nutrient broth, HIMEDIA, Mumbai, India) was approximately 4 INR/g material. The approximate yield and production medium cost was 1400 U/INR in submerged fermentation, whereas 500,000 U/INR in SSF. The media cost was more than 100 times cheap in SSF than submerged fermentation. Thus, the use of cuttle fish waste and cow dung could reduce minimum 30% of overall enzyme cost.
Downstream purification of the produced fibrinolytic enzyme
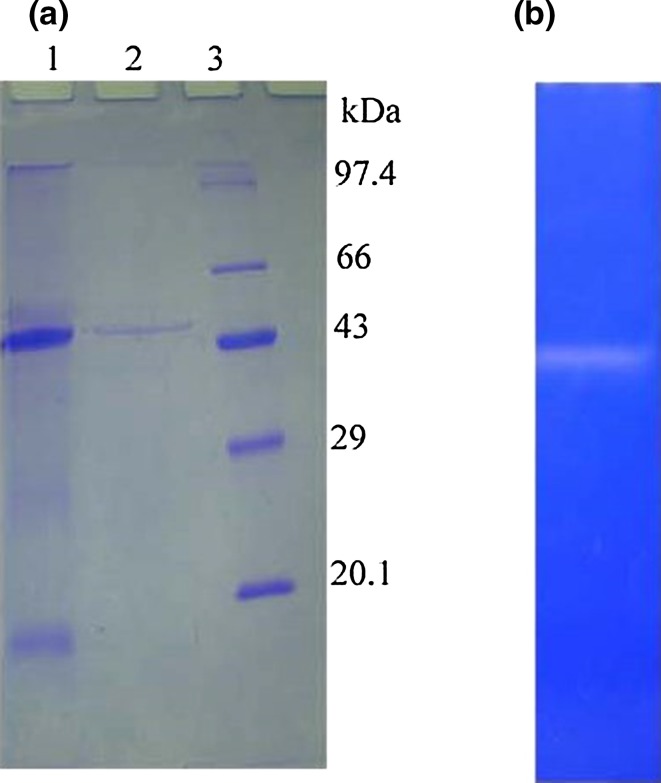
In this study, fibrinolytic enzyme was purified by the combination of ammonium sulfate precipitation, DEAE-cellulose column chromatography and affinity chromatography using casein–agarose matrix. The ammonium sulfate precipitation showed 22,040 U of fibrinolytic enzyme with 43% yield, and 1.3-fold purification was achieved. The specific activity of fibrinolytic enzyme was increased to 143 U/mg protein after DEAE-cellulose chromatography purification. The affinity chromatography purified fractions showed 364.5 U/mg protein. The highly active fraction from DEAE cellulose and casein-agarose chromatography fractions was analyzed for its homogeneity. The DEAE chromatography fractions showed partial purification of enzyme. The affinity purified enzyme migrated as a single band in SDS-PAGE gel and the molecular weight was found to be 47 kDa (Fig. 4). The molecular weight of bacterial fibrinolytic enzyme varies widely. In B. subtilis KCK-7, the molecular weight was registered as 44 kDa (Paik et al. 2004); however, the molecular weight of fibrinolytic enzyme from B. subtilis LD-8547 was reported as 30 kDa (Wang et al. 2008a).
Fig. 4.
a SDS–polyacrylamide gel electrophoresis (12%) after coomassie staining. lane 1–DEAE-cellulose fraction; lane 2–affinity chromatography purified enzyme; lane 3–molecular markers. b Fibrin zymography showing the purified fibrinolytic enzyme from B. cereus IND5
Characterization of the produced fibrinolytic enzyme
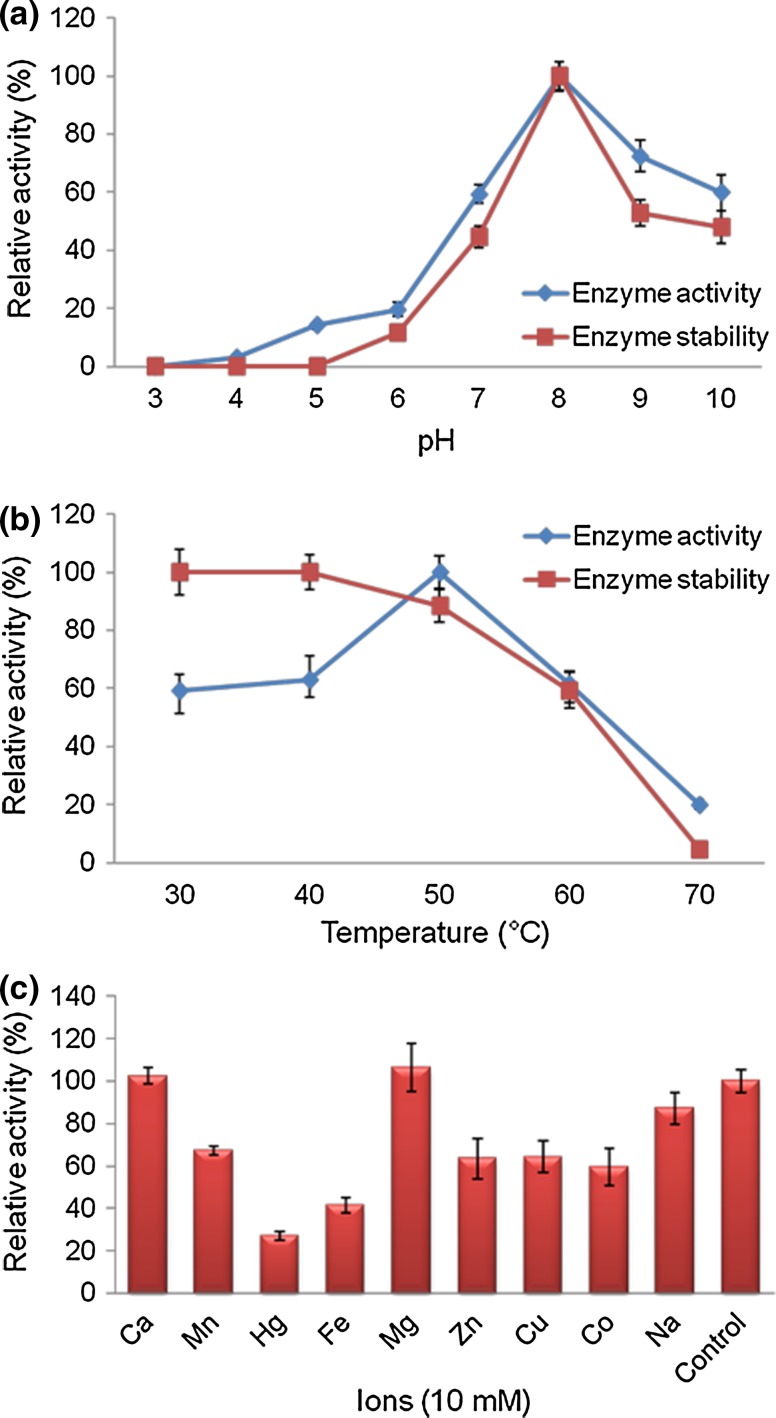
The characterization of the purified enzyme was done by analyzing the effect of temperature, pH, and ionic strength on its stability and activity. Figure 5a revealed the effect of pH on enzyme activity and stability. B. cereus IND5 fibrinolytic enzyme showed maximum activity at pH 8.0 and was stable in the range of pH from 7.0 to 9.0 which throws light on the possible applications of this enzyme in a plethora of industrial sectors. This result was acceptable with respect to the previously published results. The fibrinolytic enzyme isolated from various Bacillus sp. showed optimum activity in the range, pH 8.0–9.0 (Agrebi et al. 2010; Mahajan et al. 2012; Bajaj et al. 2013). B. cereus IND5 fibrinolytic enzyme showed maximum activity at 50 °C (Fig. 5b). Bajaj et al. (2013) reported that several Bacillus sp. fibrinolytic enzymes have shown optimum activity at 35–40 °C. However, 60 °C was optimum for the activity of fibrinolytic enzyme produced from B. subtilis A26 (Agrebi et al. 2009). B. cereus IND5 fibrinolytic enzyme activity was inhibited by the presence of tested metal ions except Mg2+ and Ca2+ (Fig. 5c). It was previously reported that the fibrinolytic enzyme of B. subtilis A26 was activated in the presence of Ba2+, Cu2+, K+, Mg2+, Mn2+ and Na+ and enzyme activity was inhibited by the addition of Zn2+ and Hg2+ (Agrebi et al. 2009). In B. subtilis ICTF-1, fibrinolytic enzyme activity was inhibited by ions like Zn2+, Hg2+ and Fe3+ (Mahajan et al. 2012).
Fig. 5.
Biochemical characterization of the purified fibrinolytic enzyme. a Effect of pH on enzyme activity and stability. Enzyme assay was carried out at 37 °C. b Effect of temperature on enzyme activity and stability. Enzyme assay was carried out at pH 8.0 with 0.1 M Tris buffer. c Effect of various ions on enzyme activity. Enzyme assay was carried out at pH 8.0 and at 50 °C
Conclusion
The present study revealed that B. cereus IND5 effectively utilized cuttle fish waste and cow dung for its growth and enzyme production. This substrate could have unprecedented potential for the production of fibrinolytic enzyme. The interactions of pH, casein, and magnesium sulfate were evaluated by response surface methodology. Fibrinolytic enzyme production was significantly increased by altering the medium pH and at increased concentration of casein. The purified enzyme was highly active at 50 °C and was stable up to pH 8.0. Considering these properties, it could be useful for various biotechnological applications.
Acknowledgements
The Deanship of Scientific Research at King Saud University is gratefully acknowledged for providing fund to this Prolific Research Group (PRG-1437-28).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Adegunloye DV, Adetuyi FC, Akinyosoye FA, Doyeni MO. Microbial analysis of compost using cowdung as booster. Pak J Nutr. 2007;6:506–510. doi: 10.3923/pjn.2007.506.510. [DOI] [Google Scholar]
- Agrebi R, Haddar A, Hajji M, Frikha F, Manni L, Jellouli K, Nasri M. Fibrinolytic enzymes from a newly isolated marine bacterium Bacillus subtilis A26: characterization and statistical media optimization. Can J Microbiol. 2009;55:1049–1061. doi: 10.1139/W09-057. [DOI] [PubMed] [Google Scholar]
- Agrebi R, Hmider N, Hajji M, Ktari N, Haddar A, Zouari NH, Nasri M. Fibrinolytic serine protease isolated from Bacillus amyloliquefaciens An6 grown on Mirabilis Jalapa tuber powders. Appl Biochem Biotechnol. 2010;162:75–88. doi: 10.1007/s12010-009-8800-z. [DOI] [PubMed] [Google Scholar]
- Ahn MY, Hahn BS, Ryu KS, Kim JW, Kim I, Kim YS. Purification and characterization of a serine protease with fibrinolytic activity from the dung beetles, Catharsius molossus. Thromb Res. 2003;112:339–347. doi: 10.1016/j.thromres.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Anson ML. The estimation of pepsin, trypsin, papain, and cathepsin with hemoglobin. J Gen Physiol. 1938;22:79–89. doi: 10.1085/jgp.22.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awarenet (2004) Handbook for the prevention and minimization of waste and valorization of by-products in European agro-food industries. Agro-food waste minimization and reduction network (AWARENET). Grow Programme, European Commission, pp 1–7
- Bajaj BK, Sharma N, Singh S. Enhanced production of fibrinolytic protease from Bacillus cereus NS-2 using cotton seed cake as nitrogen source. Biocatal Agric Biotechnol. 2013;2:204–209. [Google Scholar]
- Bajaj BK, Singh S, Khullar M, Singh K, Bhardwaj S. Optimization of fibrinolytic protease production from Bacillus subtilis I-2 using agro-residues. Braz Arch Biol Technol. 2014;57:653–662. doi: 10.1590/S1516-8913201402132. [DOI] [Google Scholar]
- Bednarski A. Identifying unknown bacteria using biochemical and molecular methods. St Louis: Washington University; 2006. [Google Scholar]
- Ben Rebah F, Frikha F, Kammoun W, Belbahri L, Gargouri Y, Miled N. Culture of Staphylococcus xylosus in fish processing by-product-based media for lipase production. Lett Appl Microbiol. 2008;47:549–554. doi: 10.1111/j.1472-765X.2008.02465.x. [DOI] [PubMed] [Google Scholar]
- Blann AD, Landray MJ, Lip GY. An overview of antithrombotic therapy. BMJ. 2002;325:762–765. doi: 10.1136/bmj.325.7367.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode C, Runge MS, Smalling RW. The future of thrombolysis in the treatment of acute myocardial infarction. Eur Heart J. 1996;17:55–60. doi: 10.1093/eurheartj/17.suppl_E.55. [DOI] [PubMed] [Google Scholar]
- Bozzano A, Sardà F. Fishery discard consumption rate and scavenging activity in the northwestern Mediterranean Sea. ICES J Mar Sci. 2002;59:15–28. doi: 10.1006/jmsc.2001.1142. [DOI] [Google Scholar]
- Bressollier P, Letourneau F, Urdaci M, Verneuil B. Purification and characterization of a keratinolytic serine proteinase from Streptomyces albidoflavus. Appl Environ Microbiol. 1999;65:2570–2576. doi: 10.1128/aem.65.6.2570-2576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CT, Fan MH, Kuo FC, Sung HY. Potent fibrinolytic enzyme from a mutant of Bacillus subtilis IMR-NK1. J Agric Food Chem. 2000;48:3210–3216. doi: 10.1021/jf000020k. [DOI] [PubMed] [Google Scholar]
- Dilipkumar M, Rajasimman M, Rajamohan N (2011) Optimization of inulinase production from garlic by Streptomyces sp. in solid state fermentation using statistical designs. Biotechnol Res Int. doi:10.4061/2011/708043 [DOI] [PMC free article] [PubMed]
- Dilipkumar M, Rajasimman M, Rajamohan N. Enhanced inulinase production by Streptomyces sp. in solid state fermentation through statistical designs. 3 Biotech. 2013;3:509–515. doi: 10.1007/s13205-012-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Nomura K, Hong K, Ito Y, Asada A, Nishimuro S. Purification and characterization of a strong fibrinolytic enzyme (nattokinase) in the vegetable cheese natto, a popular soybean fermented food in Japan. Biochem Biophys Res Commun. 1993;197:1340–1347. doi: 10.1006/bbrc.1993.2624. [DOI] [PubMed] [Google Scholar]
- Fujita M, Ito Y, Hong K, Nishimuro S. Characterization of nattokinase-degraded products from human fibrinogen or cross-linked fibrin. Fibrinolysis. 1995;9:157–164. doi: 10.1016/S0268-9499(95)80005-0. [DOI] [Google Scholar]
- Fulhage CD. Reduce environmental problems with proper land application of animal manure. Columbia: University of Missouri Extension; 2000. [Google Scholar]
- Gad RG, Nirmala S, Sivvaswamy SN. Fibrinolytic enzyme from Bacillus amyloliquefaciens: Optimisation and scale up studies. Int J Pharm Pharm Sci. 2014;6:370–378. [Google Scholar]
- Ghaly AE, Ramakrishnan VV, Brooks MS, Budge SM, Dave D. Fish processing wastes as a potential source of proteins, amino acids and oils: a critical review. J Microb Biochem Technol. 2013;5:107–129. [Google Scholar]
- Ghorbel S, Soussi N, Ellouz YT, Duffosse L, Guerard F, Nazri M. Preparation and testing of Sardinella protein hydrolysate as nitrogen source for extracellular lipase production by Rhizopus oryzae. World J Microbiol Biotechnol. 2005;21:33–38. doi: 10.1007/s11274-004-1556-2. [DOI] [Google Scholar]
- Haddar A, Fakhfakh-Zouari N, Hmidet N, Frikha F, Nasri M, Kamoun AS. Low-cost fermentation medium for alkaline protease production by Bacillus mojavensis A21 using hulled grain of wheat and Sardinella peptone. J Biosci Bioeng. 2010;110:288–294. doi: 10.1016/j.jbiosc.2010.03.015. [DOI] [PubMed] [Google Scholar]
- He J, Chen S, Gu J. Identification and characterization of Harobin, a novel fibrino(geno)lytic serine protease from a sea snake (Lapemis hardwickii) FEBS. 2007;581:2965–2973. doi: 10.1016/j.febslet.2007.05.047. [DOI] [PubMed] [Google Scholar]
- Holden RW. Plasminogen activators: pharmacology and therapy. Radiology. 1990;174:993–1001. doi: 10.1148/radiology.174.3.174-3-993. [DOI] [PubMed] [Google Scholar]
- Itoi Y, Horinaka M, Tsujimoto Y, Matsui H, Watanabe K. Characteristic features in the structure and collagen-binding ability of a thermophilic collagenolytic protease from the thermophile Geobacillus collagenovorans MO-1. J Bacteriol. 2006;188:6572–6579. doi: 10.1128/JB.00767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhample SB, Bhagwat PK, Dandge PB. Statistical media optimization for enhanced production of fibrinolytic enzyme from newly isolated Proteus penneri SP-20. Biocatal Agric Biotechnol. 2015;4:370–379. [Google Scholar]
- Joo HS, Kumar CG, Park GC, Paik SR, Chang CS. Oxidant and SDS-stable alkaline protease from Bacillus clausii I-52: production and some properties. J Appl Microbiol. 2003;95:267–272. doi: 10.1046/j.1365-2672.2003.01982.x. [DOI] [PubMed] [Google Scholar]
- Khusro A, Kaliyan BK, Al-Dhabi NA, Arasu MV, Agastian P. Statistical optimization of thermo-alkali stable xylanase production from Bacillus tequilensis strain ARMATI. Electron J Biotechnol. 2016 [Google Scholar]
- Kim W, Choi K, Kim Y, Park H, Choi J, Lee Y, Oh H, Kwon I, Lee S. Purification and characterization of a fibrinolytic enzyme produced from Bacillus sp. strain CK 11-4 screened from Chungkook-Jang. Appl Environ Microbiol. 1996;62:2482–2488. doi: 10.1128/aem.62.7.2482-2488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xing J, Chang T, Ma Z, Liu H. Optimization of nutritional conditions for nattokinase production by Bacillus natto NLSSE using statistical experimental methods. Process Biochem. 2005;40:2757–2762. doi: 10.1016/j.procbio.2004.12.025. [DOI] [Google Scholar]
- Lomate PR, Hivrale VK. Induction of leucine aminopeptidase (LAP) like activity with wounding and methyl jasmonate in pigeonpea (Cajanas cajan) suggests the role of these enzymes in plant defense in leguminosae. Plant Physiol Biochem. 2011;49:609–616. doi: 10.1016/j.plaphy.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Mahajan PM, Nayak S, Lele SS. Fibrinolytic enzyme from newly isolated marine bacterium Bacillus subtilis ICTF-1: Media optimization, purification and characterization. J Biosci Bioeng. 2012;113:307–314. doi: 10.1016/j.jbiosc.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Mahanta N, Gupta A, Khare SK. Production of protease and lipase by solvent tolerant Pseudomonas aeruginosa PseA in solid-state fermentation using Jatropha curcas seed cake as substrate. Bioresour Technol. 2008;99:1729–1735. doi: 10.1016/j.biortech.2007.03.046. [DOI] [PubMed] [Google Scholar]
- Majumdar S, Goswami S, Keppen C, Rai SK, Mukherjee AK. Statistical optimization for improved production of fibrin (Ogen) olytic enzyme by Bacillus cereus strain FF01 and assessment of in vitro thrombolytic potential of protease enzyme. Biocatal Agric Biotechnol. 2015;4:191–198. [Google Scholar]
- Mukherjee AK, Rai SK. A statistical approach for the enhanced production of alkaline protease showing fibrinolytic activity from a newly isolated Gram-negative Bacillus sp. strain AS-S20-I. N Biotechnol. 2011;28:182–189. doi: 10.1016/j.nbt.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Mukherjee AK, Adhikari H, Rai SK. Production of alkaline protease by a thermophilic Bacillus subtilis under solid-state fermentation (SSF) condition using Imperata cylindrical grass and potato peel as low cost medium: characterization and application of enzyme in detergent formulation. Biochem Eng J. 2008;39:353–361. doi: 10.1016/j.bej.2007.09.017. [DOI] [Google Scholar]
- Oh YS, Shih IL, Tzeng YM, Wang SL. Protease produced by Pseudomonas aeruginosa K-187 and its application in the deproteinization of shrimp and crab shell wastes. Enzym Microb Technol. 2000;27:3–10. doi: 10.1016/S0141-0229(99)00172-6. [DOI] [PubMed] [Google Scholar]
- Paik HD, Lee SK, Heo S, Kim SY, Lee H, Kwon TJ. Purification and characterization of the fibrinolytic enzyme produced by Bacillus subtilis KCK-7 from Chungkookjang. J Microbiol Biotechnol. 2004;14:829–835. [Google Scholar]
- Pandey A, Soccol CR, Mitchell D. New developments in solid state fermentation: I-bioprocesses and products. Process Biochem. 2000;35:1153–1169. doi: 10.1016/S0032-9592(00)00152-7. [DOI] [Google Scholar]
- Peng Y, Yang XJ, Xiao L, Zhang YZ. Cloning and expression of a fibrinolytic enzyme (subtilisin DFE) gene from Bacillus amyloliquefaciens DC-4 in Bacillus subtilis. Res Microbiol. 2004;155:167–173. doi: 10.1016/j.resmic.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Prakasham RS, Rao CS, Sarma PN. Green gram husk—an inexpensive substrate for alkaline protease production by Bacillus sp. in solid-state fermentation. Bioresour Technol. 2006;97:1449–1454. doi: 10.1016/j.biortech.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Premalatha N, Gopal NO, Jose PA, Anandham R, Kwon SW (2015) Optimization of cellulase production by Enhydrobacter sp. ACCA2 and its application in biomass saccharification. Front Microbiol. doi:10.3389/fmicb.2015.01046 [DOI] [PMC free article] [PubMed]
- Raol GG, Raol BV, Prajapati VS, Bhavsar NH. Utilization of agro-industrial waste for β-galactosidase production under solid state fermentation using halotolerant Aspergillus tubingensis GR1 isolate. 3 Biotech. 2015;5:411–421. doi: 10.1007/s13205-014-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebah FB, Miled N. Fish processing wastes for microbial enzyme production: a review. 3 Biotech. 2013;3:255–265. doi: 10.1007/s13205-012-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejiniemon TS, Hussain RR, Rajamani B. In-vitro functional properties of Lactobacillus plantarum isolated from fermented ragi malt. South Ind J Biol Sci. 2015;1:15–23. [Google Scholar]
- Salihu A, Sallau AB, Adamu A, Kudu FA, Tajo MM, Bala TF, Yashim WD. Utilization of groundnut husk as a solid substrate for cellulase production by Aspergillus niger using response surface methodology. Waste Biomass Valoriz. 2014;5:585–593. doi: 10.1007/s12649-013-9268-1. [DOI] [Google Scholar]
- Siala R, Frikha F, Mhamdi S, Nasri M, Sellami Kamoun A. Optimization of acid protease production by Aspergillus niger I1 on shrimp peptone using statistical experimental design. Sci World J. 2012 doi: 10.1100/2012/564932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Richa K, Bose H, Karthik L, Kumar G, Rao KV. Statistical media optimization and cellulase production from marine Bacillus VITRKHB. 3. Biotech. 2014;4:591–598. doi: 10.1007/s13205-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souissi N, Ellouz-Triki Y, Bougatef A, Blibech M, Nasri M. Preparation and use of media for protease-producing bacterial strains based on by-products from Cuttlefish (Sepia officinalis) and wastewaters from marine-products processing factories. Microbiol Res. 2008;163:473–480. doi: 10.1016/j.micres.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Sugimoto S, Fujii T, Morimiya T, Johdo O, Nakamura T. The fibrinolytic activity of a novel protease derived from a tempeh producing fungus, Fusarium sp. BLB. Biosci Biotechnol Biochem. 2007;71:2184–2189. doi: 10.1271/bbb.70153. [DOI] [PubMed] [Google Scholar]
- Sumi H, Hamada H, Tsushima H, Mihara H, Muraki H. A novel fibrinolytic enzyme (nattokinase) in the vegetable cheese Natto; a typical and popular soybean food in the Japanese diet. Experientia. 1987;43:1110–1111. doi: 10.1007/BF01956052. [DOI] [PubMed] [Google Scholar]
- Sumi H, Hamada H, Nakanishi K, Hiratani H. Enhancement of the fibrinolytic activity in plasma by oral administration of nattokinases. Acta Haematol. 1990;84:139–143. doi: 10.1159/000205051. [DOI] [PubMed] [Google Scholar]
- Sumi H, Nakajima N, Mihara H. Fibrinolysis relating substances in marine creatures. Comp Biochem Physiol B. 1992;102:163–167. doi: 10.1016/0300-9629(92)90029-P. [DOI] [PubMed] [Google Scholar]
- Triki-Ellouz Y, Ghorbel B, Souissi N, Kammoun S, Nasri M. Biosynthesis of protease by Pseudomonas aeruginosa MN7 grown on fish substrate. World J Microbiol Biotechnol. 2003;19:41–45. doi: 10.1023/A:1022549517421. [DOI] [Google Scholar]
- Turpie AG. Pentasaccharides. Semin Hematol. 2002;39:158–171. doi: 10.1053/shem.2002.34091. [DOI] [PubMed] [Google Scholar]
- Vázquez JA, González MP, Murado MA. Preliminary tests on nisin and pediocin production using waste protein sources: factorial and kinetic studies. Bioresour Technol. 2006;97:605–613. doi: 10.1016/j.biortech.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan P, Vincent SG. Cow dung as a novel, inexpensive substrate for the production of a halo-tolerant alkaline protease by Halomonas sp. PV1 for eco-friendly applications. Biochem Eng J. 2012;69:57–60. doi: 10.1016/j.bej.2012.08.014. [DOI] [Google Scholar]
- Vijayaraghavan P, Vincent SG (2014) Statistical optimization of fibrinolytic enzyme production by Pseudoalteromonas sp. IND11 using cow dung substrate by response surface methodology. Springerplus. doi: 10.1186/2193-1801-3-60 [DOI] [PMC free article] [PubMed]
- Vijayaraghavan P, Vijayan A, Arun A, Jenisha JK, Vincent SG. Cow dung: a potential biomass substrate for the production of detergent-stable dehairing protease by alkaliphilic Bacillus subtilis strain VV. Springerplus. 2012 doi: 10.1186/2193-1801-1-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavan P, Arun A, Al-Dhabi NA, Vincent SG, Arasu MV, Choi KC. Novel Bacillus subtilis IND19 cell factory for the simultaneous production of carboxy methyl cellulase and protease using cow dung substrate in solid-substrate fermentation. Biotechnol Biofuels. 2016 doi: 10.1186/s13068-016-0481-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavan P, Arun A, Vincent SG, Arasu MV, Al-Dhabi NA (2016b) Cow dung is a novel feedstock for fibrinolytic enzyme production from newly isolated Bacillus sp. IND7 and its application in in vitro clot lysis. Front Microbiol. doi: 10.3389/fmicb.2016.00361 [DOI] [PMC free article] [PubMed]
- Wang CT, Ji BP, Li B, Nout R, Li PL, Ji H, Chen LF. Purification and characterization of a fibrinolytic enzyme of Bacillus subtilis DC33, isolated from Chinese traditional Douchi. J Ind Microbiol Biotechnol. 2006;33:750–758. doi: 10.1007/s10295-006-0111-6. [DOI] [PubMed] [Google Scholar]
- Wang SH, Zhang C, Yang YL, Diao M, Bai MF. Screening of a high fibrinolytic enzyme producing strain and characterization of the fibrinolytic enzyme produced from Bacillus subtilis LD-8547. World J Microbiol Biotechnol. 2008;24:475–482. doi: 10.1007/s11274-007-9496-2. [DOI] [Google Scholar]
- Wang SL, Hsu WT, Yen YH, Wang CL. Purification and characterization of three novel keratinolytic metalloproteases produced by Chryseobacterium indologenes TKU014 in a shrimp shell powder medium. Bioresour Technol. 2008;99:5679–5686. doi: 10.1016/j.biortech.2007.10.024. [DOI] [PubMed] [Google Scholar]