Version Changes
Revised. Amendments from Version 1
In this revised version, we have 1) included a brief description of our MiSeq sequencing approach, 2) provided a table (Table 1) summarizing demographic, respiratory/gastrointestinal signs/symptoms, and virological information of the four patients in whom HboV was detected, and 3) included a discussion covering general issues related to our report as per reviewers’ suggestions.
Abstract
As part of an ongoing effort to generate complete genome sequences of hand, foot and mouth disease-causing enteroviruses directly from clinical specimens, two complete coding sequences and two partial genomic sequences of human bocavirus 1 (n=3) and 2 (n=1) were co-amplified and sequenced, representing the first genome sequences of human bocaviruses from Vietnam. The sequences may aid future study aiming at understanding the evolution of the virus.
Keywords: human bocavirus, HBoV, HFMD, Vietnam
Introduction
Human bocaviruses (HBoV) are non-enveloped, single stranded DNA viruses of the family Parvoviridae, subfamily Parvovirininae and genus Bocaparvovirus. The virus genome is ~5.3 Kb in length. HBoV-1 was first discovered in 2005 1. Since then three additional HBoV species, namely HBoV-2, HBoV-3 and HBoV-4, have been discovered 2, 4. While the clinical significance of HBoV remains unknown, worldwide their prevalence in respiratory/gastrointestinal tracts varies between 0–26% 5, 6. In Vietnam, the reported prevalence of HBoV was 2–17% 7– 10. Currently, there is relatively limited sequence information, especially at genome-wide level, of HBoV from Vietnam, although such knowledge may be essential for the development of sensitive, specific diagnostic PCR for the local viral strains, and may aid future investigation documenting the circulation and spread of the viruses at global scale.
Herein we report the recovery of two complete coding sequences (CDS) and two partial genomic sequences of HBoV from swabs of Vietnamese children enrolled in our ongoing hand, foot and mouth disease (HFMD) research program in Ho Chi Minh City. The research program aims to look at various disease aspects, including pathogen evolution and its potential implication for vaccine development and implementation.
Methods and results
Whole-genome sequencing of the dominant pathogens (including coxsackievirus A6 (CV-A6), CV-A10 and CV-A16) were performed on 296 RT-PCR positive swabs using an in-house MiSeq-based approach 11. In brief, 110 µl of selected swabs were centrifuged at 13,500 rpm for 10 minutes to remove host cells or large cellular components. After DNAse treatment, viral nucleic acid (NA) was then isolated from 100 µl of supernatant using QIAamp viral RNA kit (QIAgen GmbH, Hilden, Germany), and recovered in 50 µl of elution buffer (provided with the kit). Ten microliter of the isolated NA was subjected to cDNA synthesis using Super Script III kit (Invitrogen, Carlsbad, CA, USA) and FR26RV-Endoh primer (primer sequences can be found elsewhere 11). The cDNA was then converted to double-stranded DNA using exo-Klenow (Invitrogen), and subsequently pre-amplified using Platinum PCR supermix (Invitrogen) and FR20RV primer 11. PCR product was then purified and subjected to library preparation using Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, USA) and was finally sequenced using MiSeq reagent kits (Illumina) in an Illumina MiSeq platform (Illumina) 11.
After reference-based mapping 11 to generate the complete genome sequences of the targeted enteroviruses using Geneious software v 8.1.5 (Biomatters, Ltd, Auckland, New Zealand), the remaining reads were then subjected to publicly available metagenomic pipelines; Taxonomer 12 and Sequence-based Ultra-Rapid Pathogen Identification (SURPI) 13 to explore the contents of non-enteroviral sequences in the tested swabs. Evidence of bocavirus sequences were found in four swabs (including 3 throat- and 1 rectal swabs). A reference-based mapping approach using Geneious software (Biomatters) 11 was then employed to recover the HBoV genomes from the corresponding dataset. Subsequently, 2 CDS (1 from a throat swab with 4925 bp in length and the other from a rectal swab with 4898 bp; i.e. over 90% of genome coverage) were successfully assembled with a mean coverage of 1,922 and 3,745, respectively. In the other two datasets from the remaining 2 swabs only partial genomic sequences of HBoV, each with 2870 bp in length and a mean coverage of 15.4 and 448.7, were recovered.
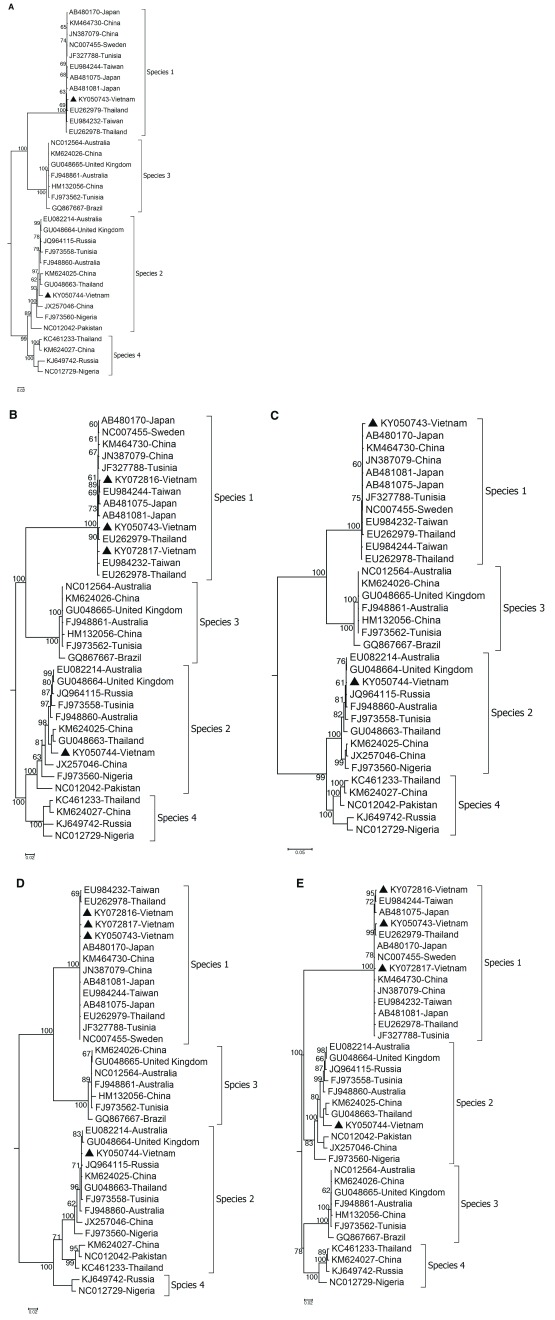
Subsequent sequence alignment and phylogenetic analysis using MUSCLE 14 and Neighbor-joining available in Geneious (Biomatters), respectively ( Figure 1) revealed that all 3 Vietnamese HBoV recovered from the throat swabs belonged to HBoV-1 and had >98% of sequence similarity at nucleotide level with other HBoV-1. The other belonged to HBoV-2 and had a close relatedness with a Thai strain CU54TH (GU048663) with a sequence similarity of 97.3% ( Figure 1). Similar results were obtained when the analyses were done for 3 individual open reading frames (ORF1, ORF2, and ORF3) of the virus genome ( Figure 1).
Figure 1. Phylogenetic trees showing the relationship between the Vietnamese bocaviruses and representative worldwide circulating strains.
A) Neighbor-joining phylogeny of CDS; B) Neighbor-joining phylogeny of partial genomic sequences spanning the region from nucleotide 1897 to 4856 of the HBoV genomes; C) Neighbor-joining phylogeny of ORF1 encoding NS1 protein; D) Neighbor-joining phylogeny of ORF2 encoding NP1 protein; E) Neighbor-joining phylogeny of ORF3 encoding VP1 and VP2 proteins. Trees were reconstructed using Neighbor-joining method available in Geneious with Tamura-Nei nucleotide substitution model, and support for individual nodes was assessed using a bootstrap procedure (1000 replicates). Bootstrap values greater than 60% are shown on the branch nodes. The Vietnamese strains from this study are indicated by solid triangles. The scale bars indicate the number of nucleotide substitution.
All the four HFMD patients (including 3 CV-A6 and 1 CV-A12, Table 1) in whom HBoV was detected had mild HFMD, and were enrolled in November 2013 – March 2014. Three had vomiting, and two presented with runny nose and cough ( Table 1).
Table 1. Demographic, clinical information, and details of MiSeq sequencing results.
| Patient | Gender | Age
(year) |
Admission
date |
HFMD grade | Respiratory/gastrointestinal signs/
symptoms |
MiSeq sequencing results | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Enterovirus | Bocavirus | ||||||||||||||
| Runny
nose |
Cough | Vomiting | Diahrrea | Serotype | Full
genome |
Mean
coverage |
Species | Complete
CDS |
Mean
Coverage |
Genbank
accession |
||||||
| 1 | Male | 0.9 | 11/3/2014 | 2a | Yes | Yes | Yes | No | Throat swab | CV-A6 | Yes | 6,024 | 1 | Yes | 1,922 | KY050743 |
| 2 | Male | 0.4 | 5/11/2013 | 2a | No | No | No | No | Rectal swab | CV-A6 | Yes | 12,303 | 2 | Yes | 3,745 | KY050744 |
| 3 | Male | 1.4 | 14/11/2013 | 1 | No | No | Yes | No | Throat swab | CV-A12 | Yes | 2,841 | 1 | No | 15.4 | KY072816 |
| 4 | Female | 0.8 | 4/12/2013 | 1 | Yes | Yes | Yes | No | Throat swab | CV-A6 | Yes | 2,493 | 1 | No | 448.7 | KY072817 |
Discussion
Herein we reported for the first time 2 complete CDS alongside two other partial genomics sequences of HBoV from Vietnam. Phylogenetically, the four HBoVs from Vietnam were closely related to other HBoV strains sampled from various countries worldwide, reflecting a wide distribution of these HBoV lineages at global scales.
All three HBoV detected in throat swabs belong to species 1, while the remaining virus detected in rectal swab was HBoV-2. This is in line with previous reports regarding the frequent detection of HBoV-1 and HBoV-2 in throat- and rectal swab, respectively 5, 6, 15– 18, albeit our sample size was small. Likewise, all the four HFMD patients in whom HBoVs were found were enrolled into our HFMD study during the seasonal peak of HBoV in southern Vietnam 8.
Although the pathogenic potential of HBoV infections remains unknown, clinical signs/symptoms such as vomiting, runny nose and cough were also commonly recorded among HFMD patients in previous reports 19– 21. HBoV has commonly been co-detected with other pathogens in respiratory and gastrointestinal tracts 5, 10, 16– 18. It was also previously detected in fecal samples of HFMD patients from Thailand 22. Clearly, further research is needed to ascribe the contribution of coinfections to clinical manifestation and pathogenesis of HFMD. Of note, previous reports showed that there might be an association between coinfections with other viral pathogens such as norovirus and rotavirus and clinical severity of HFMD patients 23.
In conclusion, to the best of our knowledge, we are the first to report the complete CDS of HBoVs from Vietnam. The contribution of HBoV to clinical manifestation of HFMD requires further research.
Data availability
Nucleotide sequence accession numbers: the HBoV sequences have been submitted to Genbank ( https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers KY050743, KY050744, KY072816 and KY072817.
Consent
The clinical samples used in this study were derived from an on-going HFMD study in three referral hospitals in Ho Chi Minh city, Vietnam. The study was reviewed and approved by the local Institutional Review Boards and the Oxford Tropical Research Ethics Committee (OxTREC), University of Oxford, Oxford, United Kingdom. Written informed consent was obtained from parent or legal guardian of each participant.
Acknowledgements
We thank Ms Le Kim Thanh from Oxford University Clinical Research Unit in Ho Chi Minh City, Vietnam for her logistic assistance and Truong Duy for his help with the resolution of the figure of the phylogenetic trees. We are indebted to patients and their parents for their participation in this study, and all the nursing and medical staff at the at Children's Hospital 1, Children's Hospital 2 and the Hospital for Tropical Diseases who provided care for the patients and helped collect clinical data.
Funding Statement
This work was supported by the Wellcome Trust [101104/Z/13/Z], [106680/B/14/Z].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 3 approved]
References
- 1. Allander T, Tammi MT, Eriksson M, et al. : Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 2005;102(36):12891–6. 10.1073/pnas.0504666102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kapoor A, Simmonds P, Slikas E, et al. : Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J Infect Dis. 2010;201(11):1633–43. 10.1086/652416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kapoor A, Slikas E, Simmonds P, et al. : A newly identified bocavirus species in human stool. J Infect Dis. 2009;199(2):196–200. 10.1086/595831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arthur JL, Higgins GD, Davidson GP, et al. : A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 2009;5(4):e1000391. 10.1371/journal.ppat.1000391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Broccolo F, Falcone V, Esposito S, et al. : Human bocaviruses: Possible etiologic role in respiratory infection. J Clin Virol. 2015;72:75–81. 10.1016/j.jcv.2015.09.008 [DOI] [PubMed] [Google Scholar]
- 6. Ong DS, Schuurman R, Heikens E: Human bocavirus in stool: A true pathogen or an innocent bystander? J Clin Virol. 2016;74:45–9. 10.1016/j.jcv.2015.11.027 [DOI] [PubMed] [Google Scholar]
- 7. Yoshida LM, Suzuki M, Yamamoto T, et al. : Viral pathogens associated with acute respiratory infections in central vietnamese children. Pediatr Infect Dis J. 2010;29(1):75–7. 10.1097/INF.0b013e3181af61e9 [DOI] [PubMed] [Google Scholar]
- 8. Tran DN, Nguyen TQ, Nguyen TA, et al. : Human bocavirus in children with acute respiratory infections in Vietnam. J Med Virol. 2014;86(6):988–94. 10.1002/jmv.23789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tran DN, Trinh QD, Pham NT, et al. : Clinical and epidemiological characteristics of acute respiratory virus infections in Vietnamese children. Epidemiol Infect. 2016;144(3):527–36. 10.1017/S095026881500134X [DOI] [PubMed] [Google Scholar]
- 10. Do AH, van Doorn HR, Nghiem MN, et al. : Viral etiologies of acute respiratory infections among hospitalized Vietnamese children in Ho Chi Minh City, 2004–2008. PLoS One. 2011;6(3):e18176. 10.1371/journal.pone.0018176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nguyen AT, Tran TT, Hoang VM, et al. : Development and evaluation of a non-ribosomal random PCR and next-generation sequencing based assay for detection and sequencing of hand, foot and mouth disease pathogens. Virol J. 2016;13:125. 10.1186/s12985-016-0580-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flygare S, Simmon K, Miller C, et al. : Taxonomer: an interactive metagenomics analysis portal for universal pathogen detection and host mRNA expression profiling. Genome Biol. 2016;17(1):111. 10.1186/s13059-016-0969-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naccache SN, Federman S, Veeraraghavan N, et al. : A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24(7):1180–92. 10.1101/gr.171934.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Li Y, Liu J, et al. : Genetic characterization of human bocavirus among children with severe acute respiratory infection in China. J Infect. 2016;73(2):155–63. 10.1016/j.jinf.2016.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schildgen O: Human bocavirus: lessons learned to date. Pathogens. 2013;2(1):1–12. 10.3390/pathogens2010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Risku M, Kätkä M, Lappalainen S, et al. : Human bocavirus types 1, 2 and 3 in acute gastroenteritis of childhood. Acta Paediatr. 2012;101(9):e405–10. 10.1111/j.1651-2227.2012.02727.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khamrin P, Malasao R, Chaimongkol N, et al. : Circulating of human bocavirus 1, 2, 3, and 4 in pediatric patients with acute gastroenteritis in Thailand. Infect Genet Evol. 2012;12(3):565–9. 10.1016/j.meegid.2012.01.025 [DOI] [PubMed] [Google Scholar]
- 19. Mirand A, le Sage FV, Pereira B, et al. : Ambulatory Pediatric Surveillance of Hand, Foot and Mouth Disease as Signal of an Outbreak of Coxsackievirus A6 Infections, France, 2014–2015. Emerg Infect Dis. 2016;22(11):1884–1893. 10.3201/eid2211.160590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee MK, Chan PK, Ho II, et al. : Enterovirus infection among patients admitted to hospital in Hong Kong in 2010: epidemiology, clinical characteristics, and importance of molecular diagnosis. J Med Virol. 2013;85(10):1811–7. 10.1002/jmv.23663 [DOI] [PubMed] [Google Scholar]
- 21. Tu PV, Thao NT, Perera D, et al. : Epidemiologic and virologic investigation of hand, foot, and mouth disease, southern Vietnam, 2005. Emerg Infect Dis. 2007;13(11):1733–41. 10.3201/eid1311.070632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Linsuwanon P, Poovorawan Y, Li L, et al. : The Fecal Virome of Children with Hand, Foot, and Mouth Disease that Tested PCR Negative for Pathogenic Enteroviruses. PLoS One. 2015;10(8):e0135573. 10.1371/journal.pone.0135573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu LJ, Xu HM, Li XJ, et al. : Co-detection in the pathogenesis of severe hand-foot-mouth disease. Arch Virol. 2012;157(11):2219–22. 10.1007/s00705-012-1396-6 [DOI] [PMC free article] [PubMed] [Google Scholar]