Summary
Pancreatic cancer is a deadly malignancy that lacks effective therapeutics. We previously reported that oncogenic Kras induced the redox master regulator Nrf2/Nfe2l2 to stimulate pancreatic and lung cancer initiation. Here, we show that NRF2 is necessary to maintain pancreatic cancer proliferation by regulating mRNA translation. Specifically, loss of NRF2 led to defects in autocrine EGFR signaling and oxidation of specific translational regulatory proteins, resulting in impaired cap-dependent and cap-independent mRNA translation in pancreatic cancer cells. Combined targeting of the EGFR effector AKT and the glutathione antioxidant pathway mimicked Nrf2 ablation to potently inhibit pancreatic cancer ex vivo and in vivo, representing a promising synthetic lethal strategy for treating the disease.
Introduction
Pancreatic ductal adenocarcinoma (PDA) is the fourth leading cause of cancer-related death in the US, with incidence nearly matching mortality. This is reflected by a short median survival of 6 months and a 5-year survival of <5% (Siegel et al., 2015). The poor prognosis of PDA relates to the advanced disease stage at the time of diagnosis and to its profound resistance to therapies (Jemal et al., 2009). PDA exhibits a wide range of genetic and epigenetic alterations including a high frequency (90–95%) of activating KRAS mutations and inactivation of the tumor suppressors TP53, P16/INK4A and SMAD4 (Jones et al., 2008). We previously found that oncogenic Kras expression induced an important regulator of redox control, the transcription factor Nuclear factor erythroid-derived 2-like 2, Nfe2l2/Nrf2 (DeNicola et al., 2011). In response to oxidative stress, Nrf2 controls the fate of cells through transcriptional upregulation of antioxidant response element-bearing genes (Hayes and Dinkova-Kostova, 2014). In the context of oncogenic Kras, Nrf2 promotes pancreatic intraepithelial neoplasia by stimulating proliferation and suppressing senescence. Thus, Nrf2 is a key player in PDA initiation through the maintenance of redox homeostasis.
Although oxidative stress may disrupt biological functions, redox reactions in a cell are often tightly regulated and play essential physiological roles (Trachootham et al., 2008). The chemical properties of cysteine thiol groups render this amino acid exquisitely sensitive to changes in cellular levels of reactive oxygen (ROS) or nitrogen (RNS) species. Certain cysteine thiols can act as redox switches, since oxidative modifications may impact properties such as catalytic activity or conformation of a protein (Barford, 2004). Although the general concepts of redox signaling have been established, the identity and function of many regulatory switches remain unclear, particularly in the context of cancer. Given the elevated levels of Nrf2 in PDA, and the ability of Nrf2 to potently lower ROS, we hypothesized that such redox switches may function as Nrf2 downstream effectors to support PDA progression.
The cellular response to stress involves regulatory changes in many processes, including transcription, mRNA processing, and translation. Historically, increased cancer cell proliferation has been shown to require increased rates of protein synthesis and ribosome number (Johnson et al., 1975; Zetterberg et al., 1995). Mutations that deregulate mRNA translation are common events in human cancers (Bilanges and Stokoe, 2007), while enforced expression of certain translation factors transforms rodent fibroblasts and promotes tumorigenesis (Wendel et al., 2004). Small molecules that target these pathways suppress mRNA translation and have antitumor effects (Choo and Blenis, 2009). Thus, deregulation of translation is an important step in oncogenic transformation and tumor maintenance. Here we show that Nrf2 directly stimulates mRNA translation by maintaining the reduced state of specific cysteine residues in multiple proteins that participate in translational regulation. In parallel, redox regulation by Nrf2 also promotes EGFR autocrine signaling through AKT in KRAS mutant cells to fuel cap-dependent translation initiation. These functions converge to promote global protein synthesis in PDA. Combined inhibition of AKT signaling and synthesis of glutathione, a vital intracellular antioxidant, synergistically hampered the survival of PDA cells in vitro and in vivo, presenting an opportunity for therapeutic intervention.
Results
Nrf2 is a critical biological dependency in pancreatic cancer
The organoid culture system supports the proliferation of normal and malignant primary pancreatic ductal cells of both mouse and human origins (Boj et al., 2015). NRF2 protein was up-regulated in human tumor (hT) organoids compared to normal counterparts (hN) (Figure S1A). While hN organoids tolerated NRF2 knockdown, this was detrimental in most hT organoids tested (3/5) (Figure 1A). Upon NRF2 knockdown (Figure S1B), the two surviving hT organoids exhibited substantial elevation of ROS (Figure 1B) and decreased cell proliferation that was largely mitigated by the antioxidant N-acetylcysteine (NAC) (Figure S1C). Similar proliferative defects were also observed in the KRAS and TP53 mutant Suit2 PDA cell line (Moore et al., 2001) (Figure S1C). When transplanted into athymic nude mice, shNRF2 Suit2 cells gave rise to smaller tumors (Figure 1C, top). Likewise, doxycyline-induced knockdown of NRF2 (Figure S1D) in established, xenografted Suit2 tumors resulted in a significant decrease in tumor growth (Figure 1C, bottom). The level of DNA damage did not correlate with NRF2 expression in these tumors (Figure S1E), nor in Suit2 cells and human T organoids in culture (Figure S1F). These data suggest that NRF2 is essential for PDA tumor maintenance through mechanisms other than prevention of ROS-induced DNA damage.
Figure 1. Nrf2 controls redox homeostasis, initiation and maintenance of pancreatic tumors.
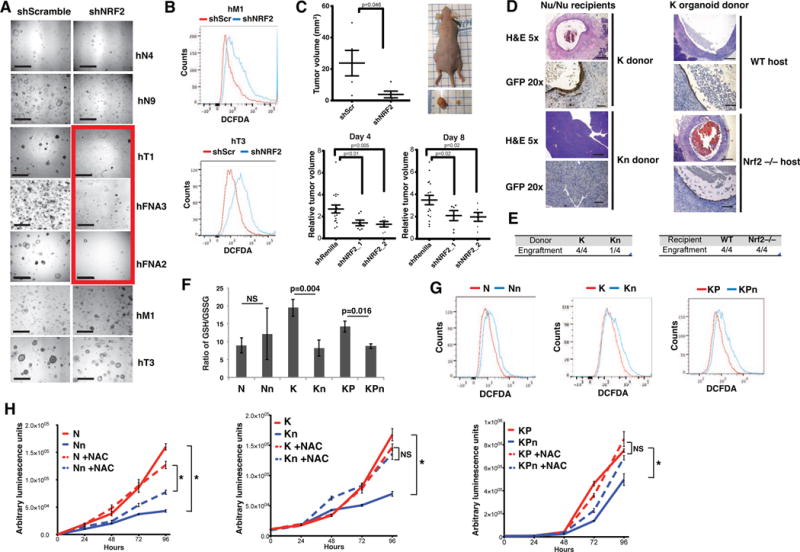
(A) Human normal (N) and tumor (T) (T= primary tumor, FNA=fine needle biopsy, M=metastasis) organoid lines following knockdown with control (shScramble) or shNRF2 hairpins. Scale bar = 1mm. Red box: human T organoid lines that did not tolerate NRF2 loss.
(B) ROS levels in human T organoids measured with CM-H2DCFDA.
(C) Tumor volume of subcutaneously xenografted Suit2 cells bearing control or shNRF2 hairpins. Top panel, constitutive knockdown (n=5) on day 28. Left flank: control tumor; right flank: shNRF2 tumor. Bottom panel, inducible knockdown 21 days post transplantation followed by doxycycline treatment for 4 or 8 days (n=17 control, n=6 mice per shNRF2 hairpin, Student’s t-test.)
(D) Hematoxylin and eosin (H&E), and immunohistochemistry (IHC) of orthotopic transplants of GFP-expressing K and Kn organoids into athymic nude mice 2 weeks after transplantation (left). H&E and IHC of orthotopic, syngeneic transplants of K organoids into B6 wildtype or Nrf2−/− mice 2 weeks after transplantation (right). 5X scale bar = 500 μm, 20X scale bar = 100 μm.
(E) Engraftment rates of orthotopic transplants.
(F) Ratio of reduced (GSH) to oxidized (GSSG) glutathione in murine organoids (n=3, Student’s t-test).
(G) ROS levels in murine organoids measured with CM-H2DCFDA, representative from 3 biological replicates.
(H) Proliferation of N, K and KP organoids grown in reduced media with or without 1.25 mM NAC. Data are mean +/− s.e.m. (n=5). *= p<0.05, NS= p>0.05, Student’s t-test.
See also Figure S1.
Given the difficulty of targeting transcription factors therapeutically, we sought to comprehensively characterize the mechanisms used by NRF2 to support PDA such that tractable approaches to counter the effects of NRF2 may be developed. To this end, we used organoids established from genetically engineered mouse models, which mimic the pathophysiological features of the human disease in a uniform genetic background (Hingorani et al., 2005). Murine pancreatic organoid cultures were grown from ductal isolates of normal “N”, KrasG12D/+ “K”, and KrasG12D/+;p53R172H/+ “KP” tissues, of either Nrf2-wild type or Nrf2-deficient “n” backgrounds. The resulting organoids appeared morphologically similar as hollow structures with a single cell layer (Figure S1G). While Nrf2-deficient organoids can be propagated in culture, orthotopic engraftment of K organoids in athymic nude mice was largely impaired in the absence of Nrf2 (Figures 1D and 1E). This defect was cell autonomous since syngeneic engraftment of K organoids was similar between a C57B6/J host and one that was deficient of Nrf2 (Figures 1D and 1E).
Pancreatic organoids are typically maintained in complete media, which contains various growth factors including EGF and NAC. Oncogenic Kras up-regulated Nrf2 and its downstream target gene Nqo1 in K and KP organoids (Figures S1H and S1I). The induction of Nrf2 and Nqo1 in KP organoids was more prominent upon exclusion of both EGF and NAC from the media (reduced) (Figures S1J and S1K). While removal of EGF and NAC limited the passaging capacity of N organoids, they were dispensable for K and KP organoids (Figure S1L). Thus, all organoids described hereafter were cultured and passaged in complete media, while experiments were all conducted in reduced media.
Oncogenic Kras activation in K and KP organoids resulted in a significant increase in intracellular glutathione (GSH) (Figure S1M), and decreased levels of ROS (Figure S1N), features reverted upon Nrf2 ablation (Figures S1M, 1F and 1G). Notably, no increases were observed in the levels of RNS (Figure S1O), or in mitochondrial superoxides (Figure S1P), either upon chronic ablation or acute knockdown of NRF2 in murine and human T organoids, respectively. This indicates that NRF2 specifically modulates cytoplasmic ROS in pancreatic ductal cells. Deficiency of Nrf2 impeded proliferation of N, K and KP organoids in culture, and was largely corrected by NAC (Figure 1H). KPn organoids did not exhibit elevated DNA damage response (Figure S1Q). Therefore, Nrf2 regulates redox homeostasis and cell proliferation in Kras mutant pancreatic epithelial cells, likely through a DNA damage-independent mechanism.
Global cysteine proteomics reveal redox-dependent regulation of the translational machinery
Cysteine residues are the most abundant cellular thiol. Alterations in cellular redox levels may cause reversible modifications on specific cysteine residues and alter the activity of their corresponding proteins (Paulsen and Carroll, 2010). Thus, reactive-cysteine-containing proteins are potential candidates to carry out redox-sensitive effector functions of Nrf2. To decipher changes in the cysteine proteome, we devised a highly sensitive proteomic method using a selectively cleavable cysteine-reactive (ICAT) reagent to enrich for and identify reduced cysteine-containing peptides (Sethuraman et al., 2004). This was combined with a parallel analysis of the total proteome by employing amine-reactive isobaric tags for relative and absolute quantification (iTRAQ) (Ross et al., 2004) (Figure 2A). By normalizing cysteine peptide changes to that of the total proteome, oxidative alterations in the cysteine proteome were quantified independent of protein expression differences, an approach not applied previously in cysteine proteomics. To establish this method, murine T organoids were treated with an inhibitor of GSH synthesis, buthionine sulfoximine (BSO). In this experiment, 88% of ICAT-enriched unique peptides contained cysteines, indicating the specificity of the technique. From a total of 4,089 proteins identified and quantified, 4,805 unique cysteine-containing peptides were detected. Upon BSO treatment, 1,192 unique cysteine-containing peptides were oxidized. Notably, cysteine residues oxidized by BSO included a previously reported redox sensitive protein, Pkm2 (Anastasiou et al., 2011). The full list of oxidized peptides is shown in Table S1.
Figure 2. Nrf2 deficiency induces cysteine oxidation of components of the translation machinery.
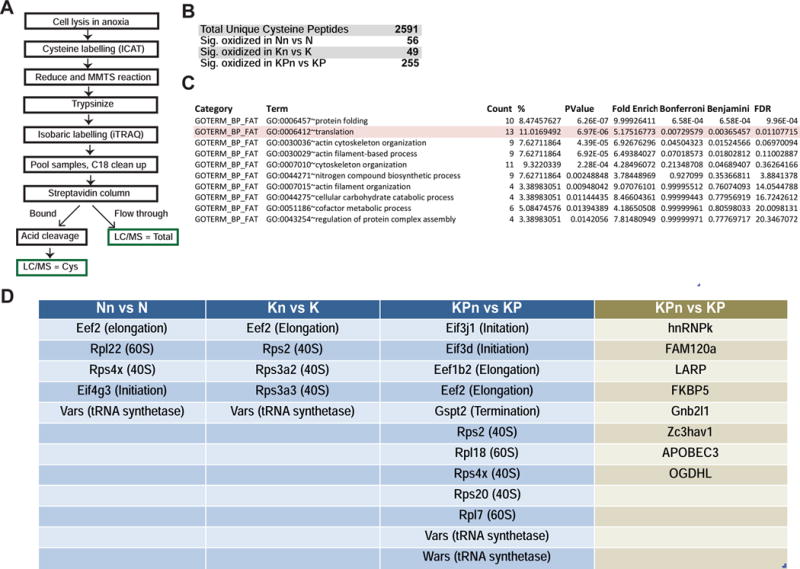
(A) Schematic of cysteine proteomics approach.
(B) Significant cysteine peptide changes identified in N, K and KP organoids compared to Nrf2-null counterparts. p<0.05, Welch’s t-test.
(C) DAVID gene ontology analysis for pathway enrichment of significantly oxidized peptides in KPn organoids compared to KP organoids.
(D) Proteins in the core translation machinery (blue) and regulators of mRNA translation (green) that were significantly oxidized (Welch’s t-test, p < 0.05) in Nrf2-deficient N, K and KP organoids.
See also Figure S2 and Tables S1–4.
To identify Nrf2-dependent, redox-sensitive effector pathways, the cysteine proteomes of 3 pairs of N, Nn, K, Kn, and 4 pairs of KP and KPn organoids were normalized and merged. A total of 2,591 unique cysteine-containing peptides corresponding to 1,446 proteins were identified in all of the four 8-plex iTRAQ runs. Of these, 56 cysteine-containing peptides were significantly oxidized when Nrf2 was deleted in N organoids, 49 in K organoids and 255 in KP organoids (Figure 2B, Table S2). As in BSO-treated tumor organoids, Pkm2C358 was also substantially oxidized in KPn organoids. Global levels of reduced cysteine peptides were similar across the different genotypes (Figure S2A), indicating the absence of widespread changes in protein oxidation upon Nrf2 ablation. To determine whether the subset of proteins specifically oxidized in KPn cells represented any particular biochemical processes, the data were analyzed using DAVID (Huang da et al., 2009) for pathway enrichment. Interestingly, proteins containing oxidized cysteines in KPn organoids were strongly enriched for participation in protein folding and in mRNA translation (Figure 2C). Nrf2 deficient T organoids also exhibited a similar enrichment (Tables S3 and S4). Besides the classic translation pathway proteins curated in the DAVID database (ribosomal proteins, translation initiation and elongation factors), we identified additional proteins implicated in translational regulation to be oxidized in KPn organoids (Figures 2D and S2B), further coupling Nrf2 activation with translational control. Translation-related peptides exhibited an average of 20–50% increase in oxidation in KPn organoids (Figure S2C), in line with studies done in yeast reporting that the majority of cysteine residues are only partially (~25%) oxidized (Brandes et al., 2011). Treatment of murine T organoids with BSO similarly led to significant oxidation of proteins in the translation machinery (Figures S2D and S2E). These genetic and pharmacological observations collectively support the redox-dependent regulation of the translation machinery through cysteine modifications, prompting us to further evaluate the functional role of Nrf2 in mRNA translation for pancreatic cancer cells.
mRNA translation is impeded in Nrf2-deficient cells
Messenger RNA translation is a vital cellular process that regulates growth and metabolism (Sonenberg and Hinnebusch, 2009). In cancer cells, hyper-activated signaling pathways influence translation, supporting uncontrolled growth and survival (Mavrakis and Wendel, 2008). When compared to N organoids, KP organoids exhibited elevated levels of mRNA associated with translationally active polysomes and decreased levels of mRNA associated with translationally inactive monosomes (Figure 3A, left), consistent with the notion that translational upregulation plays an oncogenic role in cancer. While deletion of Nrf2 from N and K organoids did not noticeably alter the distribution of monosomes and polysomes (Figure S3A), deletion of Nrf2 from KP organoids led to a measurable decrease in polysomes with a corresponding increase in monosomes (Figure 3A, right), suggesting a decrease in translation efficiency in cancer cells when Nrf2 is absent. Impaired translation in KPn organoids was not due to eIF2 mediated activation of the unfolded protein response upon elevated ROS (Figure S3B) (Ling and Soll, 2010). When pulsed with [35S]-Methionine (Met) to measure the rate of nascent protein synthesis, a significant decrease in [35S]-Met incorporation was observed in Kn and KPn organoids (Figures 3B and S3C). This translation defect was redox-dependent (Figure 3B) and was observed in organoids and Suit2 harboring shNRF2 (Figures 3B, S3D, and S3E). These effects were mitigated by NAC and other antioxidants such as trolox and glutathione ethyl ester (Figures 3B, S3D, S3E and S3F). Of note, treatment of organoids with propargylglycine, an irreversible inactivator of γ-cystathonase, did not alter the difference in [35S]-Met incorporation into proteins in KP and KPn organoids (Figure S3G). This indicates that Nrf2-dependent decrease in [35S]-Met incorporation into proteins was not due to alterations in methionine to cysteine exchange through the transsulfuration cycle in response to elevated oxidative stress (Belalcazar et al., 2014). Introduction of a heterotypic NRF2 mRNA in human T organoids expressing shRNA against endogenous NRF2 restored [35S]-Met incorporation (Figure S3H). Thus, Nrf2 regulates the activity of the translational machinery through the maintenance of redox homeostasis.
Figure 3. Nrf2 deficiency impairs protein synthesis.
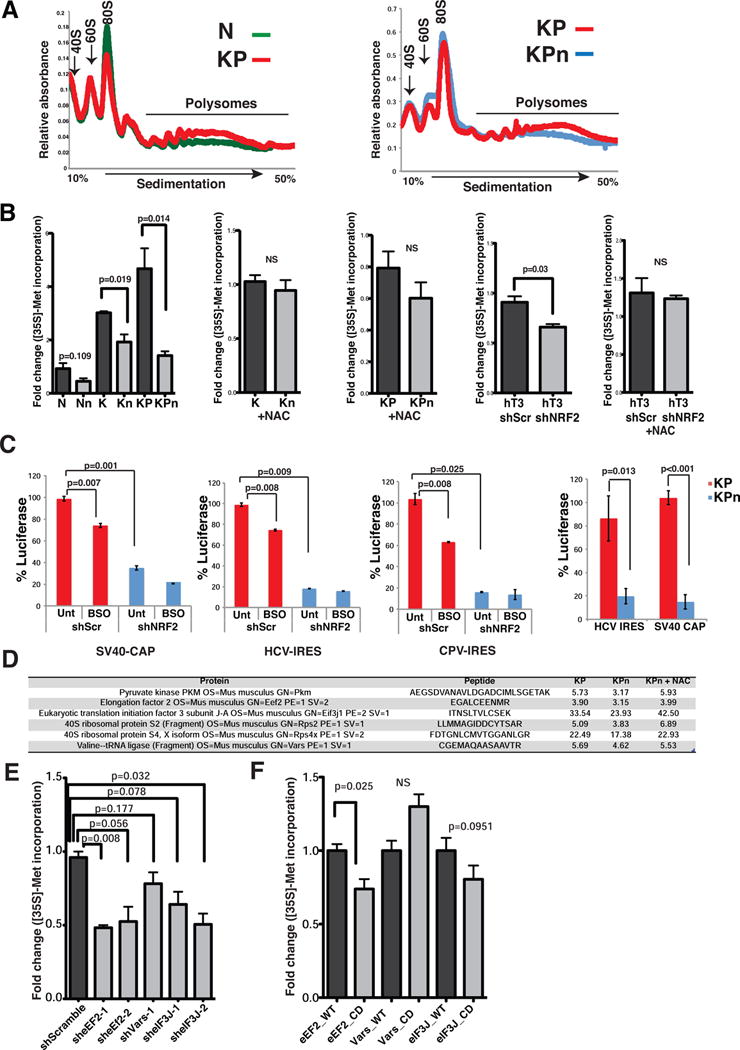
(A) Polysome profiles of N, KP and KPn organoids treated with 300 μg/ml cycloheximide for 10 min. Absorbance light at 254 nm. Representative profiles from two biological replicates.
(B) [35S]-Met incorporation into protein from murine and human organoids grown in reduced media or in media containing 1.25 mM NAC. Data are mean +/− s.e.m (n=3, Student’s t-test). NS = not significant.
(C) Activities of Renilla and firefly luciferase in Suit2 cells bearing shScr or shNRF2 (left three graphs) or KP, KPn organoids (rightmost) transfected with the biscistronic reporter plasmid 24 hrs prior. Data are percentage luciferase activity driven by SV40-CAP, HCV-IRES or CPV-IRES. Data are mean +/− s.d. (n=3, Student’s t-test).
(D) Cysteine peptide counts of translational regulatory proteins in KP, KPn and KPn organoids supplemented with 1.25 mM NAC (average of 2 biological replicates).
(E–F) [35S]-Met incorporation into protein from murine T organoids bearing shRNA (E) or V5-tagged wildtype (WT) or cysteine mutated (CD) cDNAs of indicated proteins (F). Data are mean +/− s.e.m. (n=6, Student’s t-test).
See also Figure S3.
To further analyze the steps in mRNA translation that are impacted by Nrf2 in pancreatic cancer cells, a bicistronic reporter construct encoding Renilla luciferase under the SV40 promoter and firefly luciferase under the control of the IRES sequences from the Hepatitis C Virus (HCV) was employed. Interestingly, both cap-dependent translation of Renilla luciferase and cap-independent HCV-IRES mediated translation of firefly luciferase were decreased in Suit2 cells bearing shNRF2 (Figure 3C), with no significant changes in the expression of the luciferase gene (Figure S3I). While HCV-IRES-mediated translation requires certain canonical initiation factors (Otto and Puglisi, 2004), the cricket paralysis virus IRES (CPV-IRES) (Wilson et al., 2000) enables translation of mRNAs independently of any translation initiation factors. We found that CPV-IRES-mediated translation was also impaired when NRF2 expression was suppressed (Figure 3C). These translational defects were partially phenocopied by BSO treatment (Figure 3C), and were recapitulated in KPn organoids (Figure 3C). These results suggest that NRF2 exercises redox-dependent control over multiple aspects of the translation machinery.
Since cysteine residues previously implicated as functional redox switches are reversibly oxidized, we evaluated the possibility that components of the translational machinery may also be redox switches that govern translation activity. Supplementation of KPn organoids with NAC reversed the oxidative cysteine changes in a number of translational regulatory proteins (Figure 3D). shRNA-mediated knockdown of selected initiation and elongation factors (Figure S3J) confirmed that they were independently essential for protein synthesis in PDA (Figure 3E). Ectopic expression of these factors did not enhance protein synthesis in KP organoids (Figure S3K). However, ectopic expression of the cysteine to aspartic acid oxidation mimic (Permyakov et al., 2012) of the elongation factor Eef2 led to a significant decrease in the rate of nascent protein synthesis when compared to its wildtype counterpart (Figures 3F and S3L). A similar trend was also observed for the translation initiation factor eIF3J, but not for the Valyl-tRNA synthetase, Vars (Figures 3F and S3L). These results suggest that Eef2Cys693 (and possibly additional components of the translation machinery) may function as a redox switch to modulate the activity of the elongation apparatus.
Growth factor signaling pathways upstream of cap-dependent translation are impaired in Nrf2-deficient cells
Of the four phases of protein synthesis (initiation, elongation, termination and recycling), initiation is considered to be the rate-limiting step in mRNA translation, and is often exploited by cancer cells to support tumorigenesis (Pelletier et al., 2015). Based on our observation that cap-dependent mRNA translation was compromised in NRF2-deficient cells (Figure 3C), and that the initiation factor eIF3J may also be a candidate redox switch (Figure 3F), we extended our investigation on translation initiation biochemically. eIF4E is the limiting initiation factor for mRNA translation, as it recognizes the m7GTP cap on mRNA and recruits eIF4G (Gingras et al., 1999). In the absence of mitogenic signaling, eIF4E is bound by the inhibitory 4EBPs, which sequester eIF4E from interaction with eIF4G. This process is critical for regulating mRNA translation in PDA organoids as expression of a dominant active 4EBP1-4A mutant led to substantial decrease in nascent protein synthesis (Figure S4A) and cell proliferation (Figure S4B), phenocopying Nrf2-deficiency. The levels of eIF4E, eIF4G and 4EBP1 were independent of Nrf2 status (Figures 4A and S4C), and the oxidative states of these proteins as determined through iodoacetamide enrichment for reduced cysteine peptides were also similar in KP and KPn cells (Figure S4C). Despite these, the amount of inhibitory 4EBP1 co-precipitating with eIF4E in m7GTP pull-downs was found to be elevated in Nn, Kn and KPn organoids, with a concomitant decrease in eIF4G co-precipitation (Figures 4A and S4D). This reflects a defective eIF4F complex formation that would impart decreased cap-dependent translation initiation in Nrf2-deficient KP cells.
Figure 4. Nrf2 deficiency impairs mitogenic signaling pathways governing eIF4F complex formation.
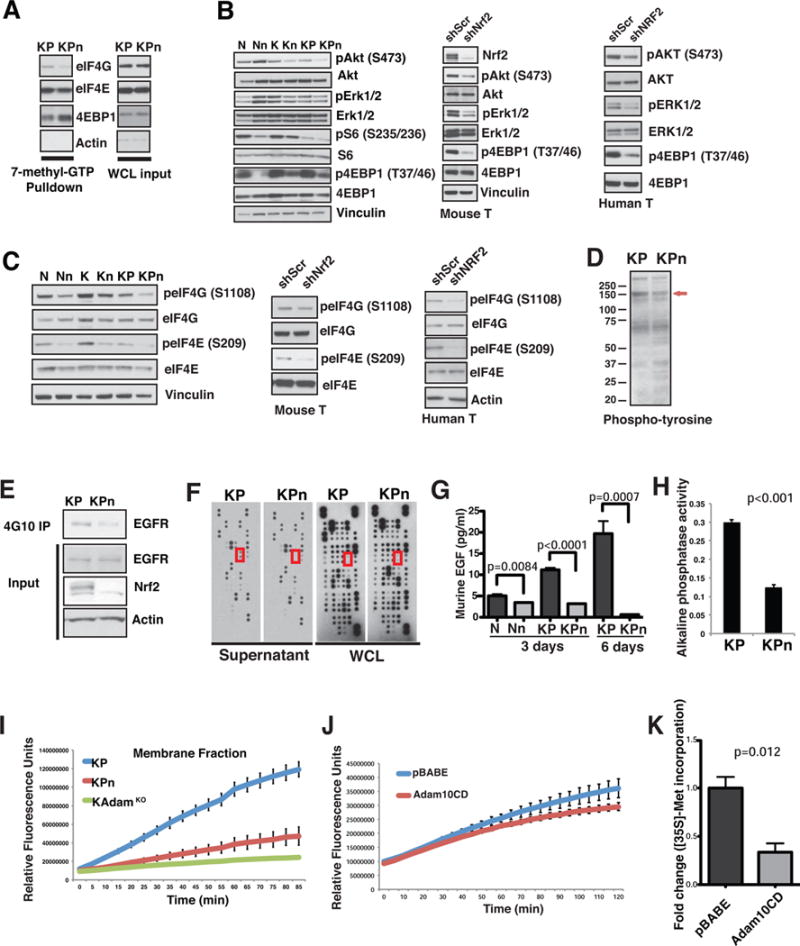
(A) Lysates from KP and KPn organoids were subjected to 7-methyl-GTP pulldowns, and analysed for indicated proteins. WCL, whole cell lysates.
(B–C) Immunoblot analysis for growth factor signaling pathway activation (B) and mRNA cap-binding proteins (C) in N, K, KP organoids as well as murine tumor (T) and human tumor (hM1, hT3) organoids with shScr or shNRF2. p=phospho.
(D–E) Immunoblot analysis of total (D) and EGFR tyrosine (Y) phosphorylation (E) in KP and KPn organoids. Red arrow, 150 kD molecular weight protein. 4G10 IP, total phospho-Y antibody immunoprecipitation.
(F) Protein array analysis for growth factors secreted into culture medium (Supernatant) and expressed in whole cell lysate (WCL) in KP and KPn organoids. Red box, murine EGF.
(G) EGF ELISA of supernatant from N, Nn, KP and KPn organoids after 3 or 6 days in culture. Data are mean +/− s.d. (n=3, Student’s t-test).
(H) Constitutive EGF shedding determined by alkaline phosphatase activity in the supernatant from KP organoids. Data are mean +/− s.d. (n=3, Student’s t-test).
(I–J) Activity of Adam10 from plasma membrane fractions of KP, KPn and K;Adam10 KO organoids (I) or KP organoids expressing control vector versus Adam10 cysteine mutant (CD) (J), measured by cleavage and increase in 5-FAM fluorescence of a FRET substrate specific to Adam10. Activity was monitored over 2 hrs at excitation excitation/emission = 490 nm/520 nm. Data are mean +/− s.d. (n=2).
(K) [35S]-Met incorporation into protein from KP organoids expressing control vector or Adam10 cysteine mutant (CD). Data are mean +/− s.d. (n=6, Student’s t-test).
See also Figure S4.
PI3K/AKT signaling has been shown to promote phosphorylation of 4EBPs at multiple sites, resulting in the liberation of eIF4E from 4EBP1 and the stimulation of cap-dependent translation initiation (Ruggero and Sonenberg, 2005). Consistent with elevated 4EBP1 at the mRNA cap, phosphorylation of Akt and 4EBP1 were both decreased in Kn and KPn organoids, as well as in murine and human T organoids expressing shNRF2 (Figure 4B). pErk was moderately decreased in Kn and KPn cells, and more clearly in T organoids expressing shNRF2 (Figure 4B). The level of pS6 was not consistently downregulated in Kn and KPn cells, suggesting that these signaling alterations operate independently of S6 kinase (Figure 4B). Consistently, phosphorylation of eIF4G and eIF4E, substrates of Akt (Raught et al., 2000), and Erk/Mnk signaling (Ueda et al., 2004), respectively, were also decreased in Kn and KPn organoids as well as in T organoids expressing shNRF2 (Figures 4C, S4E and S4F). These signaling defects were not due to differential activation of oncogenic Kras (Figure S4G), nor that of the Igf1 receptor (Figure S4H) (Molina-Arcas et al., 2013). Neither the activity nor the expression level of PTEN (Maehama and Dixon, 1998), was up-regulated in Nn and KPn organoids (Figure S4I). The signaling changes upon Nrf2 deficiency were most striking in the context of K and KP cells, reflecting critical Kras-dependency.
A hallmark of Kras mutant cells is the role of autocrine and paracrine loops in amplifying oncogenic Kras signaling through AKT and ERK (Ardito et al., 2012). Decreased activation of AKT and MAPK in Nrf2-deficient cells was not due to oxidation of these kinases (Figure S4J). Immunoblot analysis of global tyrosine phosphorylation revealed decreased phosphorylation of a protein above 150 kDa (Figure 4D), which we then confirmed to be the EGF receptor (Figure 4E). By secretome arrays (Figure 4F) and ELISA (Figure 4G), we found that the release of EGF into culture media by KPn organoids was reduced when compared to KP organoids while total intracellular levels of EGF protein (Figure 4F) and mRNA (Figure S4K) were not decreased. Indeed, induced (Figure S4L) and constitutive (Figure 4H) shedding of EGF as determined by the expression of an EGF-alkaline phosphatase (AP) construct followed by measurement of AP activity in the supernatant revealed decreased EGF shedding in KPn organoids and in Nrf2-deficient tumor cells (Figure S4M). Consistent with reports showing that EGFR signaling is coupled to activation of cap-dependent translation in EGFR wild type cells (Patel et al., 2013), EGF supplementation was able to restore defective Akt and Erk activation in KPn organoids (Figure S4N). Addition of NAC alone also partially corrected these signaling defects (Figure S4N) and restored EGF release (Figure S4O). As with NAC supplementation, EGF was sufficient to mitigate the translational defects observed in Kn and KPn organoids (Figure S4P) and improved proliferation in KPn organoids (Figure S4Q). Doxcycline-induced knockdown of NRF2 in human T organoids led to decreased pEGFR and pERRB2 (Figure S4R). Similarly, activity levels of EGFR in tumor lysates of Suit2 xenografts correlated with doxycycline induced NRF2 knockdown (Figure S4S). In one shNRF2 tumor sample where NRF2 was reactivated, pEGFR was sustained at a high level (Figure S4S), further demonstrating a role of NRF2 in EGFR activation.
Metalloproteases of the ADAM (a disintegrin and metalloprotease) family are thought to be responsible for the shedding of certain EGFR ligands, with Adam10 emerging as the main sheddase of EGF (Sahin et al., 2004). Although Adam10 was not detected in our initial cysteine proteomics experiment, mass spectrometric analysis of immunoprecipitated Adam10 revealed a substantial and specific decrease in the levels of a cysteine-containing peptide when Nrf2 was deleted (Figure S4T). Consistent with decreased EGF shedding, Adam10 was functionally impaired in KPn organoids (Figure 4I). Ectopic expression of a cysteine oxidation mimic of Adam10 (Figure S4U) inhibited the activity of endogenous Adam10 (Figure 4J) and led to a significant decrease in nascent protein synthesis in KP organoids (Figure 4K). These data suggest that redox-dependent regulation of Adam10 activity contributes to the maintenance of EGF autocrine signaling and cap-dependent translation in PDA.
Nrf2 supports cap-dependent translation of pro-survival transcripts
Enhanced eIF4F complex formation in cancer cells promotes cap-dependent mRNA translation and thereby elevates global protein synthesis rates. However, mRNAs vary widely in their inherent “translatability,” largely as a function of differences in the length and structure of their 5′UTRs. Cellular mRNAs most sensitive to alterations in eIF4F complex formation have highly structured 5′UTRs. These mRNAs often encode proteins that promote growth and transformation (Pelletier et al., 2015). In contrast, the majority of cellular mRNAs have relatively short, unstructured 5′ UTRs that enable efficient translation even when eIF4F complex levels are limiting. We found that the levels of a number of pro-survival proteins were increased in KP compared to N organoids, with no changes in housekeeping proteins (Figures 5A and S5A). Of the pro-survival proteins, Hif1α, Bcl2 and more moderately, cyclin D1, cyclin D3 and c-Myc were decreased in KPn organoids (Figure 5A). Similar observations were made in murine T organoids bearing shNrf2 (Figure 5B). This is consistent with earlier reports showing a correlation between the expression of Nrf2 and some of these proteins (Fan et al., 2014; Ke et al., 2013; Malec et al., 2010; Niture and Jaiswal, 2012; Wang et al., 2016). Quantitative PCR of polysome fractions revealed an increase in the association of the mRNAs of these proteins with the translationally inactive monosome fractions (Figure 5C), while no differences were seen for Actin, Mcl-1, and interestingly, Bcl2 mRNAs (Figure S5B). No decrease was observed in the total transcript level of these mRNAs (Figure S5C), or the half-life of the corresponding proteins (Figure S5D). Compromised Hif1α protein levels in KPn organoids corresponded with a decrease in the expression level of Hif1α target genes (Figure 5D), their inability to grow in hypoxia (Figure 5E) and heightened sensitivity to inhibition of glycolysis (Figure 5F) (Semenza, 2013). These data indicate that Nrf2-mediated protein synthesis in Kras mutant cells has a potent impact on the translation of certain proto-oncogenic mRNAs, thus supporting a role for Nrf2 in fully transformed PDA cells.
Figure 5. Nrf2 supports translation of pro-survival transcripts.
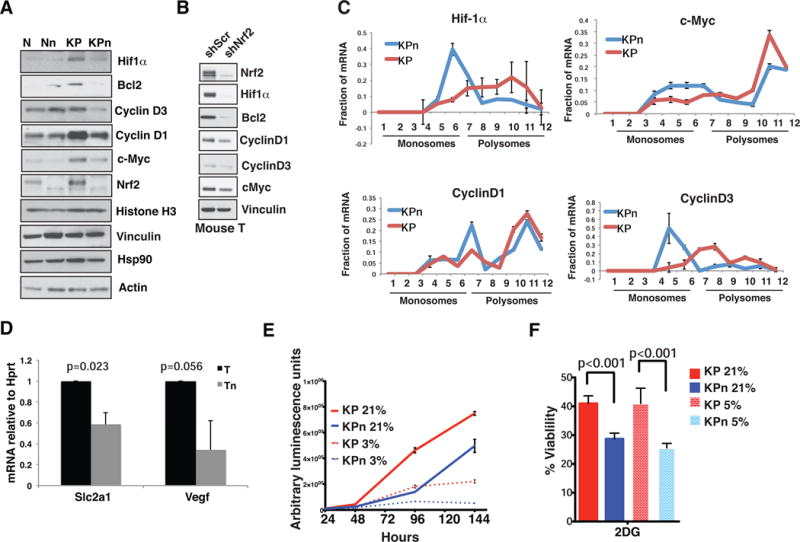
(A–B) Immunoblot analysis of oncoproteins in N and KP organoids (A) and murine T organoids (B) expressing shScramble or shNrf2.
(C) qPCR analysis of indicated mRNAs from polysome fractions of KP and KPn organoids. ΔCT calculated against 80S fraction. Data are means normalized against the sum +/− s.d. (n=2 biological replicates). Fractions 1–6: Monosomes; 7–12: Polysomes.
(D) qPCR analysis of mRNA expression of Hif1α target genes in murine T organoids. Data are mean +/− s.d. (n=3, Student’s t-test).
(E) Proliferation of organoids grown in 21% or 3% oxygen. Data are mean +/− s.e.m. (n=5).
(F) Cell viability upon treatment with 20 mM 2-deoxyglucose for 72 hours in 21% oxygen or 5% oxygen. Data are mean +/− s.e.m. (n=5, Students t-test.)
See also Figure S5.
Combined inhibition of AKT and glutathione synthesis blunts pancreatic cancer growth and survival
The PI3K/AKT pathway is believed to be a major pathway regulating global and mRNA-specific translation initiation (Ruggero and Sonenberg, 2005). We found that the basal phosphorylation status of 4EBP1 correlated strongly with the sensitivity of hT organoids to shNRF2 (Figure 6A). Inhibition of AKT using the pan-AKT inhibitor, MK2206, markedly decreased the rate of protein synthesis (Figure 6B) and increased 4EBP1-eIF4E interactions at the mRNA cap (Figure 6C) in KP organoids, the effect of which were potentiated in KPn organoids (Figures 6B and 6C). At a dose that suppressed KP intracellular GSH to a level found in KPn cells (Figure S6A), BSO bolstered the effect of MK2206 to further decrease the rate of protein synthesis (Figure 6D). Consistently, inhibition of 4EBP1 phosphorylation in both KP and human T organoids was most efficient in the presence of both inhibitors (Figure 6E).
Figure 6. Inhibition of glutathione synthesis sensitizes pancreatic cancer cells to pan AKT inhibition.
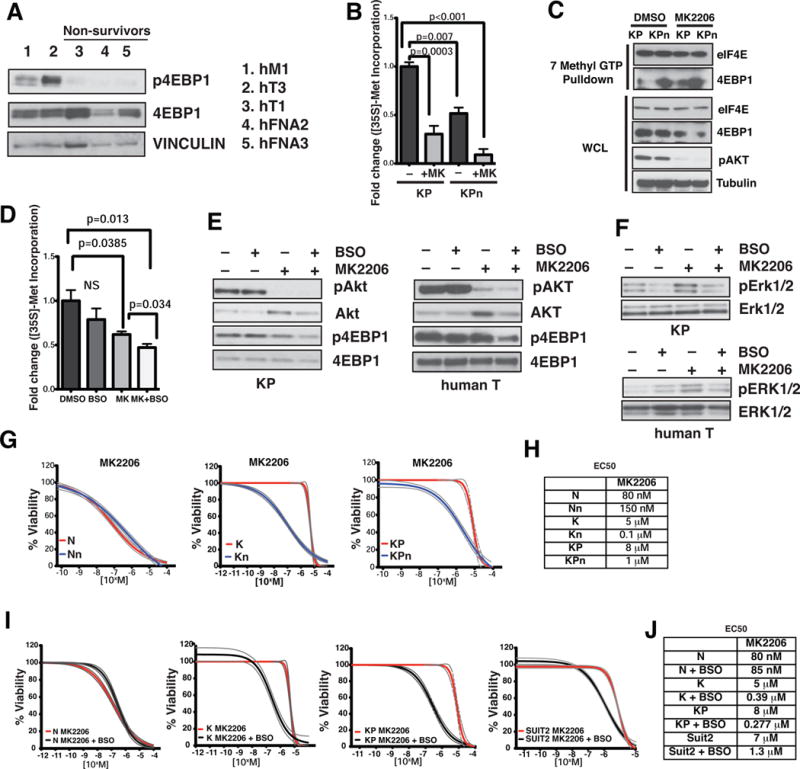
(A) Immunoblot analysis of 4EBP1 activation status in human T organoids. VINCULIN, loading control. p = phospho.
(B) [35S]-Met incorporation into protein from KP and KPn organoids treated with DMSO or 1 μM MK2206 (MK) for 48 hrs. Data are mean +/− s.e.m. (n=3, Student’s t-test.).
(C) Lysates from KP and KPn organoids treated with DMSO or 1 μM MK2206 were subjected to 7-methyl-GTP pulldowns, and analyzed for indicated proteins.
(D) [35S]-Met incorporation into protein from KP organoids treated with 1 μM MK2206 (MK), 100 μM BSO, or in combo for 48 hrs. Data are mean +/ s.e.m. (n=3, Student’s t-test.).
(E) Immunoblot analysis of AKT and 4EBP1 activation status in murine (left) and human hT1 (right) T organoids treated with 1 μM MK2206 (MK), 100 μM BSO, or in combo for 48 hrs. p = phospho.
(F) Adaptive response in KP organoids (top) and human T organoids (bottom) upon treatment with vehicle only, 1 μM MK2206, 100 μM BSO, or in combination for 48 hrs. p = phospho.
(G) Cell viability of N, K and KP and the corresponding Nrf2-deficient organoids over increasing concentrations of MK2206 for 72 hours. Dotted lines, 95% confidence intervals. (n=5).
(H) EC50 values of MK2206 in murine organoids.
(I) Cell viability of N, K, KP organoids and Suit2 cells over increasing concentrations of MK2206 in the presence or absence of BSO for 72 hours. Dotted lines, 95% confidence intervals. (n=5).
(J) EC50 values of MK2206 on pancreatic organoids and cell lines in the presence or absence of BSO.
See also Figure S6.
Reactivation of EGFR and MAPK signaling upon inhibition of AKT is an important adaptive survival response leading to drug resistance in Kras mutant cancer cells and requires an intact autocrine EGFR signaling cascade to enact (Mendoza et al., 2011). Consistent with defective EGFR signaling in Kn and KPn organoids, these adaptive responses were attenuated (Figure S6B). While BSO treatment alone had minimal effects on the activity of these signaling cascades, BSO suppressed the reactivation of EGFR (Figure S6C) and MAPK (Figure 6F) upon treatment with MK2206. Combined treatment of MK2206 with BSO led to a higher level of ROS compared to single agents alone (Figure S6D), further supporting the premise that EGFR autocrine activation in Kras mutant cells is redox-dependent.
In congruence with our biochemical observations, Kn and KPn organoids were substantially more sensitive to MK2206 (Figures 6G, 6H and S6E) and the multi-targeted PI3K inhibitor, PI-103, (Figure S6F) than K and KP counterparts. Consistently, BSO exhibited a synergistic effect with MK2206 in K and KP organoids (Figures 6I, 6J, S6G and S6H), as well as in human PDA cells (Figures 6I, 6J, S6I and S6J). Of note, sensitivity of N organoids towards MK2206 was not heightened by either BSO treatment (Figures 6I and 6J), or Nrf2 deficiency (Figures 6G and 6H), reflecting a potential therapeutic index.
Contrary to our observations with AKT inhibition, KPn organoids were not more sensitive to mTOR inhibitors such as rapamycin and Torin1 (Figures S6K and S6L). We posit this to be due to mTOR mediated growth suppression in cells that rely on extracellular proteins as an amino acid source (Palm et al., 2015), as in the case of Kras mutant cells (Bar-Sagi and Feramisco, 1986). Consistent with the notion that Nrf2 modulates Akt/mTOR activity downstream of mitogenic signaling, elevated levels of lysosomal hydrolysis were observed in KPn organoids (Figure S6M) and this process was redox dependent (Figures S6N and S6O). We posit that the proliferative advantage obtained in mTOR-inhibited Kras mutant cells may outweigh the suppressive effects from AKT and glutathione inhibition. Thus, this renders direct antagonism of AKT to be more growth suppressive than that of mTOR in this context. Sensitivity of Nrf2-deficient cells was selective towards inhibition of PI3K/AKT, as similar vulnerability was absent towards the MEK inhibitor, AZD6244 (Figures S6P and S6Q) (Davies et al., 2007), or to a panel of commonly used chemotherapeutics (Figure S6R).
Encouraged by our in vitro observations on MK2206 and BSO, we tested this strategy in the KPC mouse model (Hingorani et al., 2005). Mice were randomly assigned to treatments with vehicle, BSO, MK2206 or the combination of MK2206 and BSO. MK2206 and BSO effectively suppressed levels of pAkt (Figure S7A) and GSH (Figure S7B), respectively. While monotherapy did not markedly impact tumor growth, combination treatment was significantly growth suppressive (Figures 7A and 7B). Long-term treatment of KPC mice with this combination diminished neoplastic cell proliferation (Figure S7C), and modestly increased median survival when compared to MK2206 alone (Figure S7D). The synergistic effect of MK2206 and BSO was also observed in Suit2 xenograft models in terms of tumor kinetics (Figure 7C), and neoplastic cell proliferation (Figure 7D). Our results suggest that pro-oxidants may further augment the effectiveness of AKT inhibition in suppressing PDA growth through combined inhibition of mRNA translation and EGFR-dependent mechanisms of resistance (Figure S7E).
Figure 7. Combined inhibition of AKT and glutathione synthesis suppresses human and mouse pancreatic tumor growth in vivo.
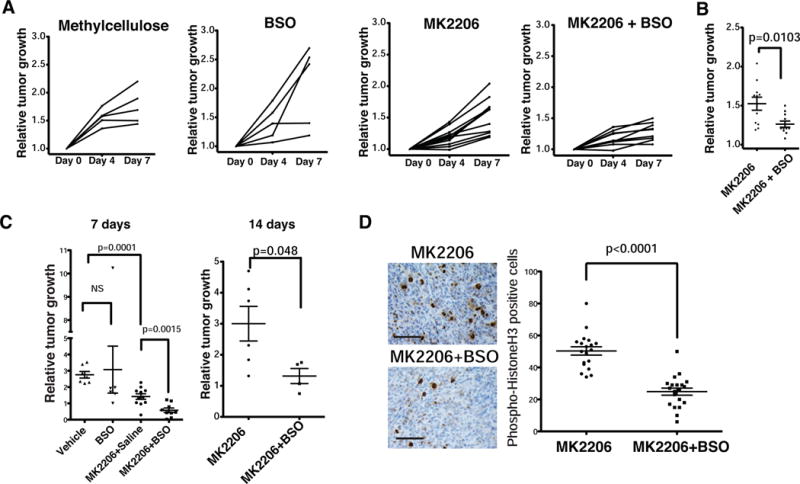
(A) Tumor volumes of KPC mice treated daily with vehicle (methylcellulose), BSO, MK2206, or the combination for 7 days. Tumor volumes were determined by ultrasound imaging on the indicated days.
(B) Relative tumor volume of KPC mice treated with MK2206 or in combination with BSO on day 7. Student’s t-test. (n=11).
(C) Relative growth of subcutaneously xenografted Suit2 tumors in mice treated with vehicle, MK2206, or MK2206+BSO for 7 days (left) or 14 days (right). Data are mean +/− s.e.m. Student’s t test.
(D) Phospho-histone H3 IHC of representative Suit2 tumors from treated mice (Left). Quantification of pH3 positivity (Right). Data are mean +/− s.e.m (n ≥ 5 fields of view). Student’s t-test.
See also Figure S7.
Discussion
The concept that elevated levels of free radicals and oxidative stress contribute to DNA damage and predisposition to cancer has led to the recommendation of antioxidant use as prophylaxis against neoplasia (Khansari et al., 2009). However, Nrf2 activators (Talalay et al., 2007) and various antioxidants have either failed to reduce cancer incidence or have instead promoted cancer in clinical and preclinical testing (Sayin et al., 2014). We present the alternative hypothesis that Nrf2’s antioxidant function stimulates pancreatic cancer in part by promoting mRNA translation and mitogenic signaling. Indeed, our findings may reflect cellular responses to a number of redox perturbants demonstrated previously to exhibit efficacy in Ras mutant cells. This includes erastin (Yang and Stockwell, 2008), dehydroascorbic acid (oxidized vitamin C) (Yun et al., 2015), as well as platinum-based cytotoxics (Socinski, 2004).
The synthetic lethal interaction we observed between Nrf2 loss and oncogenic Kras in PDA may also reflect the proto-oncogenic mRNAs that are exquisitely translationally up-regulated in Kras mutant cells. For example, Hif1α, an important transcription factor that mediates adaptive responses to intratumoral hypoxia and malignant progression in PDA (Zhao et al., 2014), was markedly decreased in Nrf2-deficient Kras-mutant cells. Hif1α likely contributes to the glycolytic phenotype of PDA (Ying et al., 2012), working in parallel with NRF2 transcriptional target genes that are metabolic enzymes governing the pentose phosphate pathway (Mitsuishi et al., 2012) to promote metabolic rewiring in cancer cells.
Regarding the potential clinical implications of this work, AKT inhibitors have already shown some preliminary activity in clinical trials (Molife et al., 2014; Yap et al., 2011), and MK2206 is currently being investigated in patients with pancreatic cancer (NCT01783171; NCT01658943). Our current study demonstrates that the combination of AKT inhibitors with oxidizing agents is active in PDA, and ongoing work will determine whether the investigational agent BSO (NCT00002730; NCT00005835; NCT00661336) or alternative agents that target different aspects of redox regulation will be most attractive for rapid clinical translation.
Experimental Procedures
Analysis of Global Protein Synthesis
Organoids or SUIT2 cells were incubated for 30 minutes in media containing 10 μCi of [35S] labeled Methionine (NEG709A005MC, Perkin Elmer). Cell lysates were prepared using standard procedures and equal amounts of total protein were separated on a 4–12% Bis-Tris polyacrylamide gel and transferred to PVDF membrane. Membranes were exposed to autoradiography film for 12–48 hours and then developed. For liquid scintillation, equal amounts of radiolabelled total protein were TCA-precipitated and washed two times in acetone and air dried at room temperature. The amount of [35S]-Met incorporated into protein was measured using a Beckman LS6500 Scintillation counter. Total protein content was determined by BCA assay (Bio-rad).
Polysome Fractionation
Organoids were treated with 300 μg/ml cyclohexamide (Sigma) in media for 10 min at 37°C and then harvested on ice in PBS containing 300 μg/ml cyclohexamide. Cells were pelleted and lysed in 10 mM Tris-HCl (pH 8), 140 mM NaCl, 1.5 mM MgCl2, 0.25% NP-40, 0.1% Triton X-100, 50 mM DTT, 150 μg/ml cyclohexamide, and 640 U/ml RNasin for 30 min. Lysates were cleared, separated on a 10%–50% sucrose gradient by ultracentrifugation, and fractionated using an ISCO gradient fractionation system.
Therapeutic intervention studies in mice
Upon detection of a mass during weekly palpation, KPC mice were subjected to high-contrast ultrasound imaging using the Vevo 2100 System with a MS250, 13–24 MHz scanhead (Visual Sonics, Inc, Amsterdam, NL). Mice with tumor diameters of 7–9 mm were randomized and enrolled one day after scanning. MK2206 (Merck) was formulated in 0.5% methylcellulose. BSO (Sigma) was formulated in saline. Mice were administered methylcellulose vehicle or 100 mg/kg MK2206 every day via oral gavage, and saline or 10 mmol/kg BSO via intraperitoneal injection. Tumor volume was monitored on days 4 and 7 after initial scan. The same dosing regimen was applied to athymic nude mice bearing Suit2 tumor cells. Drug administration was initiated three weeks post transplantation of 105 cells subcutaneously.
Supplementary Material
Acknowledgments
We thank J.R. Prigge, Montana State University, for developing the anti-Nrf2 antibody, and Dr. L. Baker for critical review of the manuscript. This work was performed with the CSHL Mass Spectrometry, Animal and Tissue Imaging, and Bioinformatics Shared Resources, which are supported by the Cancer Center Support Grant 5P30CA045508. D.A.T. is a distinguished scholar of the Lustgarten Foundation (LF) and Director of the LF-designated Laboratory of Pancreatic Cancer Research. D.A.T. is supported by the CSHL Association, the NIH (5P30CA45508-26, 5P50CA101955-07, 1U10CA180944-01, 5U01CA168409-3, 1R01CA190092-01); Carcinoid Foundation, PCUK, the David Rubinstein Center for Pancreatic Cancer Research at MSKCC, Stand Up to Cancer/KWF, the STARR foundation (I7-A718), DOD (W81XWH-13-PRCRP-IA) and the Precision Medicine Research Associates. We are also grateful for support from the following: the Damon Runyon Cancer Research Foundation (Shirley Stein fellow, DRG-2165-13, I.I.C.C.), Human Frontier Science Program (LT000190/2013, I.I.C.C.), Cancer Research Society and CCSRI (702317, N.S.), the Canadian Institute of Health Research (S.M.J.), NIH grants (AG040020 and CA152559, E.E.S.; CA159222, H.C.C.), the Swedish Research Council (537-2013-7277, D.Ö.), the Sociedad Española de Oncología Médica (SEOM, M.P.S.), the Hope Funds for Cancer Research Fellowship (W.P). This work was performed with the MSKCC molecular cytology core, which is supported by the Cancer Center Support Grant P30CA008748. CBT is a founder of Agios Pharmaceuticals and a member of its scientific advisory board. CBT also serves on the board of directors of Merck.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
I.I.C.C. and D.A.T. initiated the project and designed the research plan. S.M.J. helped with translation experiments. M.P.S. and D.Ö. helped with dose response experiments. K.R. performed mass spectrometry (MS) experiment. J.W., V.S., Y.H., M.H. performed statistical analysis of MS data. K.W., D.F., Y.P. B.D, and C.S. helped with in vivo animal study. J.E.W. performed pathological analysis. W.P., C.B.T. and I.I.C.C. designed and performed lysosomal hydrolysis experiments. E.L. and Y.P. designed and cloned cysteine mutants. H.T. and J.M.B. established human organoid lines from primary patient samples. G.M.D. provided inducible NRF2 hairpins. H.C.C. assisted in Adam10-related experiments. I.I.C.C, E.E.S., D.J.P., N.S. and D.A.T. designed the study and wrote the manuscript. The contributions of S.M.J., M.P.S. and Y.P. are equal.
References
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardito CM, Gruner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, Delgiorno KE, Carpenter ES, Halbrook CJ, Hall JC, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science. 1986;233:1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Current opinion in structural biology. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Belalcazar AD, Ball JG, Frost LM, Valentovic MA, Wilkinson JT. Transsulfuration Is a Significant Source of Sulfur for Glutathione Production in Human Mammary Epithelial Cells. ISRN biochemistry. 2014;2013:637897. doi: 10.1155/2013/637897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilanges B, Stokoe D. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene. 2007;26:5973–5990. doi: 10.1038/sj.onc.1210431. [DOI] [PubMed] [Google Scholar]
- Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes N, Reichmann D, Tienson H, Leichert LI, Jakob U. Using quantitative redox proteomics to dissect the yeast redoxome. J Biol Chem. 2011;286:41893–41903. doi: 10.1074/jbc.M111.296236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Blenis J. Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy. Cell cycle. 2009;8:567–572. doi: 10.4161/cc.8.4.7659. [DOI] [PubMed] [Google Scholar]
- Davies BR, Logie A, McKay JS, Martin P, Steele S, Jenkins R, Cockerill M, Cartlidge S, Smith PD. AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1/2 kinases: mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship, and potential for combination in preclinical models. Mol Cancer Ther. 2007;6:2209–2219. doi: 10.1158/1535-7163.MCT-07-0231. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Chen P, Tan H, Zeng H, Jiang Y, Wang Y, Wang Y, Hou X, Bi H, Huang M. Dynamic and coordinated regulation of KEAP1-NRF2-ARE and p53/p21 signaling pathways is associated with acetaminophen injury responsive liver regeneration. Drug metabolism and disposition: the biological fate of chemicals. 2014;42:1532–1539. doi: 10.1124/dmd.114.059394. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in biochemical sciences. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Johnson LF, Williams JG, Abelson HT, Green H, Penman S. Changes in RNA in relation to growth of the fibroblast. III. Posttranscriptional regulation of mRNA formation in resting and growing cells. Cell. 1975;4:69–75. doi: 10.1016/0092-8674(75)90135-x. [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke B, Shen XD, Zhang Y, Ji H, Gao F, Yue S, Kamo N, Zhai Y, Yamamoto M, Busuttil RW, et al. KEAP1-NRF2 complex in ischemia-induced hepatocellular damage of mouse liver transplants. Journal of hepatology. 2013;59:1200–1207. doi: 10.1016/j.jhep.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khansari N, Shakiba Y, Mahmoudi M. Chronic inflammation and oxidative stress as a major cause of age-related diseases and cancer. Recent patents on inflammation & allergy drug discovery. 2009;3:73–80. doi: 10.2174/187221309787158371. [DOI] [PubMed] [Google Scholar]
- Ling J, Soll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci U S A. 2010;107:4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Malec V, Gottschald OR, Li S, Rose F, Seeger W, Hanze J. HIF-1 alpha signaling is augmented during intermittent hypoxia by induction of the Nrf2 pathway in NOX1-expressing adenocarcinoma A549 cells. Free Radic Biol Med. 2010;48:1626–1635. doi: 10.1016/j.freeradbiomed.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Mavrakis KJ, Wendel HG. Translational control and cancer therapy. Cell cycle. 2008;7:2791–2794. doi: 10.4161/cc.7.18.6683. [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends in biochemical sciences. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Molife LR, Yan L, Vitfell-Rasmussen J, Zernhelt AM, Sullivan DM, Cassier PA, Chen E, Biondo A, Tetteh E, Siu LL, et al. Phase 1 trial of the oral AKT inhibitor MK-2206 plus carboplatin/paclitaxel, docetaxel, or erlotinib in patients with advanced solid tumors. Journal of hematology & oncology. 2014;7:1. doi: 10.1186/1756-8722-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS, Downward J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer discovery. 2013;3:548–563. doi: 10.1158/2159-8290.CD-12-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PS, Sipos B, Orlandini S, Sorio C, Real FX, Lemoine NR, Gress T, Bassi C, Kloppel G, Kalthoff H, et al. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Archiv: an international journal of pathology. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- Niture SK, Jaiswal AK. Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J Biol Chem. 2012;287:9873–9886. doi: 10.1074/jbc.M111.312694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell. 2015;162:259–270. doi: 10.1016/j.cell.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, Jay-Dixon J, Sadiq AA, Jacobson BA, Kratzke RA. Resistance to EGFR-TKI can be mediated through multiple signaling pathways converging upon cap-dependent translation in EGFR-wild type NSCLC. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2013;8:1142–1147. doi: 10.1097/JTO.0b013e31829ce963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permyakov SE, Zernii EY, Knyazeva EL, Denesyuk AI, Nazipova AA, Kolpakova TV, Zinchenko DV, Philippov PP, Permyakov EA, Senin II. Oxidation mimicking substitution of conservative cysteine in recoverin suppresses its membrane association. Amino acids. 2012;42:1435–1442. doi: 10.1007/s00726-011-0843-0. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Gygi SP, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. The EMBO journal. 2000;19:434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & cellular proteomics : MCP. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Sonenberg N. The Akt of translational control. Oncogene. 2005;24:7426–7434. doi: 10.1038/sj.onc.1209098. [DOI] [PubMed] [Google Scholar]
- Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. The Journal of cell biology. 2004;164:769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Science translational medicine. 2014;6:221ra215. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. The Journal of clinical investigation. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman M, McComb ME, Huang H, Huang S, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. Journal of proteome research. 2004;3:1228–1233. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Socinski MA. Cytotoxic chemotherapy in advanced non-small cell lung cancer: a review of standard treatment paradigms. Clin Cancer Res. 2004;10:4210s–4214s. doi: 10.1158/1078-0432.CCR-040009. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talalay P, Fahey JW, Healy ZR, Wehage SL, Benedict AL, Min C, Dinkova-Kostova AT. Sulforaphane mobilizes cellular defenses that protect skin against damage by UV radiation. Proc Natl Acad Sci U S A. 2007;104:17500–17505. doi: 10.1073/pnas.0708710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Lu W, Ogasawara MA, Nilsa RD, Huang P. Redox regulation of cell survival. Antioxid Redox Signal. 2008;10:1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu X, Long M, Huang Y, Zhang L, Zhang R, Zheng Y, Liao X, Wang Y, Liao Q, et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Science translational medicine. 2016;8:334ra351. doi: 10.1126/scitranslmed.aad6095. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chemistry & biology. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TA, Yan L, Patnaik A, Fearen I, Olmos D, Papadopoulos K, Baird RD, Delgado L, Taylor A, Lupinacci L, et al. First-in-man clinical trial of the oral pan-AKT inhibitor MK-2206 in patients with advanced solid tumors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:4688–4695. doi: 10.1200/JCO.2011.35.5263. [DOI] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, Roper J, Chio II, Giannopoulou EG, Rago C, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015 doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg A, Larsson O, Wiman KG. What is the restriction point? Current opinion in cell biology. 1995;7:835–842. doi: 10.1016/0955-0674(95)80067-0. [DOI] [PubMed] [Google Scholar]
- Zhao X, Gao S, Ren H, Sun W, Zhang H, Sun J, Yang S, Hao J. Hypoxia-inducible factor-1 promotes pancreatic ductal adenocarcinoma invasion and metastasis by activating transcription of the actin-bundling protein fascin. Cancer Res. 2014;74:2455–2464. doi: 10.1158/0008-5472.CAN-13-3009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


