Abstract
Toll-like receptor (TLR) activation is central to immunity, wherein the activation of the TLR9 subfamily members TLR9 and TLR7 results in the robust induction of type I IFNs (IFN-α/β) by means of the MyD88 adaptor protein. However, it remains unknown how the TLR signal “input” can be processed through MyD88 to “output” the induction of the IFN genes. Here, we demonstrate that the transcription factor IRF-7 interacts with MyD88 to form a complex in the cytoplasm. We provide evidence that this complex also involves IRAK4 and TRAF6 and provides the foundation for the TLR9-dependent activation of the IFN genes. The complex defined in this study represents an example of how the coupling of the signaling adaptor and effector kinase molecules together with the transcription factor regulate the processing of an extracellular signal to evoke its versatile downstream transcriptional events in a cell. Thus, we propose that this molecular complex may function as a cytoplasmic transductional-transcriptional processor.
The family of Toll-like receptors (TLRs) consists of germline-encoded receptors that recognize various pathogen-associated molecular patterns, and the activation of each receptor results in overlapping and distinct cellular responses in antigen-presenting cells, such as dendritic cells (DCs), macrophages, and B cells (1, 2). In fact, the outcome of the responses of antigen-presenting cells to a given TLR signaling is critical to determining the nature of the innate and adaptive immune responses, and the versatility of the response may be mediated at least in part by adaptor proteins that interact with TLRs to link the receptor activation to distinct downstream signaling pathways (3, 4). Whereas all TLRs use the adaptor MyD88 for the signaling, some TLRs also use additional adaptors, such as Toll/IL-1 receptor (TIR) domain-containing adaptor inducing IFN-β (TRIF; also known as TICAM-1), resulting in the activation of divergent signaling cascades (5, 6). Typically, TLR4 utilizes two adaptor pathways, the MyD88/TIRAP (MAL) and TRAM (TICAM-2)/TRIF (TICAM-1) pathways (4, 7-9); the latter pathway is known to be critical to the induction of the maturation of DCs (10, 11). Conversely, TLR9 subfamily members TLR9 and TLR7 transmit signals solely by using MyD88 (12), raising an interesting issue of how signaling “input” initiated by TLR9 family activation can be “processed” by means of MyD88 alone to activate diverse downstream signaling pathways to ensure the proper “output,” such as the induction of various cytokines and maturation in DCs.
Among many cytokines induced by TLR activation, type I IFNs (IFN-α/β) have been the new focus of attention in the context of linking the innate and adaptive immune responses (4, 13-15). Particularly interesting is the high-level induction of IFN-α/β upon the activation of TLR9 subfamily members in a small subset of DCs, termed plasmacytoid DCs (pDCs) (16-18). Such induction by the same ligands, namely, unmethylated DNAs, is not observed in conventional DCs or other immune cells that also express and respond to TLR9 (12). It has been well studied that TLR4 induces the MyD88-independent, low-level IFN-β gene transcription by IRF-3 via the TRAM (TICAM-2)/TRIF (TICAM-1) pathway (4). In contrast, the underlying mechanism remains elusive regarding how the TLR9 family members activate the MyD88-dependent IFN induction pathway, particularly in pDCs. Also unknown is how the IFN induction pathway operates in conjunction with the MyD88-dependent IL-1 receptor-associated kinase (IRAK)/tumor necrosis factor receptor-associated factor 6 (TRAF6)/NF-κB activation pathway that is critical for the induction of other cytokines (19-23).
We conjectured that the MyD88 signaling pathway can be dissected in terms of its cell-type-dependent and -independent aspects, both of which would be critical to our understanding of the specialized IFN induction pathway operating in pDCs. Thus, to gain insights into the latter aspect, we examined in this study the physical and functional interactions between MyD88 and two of the transcription factors of the IRF family (24), IRF-3 and IRF-7, in nonimmune cell types. We demonstrate the presence of a unique cytoplasmic complex that involves the MyD88 adaptor and IRF-7. In addition, we provide evidence that this complex also involves IRAK4 kinase and TRAF6 molecules and is critical for MyD88 signaling, which activates the IFN-α/β gene. We discuss this molecular complex, termed cytoplasmic transductional-transcriptional processor (CTTP), in the context of how the signal initiated by TLR activation is processed to evoke the downstream transcriptional events.
Materials and Methods
Plasmid Construction. The hemagglutinin (HA)-tagged mouse IRF-7 expression vector is described in ref. 25. The fragments of the full-length mouse MyD88 and the series of deletion mutants of MyD88 were obtained by RT-PCR and ligated into the pCXN2-FLAG vector. To construct the fluorescent proteintagged expression vectors, the fragments of IRF-3, IRF-7, TRIF (provided by T. Seya, Hokkaido University, Sapporo, Japan), and MyD88 cDNA were cloned into the XhoI and NotI sites of pCAGGS-YFP or pCAGGS-CFP. The expression vectors for FLAG-tagged TRAF6 and Myc-tagged IRAK-4 were kindly provided by J. Inoue (University of Tokyo) and H. Wesche (Tularik, South San Francisco, CA), respectively.
Confocal and Time-Lapse Microscopic Analysis. Cells were cultured on glass-bottom 35-mm tissue-culture dishes (Matsunami Glass, Osaka) and were transfected with expression vectors for fluorescent-protein-tagged fusion proteins. Confocal microscopic analysis was performed by using an Olympus FV-1000 confocal microscope equipped with a multi-Argon laser, from which 458- and 514-nm beams could emanate. To avoid the bleed-through of cyan fluorescent protein (CFP) fluorescence to the yellow fluorescent protein (YFP) channel, the sequential acquisition mode was used. For time-lapse imaging, the fluorescence imaging workstation consisted of an Olympus IX71 inverted microscope equipped with a cooled charge-coupled device (Cool-SNAPHQ, Roper Scientific, Trenton, NJ), excitation and emission filter wheels (MAC 5000, Ludl Electronic Products, Hawthorne, NY), and a Xenon 75-W light source, all controlled by metamorph software (Universal Imaging, Downingtown, PA). Images were recorded every minute for up to 2 h. Starting after 20 min, cells were stimulated with 1 μM CpG-A with 3% 1,2-dioleoyloxy-3-trimethylammonium-propane (DOTAP) (Boehringer Mannheim). Synthesized unmethylated CpG-A oligodeoxynucleotide (CpG-A ODN) was purchased from Hokkaido System Science (Sapporo, Japan). The sequence of CpG-A ODN is ggTGCATCGATGCAgggggG (16). Capital and lowercase letters indicate bases with phosphodiester- and phosphorothioate-modified backbones, respectively. Complexation of CpG-A with DOTAP was performed according to the manufacturer's recommendations.
Fluorescence Resonance Energy Transfer (FRET) Measurement. The method of sensitized FRET measurement is described in ref. 26. Fluorescent images were acquired sequentially through YFP, CFP, and FRET filter channels. Filter sets used were YFP (excitation, 500/25 nm; emission, 535/26 nm), CFP (excitation, 440/21 nm; emission, 480/30), and FRET (excitation, 440/21 nm; emission, 535/26 nm). An XF2034 (455 DRLP) dichroic mirror (Omega Optical, Brattleboro, VT) was used. Images were acquired by using the 4 × 4 binning mode and 100- to 200-ms integration times. The background was subtracted from raw images before FRET calculations. Corrected FRET (FRETC) was calculated on a pixel-by-pixel basis for the entire image by using the following equation: FRETC = FRET - (0.5 × CFP) - (0.02 × YFP), where FRET, YFP, and CFP correspond to background-subtracted images of cells coexpressing CFP and YFP acquired through the FRET, YFP, and CFP channels, respectively. The fractions of the bleed-through of CFP and YFP fluorescence were 0.5 and 0.02, respectively, through the FRET channel. FRETC images are presented in the pseudocolor mode.
Reporter Assay. HEK293T cells seeded on six-well plates were transiently cotransfected with 50 ng of reporter plasmid [pNF-κB-Luc (Stratagene) or p125-Luc (27)] together with expression plasmids for IRFs and MyD88 by lipofection. The total amounts of DNA were kept constant by supplementation with an empty vector (pEF-BOS). After 24 h of transfection, cells were harvested, and luciferase activity was measured by using the Dual-Luciferase reporter assay system (Promega). In all cases, the data were normalized for transfection efficiency by dividing firefly luciferase activity by Renilla luciferase activity.
Immunoprecipitation and Immunoblotting. Immunoprecipitation and immunoblotting were carried out as described in ref. 28. Monoclonal antibodies against the following epitopes were purchased from the indicated manufacturers: HA, Sigma-Aldrich; FLAG, Santa Cruz Biotechnology; and Myc, Santa Cruz Biotechnology.
Measurement of IFN-α Production by Splenic pDCs. The generation of IRAK4-/- mice is described in ref. 23. MyD88-/- mice were kindly provided by S. Akira (Osaka University, Osaka). Spleens from mutant or WT littermates were digested with 1 mg/ml collagenase A (Roche Biochemicals) and EDTA (20 mM, final concentration) and subjected to the negative selection of T and B cells with anti-CD5 and anti-CD19 antibodies (BD Biosciences) and anti-rat IgG-coated Dynabeads (Dynal Biotech, Oslo) according to the manufacturer's instructions. Recovered cells were incubated with anti-B220 and anti-CD11c antibodies (BD Biosciences), and B220+/CD11c+ pDCs were sorted by using a FACSDiva (BD Biosciences). Sorted cells were seeded into 96-well plates at 2 × 105 cells per ml and stimulated with 3 μM CpG-A or a synthetic single-stranded RNA, poly(uridylic acid) (Sigma; 5 μg/ml in 3% DOTAP), for 24 h. IFN-α induction levels then were determined by ELISA (PBL Biomedical Laboratories, Piscataway, NJ).
Results
Cytoplasmic Colocalization of IRF-7 and MyD88. In view of the fact that MyD88 and the IRF-3 and IRF-7 transcription factors all reside in the cytoplasm, we examined whether there is any intermolecular association among MyD88 and the IRF-3 and IRF-7 transcription factors. We first expressed IRF-7 and IRF-3, each tagged with the YFP (hereafter called IRF-7YFP and IRF-3YFP, respectively), in HEK293T cells and subjected these cells to confocal microscopic analysis. These IRF fusion proteins are functional as assessed by the activation of an IFN-β promoter-driven luciferase reporter gene (p125-Luc) (27) (data not shown). As shown in Fig. 1a, both IRFs were expressed in the cytoplasm; however, granular structures were observed for IRF-7YFP but not for IRF-3YFP. In addition, MyD88 tagged with CFP (MyD88CFP) also formed similar structures, whereas CFP-tagged adaptor TRIF was expressed diffusely (Fig. 1a). Interestingly, when IRF-7YFP was coexpressed with MyD88CFP, a significant fraction of YFP-IRF-7 merged with MyD88CFP (Fig. 1b). Conversely, the diffusely expressed IRF-3YFP did not merge with MyD88CFP.
Fig. 1.
Subcellular localization of IRFs and MyD88. (a) Confocal images of HEK293T cells transiently expressing IRF-3YFP, IRF-7YFP, MyD88CFP, or TRIFCFP. (b) Confocal images of HEK293T cells coexpressing MyD88CFP with IRF-3YFP or IRF-7YFP. (c) HeLa cells were transfected with expression plasmids for IRF-7YFP or IRF-3YFP and MyD88CFP. FRET, YFP, and CFP images were collected by using an inverted fluorescence microscope equipped with a cooled charge-coupled device camera. FRETC was determined as described in the text and presented in the pseudocolor mode as indicated below the photographs.
FRET Analysis of IRFs and MyD88. The above results suggest a previously unidentified molecular interaction between the adaptor MyD88 and the transcription factor IRF-7 in the cytoplasm. To gain further insights into the interaction between IRF-7 and MyD88, these tagged molecules were coexpressed in HeLa cells and subjected to FRET analysis. As shown in Fig. 1c, FRETC images revealed a strong energy transfer from MyD88CFP to IRF-7YFP, but not to IRF-3YFP, indicating that MyD88 and IRF-7 selectively form a cytoplasmic complex and are in direct contact with each other.
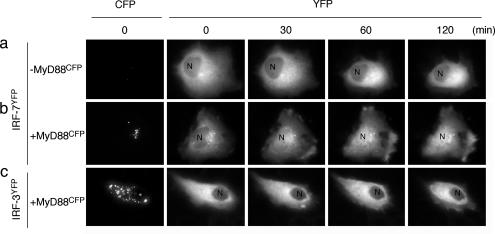
MyD88-Dependent Nuclear Translocation of IRF-7. We detected in HeLa cells the expression of mRNA for TLR9 that recognizes unmethylated DNA oligonucleotides (CpG ODNs) (data not shown). It has been known that A/D type CpG (CpG-A) ODN, containing a stretch of G residues at the 3′ end, strongly induces IFN-α/β genes upon TLR9 activation in pDCs (16, 29). We therefore asked whether the CpG-A ODN stimulation of HeLa cells causes the nuclear translocation of IRF-7. When IRF-7YFP was solely expressed and stimulated by CpG-A ODN, the nuclear translocation of IRF-7YFP was not observed (Fig. 2a). However, when MyD88CFP and IRF-7YFP were coexpressed, a notable fraction of IRF-7YFP underwent nuclear translocation after CpG-A ODN stimulation (Fig. 2b), whereas IRF-3YFP remained mostly in the cytoplasm (Fig. 2c). Thus, these results further lend support to the notion that IRF-7, not IRF-3, is activated by CpG DNA via the TLR9-dependent MyD88 signaling pathway. It is currently unknown how IRF-7 is activated by MyD88, but on the basis of previous works demonstrating the importance of IRF-7 phosphorylation for its activation (24, 25), we infer that MyD88 may recruit a kinase(s).
Fig. 2.
Nuclear translocation of IRF-7 induced by CpG-A ODN stimulation. HeLa cells expressing IRF-7YFP alone (a), MyD88CFP and IRF-7YFP (b), or MyD88CFP and IRF-3YFP (c) were placed on a time-lapse microscope and imaged every minute. Cells were stimulated with 1 μM CpG-A plus DOTAP and incubated for up to 2 h. Fluorescent images of YFP at the indicated periods are shown. CFP images indicate the expression level of MyD88. N, nucleus.
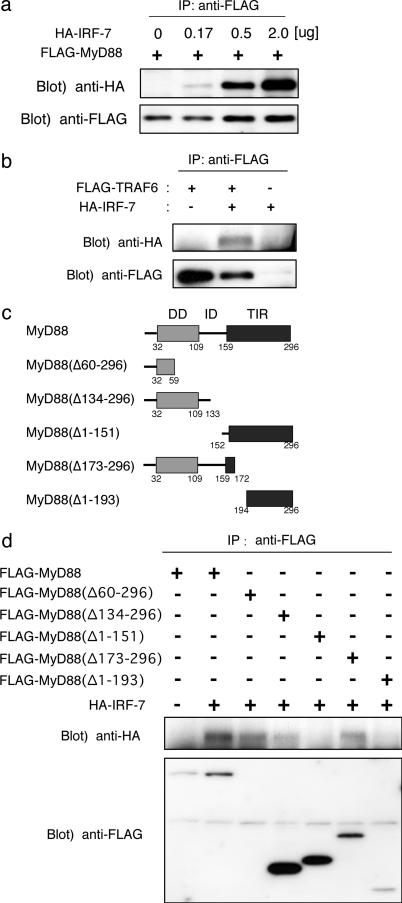
Association of IRF-7 with MyD88 and TRAF6. To study further the structure-function relationship between MyD88 and IRF-7, we examined the association of IRF-7 with MyD88 and another adaptor, TRAF6, which functions downstream of MyD88 (19). As shown in Fig. 3a, HA-tagged IRF-7 was coimmunoprecipitated with FLAG-tagged MyD88 when they were coexpressed in HEK293T cells. In addition, the coimmunoprecipitation of HA-tagged IRF-7 and FLAG-tagged TRAF6 was observed (Fig. 3b).
Fig. 3.
Association of IRF-7 with MyD88 and TRAF6. (a) Coimmunoprecipitation of IRF-7 with MyD88. HEK293T cells were transfected transiently with various concentrations of pEF-HA-IRF-7 (0, 0.17, 0.5, and 2.0 μg) together with pCXN2-FLAG-MyD88 (2.0 μg) in six-well plates, and cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody. IRF-7 was immunoblotted with a rabbit polyclonal anti-HA antibody. MyD88 was immunoblotted with anti-FLAG antibody. (b) HEK293T cells were transfected transiently with the indicated combinations of pEF-HA-IRF-7 (2.0 μg) and pME-FLAG-TRAF6 (2.0 μg), and cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody. (c) Schematic structures of the deletion mutants of MyD88. (d) HEK293T cells were transfected transiently with the indicated combinations of HA-tagged IRF-7 (2.0 μg) and FLAG-tagged full-length or deletion mutants of MyD88 (2.0 μg) and subjected to immunoprecipitation with the anti-FLAG antibody.
We further examined the association region of MyD88 that is responsible for its interaction with HA-tagged IRF-7 by generating deletion mutants, each tagged by the FLAG epitope (Fig. 3c). As shown in Fig. 3d, MyD88(Δ173-296), MyD88(Δ134-296), or MyD88(Δ60-296) lacking the TIR domain responsible for interaction with TLRs (21) still interacted with IRF-7, but MyD88(Δ1-193) and MyD88(Δ1-151), both lacking the “death domain” that interacts with IRAK1/4 kinases (20), failed to interact with IRF-7. We presume that the interaction with IRF-7 occurs at the N-terminal region of MyD88, because of the coimmunoprecipitation of IRF-7 and MyD88(Δ60-296) (Fig. 3d).
Activation of IFN Promoter by MyD88 and TRAF6. We next examined the effect of MyD88 and TRAF6 on activation of the IFN promoter by IRF-3 and IRF-7, by using a p125-luc; this promoter responds to both IRF-3 and IRF-7 in virally infected cells (24, 27, 30). As shown in Fig. 4a, the expression of IRF-7 alone in these cells did not activate the gene, but a strong activation was observed upon the coexpression of WT MyD88 in a dose-dependent manner. Conversely, IRF-3 that failed to interact with MyD88 also failed to activate the gene (Fig. 4a). Interestingly, the coexpression of IRF-7 and TRAF6 also caused the activation of the IFN promoter (Fig. 4b). Furthermore, the activation of the promoter by IRF-7 and MyD88 was enhanced further by TRAF6 in a dose-dependent manner, supporting the notion that IRF-7 is integrated fully to the complex of the MyD88-TRAF6-signaling pathway (Fig. 4c).
Fig. 4.
IRF-7 activation by MyD88 and TRAF6. HEK293T cells were cotransfected transiently with p125-Luc (50 ng) and the expression vector for the indicated combinations of MyD88 (10 ng), MyD88 mutants (10 ng), IRF-3 (0, 1, or 10 ng), IRF-7 (10 ng in b; 0, 1, or 10 ng in a and d), and TRAF6 [10 ng in b; 0, 1, 5 or 10 ng in c]. After 24 h of transfection, cells were harvested, and luciferase activity was measured.
MyD88 Mutants and Activation of IRF-7. To characterize further the role of MyD88 in IRF-7-mediated transcriptional activation, we assessed the effects of MyD88 mutants on the transcriptional activation of the p125-Luc reporter gene. MyD88(Δ134-296), which fails to interact with TLRs (21), still interacted with IRF-7 and activated the reporter gene even more strongly than full-length MyD88 in an IRF-7-dependent manner in this assay (Figs. 3d and 4d). Conversely, MyD88(Δ1-151) and MyD88(Δ1-193), both of which failed to interact with IRF-7, also failed to activate the reporter gene (Figs. 3d and 4d). These observations collectively suggest that the MyD88-IRF-7 complex formation is indeed critical for IRF-7 activation. It is worth noting that MyD88(Δ60-296), which still interacted with IRF-7 (Fig. 3d), failed to activate the IFN promoter (Fig. 4d), indicating that the deleted region of MyD88 is necessary for the associated IRF-7 to activate this transcription factor (see below).
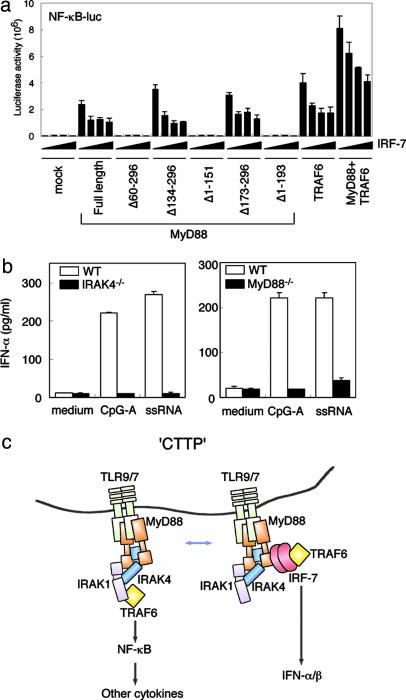
We also examined the effects of IRF-7 on MyD88- and TRAF6-dependent activation of NF-κB by using the pNF-κB-Luc reporter gene in HEK293T cells. As reported in refs. 21 and 23, the NF-κB reporter gene was activated by expressing MyD88 or TRAF6, and the activation was enhanced further by coexpressing these two adaptors (Fig. 5a). It is worth noting that the MyD88- and TRAF6-mediated reporter gene activations all were inhibited when IRF-7 was coexpressed (Fig. 5a). These results suggest that the MyD88-dependent IRF-7 and NF-κB pathways may operate in parallel downstream of TRAF6 (see Discussion).
Fig. 5.
Involvement of IRAK4 in the TLR9/7-dependent induction of IFN-α in pDCs. (a) The effect of IRF-7 expression on MyD88- or TRAF6-mediated NF-κB activation was monitored by using NF-κB-luc. HEK293T cells were cotransfected transiently with NF-κB-luc (50 ng) and the expression vectors for the indicated combinations of full-length MyD88, mutant MyD88 (10 ng), TRAF6 (100 ng), and IRF-7 (0, 1, 5, or 10 ng). Luciferase activity was measured 24 h after transfection. (b) IFN-α induction in pDCs stimulated by CpG-A or single-stranded RNA depends on MyD88 and IRAK4. Splenic pDCs (B220+CD11cint cells) were sorted and stimulated with 3 μM CpG-A ODN or 5 μg/ml poly(uridylic acid) plus DOTAP for 24 h. The concentration of IFN-α in culture supernatants was measured by ELISA. (c) Schematic illustration of the CTTP, providing a model of the TLR-dependent MyD88 signaling that potentially processes the signal to activate distinct transcription factors by means of the identified CTTP complex. For consistency with a previous report (19-21), IRAK1 also is included, although this molecule was not examined in this study. The complex may be in dynamic state depending on the expression levels of these molecules in this complex, as demonstrated in a. In normally growing cells, in which the IRF-7 level is very low, the CTTP complex may be shifted to the NF-κB pathway, but it may change in favor of the IRF-7 pathway when its expression level increases, owing to the capability of IRF-7 to interact with both MyD88 and TRAF6. Additionally, the CTTP complex may be regulated by their distinct compartmentalization in a cell. Although this model is consistent with our present data, it needs to be clarified further in the physiological context, for example, by examining DCs lacking IRF-7.
IRAK4 Kinase Participation in IRF-7 Pathway. The death domain of MyD88 is responsible for the interaction with IRAK4 kinase that is critical to MyD88 signaling (20, 23). In this regard, it is particularly interesting that MyD88(Δ60-296) failed to activate both the IFN and NF-κB reporter genes (Figs. 4d and 5a); the deleted region of MyD88 may be required for the interaction and/or activation of IRAK4, which is in turn required for the IFN gene activation pathway. We then examined the effect of IRAK4 on MyD88-IRF-7-mediated IFN promoter activation by transient expression analysis, but we found that coexpression of IRAK4 inhibited the expression level of MyD88 and IRF-7 proteins for unknown reasons (data not shown).
To this end, we examined splenic pDCs from IRAK4-deficient mice along with the pDCs from MyD88-deficient mice in terms of IFN-α production. As shown in Fig. 5b, IFN-α production induced by CpG-A ODN or the poly(uridylic acid) single-stranded RNA was abolished totally in mutant pDCs, collectively supporting the view that IRAK4 is the integral member not only for the MyD88-dependent induction pathway of other cytokines (such as IL-6 and TNF-α) but also for the complex of the MyD88-TRAF6-IRF-7 pathway activated by TLR9 and TLR7. Although it remains to be studied further how IRAK4 contributes to the activation of IRF-7, these observations collectively suggest the bifurcation of the IRF-7 and NF-κB pathways downstream of the TLR9/7-dependent MyD88-IRAK4-TRAF6 signaling and that these two pathways may interfere with each other (see Discussion).
Discussion
Our present work may have revealed an aspect of TLR9-MyD88 signaling that activates IRF-7 by means of TRAF6 and IRAK4, through the formation of a complex structure. Our results in toto suggest that the complex is critical to regulating the proper processing of the TLR signal to evoke downstream transcriptional events; we therefore propose to name this complex “the cytoplasmic transductional-transcriptional processor” (CTTP) (Fig. 5c). In analogy to computing terminology, in which the processor is central to converting the input to generate the output, CTTP may serve as a general cytoplasmic point of an organized array of signaling molecules and transcription factors that dynamically determines the specificity, strength, and longevity of the input signal to the output of transcriptional events. Our results suggest that the MyD88-dependent IRF-7 and NF-κB activation pathways diverge and may interfere with each other if the function of the CTTP complex is skewed, for example, by the overexpression of an integral molecule, as evidenced by the inhibition of the NF-κB reporter gene by IRF-7. It also is possible that the compartmentalization of CTTP in distinct vesicular structures in a given cell may affect down-stream gene expression machineries. In other words, the function of CTTP may be in dynamism in terms of its subcellular localization, and this issue of compartmentalization of the CTTP complex needs to be clarified further, particularly in DCs.
After the completion of this work, the association of IRF-7 with MyD88 and TRAF6 in HEK293T cells was reported (31), although the involvement of IRAK4 and the definition of CTTP complex have not been studied. Both our present study and this recent study leave unanswered the question of whether these experiments faithfully reflect the physiological pathways of MyD88 signaling, because the formation of the complexes was examined in transient expression assays by using nonimmune cells. Therefore, it remains to be determined as to what extent these interactions contribute to the TLR-dependent induction of IFN and other cytokines in physiological immune responses.
Acknowledgments
We thank Holger Wesche for IRAK4 expression vector; Jun-ichiro Inoue for FLAG-tagged TRAF6 expression vector; Tsukasa Seya for TRIF expression vector; Shizuo Akira for MyD88-deficient mice; and Edward Barsoumian, Hugh Rosen, and Jan Vilcek for invaluable advice. This work was supported in part by a grant for Advanced Research on Cancer and Grant-In-Aid 16017220 for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and grants from the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim, the Mochida Memorial Foundation and Pharmaceutical Research, the Nakajima Foundation, the Sumitomo Foundation, and the Mitsukoshi Foundation. H.N. was supported by an Ishidu Shun Memorial Scholarship.
Author contributions: K.H., H.Y., Y.O., A.T., and T.T. designed research; K.H., H.Y., T.M., H.N., N. Shimada, Y.O., and A.T. performed research; N. Suzuki and W.-C.Y. contributed new reagents/analytical tools; K.H., H.Y., Y.O., and T.T. analyzed data; and K.H., Y.O., and T.T. wrote the paper.
Abbreviations: CFP, cyan fluorescent protein; CTTP, cytoplasmic transductional-transcriptional processor; DC, dendritic cell; DOTAP, 1,2-dioleoyloxy-3-trimethylammonium-propane; FRET, fluorescence resonance energy transfer; FRETC, corrected FRET; IRAK, IL-1 receptor-associated kinase; IRF, IFN regulatory factor; pDC, plasmacytoid DC; TLR, Toll-like receptor; TRAF6, tumor necrosis factor receptor-associated factor 6; TRIF, Toll/IL-1 receptor domain-containing adaptor inducing IFN-β; YFP, yellow fluorescent protein.
References
- 1.Janeway, C. A., Jr., & Medzhitov, R. (2002) Annu. Rev. Immunol. 20, 197-216. [DOI] [PubMed] [Google Scholar]
- 2.Takeda, K., Kaisho, T. & Akira, S. (2003) Annu. Rev. Immunol. 21, 335-376. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill, L. A., Fitzgerald, K. A. & Bowie, A. G. (2003) Trends Immunol. 24, 286-290. [DOI] [PubMed] [Google Scholar]
- 4.Akira, S. & Takeda, K. (2004) Nat. Rev. Immunol. 4, 499-511. [DOI] [PubMed] [Google Scholar]
- 5.Oshiumi, H., Matsumoto, M., Funami, K., Akazawa, T. & Seya, T. (2003) Nat. Immunol. 4, 161-167. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., Takeuchi, O., Sugiyama, M., Okabe, M., Takeda, K. & Akira, S. (2003) Science 301, 640-643. [DOI] [PubMed] [Google Scholar]
- 7.Horng, T., Barton, G. M. & Medzhitov, R. (2001) Nat. Immunol. 2, 835-841. [DOI] [PubMed] [Google Scholar]
- 8.Hoebe, K., Du, X., Georgel, P., Janssen, E., Tabeta, K., Kim, S. O., Goode, J., Lin, P., Mann, N., Mudd, S., et al. (2003) Nature 424, 743-748. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald, K. A., Rowe, D. C., Barnes, B. J., Caffrey, D. R., Visintin, A., Latz, E., Monks, B., Pitha, P. M. & Golenbock, D. T. (2003) J. Exp. Med. 198, 1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaisho, T., Takeuchi, O., Kawai, T., Hoshino, K. & Akira, S. (2001) J. Immunol. 166, 5688-5694. [DOI] [PubMed] [Google Scholar]
- 11.Hoebe, K., Janssen, E. M., Kim, S. O., Alexopoulou, L., Flavell, R. A., Han, J. & Beutler, B. (2003) Nat. Immunol. 4, 1223-1229. [DOI] [PubMed] [Google Scholar]
- 12.Wagner, H. (2004) Trends Immunol. 25, 381-386. [DOI] [PubMed] [Google Scholar]
- 13.Biron, C. A. (2001) Immunity 14, 661-664. [DOI] [PubMed] [Google Scholar]
- 14.Hertzog, P. J., O'Neill, L. A. & Hamilton, J. A. (2003) Trends Immunol. 24, 534-539. [DOI] [PubMed] [Google Scholar]
- 15.Honda, K., Sakaguchi, S., Nakajima, C., Watanabe, A., Yanai, H., Matsumoto, M., Ohteki, T., Kaisho, T., Takaoka, A., Akira, S., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 10872-10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemmi, H., Kaisho, T., Takeda, K. & Akira, S. (2003) J. Immunol. 170, 3059-3064. [DOI] [PubMed] [Google Scholar]
- 17.Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S. & Reis e Sousa, C. (2004) Science 303, 1529-1531. [DOI] [PubMed] [Google Scholar]
- 18.Krug, A., Luker, G. D., Barchet, W., Leib, D. A., Akira, S. & Colonna, M. (2004) Blood 103, 1433-1437. [DOI] [PubMed] [Google Scholar]
- 19.Cao, Z., Xiong, J., Takeuchi, M., Kurama, T. & Goeddel, D. V. (1996) Nature 383, 443-446. [DOI] [PubMed] [Google Scholar]
- 20.Wesche, H., Henzel, W. J., Shillinglaw, W., Li, S. & Cao, Z. (1997) Immunity 7, 837-847. [DOI] [PubMed] [Google Scholar]
- 21.Medzhitov, R., Preston-Hurlburt, P., Kopp, E., Stadlen, A., Chen, C., Ghosh, S. & Janeway, C. A., Jr. (1998) Mol. Cell 2, 253-258. [DOI] [PubMed] [Google Scholar]
- 22.Li, S., Strelow, A., Fontana, E. J. & Wesche, H. (2002) Proc. Natl. Acad. Sci. USA 99, 5567-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki, N., Suzuki, S., Duncan, G. S., Millar, D. G., Wada, T., Mirtsos, C., Takada, H., Wakeham, A., Itie, A., Li, S., et al. (2002) Nature 416, 750-756. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi, T., Ogasawara, K., Takaoka, A. & Tanaka, N. (2001) Annu. Rev. Immunol. 19, 623-655. [DOI] [PubMed] [Google Scholar]
- 25.Sato, M., Hata, N., Asagiri, M., Nakaya, T., Taniguchi, T. & Tanaka, N. (1998) FEBS Lett. 441, 106-110. [DOI] [PubMed] [Google Scholar]
- 26.Sorkin, A., McClure, M., Huang, F. & Carter, R. (2000) Curr. Biol. 10, 1395-1398. [DOI] [PubMed] [Google Scholar]
- 27.Sato, M., Suemori, H., Hata, N., Asagiri, M., Ogasawara, K., Nakao, K., Nakaya, T., Katsuki, M., Noguchi, S., Tanaka, N. & Taniguchi, T. (2000) Immunity 13, 539-548. [DOI] [PubMed] [Google Scholar]
- 28.Takaoka, A., Mitani, Y., Suemori, H., Sato, M., Yokochi, T., Noguchi, S., Tanaka, N. & Taniguchi, T. (2000) Science 288, 2357-2360. [DOI] [PubMed] [Google Scholar]
- 29.Verthelyi, D. & Zeuner, R. A. (2003) Trends Immunol. 24, 519-522. [DOI] [PubMed] [Google Scholar]
- 30.Lin, R., Genin, P., Mamane, Y. & Hiscott, J. (2000) Mol. Cell. Biol. 20, 6342-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai, T., Sato, S., Ishii, K. J., Coban, C., Hemmi, H., Yamamoto, M., Terai, K., Matsuda, M., Inoue, J. I., Uematsu, S., et al. (2004) Nat. Immunol. 5, 1061-1068. [DOI] [PubMed] [Google Scholar]