Abstract
By analyzing the gene expression profile between tumor cells and revertant counterparts that have a suppressed malignant phenotype, we previously reported a significant down-regulation of translationally controlled tumor protein (TCTP) in the revertants. In the present study, we derived, by using the H1 parvovirus as a selective agent, revertants from three major solid cancers: colon, lung, and melanoma cell lines. These cells have a strongly suppressed malignant phenotype both in vitro and in vivo. The level of TCTP is decreased in most of the revertants. To verify whether inhibition of TCTP expression induces changes in the malignant phenotype, in the classical, well established model of “flat reversion,” v-src-transformed NIH3T3 cells were transfected with antisense TCTP. By inhibiting the expression of TCTP, the number of revertant cells was raised to 30%, instead of the reported rate for spontaneous flat revertants of 10-6. Because TCTP encodes for a histamine-releasing factor, we tested the hypothesis that inhibitors of the histaminic pathway could be effective against tumor cells. We show that some antihistaminic compounds (hydroxyzine and promethazine) and other pharmacological compounds with a related structure (including thioridazine and sertraline) kill tumor cells and significantly decrease the level of TCTP. All together, these data suggest that, with tumor reversion used as a working model, TCTP was identified as a target and drugs were selected that decrease its expression and kill tumor cells.
A “target” in cancer can be defined as a protein whose expression or biological function is different between normal and tumor cells. Such a modification will be harmful to the normal cell, leading to transformation, and thus targeting it and changing its activity could lead to the suppression and/or reversion of the malignancy.
The general approach used so far to identify such target proteins was to analyze the difference between normal and cancer cells, thus answering the question of how a normal cell becomes malignant. We have suggested a different approach, namely to analyze what causes a malignant cell to revert (1–4). One of the advantages of such a strategy is that the revertant cell has acquired the molecular knowledge of how to escape malignancy. We suggested that in such revertant cells the molecular mechanisms to override cancer are present (1–4). The understanding of how this reversion happens may lead to the identification of targets that were not disclosed by comparing normal and tumor cells.
The premises of tumor reversion were discovered in the mid-1960s, when investigators established a cell line of normal mouse fibroblasts, NIH3T3, and the first studies pointed toward a sensitivity to contact inhibition in culture. This sensitivity was caused by a reversible arrest of growth in G1 (5). After infecting this cell line with polyoma virus, or simian virus 40 (SV40), there was a loss of sensitivity to contact inhibition, and the NIH3T3 grew in clusters and multilayers. In 1968, Pollack, Green, and Todaro (6) described for the first time the selection of sublines of NIH3T3 infected with polyoma or SV40 that had regained an increased sensitivity to contact inhibition and, most importantly, a decreased tumor-producing ability. They called these cells “revertants.” Later studies led to the discovery of K-rev, a 21-kDa protein with revertant-inducing activity on Kirsten sarcoma virus-transformed NIH3T3 cells (7, 8). These experiments focused at obtaining revertants by interfering specifically with a single oncogene-induced tumor. In contrast, our studies in which revertants were derived from human tumor cell lines originating from patients' material harboring the full range of abnormalities present in the tumor cell provide a more comprehensive approach of tumor reversion.
We have previously described translationally controlled tumor protein (TCTP) as a gene down-regulated in tumor reversion (4). It was initially identified (9) as one of four mRNA that occur predominantly as untranslated, partially suppressed messenger ribonucleoprotein particles in mouse sarcoma ascites cells and further characterized as p23/p21 (10, 11). TCTP is a house-keeping gene expressed in several nontumoral cells, including erythrocytes (12). The first overexpression experiments in the analysis of TCTP showed its binding to tubulin (13, 14).
Besides these intracellular functions for TCTP, this molecule has been identified as a histamine-releasing factor (HRF) (15). More recently, the solution structure of TCTP suggests a strong homology with Mss4, a chaperone binding GDP/GTP free G protein (16). TCTP also interacts with TSAP6, translation elongation factor eEF1A, and its guanine nucleotide exchange factor eEF1B-β (17–19).
In the present study we extend the analysis of reversion to some of the major cancers. We further demonstrate that down-regulation of TCTP can, by itself, induce tumor reversion, and we describe drugs that decrease the level of TCTP and kill tumor cells.
Materials and Methods
Revertant Cells. The tumor cells lines DLD-1, A549, WM115 and 266-4, SK-MEL28, and Hs852T were obtained from the American Type Culture Collection. For the isolation of revertants we applied the same technology as described (4). Different concentrations of H1 parvovirus were used to infect tumor cells with a multiplicity of infection of 10–1,000 plaque-forming units per cell. Surviving colonies were isolated by using collagenase/dispase (Roche Diagnostics). Growth of isolated colonies and parental tumor cell lines was tested in soft agar (agar-noble, Difco). A total of 107 cells per site were injected in scid/scid mice for in vivo tumorigenicity tests, and statistical analysis on the growth was performed as described (3). H1 parvovirus DNA was amplified by using the following primers: 5′-CTAGCAACTCTGCTGAAGGAACTC-3′ and 5′-TAGTGATGCTGTTGCTGTATCTGATG-3′.
Antibodies. For Western blot analysis the following antibodies were used: antihistamine-releasing factor (TCTP) (Medical and Biological Laboratories, Nagoya, Japan) and antiactin (Santa Cruz Biotechnology). For immunofluorescence analysis, phalloidin-FITC (Sigma), anti-α-tubulin (Sigma), anti-mouse CY3, and 4′,6-diamidino-2-phenylindole were used. Anti-H1-parvovirus virions antibodies were generated in rabbits by injecting 5 × 109 plaque-forming units per cell per rabbit of UV-inactivated H1 parvovirus (Agro-Bio, La Ferté St. Aubin, France).
Flat Revertant Cells. NIH3T3 cells were transformed with v-src. Foci were isolated and maintained in culture for 4 weeks before the isolation of flat revertants. The antisense TCTP was generated by cloning the cDNA corresponding to the coding region of tpt1/TCTP 3′-5′ in pBK-RSV (Stratagene). Transfection of v-src-transformed NIH3T3 cells with antisense TCTP was performed by using Lipofectamine 2000 (Invitrogen), clones were selected with G418 (600 μg/ml), and further subclones were isolated.
Pharmacological Compounds. All pharmacological compounds were purchased from Sigma, except sertraline and paroxetine, which were purchased from Sequoia Research Products (Oxford, U.K.) and Apin Chemicals (Oxon, U.K.), respectively.
Cell Viability Assays. The cytotoxicity of the pharmacological compounds was measured on U937 cells by treating the cells for 6 days with various concentrations of the compounds. ATP level was measured by a luminescent cell viability assay. Celltiter-glo (Promega) was used following the manufacturer's instructions and read on a Victor2 plate reader (PerkinElmer).
In Vivo Evaluation of Antitumor Effect of Pharmacological Compounds. Subcutaneous tumors from MDA-MB231 and U937 cells were induced by injecting 107 cells into the right flank of scid/scid mice. Treatment (promethazine at 22.5 mg/kg, sertraline at 18.0 mg/kg, and thioridazine at 6.75 mg/kg) started 2 days before the injection of the tumor cells in the early-stage protocol, or when tumors reached a palpable volume (4 mm3) for late-stage treatment. For U937-derived tumors, mice were treated once a day with an i.p. injection over a period of 28 days and monitored over a period up to 80 days. For the MDA-MB231 tumor-bearing animals the same treatment lasted 60 days, and they were monitored for 80 days.
Results
Identification of Revertants from Colon, Lung, and Melanoma Cancer Cell Lines. Our previous conclusion on revertants was based on cells derived from two leukemia cell lines (K562 and U937) and three breast cancer cell lines (BT20, T47D, and MDA-MB231). To consolidate the concept of tumor reversion it was important to have other models of reversion, especially for solid tumors. We then derived revertants from colon, lung, and melanoma tumor cells. DLD-1 is a colorectal adenocarcinoma cell line, A549 is a lung carcinoma cell line, and WM-266-4, WM-115, SK-MEL-28, and Hs852T are melanoma cell lines.
To obtain these revertants, we used the approach as described (1, 2, 4): the tumor cells were infected with H1 parvovirus that kills preferentially the malignant cells (20, 21), while sparing their normal counterparts and cells that are resistant to the cytopathic effect of the virus. Resistant clones were isolated, and their phenotype was assessed by measuring anchorage dependence in a soft agar assay (Fig. 1A) and their tumorigenicity in scid/scid mice (Fig. 1B).
Fig. 1.
Tumor revertants for colon, lung, and melanoma cell lines. (A) In vitro, soft agar growth for colon (DLD-1), lung (A549), melanoma (WM266.4), and corresponding revertants reported as number of colonies. (B) In vivo tumorigenicity after injection of 107 tumor or revertant cells, measured as mean tumor volume. (C) PCR analysis to detect the presence of H1 parvovirus. (D) Western blot analysis for TCTP expression in tumor (control cell lines are boxed) and revertant cells. Actin is used as loading control.
The colon cancer cells DLD-1 form a large number of colonies in soft agar, whereas the two revertant clones, CL-4 and CL-16, display anchorage dependence and hardly grew under the same conditions (Fig. 1 A). In vivo tumorigenicity in scid/scid mice showed a three-time reduced tumor volume at the end of the study compared with the parental cell line. The two revertants from the lung carcinoma A549 were isolated; CL-46 showed strong anchorage dependence in soft agar (Fig. 1 A) and strongly reduced growth in scid/scid mice (Fig. 1B). The other revertant, CL-1, did not grow at all under the same conditions (Fig. 1 A).
These CL-1 cells totally reverted, showing a complete suppression of their malignant growth. Four melanoma cell lines were used. WM-115 and WM-266-4 both were derived from the same patient; WM-115 was from the primary tumor and WM-266-4 was from a metastasis. SK-MEL-28 and Hs852T were the other melanoma cell lines. All four melanomas were highly susceptible to the killing effect of the H1 parvovirus. We show the reduced tumorigenicity of the revertant clone obtained from WM-266-4 (Fig. 1 A and B).
Persistent Infection and Expression of H1 Parvovirus Is Not Necessary for Maintaining the Suppressed Malignant Phenotype. An important question to answer was whether the suppressed malignant phenotype in the revertants is caused by continuous production of the H1 parvovirus. Our previous experiments indicated that both revertants derived from the leukemia cell lines (K562 and U937) continue to be H1 parvovirus-positive for years after the initial infection (1, 2). For the breast cancer cell lines, the revertants derived from BT20 and T47D do not produce any virus at all anymore, whereas the revertants derived from MDA-MB231 continue to produce parvovirus (4). These results suggested that persistent H1 parvovirus infection is not necessary for maintaining the suppressed malignant phenotype in all of the revertants. The PCR analysis shows that both revertants of DLD-1, one of the two revertants from A549, and none of the melanoma revertants are H1-positive (Fig. 1C). The tumor cells DLD-1, A549, and WM-266-4 and their isolated revertants were thus analyzed for the presence of H1 virions by fluorescence-activated cell sorting (Fig. 4A, which is published as supporting information on the PNAS web site). Low amounts of the revertant cells from DLD-1 and A549 tumor cells stained positive for H1 viral particles, suggesting that only a small percentage of these revertants produces infectious virus. Interesting to note is that the DLD-1 revertant CL-16 produces infectious virus capable of replicating in SV40-transformed newborn human kidney cells, but cannot induce plaque formation on the same indicator cells. Conversely, the DLD-1 revertant CL-4 harbors a host range mutant of the virus that lost its ability to induce lysis of the indicator cell line NBE (Fig. 4B).
The limiting dilution assay of A549 parental and revertant clone CL-1 shows that 32% of the revertant cells produce infectious H1 particles (Fig. 4B). The melanoma revertant WM266-4-Cl4 was found to be negative in fluorescence-activated cell sorting analysis for viral particles (Fig. 4A) and for producing infectious virus (Fig. 4B).
These data confirm that the virus by itself is not sufficient for maintaining the suppressed malignant phenotype.
TCTP Expression in the Revertant Cells. In the study of reversion, we observed that the gene that is the most strongly down-regulated is TCTP, and we partially correlated the process of reversion to inhibition of TCTP expression (4). We found that decreasing TCTP by antisense cDNA in U937 cells promotes apoptosis, whereas knocking down its expression in breast carcinoma cell lines induces a reorganization by forming ductal/acinar structures similar to those structures described by Bissell's group (22). In the present study, we checked the level of TCTP expression. In one of the two colon revertant models (DLD-1 CL-16), one of the lung (A549 CL-46) revertant models, and all of the melanoma revertant models, the level of the protein is significantly reduced (Fig. 1D). These results, taken together with the previous data (4) obtained from revertants, suggest that in the majority of cases there is a down-regulation of TCTP expression during the process of tumor reversion.
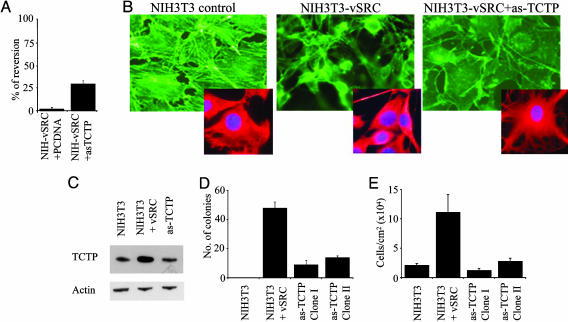
Induction of Flat Revertants in v-src-Transformed NIH3T3 by Inhibition of TCTP Expression. To position our data on TCTP and its role in reversion into the classical framework of “flat reversion” carried out in the late 1960s, we investigated how v-src-induced transformation could be influenced by the level of expression of TCTP. The transformation of NIH3T3 by v-src induces foci formation on cell monolayer and colony formation in soft agar. The flat revertant cells are generally defined as a variant of a malignant cell in which the characteristic high-saturation density and piled-up morphology have reverted to the flatter morphology associated with nontransformed cells.
Of relevance is the fact that in NIH3T3 transformed by v-src, the expression of TCTP is highly increased and the use of a TCTP antisense cDNA, although reducing the TCTP expression, strongly increases the percentage of reversion up to 30% (Fig. 2 A and C). This increase is extremely high when compared with the spontaneous rate of reversion that was previously established as being one upon 106 (6). The morphology of v-src-transformed NIH3T3 is typically piled up, whereas the antisense TCTP gives rise to flat revertants (Fig. 2B) and is able to restore contact inhibition and anchorage-dependent growth (Fig. 2D). The cell density at confluence is another parameter we tested to better define the flat revertants derived by inhibition of the expression of TCTP. This density is also strongly decreased when the expression of TCTP is inhibited (Fig. 2E). These results suggest that targeting TCTP and decreasing its level of expression in v-src-transformed NIH3T3 leads to strong reversion of the malignant phenotype.
Fig. 2.
Flat revertants induced by antisense TCTP in v-src-transformed NIH3T3 cells. (A) Percentage of tumor reversion in NIH3T3 v-src and NIH3T3 v-src transfected with antisense TCTP. (B) Phalloidin (green) and α-tubulin (red) staining in the indicated cell lines. (C) Western blot to measure TCTP expression. (D) Soft agar analysis of NIH3T3, NIH3T3 v-src, and two flat revertants. (E) Maximum reachable cell density at confluence.
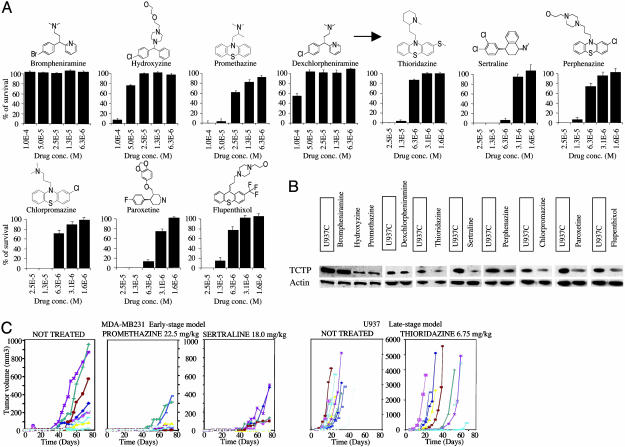
Identification of Chemical Compounds Killing Tumor Cells and Down-Regulating the Expression of TCTP. Once we established that TCTP is a valid target in tumors and an important molecule in tumor reversion, we tried to find molecules that could inhibit its expression. TCTP is also known as a HRF (15), so we hypothesized that compounds inhibiting the histaminic pathway also could inhibit the function of HRF and have an anticancer effect. We then searched for chemical compounds already known to antagonize the histaminic pathway. Brompheniramine, hydroxyzine, promethazine, and dexchlorpheniramine are known to be effective antihistaminic drugs. We tested the in vitro effect of these drugs on human leukemia U937 (Fig. 3A). Incubation of the cells with the drugs at different concentrations for 6 days revealed that hydroxyzine and promethazine had a significant cytophatic effect. Conversely, brompheniramine and dexchlorpheniramine, both part of the same chemical group of alkylamines, did not have any significant cytopathic effect. We then investigated whether structurally related molecules would have a cytopathic effect on cancer cells. Thioridazine, sertraline, perphenazine, chlorpromazine, paroxetine, and flupenthixol, generally with no pronounced antihistaminic properties, showed higher cytotoxicity at lower concentrations (Fig. 3A).
Fig. 3.
Pharmacological compounds killing tumor cells and decreasing TCTP level. (A) U937 cell plus control solvent viability at different concentrations of the indicated drug, expressed as percentage of survival after 6 days of treatment. The arrow indicates a second generation of compounds. (B) Western blots showing TCTP expression in U937 cells after drug incubation (U937 plus control are boxed). (C) In vivo tumor formation in scid/scid mice during drug treatment. (Left) MDA-MB231 treated with promethazine or sertraline by using an early-stage protocol. (Right) U937 treated with thioridazine by using a late-stage protocol. (Each curve represents the tumor formation in a different mouse.) Treatment with promethazine or sertraline for the early stage models and thioridazine for the late-stage models results in a highly significant tumor-growth delay, with some of the animals treated displaying no tumor growth at all anymore.
We checked the effect of these drugs on TCTP expression (Fig. 3B and Fig. 5, which is published as supporting information on the PNAS web site). A correlation between the cytopathic effect and inhibition of the expression of TCTP can be observed. The down-regulation of the protein does not seem to be caused by inhibition of transcription or protein degradation caused by cell death. On the contrary, the results indicate an increase in the expression level of TCTP mRNA after drug treatment (Fig. 6, which is published as supporting information on the PNAS web site). The compounds also were used in vivo on breast cancer MDA-MB231 and monocytic leukemia U937 cell lines (Fig. 3C). The volume of the tumors generated by injection of the cell lines into scid/scid mice was consistently reduced by drug treatment. Administration of drugs 2 days before the injection of the tumor cells (early-stage model) highly reduced the growth of the tumor in the animal (Fig. 3C). Administration of drugs when tumors reached a palpable volume (4 mm3) (late-stage treatment) also inhibited growth (Fig. 3C). For all drugs tested in the animals at the concentrations described here, there was no weight loss or any other general signs of toxicity present. These results suggest that these drugs have an anticancer effect by reducing directly or indirectly the level of TCTP.
Discussion
The aim of this study was to provide a stronger basis for tumor reversion as a biological process that ultimately would lead to the discovery of additional targets for the treatment of cancer. We described three sets of experiments. In the first one, biological models of tumor reversion were derived from three solid tumor cell lines. In the second, we asked the question of whether TCTP is a target protein by analyzing its capacity to modify the malignant phenotype in the framework of the experiments describing flat revertants, which initiated the whole field of tumor reversion (6). In the last set of experiments, we showed pharmaceutical agents reducing the level of TCTP and killing tumor cells.
We initiated our work on tumor reversion by asking the question of whether human leukemia cells would have the capacity to revert. In an earlier paper (1), we described these revertants, derived from the human erythroleukemia cell line K562. These KS cells (K562 Suppressed) were selected by using as a tool the H1 parvovirus that kills preferentially the tumor cells while sparing their “normal counterparts” (21, 23). These normal counterparts were unique cells among the leukemia cells (a frequency of 10-5 to 10-6 in further experiments). There was not a single sign of terminal differentiation and almost no spontaneous apoptosis (<1%) in the KS clones analyzed. However, they were not normal cells at all, the KS having an abnormal number of chromosomes and other molecular abnormalities found in this kind of tumor (1). They just lost their tumorigenicity. These experiments later were reproduced by deriving other revertants from the monocytic leukemia cell line U937. Here, too, the rate of reversion was very low, and no terminal differentiation was detected (2). Later, we derived revertants from solid tumors in the breast cancer cell lines, BT20, T47D, and MDA-MB231 (4).
We suggested then that tumor reversion is a biological process, with the capacity to override the oncogenic events to yield revertant cells. Some of these revertants lost almost their entire capacity to grow in soft agar and to form tumors after injection into scid/scid mice. In the revertants described in the present study and derived from colon, lung, and melanoma cell lines, we found the same characteristic loss of the malignant phenotype, with some of the clones barely growing. Because both the K562- and U937-derived revertants (KS and US) continued to produce H1 parvovirus, we thought that expression of the viral proteins was necessary for maintaining pressure on the cells, which would lead to a suppressed malignant phenotype. We were surprised to observe that in the revertants derived from the breast cancer cell lines (4) only one (MDA-MB231S) continued to produce the virus. This finding also seems true for the colon and lung revertants, where only one of the two revertant clones from lung and both colon revertants continued to produce the H1 parvovirus, whereas the melanoma-derived revertants did not produce it at all. These experiments led us to conclude that continuous infection with H1 parvovirus is not mandatory for maintaining the suppressed phenotype. H1 parvovirus functions as a selective agent with some clones continuing to produce it, whereas others did not produce it at all; this presence of H1 parvovirus does not change the parameters of the reversion. It remains possible, however, that during the selection procedure the virus induces the reversion process.
With a large-scale screening analysis (4) for the differentially expressed genes between the parental tumor cells and the revertants, among the 263 candidates, TCTP had the most striking differential expression. In megasort analysis, the results showed 248 signals for TCTP detected in the malignant cell line U937, but only two signals in the revertant US4 cells. This differential expression for a gene is the strongest we have ever detected. Further experiments showed that the down-regulation of TCTP by antisense cDNA or RNA interference induces an increase of apoptosis in U937 cells (up to 15%) but, most strikingly, leads to the reorganization of the breast cancer cell into ductal/acinar structures (22).
The analysis of the primary sequence of TCTP, so far, does not lead to any possible interpretation of its function. The NMR structure of the Schizosaccharomyces pombe TCTP has been reported (16). The dali search for homologous protein folds detected mainly two structural homologs, the guanine nucleotide exchange factor Mss4 and the MsrB peptide methionine sulfoxide reductase (Msr) fragment pilB (24). The guanine nucleotide-free chaperone Mss4 and its yeast homologue, Dss4, interact with the transient GDP/GTP-free form of the Rab. The Rab binding site on MSS4 coincides with the regions of highest sequence conservation in the TCTP family. The structural homology with the methionine–R-sulfoxide reductase (MsrB) is at the level of two antiparallel β sheets. Msrs protect against oxidative damage: Msr domains (MsrA and MsrB) of the pilB protein from Neisseria gonorrhoeae each reduce different epimeric forms of methionine sulfoxide. The structural homology between TCTP and pilB allows us to propose that the surface-exposed face of the four-stranded β sheets may constitute the site of interaction between TCTP and its biological partners. However, none of the amino acids located in pilB putative active site are structurally conserved in TCTP, and therefore no further biological insight can be revealed.
We confirmed that TCTP is down-regulated in one of the two colon cancer revertants, one of the two lung cancer revertants, and all of the melanoma revertants. Together with the KS (4) there are thus only three revertant clones where TCTP is not down-regulated. This finding makes sense because the analysis of tumors directly derived from patients also shows an increase of TCTP in most of the cancers analyzed, although not in all of them (4). These results suggest that all of the pathways of reversion would not involve the inhibition of TCTP. The decrease in TCTP level in most of the revertants together with the antisense experiments in U937 cells and the reorganization experiments with RNA interference in breast cancer cells argue in favor of TCTP being a reasonable target for reversion.
To validate TCTP as a target we went back to similar experiments as the ones carried out in the mid-1960s where flat revertants of NIH3T3-transformed cells were, for the first time, identified. The data presented in this article suggest that down-regulation of TCTP by antisense induces a high amount of flat revertants in v-src-transformed NIH3T3 cells and strengthens the idea that TCTP is a valuable target for tumor reversion.
We have described the effect of pharmacological compounds on decreasing the level of TCTP and killing cancer cells. Because TCTP corresponds to HRF and because it has to be down-regulated to induce tumor reversion, we hypothesized that this effect could be mimicked by using antihistaminic drugs. Hydroxyzine is part of the piperazines, and promethazine is part of the phenotiazines. They both are antihistaminic by antagonizing the H1 receptor and also act on the CNS. The role of histamine in its antitumor activity is controversial. Promethazine inhibits the effect of histamine on endotoxins against certain solid tumors (25). Another report suggests that certain H1 antagonists increase the growth rate of melanoma and fibrosarcoma cell lines (26). In any case it is now well documented by animal studies and clinical data that H1 receptor antagonists do not promote tumor formation (27). We found that structurally related drugs, whether neuroleptics or antidepressive agents, have a cytophatic effect on U937 and MDA-MB231 cells. Antihistaminics and neuroleptics are widely used in patients with cancer, as antiallergic, antidepressive, or antiemetic agents. Their use, as a potentially effective single anticancer agent, is not well established in clinical assays. In vitro studies have revealed that some phenotiazines, including promethazine, thioridazine, perphenazine, and chlorpromazine, had an antiproliferative effect (28–30).
Thioridazine inhibits cell growth of MCF7 and MDA-MB231 tamoxifen-resistant cells (31), by blocking p170mdr-1 (32). Phenotiazines act either on H1 or dopamine receptors. Here, we propose that, at least in part, these drugs act through down-regulation of TCTP at the protein level. The fact that the mRNA level of TCTP is up-regulated by sertraline and thioridazine indicates that the transcription machinery is unaffected and may compensate for the diminution of TCTP. The drugs used at the concentration described here, which is high, do not induce in the mice any general signs of toxicity but kill the cancer cells and delay tumor formation. The fact that in one of the protocols we started the treatment 2 days before inoculating the tumor cells and that such an approach results in decreased tumor formation may suggest that these drugs could have a preventive effect on tumor formation. Whether TCTP is a direct or an indirect target of these drugs remains to be investigated. The most simple explanation would be provided if the drugs directly bind TCTP. However, it is equally important to know whether these drugs affect TCTP indirectly by, for example, acting on some of the TCTP interactor proteins. In such a situation the function or subcellular distribution of TCTP would be modified, which could ultimately lead to a significant decrease in TCTP levels.
In conclusion, our approach, which identified TCTP as a key player in the process of tumor reversion, also leads to the identification of a series of pharmaceutical compounds able to diminish its expression and kill malignant cells. This strategy may contribute to new alternatives in cancer treatment.
Supplementary Material
Acknowledgments
We thank Françoise Rohfritsch for technical assistance; David Hangauer and Werner Bollag for precious advice and providing an additional list of compounds after our first experiments with antihistaminic drugs; Franck Sturtz for his help and support during the entire study; Philippe Genne for carrying out the animal studies; and Pierre Chardin for helpful discussion and the v-src plasmid.
Author contributions: R.A. and A.T. designed research; M.T., G.F., S.P., A.L., A.G., S. Beaucourt, and D.D. performed research; M.T., G.F., S.P., A.L., A.G., S. Beaucourt, D.D., S. Besse, L.S., J.C., D.M., R.A., and A.T. analyzed data; and R.A. and A.T. wrote the paper.
Abbreviations: TCTP, translationally controlled tumor protein; HRF, histamine-releasing factor.
References
- 1.Telerman, A., Tuynder, M., Dupressoir, T., Robaye, B., Sigaux, F., Shaulian, E., Oren, M., Rommelaere, J. & Amson, R. (1993) Proc. Natl. Acad. Sci. USA 90, 8702-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemani, M., Linares-Cruz, G., Bruzzoni-Giovanelli, H., Roperch, J. P., Tuynder, M., Bougueleret, L., Cherif, D., Medhioub, M., Pasturaud, P., Alvaro, V., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 9039-9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roperch, J. P., Alvaro, V., Prieur, S., Tuynder, M., Nemani, M., Lethrosne, F., Piouffre, L., Gendron, M. C., Israeli, D., Dausset, J., et al. (1998) Nat. Med. 4, 835-838. [DOI] [PubMed] [Google Scholar]
- 4.Tuynder, M., Susini, L., Prieur, S., Besse, S., Fiucci, G., Amson, R. & Telerman, A. (2002) Proc. Natl. Acad. Sci. USA 99, 14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilausen, K. & Green, H. (1965) Exp. Cell Res. 40, 166-168. [DOI] [PubMed] [Google Scholar]
- 6.Pollack, R. E., Green, H. & Todaro, G. J. (1968) Proc. Natl. Acad. Sci. USA 60, 126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noda, M., Kitayama, H., Matsuzaki, T., Sugimoto, Y., Okayama, H., Bassin, R. H. & Ikawa, Y. (1989) Proc. Natl. Acad. Sci. USA 86, 162-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitayama, H., Sugimoto, Y., Matsuzaki, T., Ikawa, Y. & Noda, M. (1989) Cell 56, 77-84. [DOI] [PubMed] [Google Scholar]
- 9.Yenofsky, R., Bergmann, I. & Brawerman, G. (1982) Proc. Natl. Acad. Sci. USA 79, 5876-5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohm, H., Gross, B., Gaestel, M., Bommer, U. A., Ryffel, G. & Bielka, H. (1991) Biomed. Biochim. Acta 50, 1193-1203. [PubMed] [Google Scholar]
- 11.Chitpatima, S. T., Makrides, S., Bandyopadhyay, R. & Brawerman, G. (1988) Nucleic Acids Res. 16, 2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez, J. C., Schaller, D., Ravier, F., Golaz, O., Jaccoud, S., Belet, M., Wilkins, M. R., James, R., Deshusses, J. & Hochstrasser, D. (1997) Electrophoresis 18, 150-155. [DOI] [PubMed] [Google Scholar]
- 13.Gachet, Y., Tournier, S., Lee, M., Lazaris-Karatzas, A., Poulton, T. & Bommer, U. A. (1999) J. Cell Sci. 112, 1257-1271. [DOI] [PubMed] [Google Scholar]
- 14.Bommer, U. A. & Thiele, B. J. (2004) Int. J. Biochem. Cell Biol. 36, 379-385. [DOI] [PubMed] [Google Scholar]
- 15.MacDonald, S. M., Rafnar, T., Langdon, J. & Lichtenstein, L. M. (1995) Science 269, 688-690. [DOI] [PubMed] [Google Scholar]
- 16.Thaw, P., Baxter, N. J., Hounslow, A. M., Price, C., Waltho, J. P. & Craven, C. J. (2001) Nat. Struct. Biol. 8, 701-704. [DOI] [PubMed] [Google Scholar]
- 17.Amzallag, N., Passer, B. J., Allanic, D., Segura, E., Thery, C., Goud, B., Amson, R. & Telerman, A. (September 21, 2004) J. Biol. Chem., www.jbc.org/cgi/reprint/M404850200v2. [DOI] [PubMed]
- 18.Cans, C., Passer, B. J., Shalak, V., Nancy-Portebois, V., Crible, V., Amzallag, N., Allanic, D., Tufino, R., Argentini, M., Moras, D., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 13892-13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langdon, J. M., Vonakis, B. M. & MacDonald, S. M. (2004) Biochim. Biophys. Acta 1688, 232-236. [DOI] [PubMed] [Google Scholar]
- 20.Toolan, H. W. & Ledinko, N. (1968) Virology 35, 475-478. [DOI] [PubMed] [Google Scholar]
- 21.Mousset, S. & Rommelaere, J. (1982) Nature 300, 537-539. [DOI] [PubMed] [Google Scholar]
- 22.Weaver, V. M., Petersen, O. W., Wang, F., Larabell, C. A., Briand, P., Damsky, C. & Bissell, M. J. (1997) J. Cell Biol. 137, 231-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toolan, H. W., Rhode, S. L., III, & Gierthy, J. F. (1982) Cancer Res. 42, 2552-2555. [PubMed] [Google Scholar]
- 24.Holm, L. & Sander, C. (1996) Science 273, 595-603. [DOI] [PubMed] [Google Scholar]
- 25.Bloksma, N., van de Wiel, P., Hofhuis, F., Kuper, F. & Willers, J. (1984) Cancer Immunol. Immunother. 17, 33-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandes, L. J., Warrington, R. C., Arron, R. J., Bogdanovic, R. P., Fang, W., Queen, G. M., Stein, D. A., Tong, J., Zaborniak, C. L. & LaBella, F. S. (1994) J. Natl. Cancer Inst. 86, 770-775. [DOI] [PubMed] [Google Scholar]
- 27.Hardman, J. G. & Limbird, L. E., eds. (1996) Goodman and Gilman's The Pharmacological Basis of Therapeutics (McGraw–Hill, New York).
- 28.Strobl, J. S., Kirkwood, K. L., Lantz, T. K., Lewine, M. A., Peterson, V. A. & Worley, J. F., III (1990) Cancer Res. 50, 5399-5405. [PubMed] [Google Scholar]
- 29.Gil-Ad, I., Shtaif, B., Levkovitz, Y., Dayag, M., Zeldich, E. & Weizman, A. (2004) J. Mol. Neurosci. 22, 189-198. [DOI] [PubMed] [Google Scholar]
- 30.Zhelev, Z., Ohba, H., Bakalova, R., Hadjimitova, V., Ishikawa, M., Shinohara, Y. & Baba, Y. (2004) Cancer Chemother. Pharmacol. 53, 267-275. [DOI] [PubMed] [Google Scholar]
- 31.Strobl, J. S. & Peterson, V. A. (1992) J. Pharmacol. Exp. Ther. 263, 186-193. [PubMed] [Google Scholar]
- 32.Yu, D., Liu, B., Jing, T., Sun, D., Price, J. E., Singletary, S. E., Ibrahim, N., Hortobagyi, G. N. & Hung, M. C. (1998) Oncogene 16, 2087-2094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.