Abstract
1-Deoxysphingolipids (deoxySLs) are atypical sphingolipids that are elevated in the plasma of patients with type 2 diabetes and hereditary sensory and autonomic neuropathy type 1 (HSAN1). Clinically, diabetic neuropathy and HSAN1 are very similar, suggesting the involvement of deoxySLs in the pathology of both diseases. However, very little is known about the biology of these lipids and the underlying pathomechanism. We synthesized an alkyne analog of 1-deoxysphinganine (doxSA), the metabolic precursor of all deoxySLs, to trace the metabolism and localization of deoxySLs. Our results indicate that the metabolism of these lipids is restricted to only some lipid species and that they are not converted to canonical sphingolipids or fatty acids. Furthermore, exogenously added alkyne-doxSA [(2S,3R)-2-aminooctadec-17-yn-3-ol] localized to mitochondria, causing mitochondrial fragmentation and dysfunction. The induced mitochondrial toxicity was also shown for natural doxSA, but not for sphinganine, and was rescued by inhibition of ceramide synthase activity. Our findings therefore indicate that mitochondrial enrichment of an N-acylated doxSA metabolite may contribute to the neurotoxicity seen in diabetic neuropathy and HSAN1. Hence, we provide a potential explanation for the characteristic vulnerability of peripheral nerves to elevated levels of deoxySLs.
Keywords: lipids/chemistry, sphingolipids, chemical synthesis, inborn errors of metabolism, neurons, diabetes, metabolic syndrome, mitotoxicity, peripheral neuropathy, ES-285
1-Deoxysphingolipids (deoxySLs) are a recently discovered class of atypical sphingolipids that are generated when serine palmitoyltransferase (SPT) uses l-alanine instead of l-serine in the condensation reaction with palmitoyl-CoA. This leads to the generation of 1-deoxysphinganine (doxSA) instead of canonical sphinganine (SA) (1, 2). Specific point mutations in SPT, which induce a pronounced shift in the preference of the enzyme toward l-alanine, were found to be causal of the rare genetic peripheral neuropathy hereditary sensory and autonomic neuropathy type 1 (HSAN1) (2). This autosomal-dominant inherited disease is characterized by slow and length-dependent dying-back of sensory axons, with variable damage also to motor and autonomic neurons (3). To date, eight different mutations in SPT have been reported to cause HSAN1 (4–9), and the severity of the disease correlates with the type of mutation (10) as well as with the deoxySL levels in the patients’ plasma (11). DoxSA is neurotoxic in vitro (2), and suppression of doxSA generation by overexpression of the wild-type enzyme or by l-serine supplementation prevented the onset and improved the clinical signs of the disease in HSAN1 mutant mice (12, 13). More recently, similar findings have been reported from an HSAN1 Drosophila model (14). Hence, the increased doxSA levels in HSAN1 patients are thought to be causal of neuronal degeneration in this disease.
Total plasma levels of deoxySLs can be as high as 1.2 µM in HSAN1 patients (2) compared with 0.1–0.3 µM found in healthy individuals (15). Plasma levels are also significantly elevated in patients suffering from metabolic syndrome and type 2 diabetes (15–17). Furthermore, the neuropathological characteristics of HSAN1 closely resemble those of diabetic peripheral neuropathy, and the reduction of plasma deoxySL levels by dietary l-serine supplementation significantly improves clinical signs of neuropathy in diabetic rats (18). Taken together, these findings suggest that doxSA, or one of its downstream metabolites, also contributes to neuronal degeneration in diabetic neuropathy. The exact pathomechanism of doxSA-mediated neurotoxicity, however, still remains unknown.
So far, little is known about the metabolism of deoxySLs (supplemental Fig. S1). DoxSA is N-acylated to 1-deoxydihydroceramides (doxDHCers) by ceramide synthases (CerSs) (19, 20), and desaturated downstream metabolites of doxDHCers, i.e., 1-deoxyceramides (doxCers) (2, 21–23) and 1-deoxysphingosine (doxSO) (24), have been detected. Because deoxySLs lack the hydroxyl group at the C1 position, it appears unlikely that they are used for the synthesis of complex sphingolipids or that they are degraded via sphingosine-1-phosphate (S1P) by S1P lyase (25), although this has never been experimentally addressed.
In contrast to canonical sphingolipids, the transport and/or localization of deoxySLs in cells remains unknown. The localization, transport routes, and protein-lipid interactions of deoxySLs could be the same as for canonical sphingolipids, but this has not been shown yet. Indeed, the study of the localization and trafficking of lipids harbors some experimental challenges. Their subcellular localization is most often detected by directly fluorescently labeling the lipids. However, the conjugation of a fluorophore can alter the lipid characteristics substantially, affecting its downstream transport and metabolism. For example, nitrobenzoxadiazole- or BODIPY-ceramides are trapped in the Golgi and do not show the expected distribution in the endoplasmic reticulum (ER) and plasma membrane (26). Thus, to reduce the risk of artifacts, the structural modification of the lipid should be as minimal as possible to avoid changing its size and polarity.
Here, we developed a novel doxSA probe, which is highly similar to its natural counterpart, with the aim of answering the following fundamental questions: Can deoxySLs be recycled to canonical sphingolipids or fatty acids? Are there major, so far unknown, downstream doxSA metabolites? Where do deoxySLs localize in cells? And can these results provide valuable insights into the biology of deoxySL-mediated neurotoxicity? Our data show that deoxySLs cannot be recycled to canonical sphingolipids or fatty acids and that the class of deoxySLs is restricted to only a few lipid species. Exogenously added (2S,3R)-2-aminooctadec-17-yn-3-ol (alkyne-doxSA) prominently localizes to mitochondria, inducing changes in the organelle morphology such as swelling, rounding up, and increased fragmentation. Exogenous treatment with natural doxSA, but not SA, also induced the observed changes in the mitochondrial phenotype, in particular, loss of internal cristae structures, and inhibited normal mitochondrial function in situ. Cotreatment with doxSA and the CerS inhibitor Fumonisin B1 (FB1) fully rescued mitotoxicity caused by doxSA treatment in cultured fibroblasts. In adult sensory neurons, doxSA-induced changes in mitochondrial phenotype preceded axonal degeneration, arguing that mitotoxicity could underlie deoxySL-induced neurotoxicity in HSAN1, but potentially also in diabetic neuropathy.
MATERIALS AND METHODS
Lipid and chemical probes
The synthesis of alkyne-doxSA is described in detail in the supplemental material to this study and depicted in supplemental Fig. S2. Synthesis of (2S,3R)-2-aminooctadec-17-yn-1,3-diol (alkyne-SA) and alkyne-oleate are published in Refs. 27 and 28, respectively. Synthesis of azido-sulfo-tetramethyl-BODIPY (ASTM-BODIPY) was published in Ref. 29. The sphingoid base probes d3-doxSA, d7-SA, and d7-SO and the ceramide probes d18:1/12:0 and m18:0/12:0 were purchased from Avanti Polar Lipids.
Cell lines and general cell culture procedures
The following cell lines were used in this study: HuH7 (human hepatocarcinoma; JCRB0403), HepG2 (human hepatocarcinoma; ATCC-HB8065), HCT116 (human colorectal carcinoma; ATCC-CCL247), A431 (human epidermoid carcinoma; ATCC-CRL1555), B104 (mouse neuroblastoma; ATCC-CRL1887), and A172 (human glioblastoma; ATCC-CRL1620). Generation of the mouse embryonic fibroblast (MEF) cell lines, MEF wild-type and MEF S1P lyase-deficient (MEF SGPL1−/−), were described previously (30). HuH7 cells were cultured in RPMI medium (Gibco) containing 10% fetal calf serum (Gibco) and 1% penicillin/streptomycin (Gibco). All other cell lines were cultured in DMEM medium (Gibco) containing 10% fetal calf serum (Gibco) and 1% penicillin/streptomycin (Gibco).
Cell culture for LC/MS analysis
MEF cells were cultured until ∼70% confluent and then treated with 1 µM labeled lipid probes (alkyne-doxSA, d3-doxSA, alkyne-SA, or d7-SA) from ethanolic stock solutions. Cells were harvested after various incubation times. If needed, the medium was collected, lyophilized, and then stored at −20°C until lipid extraction. Upon harvesting, cells were first washed with PBS and then trypsinized. Cells were counted and washed with PBS, and cell pellets were stored at −20°C until lipid extraction.
Analysis of sphingoid and deoxy-sphingoid backbones by LC/MS
Frozen cell pellets and lyophilized medium were first resuspended in 100 µl of PBS. Then, 500 µl methanol, including 200 pmol internal standard (d7-SA and d7-SO), was added to each sample. Samples were agitated for 1 h at 37°C and centrifuged at 16,000 g for 5 min to pellet precipitated protein, and the supernatant was transferred to a new tube before acid-base hydrolysis was performed as described earlier (2, 31).
Lipids were redissolved in 80 µl of derivatization mix [75 µl 56.7% methanol, 33.3% ethanol, and 10% water + 5 µl of ortho-phthaldialdehyde working solution (990 µl 3% borate + 10 µl of 50 mg/ml o-phthalaldehyde in ethanol, + 1.5 µl 2-mercaptoethanol)] for analysis by LC/MS. A C18 column (Uptisphere 120 Å, 5 µm, 125 × 2 mm; Interchim, Montluçon, France) and a binary solvent system were used, with solvent A as 1:1 methanol/5 mM ammonium acetate in water and solvent B as methanol. The column was equilibrated by using a mixture of 50% solvent A and 50% solvent B, and 25 µl sample was injected. The gradient then ramped to 100% solvent B over 32 min, followed by a ramping back down to 50% solvent A/50% solvent B for 1 min, and column equilibration for 2 min with the same mixture before loading of the next sample. An atmospheric pressure chemical ionization (APCI) source was used for ionization, and detection of ions was performed in full scan mode by using a Q-Exactive hybrid quadrupole Orbitrap mass spectrometer (Thermo, Reinach, BL, Switzerland). The detection parameters were as follows: scan range of m/z 120–1,200, mass resolution of 140,000, maximum injection time 512 ms.
Labeled 1-deoxysphingoid and sphingoid base backbones were identified based on their accurate mass (within 5 ppm) and retention times as compared with external standards (except for alkyne-doxSO, for which a standard does not exist). Peaks were quantified in XCalibur (Thermo) by integrating the area under the peak and normalizing to the added internal standards.
Extraction and analysis of ceramides and doxCers by LC/MS
Frozen cell pellets were resuspended in 100 µl of PBS. Then, 666 µl methanol and 333 µl chloroform, including 200 pmol internal standards (d18:1/12:0; m18:0/12:0), were added. The samples were then agitated at 1,000 rpm and 37°C for 1 h, followed by the addition of 500 µl chloroform and 200 µl alkaline water (20 mM ammonia in water). The samples were centrifuged at 16,000 g and the water phase was removed. The organic phase was then washed three times with 1 ml of alkaline water. Lipids were dried under a steady stream of nitrogen, resuspended in methanol, and analyzed by LC/MS.
The same binary solvent system and LC column as above were used for the liquid chromatography. However, in this case, a ratio of 70% solvent A and 30% solvent B was used for loading, followed by the gradient ramping to 100% solvent B until 28 min, which remained until 69 min. The gradient then ramped down to 70% solvent A and 30% solvent B for the next 3 min, and remained at this ratio for the last 3 min for column equilibration. An APCI source was used for ionization, and the MS was run in full scan and all ion fragmentation modes. All other method parameters were the same as for the acid-base hydrolyzed sample method.
Identification of alkyne- and d3-labeled doxDHCer and doxCer species
The alkyne and deuterium labeled N-acyl-1-deoxySL-derivatives were identified, and the findings confirmed, in multiple ways (supplemental Fig. S3, supplemental Table S1). Identification was performed based on accurate mass (within 5 ppm) of the nonfragmented molecular ions, as well as the appropriate fragments, including the loss of one water molecule, but not two, and the 1-deoxysphingoid base backbone. For the alkyne-ceramide species, identification was based on full mass, the loss of two water molecules, and the labeled sphingoid base backbone. The mass of the most abundant fragment of alkyne-doxSA (alkyne-doxSA – water = 264.26837 Da) is equal to the mass of a sphingosine minus two water molecules, whereas the mass of the most abundant alkyne-doxSO fragment (alkyne-doxSO – water = 262.25256 Da) is equal to the mass of a SA backbone minus two water molecules. Therefore, in order to distinguish exactly which species arose from the alkyne-doxSA, the selected ion chromatogram for the masses of 264.26837 and 262.25256 Da for the alkyne-doxSA-treated cells was compared with that of untreated cells and unlabeled doxSA-treated cells. If a peak only existed in the alkyne-doxSA-treated cells, then this confirmed that this species was an alkyne-doxDHCer or alkyne-doxCer species. The alkyne-deoxySL species were further confirmed through checking the mass spectrum of each peak. If a peak showed the loss of two waters in the spectrum, this indicated that it was a ceramide, and not an alkyne-doxDHCer or alkyne-doxCer.
Quantification was performed in XCalibur (Thermo) and was based on spiking the cell extracts with internal standards (d18:1/12:0; m18:0/12:0). This quantification is only relative; for absolute quantification, one would need an internal standard for each species, including all of the alkyne species.
Analysis of the metabolism of alkyne lipids by TLC
This method has been described previously (28). In brief, cells were incubated with alkyne lipids added to the growth medium from ethanolic stock solutions. For pulse-chase experiments, the alkyne lipid-containing medium was removed, and cells were washed once with growth medium and then further cultured in fresh growth medium. Harvested cells were washed once with PBS and subjected to methanol/chloroform extraction. Dried lipids were redissolved in chloroform and 30 µl click-reaction mix (10 µl of 2 mg/ml 3-azido-7-hydroxycoumarin, 250 µl of 10 mM [acetonitrile]4 CuBF4 in acetonitrile, and 850 µl of ethanol) was added. The click reaction was performed at 43°C for 3 h, and then the click-reacted lipids were applied to TLC silica plates and separated in solvent 1 (chloroform/methanol/water/acetic acid 65/25/4/1, two-thirds of the plate). TLCs were dried and then developed in solvent 2 (hexane/ethyl acetate 1/1, whole plate). Saturated versus desaturated lipids such as SA/SO, doxSA/doxSO, or dihydroceramide (DHCer)/ceramide were not separated from each other by using this TLC method. Directly before fluorescence detection, TLCs were briefly soaked in 4% (v/v) N,N,-diisopropylethylamine in hexane. Fluorescent images of the TLC plates were acquired by using a 420 nm LED (Roithner Lasertechnik, Vienna, Austria) filtered through a colored glass filter (HEBO V01, Hebo Spezialglas) for excitation and an electron multiplying charge coupled device camera (Rolera-MGi Plus Fast 1394, Q-imaging), equipped with 494/20 (channel for detection of the coumarin signal) and 572/28 (channel for detection and correction of background fluorescence) bandpass emission filters. Lipid bands were identified according to their running heights compared with chemically synthesized alkyne standards of different lipid classes. Synthesis of alkyne lipid TLC standards has been previously described (28). Quantification was performed by using the GelPro analyzer software.
Analysis of subcellular localization of alkyne lipids by fluorescence microscopy
Cells were incubated with alkyne lipids added to the growth medium from ethanolic stock solutions. Cells were washed once with PBS and fixed in 3.7% formalin in PBS for 10 min. They were then washed sequentially with 155 mM ammonium acetate, PBS, and 100 mM Hepes-KOH pH 7.4. For the click reaction, 1,000 µl prewarmed 100 mM Hepes-KOH, pH 7.4, containing 25 nmol ASTM-BODIPY, was added per well. The reaction was initiated by addition of 20 µl of 100 mM Cu(I)TFB in acetonitrile, and the dish was incubated for 1 h at 43°C. The samples were then washed sequentially with 20 mM EDTA, 155 mM ammonium acetate, and PBS. Coverslips were mounted in Mowiol 4-88 (Calbiochem).
Mitotracker CMXRos (Molecular Probes M7512) staining was performed according to the manufacturer’s instructions. In brief, cells were incubated with 50 nM Mitotracker probe in growth medium for 15 min before cell fixation. For immunofluorescence, antibody staining was performed before submitting the samples to the click reaction. The antibodies used in this study were as follows: anti-Tom20 (Santa Cruz, sc-11415; 1:1,000), anti-Giantin (Abcam 24586; 1:200), anti-protein-disulfide-isomerase (anti-PDI; Stressgen SPA891; 1:200), and anti-Lamp1 (Abcam ab25245; 1:1,000). All antibodies were diluted in 1% cold fish gelantin/0.01% saponin in PBS, except anti-PDI, which was diluted in 1% cold fish gelantin/0.1% saponin in PBS. Secondary antibodies were labeled with Alexa 350, Alexa 555, or Alexa 647 (Invitrogen). Actin filaments were stained with 1 µM Phalloidin-tetramethylrhodamine isothiocyanate (Phalloidin-TRITC) (Sigma P1951) and the cell nuclei with 4’,6-diamidino-2-phenylindole (DAPI).
Epifluorescence microscopy was performed by using a Zeiss Observer.Z1 microscope equipped with a Plan-Apochromat 63× [1.40 numerical aperture (NA)] and a Photometrics Coolsnap K4 camera. Images were processed by using Fiji software (32).
Quantification of mitochondrial fragmentation in fibroblasts
MEF cells were treated for the indicated times with lipid (doxSA/SA; Avanti Polar Lipids) and inhibitor (FB1; Sigma Aldrich) from ethanolic stock solutions. Cells were fixed, and mitochondria were stained by using anti-Tom20 antibody. Cells were additionally stained with DAPI and Phalloidin-TRITC. Four images of 250 µm × 250 µm were acquired per condition and randomly numbered. In a blinded fashion, cells were counted and assigned as either displaying tubular or fragmented mitochondria or judged to be close to detachment and cell death.
X-box binding protein 1 splicing assay
X-box binding protein 1 (XBP1) splicing assay was performed according to the protocol of Iwakoshi et al. (33). In brief, RNA was isolated with QIAzol Reagent (Qiagen), and cDNA was synthetized by using random primer (Invitrogen, 48190011) and Moloney murine leukemia virus Reverse Transcriptase (Promega). Qualitative PCR was performed by using primers surrounding the XBP1 splicing site (5′-ACACGCTTGGGAATGGACAC-3′ and 5′-CCATGGGAAGATGTTCTGGG-3′). Products were separated by a 3% agarose gel electrophoresis and visualized by SYBR-Green staining (Thermo Fisher Scientific). Quantification was performed by using the GelPro analyzer software.
Quantification of cellular ATP levels
MEF cells were treated for the indicated times with lipid (doxSA/SA, Avanti Polar Lipids) and inhibitor (FB1; Sigma-Aldrich) from ethanolic stock solutions. Cells were lysed in buffer containing 100 mM Hepes, pH 7.4, 4 mM EDTA, and 0.1% Triton X-100. The ATP content was quantified by using the CellTiter-Glo Luminescent Viability Assay (Promega), according to the manufacturers’ instructions. The protein content was determined by using Bradford Reagent (Sigma-Aldrich). ATP levels were normalized to protein content.
Quantification of oxygen consumption rate and glycolytic flux evaluation
Real-time oxygen-consumption rate and extracellular acidification rate were determined by using a XF-96 Extracellular Flux Analyzer (Seahorse Bioscience). A total of 1 × 104 cells were seeded per well. After attachment, cells were treated for the indicated times with lipid and inhibitor from ethanolic stock solutions. The assay was performed in triplicate measurements in bicarbonate-free RPMI medium supplemented with 11 mM glucose, 1 mM pyruvate, and 2 mM glutamine. Three consecutive measurements were performed under basal conditions and after the sequential addition of 1 µM oligomycin, 2 µM fluoro-carbonyl cyanide phenylhydrazone (FCCP), 1 µM rotenone, and 1 µM antimycin, respectively. All reagents were purchased from Sigma, except FCCP, which was purchased from Tocris.
Ultrastructural analysis of mitochondria by transmission electron microscopy
MEF cells were seeded onto poly-l-lysine-coated glass bottom dishes. After attachment, cells were incubated for 24 h with 1 µM doxSA added to the growth medium from an ethanolic stock solution. Control cells were treated with the same amount of pure ethanol (final concentration 0.2%). Fixation was performed by addition of the fixative [8% paraformaldehyde, 5% glutaraldehyde in CB buffer (66 mM cacodylate buffer, pH 7.2)] to the growth medium at a 1:1 vol ratio. After primary fixation (room temperature, 3 h), washing with CB, postfixation (2.5% glutaraldehyde in CB, 4°C, 16 h), twice washing with CB containing 200 mM sucrose and twice with CB, the samples were contrasted by using 1% osmiumtetroxide in CB on ice for 2 h. After washing with CB and water, the samples were incubated with 2% uranium acetate in water at 4°C for 2 h before final water washes. Sample dehydration using ethanol and Epon embedding and polymerization followed. Coverslips were removed, and sections (80 nm thickness) oriented in-plane to the cell growth support were produced and mounted onto slot grids. Electron micrographs and tomography tilt series were acquired on a JEOL JEM-2200FS microscope operated at 200 kV, equipped with a TemCam-F416 camera (TVIPS, Munich, Germany) and using a 20 eV energy filter slit. The microscope was controlled by the SerialEM software. Under focus was adjusted to 3,000 nm. Micrographs were acquired 2-fold binned. Before tomography data acquisition, areas of interest were preexposed for at least 10 min. Tilt series, collected over a total angular tilt range from −72° to +72° at 2.0° increments, were first aligned with one another by cross-correlation and subsequently by patch tracking using the IMOD software package (ver. 4.7; http://bio3d.colorado.edu/imod). A single reconstructed volume was computed from each tilt series by radially weighted back projection.
Isolation of mitochondria and analysis of mitochondrial membrane potential
MEF cells were grown to 85–95% confluence in 150 × 20 mm culture plates. The medium was aspirated, and cells were then washed two times with PBS. The cell monolayer was resuspended in 5 ml of cold isolation buffer (IB; 200 mM mannitol, 50 mM sucrose, 5 mM KH2PO4, 5 mM MOPS, 0.1% fatty acid free BSA, and 1 mM EGTA, pH 7.15) and transferred to a Sorvall centrifuge tube. The plate was then washed with another 5 ml cold IB, which was also transferred to the centrifuge tube. Three plates were pooled together. The cells were then centrifuged for 10 min at 4°C and 478 g in an SS-34 rotor (Sorvall, Thermo Fisher Scientific). The supernatant was removed, and the cell pellet was retained.
Cell pellets were resuspended in 3.5 ml cold IB and homogenized by 40 strokes with the tight plunger at 1,600 rpm, and the homogenate was centrifuged for 3 min at 1,075 g and 4°C. The supernatant was collected and spun for 10 min at 11,950 g and 4°C. Next, the supernatant was decanted, and the inner walls of the centrifuge tube were dried. The mitochondrial pellet was resuspended in 200 µl of cold respiration buffer (RB; 130 mM KCl, 5 mM K2HPO4·3 water, 20 mM MOPS, 2.5 mM EGTA, 3 mM succinate, 0.1% BSA, pH 7.15) and spun at 9,500 g for 10 min. The supernatant was again decanted, and the mitochondrial pellet was resuspended in 50 µl RB. Mitochondria and all equipment were kept on ice for the duration of the isolation and the assay.
A solution of RB containing 10 µM rhodamine 123 was prepared, and mitochondria were added to a working density of 0.2 mg/ml. The fluorescence values were read at excitation 470–495 nm and emission 520–550 nm (Varian Cary Eclipse fluorescence spectrophotometer, Agilent) upon addition of doxSA (Avanti Polar Lipids), SA (Avanti Polar Lipids), and FB1 (Sigma-Aldrich) from ethanolic stock solutions. Sodium salicylate (5 mM) was used as a positive control. Fluorescence values were read every 5 s up to 2.5 min.
Analysis of doxSA-mediated toxicity on primary dorsal root ganglia neurons
All animal experiments were performed in accordance to the Animal Welfare Act and the guidelines of the Landesamt für Natur, Umwelt und Verbraucherschutz. Female C57BL/6J mice (8 weeks old, Charles River) were used for all primary neuronal culture experiments.
Adult lumbar dorsal root ganglia (DRG) were dissected and collected in ice-cold Hank’s balanced salt solution (HBSS; Gibco). The ganglia were transferred into a sterile tube, washed twice with HBSS, and incubated in Neurobasal-A medium (Gibco) containing collagenase type I (3,000 U/ml; Worthington) at 36.5°C for 15 min, followed by 30 min with trypsin (0.25%; Gibco). Serum was then added to stop trypsin digestion. Ganglia were dissociated by gently pipetting up and down. The cell suspension was filtered by using a nylon cell strainer (70 μm; BD Falcon) and centrifuged at 76 g for 5 min. Dissociated neurons were resuspended in Neurobasal-A medium supplemented with B-27 (Gibco) and were plated at low density on laminin-coated (5 μg/ml; Roche) coverslips.
The culture was maintained in a humidified atmosphere containing 5% CO2 in air at 36.5°C. Vehicle (100% ethanol) or sphingolipids in ethanol were added 2 h after plating, when neurons were already attached to the substrate.
DRG neurons were fixed with 4% paraformaldehyde and 4% sucrose. Cells were immunostained for Tuj1 (Sigma, T0198; 1:1,000) and Tom20 (Santa Cruz, sc-11415; 1:1,000). When necessary, the alkyne-moiety was subsequently reacted with ASTM-BODIPY, as described above for the staining of fibroblasts. Secondary antibodies were labeled with Alexa 350, Alexa 488, or Alexa 555 (Invitrogen). Epifluorescence microscopy was performed by using a Zeiss Observer.Z1 microscope equipped with a Plan-Apochromat 63× (1.40 NA) and a Photometrics Coolsnap K4 camera. Optical sectioning was performed by using the apotome mode. If applying, maximum projections of z-stacks were calculated by summarizing corresponding pixel values. Images were processed by using Fiji software.
Quantification was carried out for five independent experiments. For quantification of mitochondrial morphology, images were randomly taken with an Axiovert microscope (Zeiss). In a blinded setup, cells with neurite outgrowth were assigned to display normal or disturbed mitochondrial morphology. Disturbed mitochondrial morphology was assigned for swollen or irregularly distributed mitochondria in at least three neurites. For quantification of neurite length, images were randomly taken with an Axiovert microscope (Zeiss) and analyzed by using ImageJ software (NIH). More than 200 neurons for each condition were quantified for mitochondrial morphology and neurite length, respectively.
RESULTS
Uptake, N-acylation, and desaturation of doxSA differs greatly from that of the canonical variant SA
To answer basic questions regarding deoxySL biology, we synthesized an alkyne analog of doxSA that structurally differs from the natural version only by the presence of an alkyne moiety (triple bond) between C17 and C18. All alkyne lipids used in this study are depicted in supplemental Fig. S4A. The alkyne group does not alter the polarity and only minimally changes the size of the lipid, while facilitating a highly specific and sensitive tracking of the probe via a copper(I)-catalyzed azide alkyne cycloaddition click reaction (34). The click reaction covalently attaches an azido-reporter molecule (most often an azido-fluorophore) to the alkyne group of the probe, and, in recent years, robust protocols have been developed that allow the use of alkyne lipids to answer biological questions (35). These methods include tracking the metabolism of alkyne lipids by fluorescence TLC (28) and the determination of their localization within cells by fluorescence microscopy (29, 36–38) (supplemental Fig. S4B). Here, we chemically synthesized alkyne-doxSA (supplemental Fig. S2). Its canonical sibling, alkyne-SA, was previously used for in vitro CerS assays (27) and localization studies (36).
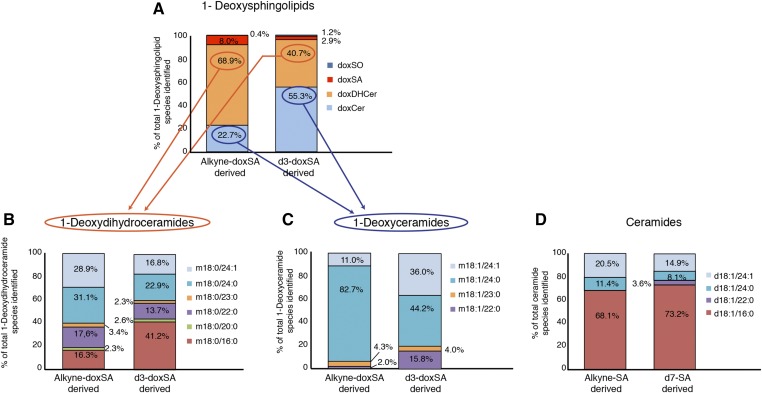
To verify the suitability of the alkyne probes, we first compared the uptake, desaturation (supplemental Fig. S5) and N-acylation profiles (Fig. 1) of alkyne-doxSA and alkyne-SA with that of their deuterated variants d3-doxSA and d7-SA, respectively, in MEFs. Our data showed a comparable rate of uptake for the alkyne compared with the related deuterated probes and a faster cellular uptake of doxSA compared with SA (supplemental Fig. S5A, B). Desaturation of doxSA was slower than that for SA, and the rate of desaturation for both alkyne probes lagged behind that of the deuterated ones (supplemental Fig. S5C, D). We also analyzed the downstream N-acylation patterns of all probes after an incubation time of 24 h. Labeled deoxySLs were found in substantial quantities as free sphingoid bases, doxDHCers, and doxCers (Fig. 1A). We further observed great differences in the N-acylation pattern of 1-deoxySLs versus canonical sphingolipids. Whereas labeled doxDHCers were found with a variety of N-acyl fatty acids (C16:0-C24:1) attached (Fig. 1B), the majority of labeled doxCers contained lignoceric or nervonic acid (C24:0 and C24:1) (Fig. 1C). In contrast, the majority of labeled ceramides were N-acylated with palmitic acid (C16:0) (Fig. 1D). Notably, these N-acylation profiles were obtained from MEF cells, and, as established for canonical sphingolipids, the produced ceramide species will most likely depend on the expression levels of the different CerS enzymes in the specific cell type under investigation. However, our data and earlier measurements of 1-deoxySLs compared with canonical ceramide species in LLC-PK (1) and MEF cells (21) clearly suggest that mammalian CerSs are discriminatory, not only for the accepted acyl-CoA, but also for the preferred sphingoid base substrate. This notion is in concordance with the fact that we observed a reduced amount of desaturated metabolites and a slight tendency for longer fatty acids to be attached to the alkyne sphingoid bases compared with the respective deuterated sphingoid bases. Hence, the responsible dihydroceramide desaturase and CerSs seem to be sensitive to the slightly shorter sphingoid backbone [triple bonds between two carbons are 34 pm shorter than single bonds (39)], emphasizing the importance that the structural modification of a probe should be as minimal as possible.
Fig. 1.
Alkyne-(deoxy)sphingoid bases are converted to the same downstream metabolites as their deuterated counterparts. LC/MS analysis of the downstream metabolism of the sphingoid base probes is shown. MEF cells were treated for 24 h either with alkyne-doxSA and d3-doxSA or with alkyne-SA and d7-SA. The proportion of each labeled downstream lipid is shown as the percentage of the total amount of the labeled lipid species identified. A: Alkyne- and d3-labeled deoxySL-classes detected. Please note that doxCers comprise only part of the deoxySLs made downstream of the doxSA precursors after a 24 h incubation. The rest of the label is present in the saturated forms doxDHCer and the free doxSA and doxSO sphingoid bases. B: N-acylation patterns of alkyne-doxSA and d3-doxSA-derived doxDHCer species. C: N-acylation patterns of alkyne-doxSA and d3-doxSA-derived doxCer species. D: N-acylation patterns of alkyne-SA and d7-SA-derived ceramide species. Please note that no labeled DHCer species were detectable after a 24 h incubation with either alkyne-SA or d7-SA. Ceramide nomenclature: d (dihydroxy-sphingoid base) or m (monohydroxy-sphingoid base); number of C-atoms in sphingoid base; :number of double bonds in sphingoid base; / number of C-atoms in N-acyl fatty acid; : number of double bonds in N-acyl fatty acid.
DeoxySLs are not converted to canonical sphingolipids or fatty acids
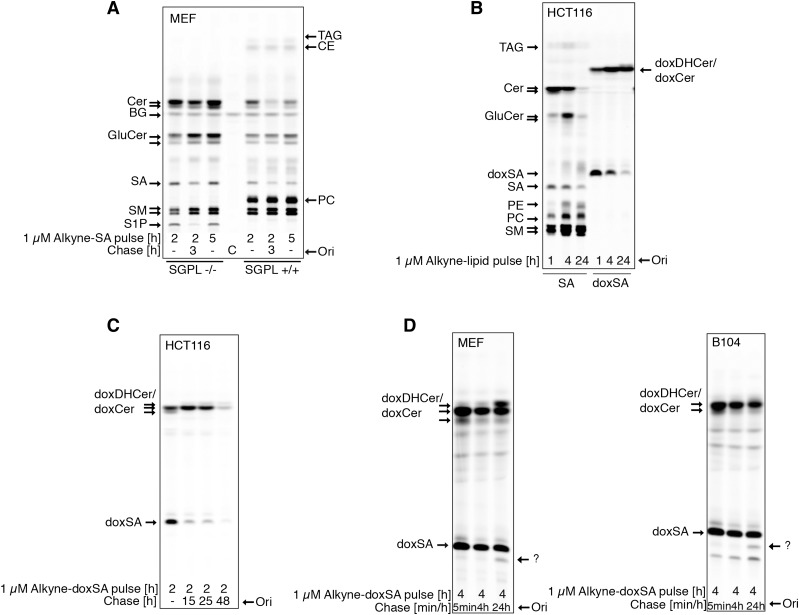
Canonical sphingolipids can be degraded via S1P by S1P lyase (SGPL1) to fatty aldehydes and, consequently, fatty acids. This fact was recapitulated by treatment of MEF cells with alkyne-SA that resulted in the appearance of the alkyne label in ceramides and more complex sphingolipids, such as sphingomyelin and glycosphingolipids (Fig. 2A, right lanes). In addition, the alkyne label was detectable in phosphatidylcholine (PC), phosphatidylethanolamine (PE), and triacylglycerol (TAG), as the alkyne label is transferred from SA to fatty acids. In concordance with this, fibroblasts without functional SGPL1 cannot convert alkyne-SA to alkyne fatty acids, as concluded from a lack of labeling in the PC, PE, or TAG bands for these cells (Fig. 2A, left lanes).
Fig. 2.
DeoxySLs are not recycled to canonical sphingolipids or fatty acids. Metabolic tracing of alkyne-doxSA and alkyne-SA by click fluorescence TLC. A: MEF WT or MEF S1P lyase−/− cells were treated with 1 μM alkyne-SA. Note that the alkyne signal is restricted to sphingolipids in MEF S1P lyase−/− cells, but also appears in glycerolipids in MEF WT cells. B. HCT116 cells were treated with 1 μM alkyne-SA or 1 μM alkyne-doxSA. Note that the alkyne signal of alkyne-doxSA was not detected in any canonical sphingolipids or glycerolipids. C: HCT116 cells were given a short pulse of 1 μM alkyne-doxSA for 2 h, followed by a chase up to 48 h in nonsupplemented growth medium. Note that the total alkyne signal decreased with prolonged chase times. D: MEF and B104 cells were treated with 1 μM alkyne-doxSA for 4 h, followed by a chase up to 24 h in nonsupplemented medium. The arrows indicate an alkyne signal that runs in a band at the height of SA after a 24 h chase, indicating a possible hydroxylated derivate of doxSA. Because of the lack of alkyne signal appearing in canonical sphingolipids or glycerolipids, it can be excluded that the hydroxylation would be at the C1 position. Please note that sphingolipid classes such as ceramides, glucosylceramides, and sphingomyelins run on the TLC according to the N-acyl fatty acid attached. Hence, most often, two bands (long-chain versus very-long-chain N-acyl metabolites) can be separated on the TLC. BG, background; C, control; CE, cholesterolester; Cer, ceramide; GluCer, glucosylceramide; Ori, origin; SM, sphingomyelin.
Hence, tracking the metabolism of lipids by using their alkyne-probes and detection by click-fluorescence TLC provides a good overview of the downstream metabolism of the respective lipid. We therefore screened different cell types with a particularly interesting canonical sphingolipid metabolism for potential cell type-specific doxSA metabolism. The screen included a glia cell line (A172) and a neuronal cell line (B104), because of the strong peripheral neurodegeneration seen in HSAN1 and diabetic neuropathy, but also because of the reported neurotoxic effects of doxSA on CNS neurons in vivo (40–43). We further tested liver cell lines (Huh7 and HepG2) because deoxySLs are mostly transported on VLDL and LDL (16) and therefore may be of hepatic origin (22). Fibroblasts (MEFs), epidermal (A431), and epithelial (HCT166) cells were chosen because the skin is also affected in HSAN1, and the dermal barrier strongly depends on a well-balanced sphingolipid synthesis. None of the investigated cell lines showed any transfer of the alkyne label of alkyne-doxSA to canonical sphingolipids or glycerolipids. The results from human epithelial cells are shown as an example (Fig. 2B). In detail, whereas incubation with alkyne-SA always resulted in labeling of diverse sphingolipids and glycerolipids, after treatment with alkyne-doxSA, doxDHCer/doxCer were the only lipids labeled, even after prolonged incubation (Fig. 2B) or chase times (Fig. 2C). Hence, although suspected previously, our tracing data allow us to conclude that doxSA is not subjected to hydroxylation at C1 for conversion to canonical sphingolipids in all cell types tested. The absence of labeled glycerolipids such as PC, PE, or TAG also suggests that no other metabolic pathway converts deoxySLs directly to fatty acids in appreciable amounts. In addition, the detailed analysis of pulse-chase experiments with alkyne-doxSA revealed a continuous reduction of the total alkyne signal during the chase times for all cell types tested. The loss of the label was fastest in HCT116 cells, where, after a 48 h chase, only 18% of the signal remained (Fig. 2C). In these experiments, we observed no apparent cell toxicity or detachment of the cells and excluded a signal loss due to secretion by analyzing the culture medium used during the chase. In addition, we searched for cell type-specific doxSA metabolites in the various cell lines (MEF, HCT116, A431, HuH7, HepG2, B104, and A172). In some cell lines, a minor metabolite of alkyne-doxSA was detected running on the TLCs at the position of SA (Fig. 2D). This indicates a doxSA hydroxylation at a position different from C1 in these cells. A C1 hydroxylation was ruled out because no canonical alkyne sphingolipids or alkyne glycerolipids were generated from alkyne-doxSA via the SGPL1 pathway.
Alkyne deoxySLs localize to mitochondria
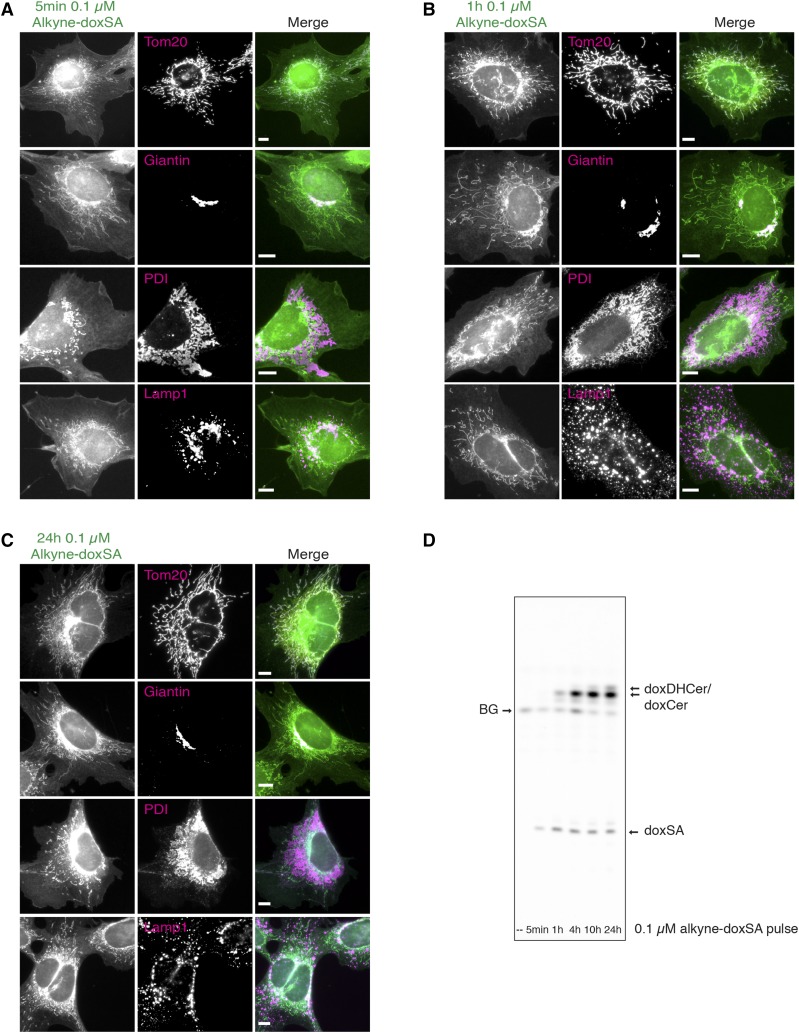
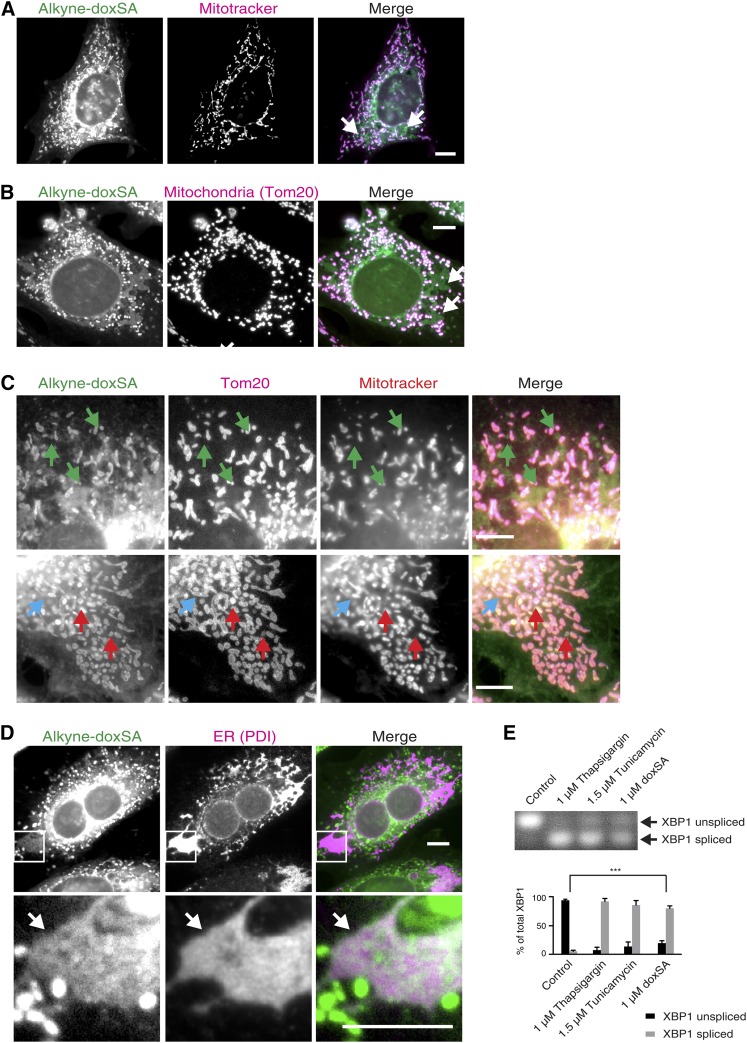
To study the subcellular localization of deoxySLs, we treated MEF cells with a nontoxic concentration of 0.1 µM alkyne-doxSA for up to 24 h and analyzed the distribution of alkyne lipids by fluorescence microscopy. Five minutes after treatment, we observed prominent labeling of mitochondria, as confirmed in costaining experiments using an antibody against the mitochondrial protein Tom20 (Fig. 3A). In addition, our data showed colocalization of perinuclear alkyne staining with immunofluorescence staining against the Golgi protein marker giantin. After 1 h (Fig. 3B) and 24 h of treatment (Fig. 3C), mitochondrial labeling remained the most prominent, whereas the intensity of the labeling increased over time. Apart from mitochondria, the alkyne deoxySLs remained detectable in Golgi and, to a minor degree, were detectable in other cellular structures such as the ER (1 and 24 h), coinciding with the conversion of alkyne-doxSA to doxDHCer and doxCer species (Fig. 3D). This was expected, because the enzymes responsible for the synthesis of doxDHCer and doxCer are located in the ER, although a CerS activity has also been reported for mitochondria (44). As shown by costaining against a lysosomal marker protein (Lamp1), no prominent colocalization of the alkyne-signal was seen for lysosomes within the analyzed treatment time (5 min to 24 h). In contrast to the exogenous treatment with alkyne-doxSA, exogenous treatment of cells with alkyne-oleate, a fatty acid probe that is mostly incorporated into PC (28), does show a very broad staining of all cellular membranes (supplemental Fig. S6A). In addition, incubation of cells with alkyne-doxSA after fixation did not lead to any appreciable cellular staining (supplemental Fig. S6B).
Fig. 3.
Exogenous alkyne-doxSA localizes to mitochondria. Subcellular tracking of alkyne-doxSA by fluorescence microscopy. MEF cells were treated with a nontoxic concentration of 0.1 μM alkyne-doxSA for 5 min (A), 1 h (B), or 24 h (C). After treatment for the indicated times, cells were fixed and immunostained for organelle protein markers (magenta), the alkyne moiety was reacted with ASTM-BODIPY (green), and the cells analyzed by fluorescence microscopy. Scale bars, 10 μm. D: Analysis of the metabolic fate of the alkyne probe by click-fluorescence TLC. BG, background.
DoxSA induces mitochondrial fragmentation
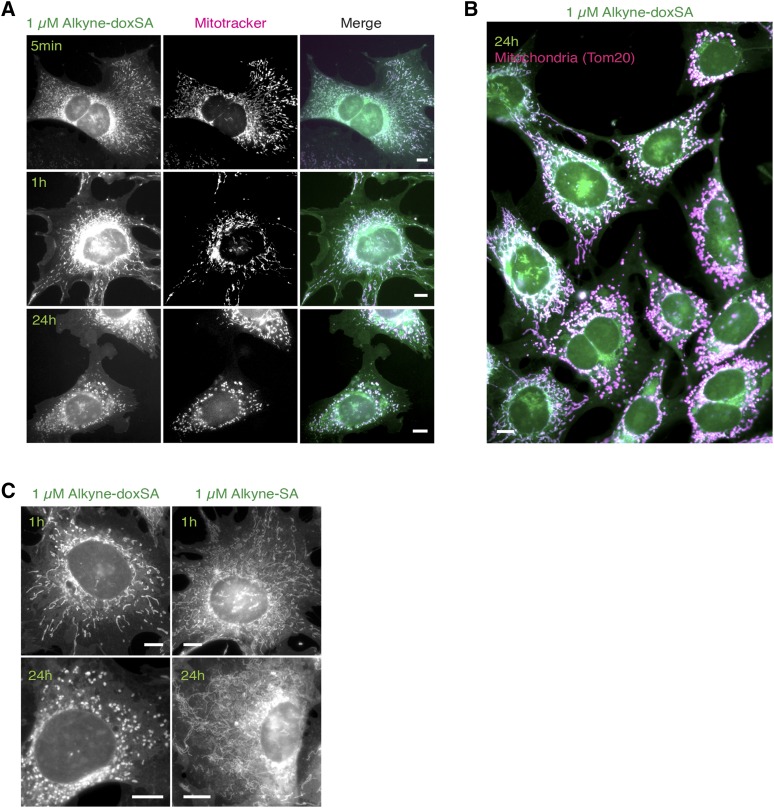
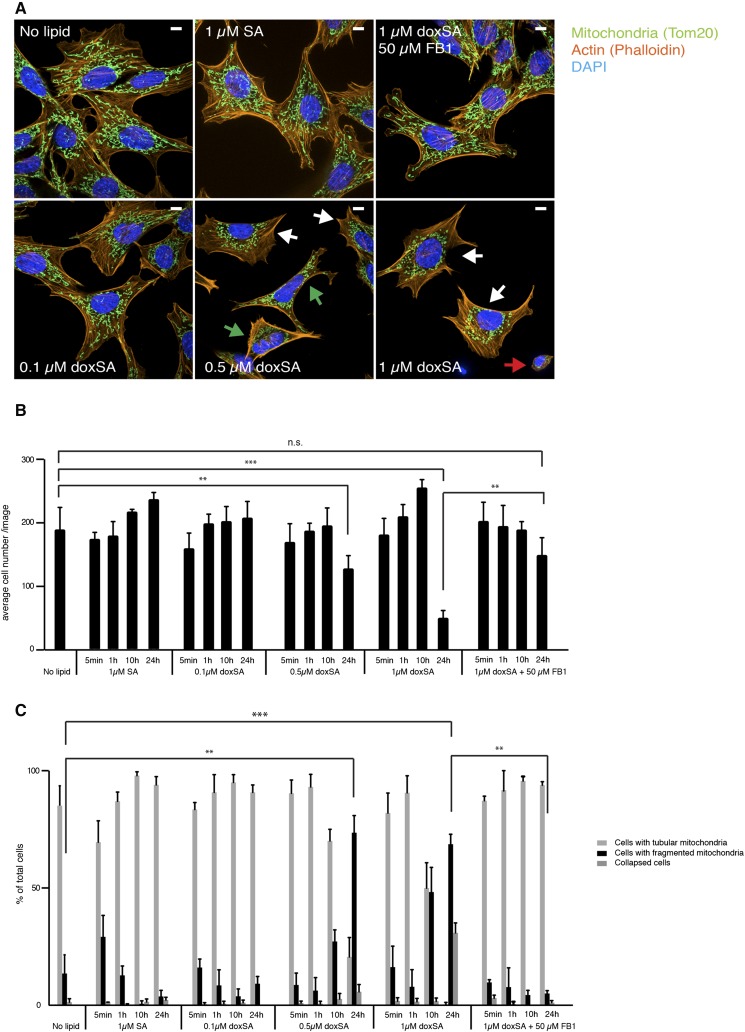
Next, we evaluated the cytotoxic effects of doxSA treatment. Treatment of MEF cells with a cytotoxic concentration of 1 µM alkyne-doxSA for only 5 min also allowed for the detection of the lipid in mitochondria, and the staining intensity increased with incubation time (Fig. 4A). However, in contrast to treating cells with a low alkyne-doxSA concentration (i.e., 0.1 µM; Fig. 3), treatment of MEF cells with 1 µM alkyne-doxSA resulted in changes in mitochondrial morphology. Whereas mitochondria of nontreated cells were mostly tubular with only some fissioning mitochondria, 1 µM alkyne-doxSA treatment led to swollen and hyperfused mitochondria (1 h), and prolonged presence resulted in fragmented mitochondria in approximately 50% of all cells (Fig. 4B). To validate the specificity of the effect, we compared the mitochondrial morphology upon alkyne-doxSA or alkyne-SA treatment (Fig. 4C). Treatment with 1 µM alkyne-SA also led to labeling of mitochondria, but with a comparatively stronger staining of other organelles. Most importantly, we did not observe any increase in mitochondrial fragmentation upon treatment with alkyne-SA. To corroborate these findings and to exclude any potential effect of the alkyne moiety, additional experiments using untagged natural doxSA and SA were performed (Fig. 5). Here, cells were treated for the indicated times with 1 µM SA, with increasing concentrations of doxSA (0.1, 0.5, and 1 µM), or with 1 µM doxSA in the presence of a CerS inhibitor (FB1), respectively. Cells were then fixed, stained, and assigned as displaying tubular-shaped mitochondria, fragmented mitochondria, or being collapsed, meaning being close to detachment and cell death (see Fig. 5A for representative pictures). Continuous treatment with 0.5 or 1 µM doxSA for 24 h was cytotoxic to the cells, as indicated by a significant reduction in average cell number per image (Fig. 5B). Mitochondrial fragmentation was significantly increased after 24 h with 0.5 or 1 µM doxSA, but not with 0.1 µM doxSA or 1 µM SA treatment (Fig. 5C). Also, cotreatment with 50 µM FB1 in combination with 1 µM doxSA fully prevented mitochondrial fragmentation. To further decipher this finding, we quantified the cellular uptake of alkyne-doxSA in the presence or absence of FB1 (supplemental Fig. S7). Cotreatment with FB1 did not change the uptake of alkyne-doxSA, whereas it significantly inhibited its N-acylation by CerSs. Hence, we conclude that an N-acyl downstream product, and not the 1-deoxysphingoid base itself, is responsible for the observed mitochondrial changes upon doxSA-mediated toxicity.
Fig. 4.
Exogenous alkyne-doxSA, but not alkyne-SA, treatment induces mitochondrial fragmentation. Subcellular tracking of toxic concentrations of alkyne-doxSA by fluorescence microscopy is shown. Scale bars, 10 μm. A: MEF cells were treated with 1 μM alkyne-doxSA for 5 min, 1 h, or 24 h. Mitochondria were stained by using mitotracker CMXRos (magenta); cells were fixed, and the alkyne moiety was reacted with ASTM-BODIPY (green). B: MEF cells were incubated for 24 h with 1 μM alkyne-doxSA. Cells were fixed and costained for the mitochondrial marker Tom20 (magenta) and the alkyne moiety was reacted with ASTM-BODIPY (green). C: MEF cells were treated for 1 or 24 h with either 1 μM alkyne-doxSA or 1 μM alkyne-SA before fixation and detection of the alkyne label with ASTM-BODIPY.
Fig. 5.
Mitochondrial fragmentation upon doxSA treatment depends on functional CerSs. MEF cells were treated for the indicated times with different concentrations of unlabeled doxSA or SA before fixation and staining of mitochondria (Tom20; green), actin filaments (Phalloidin; orange), and nucleus (DAPI; blue). Data are presented as average ± SD. * P < 0.05; ** P < 0.01; *** P < 0.001. A: Representative images of cells showing normal mitochondrial morphology, fragmenting (green arrows) and fragmented (white arrows) mitochondria, or a collapsed cellular phenotype with little visible cytoplasm around the nucleus (red arrow). Scale bar, 10 μm. B: Average cell number per image. C: Quantification of cells with normal mitochondria, fragmented mitochondria, or cells with collapsed actin filaments. n.s., not significant.
Prolonged incubation with alkyne-doxSA at toxic concentrations causes the appearance of dense alkyne-positive ER structures
We further noticed that prolonged exposure of cells to toxic concentrations of alkyne-doxSA not only resulted in fragmented mitochondria, but also led to changes in alkyne-doxSA localization and in the morphology of other organelles in the cultured MEF cells. In detail, large, intensely stained structures became visible upon long incubation with 1 µM alkyne-doxSA (Fig. 6A, white arrows). These structures only appeared after prolonged incubation, and the alkyne labeling did not coincide with either the membrane potential-sensitive staining of Mitotracker CMXRos (Fig. 6A) or the mitochondrial marker Tom20 (Fig. 6B), hence excluding a mitochondrial origin for most of these structures. However, a small fraction may represent mitochondria that have lost membrane potential, as triple staining revealed (Fig. 6C). Further costainings suggested that the majority of the observed structures are enlarged ER domains, as concluded from the overlapping signal of the alkyne staining with the ER protein PDI (Fig. 6D, white arrow). These findings indicate that prolonged treatment with alkyne-doxSA at toxic concentrations leads to morphological changes of the ER, from a net-like appearance to enlarged and dense-appearing sheets. To determine whether ER stress is occurring upon doxSA treatment, we analyzed the alternative splicing product of the XBP1, that is known to occur when ER stress triggers the unfolded protein response (45). Our data showed that XBP1 splicing was significantly induced after 24 h of exogenous treatment with doxSA (Fig. 6E).
Fig. 6.
Long-term incubation with alkyne-doxSA at toxic concentrations leads to prominent alkyne-positive ER structures and ER stress. MEF cells were treated for 24 h with 1 μM alkyne-doxSA. After fixation, the alkyne moiety was reacted with ASTM-BODIPY (green). Scale bars, 10 μm. Please note that unidentified alkyne-positive structures (white arrows) appear (after the long doxSA incubation at a toxic concentration), which do not colocalize with mitochondrial membrane-dependent Mitotracker staining (magenta) (A) or with the mitochondrial marker protein Tom20 (magenta) (B). C: Triple stainings show some mitochondria (Tom20-positive; magenta) that were alkyne-positive (green), but did not take up the Mitotracker stain (red), indicating total loss of the membrane potential (red arrows). In contrast, a blue arrow marks a mitochondrium (Tom20-positive; magenta), that took up neither the alkyne lipid (green) nor the Mitotracker stain (red), whereas green arrows indicate alkyne-positive structures of unknown origin that are not hyperfragmented mitochondria. D: Further immunofluorescence costainings show colocalization of the unidentified alkyne-positive (green) structures (white arrow) with the ER protein PDI (magenta). Please note that, because of the very prominent alkyne staining in mitochondria, alkyne staining in ER appears less bright, although highly specific. E: To probe for induction of ER stress, MEF cells were treated for 24 h with 1 μM doxSA, and alternative splicing of the ER stress marker XBP1 was analyzed, showing that, apart from mitochondrial hyperfragmentation, doxSA toxicity leads to ER stress. Data are presented as average ± SD. *** P < 0.001.
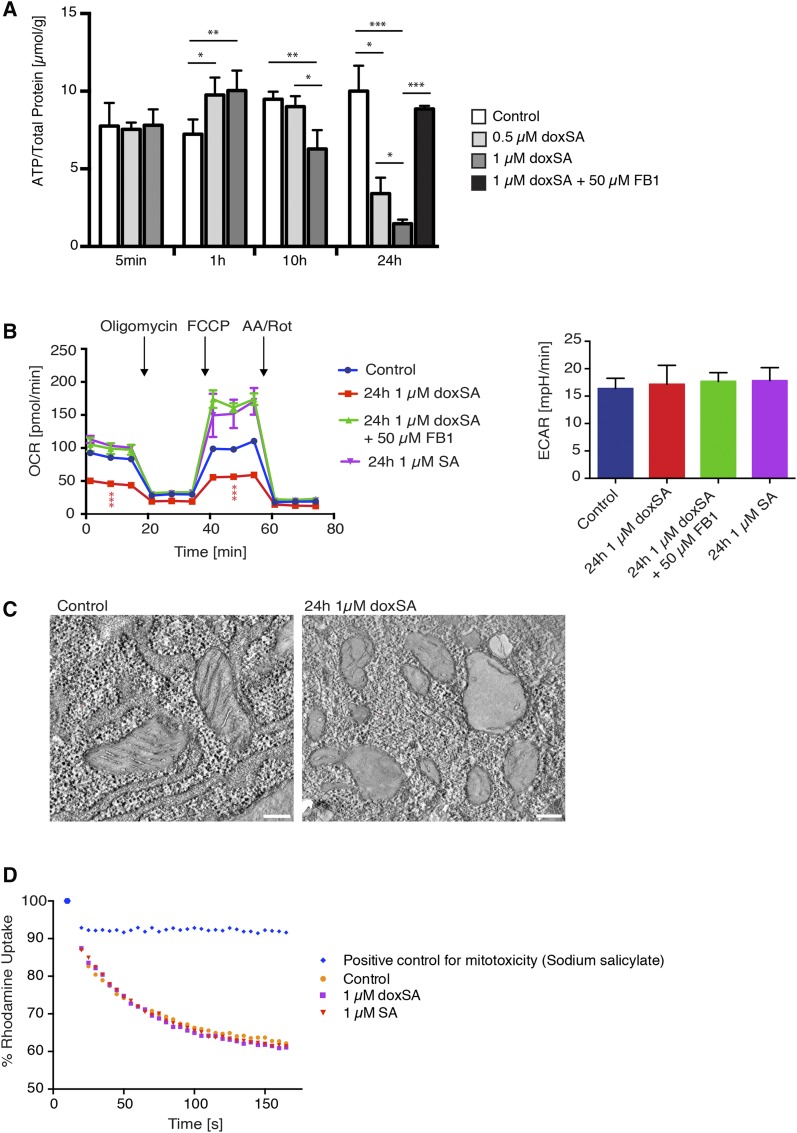
DoxSA treatment induces mitochondrial dysfunction in situ
Hyperfragmentation of mitochondria leads to a reduced capacity for ATP production by mitochondrial respiration (46). We therefore hypothesized that doxSA treatment could lead to a reduction of total cellular ATP levels. Indeed, doxSA treatment of MEF cells at toxic concentrations led to a significant increase in cellular ATP after 1 h and a pronounced reduction in cellular ATP levels after 10 and 24 h of treatment (Fig. 7A). This drop in cellular ATP was, like the mitochondrial fragmentation (Fig. 5C), rescued by cotreatment with 50 µM FB1. Measurement of mitochondrial function by using a Seahorse Analyzer showed an approximately 50% reduction in basal as well as maximal respiration in MEF cells treated for 24 h with 1 µM doxSA (Fig. 7B). This effect was again rescued by cotreatment with FB1, whereas no differences in the extracellular acidification rate, as a function of glycolysis, were detectable. Ultrastructural analysis of the hyperfragmented mitochondria by electron microscopy showed mitochondria that were spherical and had largely lost internal cristae structures (Fig. 7C). In contrast to the observed mitochondrial dysfunction after doxSA treatment in situ, direct doxSA treatment of isolated mitochondria did not lead to loss of mitochondrial membrane potential in vitro (Fig. 7D). Here, functional mitochondria from MEF cells were incubated with 1 µM doxSA and subsequently the membrane potential depended uptake of the fluorescent dye rhodamine (RH-123) (47) was analyzed.
Fig. 7.
Mitochondrial fragmentation upon doxSA treatment is characterized by a reduction of cellular ATP levels, reduced mitochondrial respiratory capacity, and loss of internal mitochondrial cristae structures. A: MEF cells were treated for the indicated times with different doxSA concentrations and FB1 before lysis and measurement of cellular ATP levels. Data are presented as average ± SD. B: MEF cells were treated for 24 h with 1 μM doxSA, 1 μM doxSA + 50 μM FB1, or 1 μM SA, respectively, before analysis of glycolysis rate, mitochondrial respiration, and maximal respiration capacity. OCR, oxygen consumption rate. Data are presented as average ± SD. C: MEF cells were treated for 24 h with 1 μM doxSA, before fixation and analysis of mitochondrial structures by electron microscopy. Scale bars, 200 nm. D: Mitochondria were isolated from MEF cells and incubated in vitro with 1 μM doxSA, 1 μM doxSA + FB1, or 1 μM SA. Uptake of rhodamine 123 was analyzed to measure mitochondrial membrane potential. * P < 0.05; ** P < 0.01; *** P < 0.001.
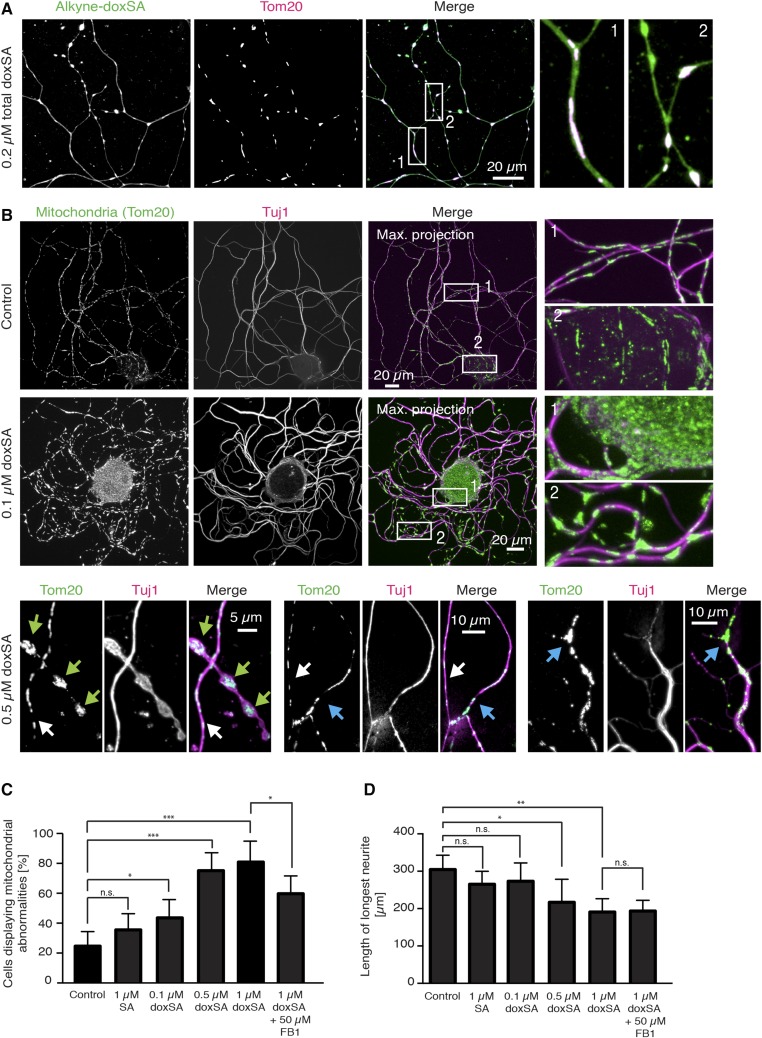
Exogenous doxSA treatment leads to swollen spherical mitochondria in primary sensory neurons
Although changes in ER morphology appeared only at late stages of doxSA-induced cellular toxicity, when the cell was close to cell death, we observed that the changes in mitochondrial morphology, such as swelling, were an early characteristic after doxSA treatment and did not per se result in cell death. To evaluate whether mitotoxicity could be a common mechanism underlying the known doxSA-induced neuronal degeneration, we treated primary DRG neurons with a mixture of doxSA and alkyne-doxSA (ratio 1:1) and confirmed uptake and mitochondrial localization of the lipids also in these cells (Fig. 8A). Alkyne-doxSA prominently stained the mitochondria within the cell body and along the neurites. To probe for the effect of doxSA on mitochondria in neurons, we then treated DRG neurons for 22 h with doxSA and stained for mitochondria (Tom20) and the neuron-specific class III β-tubulin (tuj1) (48) (Fig. 8B). Whereas mitochondria in control samples were evenly distributed along the neurites and appeared fine and streak-like, in doxSA-treated cells, we noticed the occurrence of swollen, more spherical, and less evenly distributed mitochondria in neurites and accumulation of mitochondria in the cell body already at the lowest concentration tested (0.1 µM). This phenotype was more pronounced at higher concentrations. Specifically, after treating neurons with 0.5 µM doxSA, we observed grossly swollen mitochondria (green arrows) and irregularly distributed mitochondria (blue arrows) in degenerating neurites compared with unaffected neurites (white arrows).
Fig. 8.
Exogenous doxSA-treatment leads to swollen spherical mitochondria in primary DRG neurons. A: DRG neurons were cultured for 2 h and treated for further 22 h with a mixture of alkyne-doxSA and unlabeled doxSA (total 0.2 μM). After fixation, the cells were stained for mitochondria (Tom20; magenta) and the alkyne moiety was reacted with ASTM-BODIPY (green). Alkyne-doxSA was taken up by the neurons and localized to mitochondria. B: DRG neurons were cultured for 2 h and treated for further 22 h with the indicated concentrations of unlabeled doxSA. Cells were fixed and stained for mitochondria (green) and the neuron-specific β III-tubulin variant tuj1 (magenta). Whereas mitochondria in control-treated DRG neurites appeared fine and “streak-like,” treatment with as little as 0.1 μM doxSA induced swelling of the mitochondria. Higher concentrations led to grossly swollen spherical mitochondria (green arrows) and irregularly distributed mitochondria and axonal degeneration in affected neurites (blue arrows) versus not-affected neurites (white arrows). Please note that images in this figure are optical sections performed using the apotome mode or, if indicated, maximum projections of z-stacks of optical sections. C and D: DRG neurons were cultured for 2 h and treated for a further 22 h with the indicated lipids. Cells were fixed and stained for mitochondria and the neuron-specific β III-tubulin variant tuj1 and quantified for mitochondrial morphology (C) and axonal outgrowth (D). Data are presented as average ± SD. * P < 0.05; ** P < 0.01; *** P < 0.001. n.s., not significant.
To quantify our observations, we isolated DRG neurons from five different mice and treated them separately for 22 h with SA, different concentrations of doxSA, or a combination of doxSA and FB1, respectively. Although the initially observed apparent accumulation of mitochondria in cell bodies was not statistically significant, differences in the number of neurons showing mitochondrial abnormalities in shape (swollen or spherical) or distribution along the neurites (nonregular distribution with gaps in between) were significant already at the lowest concentration tested (0.1 µM doxSA) and increased with higher concentrations (Fig. 8C). Treatment with 1 µM SA led to a slight, albeit not significant, increase in neurons showing mitochondrial abnormalities. In contrast to fibroblasts, cotreatment of 1 µM doxSA with 50 µM FB1 only partially rescued the effect of doxSA treatment on mitochondrial morphology (Fig. 8C). Quantification of neurite length revealed significant differences for treatment with higher doxSA concentrations (0.5 and 1 µM doxSA), but no significant effect on neurite outgrowth for treatment with 1 µM SA or 0.1 µM doxSA (Fig. 8D), indicating that the observed mitochondrial disturbances are likely to precede axonal degeneration. However, cotreatment of 1 µM doxSA with FB1 did not rescue the inhibitory effect of doxSA on neurite length, pointing toward an additional toxic effect that the doxSA sphingoid base exhibits on axonal outgrowth.
DISCUSSION
Because of their only recent identification in humans, very little is known about effects and functions of deoxySLs (49). In this study, we addressed some fundamental questions about the cellular metabolism and localization of deoxySLs, potentially leading to novel insights about their pathomechanism in diseases like diabetic neuropathy and HSAN1. To enable these experiments, we synthesized a novel traceable doxSA probe, alkyne-doxSA, with superior similarity to naturally formed doxSA and allowing for easy detection via click-chemistry methods (27–29, 37).
First, by comparing the uptake and metabolism of the novel and chemically synthesized alkyne-doxSA with that of its deuterated counterpart, we could verify it being a highly suitable probe for the study of deoxySLs in mammalian cells. The acquired data also underline that, whereas canonical sphingolipids are mainly desaturated and hence rarely present as DHCers, a smaller proportion of deoxySLs are desaturated. This notion was also depicted in earlier studies that found approximately one-third of the total deoxySLs in human plasma to be doxDHCer and two-thirds as doxCer (2). In agreement with a previous study (21), our results also indicate profound differences in the N-acylation pattern of doxDHCers versus doxCers. In detail, whereas in the MEF cells studied here, doxDHCers are found with a variety of N-acyl fatty acids attached (C16-C24), doxCer were detectable only with very long chain N-acyl fatty acids (C22–C24). This argues for a selectivity of the responsible dihydroceramide desaturase for very long-chain N-acylated doxDHCers or for an exchange of N-acyl fatty acids subsequent to desaturation. Although, just like for canonical sphingolipids, CerS expression levels in a given cell will likely affect the specific N-acyl pattern of the deoxySLs, but the presence of different N-acylation patterns between deoxySLs and canonical ceramides also highly indicates that CerSs may discriminate between their sphingoid base substrates. According to the Genevestigator database (50), which is based on a large set of quality-controlled microarrays, CerS5, CerS6, and CerS2 are the most highly expressed CerS isoforms in MEFs. In concordance with their known substrate specificity, we detected the most abundant doxDHCers to be N-acylated with C16:1, C24:0, and C24:1 fatty acids. Furthermore, CerS2 is also one of the most highly expressed CerS enzymes in the DRG of both humans and mice (along with CerS6 and CerS5, respectively) (50), indicating that ceramide and dox(DH)Cer species found in peripheral nerves are likely be similar to those found in the fibroblasts. However, to clearly decipher the activity of the different CerSs with deoxySLs, in vitro assays with single CerSs present in the assay buffer would be needed.
In addition to the metabolic data and, although often assumed, this study is the first to show that there is no conversion of deoxySLs to canonical sphingolipids (via a C1-hydroxylase) or directly to fatty acids in any of the cell types tested. Our tracing studies further confirm that deoxySLs are metabolized to only few downstream lipid species, when compared with canonical sphingolipids.
Our experiments with exogenous alkyne-doxSA treatment of cultured cells showed a very prominent localization of the alkyne signal in mitochondria as early as 5 min after administration. Although also colocalizing with Golgi and ER markers, the signal in mitochondria remained the most prominent at all time points analyzed. Furthermore, we found a striking change in mitochondrial morphology upon cellular treatment with alkyne-doxSA or doxSA, but not with alkyne-SA or SA, although exogenously added alkyne-SA also partially localized to mitochondria in cultured fibroblasts. In detail, upon long incubations (10–24 h), at toxic levels (0.5–1.0 µM doxSA), the majority of cells showed completely spherical, hyperfragmented mitochondria. At toxic concentrations and long incubation times, we also observed a prominent staining of alkyne-deoxySLs in the ER that had undergone morphological changes, leading to enlarged and highly visible perinuclear ER structures. Furthermore, our analysis showed that the cells were undergoing ER stress, and, indeed, ER stress has already been linked to doxSA toxicity in a former study that analyzed doxSA under its pharmaceutical code ES-285 as a potential anticancer drug (51). It was reported that doxSA induced apoptosis in cancer cells via an ER-stress mediated pathway and caspase 3 and 12 activation. Hence, induction of ER stress may be part of the general cytotoxicity mechanism of doxSA and may be what finally leads to apoptosis.
In addition to the ER stress, we observed that doxSA concentrations, which are not high enough to induce cell death, still resulted in changes in mitochondrial morphology. Most importantly, we found that primary DRG neurons displayed changes toward mitochondrial swelling and abnormal mitochondrial distribution already at the lowest doxSA concentration (0.1 µM) tested. In line with this, changes in mitochondrial morphology have been observed before in lymphoblasts from HSAN1 patients that have an increased endogenous doxSA generation (52). The authors detected swollen, perinuclear mitochondria with enlarged cristae and a discontinuous outer membrane in these cells by electron microscopy. In the present study, we show that the same mitochondrial disturbances are also induced by exogenous treatment with doxSA and that doxSA causes mitochondrial dysfunction in respiration and ATP generation in situ. Interestingly, in fibroblasts, inhibition of CerS activity with FB1 completely rescued doxSA-induced mitotoxicity, indicating that not the presence of the free sphingoid base, but of one of its N-acylated downstream products, is responsible for the mitochondrial dysfunction. This is also supported by a recent paper showing that 24 h treatment with 0.5 µM doxSA reduced cellular metabolic activity [3-(4,5-dimethylthiazol-2-yl)2,5-diphenyltetrazolium bromide (MTT test)] to approximately 50% in cortical neurons, but did not lead to a significant increase of cell death (lactate dehydrogenase test) (53). Here, reduced metabolic activity (MTT test) upon doxSA treatment was also rescued in the cortical neurons by preincubation of the cells with FB1, again indicating that doxSA needs to be N-acylated to dox(DH)Cers to affect mitochondrial function. In our hands, FB1 cotreatment had no positive effect on length of axonal outgrowth, implying that the sphingoid base itself possesses an additional toxic effect that limits axonal growth in peripheral neurons. However, in line with our results in fibroblasts, FB1 cotreatment significantly reduced the percentage of peripheral neurons that showed disturbed mitochondrial morphology.
Together, our results suggest that elevated deoxySL levels impair mitochondrial function and lead to an energy deficit in affected cells, especially in those that highly depend on respiration for ATP production. Indeed, peripheral nerves have very high and specific energy requirements because of the substantial length of the axons. This makes them particularly vulnerable to energy deficits. A chronic energy deficit due to mitotoxicity has been proposed as the underlying cause for a wide range of distal painful symmetrical peripheral neuropathies, including diabetic neuropathy (54). Furthermore, several inherited peripheral neuropathies are caused by mutations in genes responsible for mitochondrial fusion, fission, or transport (55). Specifically, it is known that mutations in mitofusin 2 (MFN2), a protein that controls mitochondrial fusion, cause hereditary motor and sensory neuropathy Charcot-Marie-Tooth disease type 2A (56), a disease with similar symptoms to HSAN1. Baloh et al. showed that expression of the MFN2 mutants in cultured DRG neurons resulted in fragmented and abnormally clustered mitochondria (57). Hence, although speculative, a decreased mitochondrial function may also be the underlying cause of the neurodegeneration seen in HSAN1. A pathomechanism based on doxSA-induced mitotoxicity could furthermore explain the lancinating pain attacks seen in HSAN1. Here, the chronic energy deficit would lead to inefficient repolarization of the neuronal membrane and spontaneous discharges that are experienced as painful attacks (58).
In conclusion, we have developed and validated a novel and useful probe to address deoxySL localization and metabolism. Using this probe, we have gathered data that sheds light onto the enigmatic biology of deoxySLs, showing that localization of deoxySLs to mitochondria disrupt mitochondrial integrity, whereas the subsequent reduction in mitochondrial function may likely provide an explanation for the observed particular vulnerability of peripheral nerves to deoxySLs in diseases such HSAN1 or diabetes.
Supplementary Material
Acknowledgments
A.P. thanks Katharina Meyer (DZNE, Germany) and J. Thomas Hannich (University of Geneva, Switzerland) for very helpful discussions on mitochondrial dynamics and doxSA biology, respectively. I.A. thanks Michele Visentin (University Hospital Zurich, Switzerland) for his help with the mitochondrial membrane potential assay and for the helpful discussions.
Footnotes
Abbreviations:
- alkyne-doxSA
- (2S,3R)-2-aminooctadec-17-yn-3-ol
- alkyne-SA
- (2S,3R)-2-aminooctadec-17-yn-1,3-diol
- ASTM-BODIPY
- azido-sulfo-tetramethyl-BODIPY
- CerS
- ceramide synthase
- DAPI
- 4’,6-diamidino-2-phenylindole
- DHCer
- dihydroceramide
- deoxySL
- 1-deoxysphingolipid
- doxCer
- 1-deoxyceramide
- doxDHCer
- 1-deoxydihydroceramide
- doxSA
- 1-deoxysphinganine
- doxSO
- 1-deoxysphingosine
- DRG
- dorsal root ganglia
- ER
- endoplasmic reticulum
- FB1
- Fumonisin B1
- HSAN1
- hereditary sensory and autonomic neuropathy type 1
- IB
- isolation buffer
- MEF
- mouse embryonal fibroblast
- PC
- phosphatidylcholine
- PDI
- protein-disulfide-isomerase
- PE
- phosphatidylethanolamine
- RB
- respiration buffer
- SA
- sphinganine
- SPT
- serine palmitoyltransferase
- S1P
- sphingosine-1-phosphate
- TAG
- triacylglycerol
- XBP1
- X-box binding protein 1
M.A.R.L. was supported by Deutsche Forschungsgemeinschaft (DFG) Grant SFB1123. I.A., T.H., D.E., and A.O. were supported by the 7th Framework Program of the European Commission “RESOLVE” Project 305707; Swiss National Foundation SNF Project 31003A_153390/1; the Hurka Foundation; the Novartis Foundation; and the Rare Disease Initiative Zurich (“radiz,” Clinical Research Priority Program for Rare Diseases, University of Zurich). F.B. is supported by the DFG, International Research in Paraplegia, and Wings for Life. C.T., A.G., L.K., K.W., E.L., and A.P. were supported by DFG Grants SFB645 and TRR83, and A.A.-A. and C.L. also were supported by DFG Grant TRR83.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Zitomer N. C., Mitchell T., Voss K. A., Bondy G. S., Pruett S. T., Garnier-Amblard E. C., Liebeskind L. S., Park H., Wang E., Sullards M. C., et al. . 2009. Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxysphinganine: a novel category of bioactive 1-deoxysphingoid bases and 1-deoxydihydroceramides biosynthesized by mammalian cell lines and animals. J. Biol. Chem. 284: 4786–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Penno A., Reilly M. M., Houlden H., Laura M., Rentsch K., Niederkofler V., Stoeckli E. T., Nicholson G., Eichler F., Brown R. H. Jr., et al. . 2010. Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J. Biol. Chem. 285: 11178–11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houlden H., King R., Blake J., Groves M., Love S., Woodward C., Hammans S., Nicoll J., Lennox G., O’Donovan D. G., et al. . 2006. Clinical, pathological and genetic characterization of hereditary sensory and autonomic neuropathy type 1 (HSAN I). Brain. 129: 411–425. [DOI] [PubMed] [Google Scholar]
- 4.Bejaoui K., Wu C., Scheffler M. D., Haan G., Ashby P., Wu L., de Jong P., and Brown R. H. Jr. 2001. SPTLC1 is mutated in hereditary sensory neuropathy, type 1. Nat. Genet. 27: 261–262. [DOI] [PubMed] [Google Scholar]
- 5.Dawkins J. L., Hulme D. J., Brahmbhatt S. B., Auer-Grumbach M., and Nicholson G. A.. 2001. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat. Genet. 27: 309–312. [DOI] [PubMed] [Google Scholar]
- 6.Auer-Grumbach M., Bode H., Pieber T. R., Schabhuttl M., Fischer D., Seidl R., Graf E., Wieland T., Schuh R., Vacariu G., et al. . 2013. Mutations at Ser331 in the HSN type I gene SPTLC1 are associated with a distinct syndromic phenotype. Eur. J. Med. Genet. 56: 266–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotthier A., Penno A., Rautenstrauss B., Auer-Grumbach M., Stettner G. M., Asselbergh B., Van Hoof K., Sticht H., Levy N., Timmerman V., et al. . 2011. Characterization of two mutations in the SPTLC1 subunit of serine palmitoyltransferase associated with hereditary sensory and autonomic neuropathy type I. Hum. Mutat. 32: E2211–E2225. [DOI] [PubMed] [Google Scholar]
- 8.Murphy S. M., Ernst D., Wei Y., Laura M., Liu Y. T., Polke J., Blake J., Winer J., Houlden H., Hornemann T., et al. . 2013. Hereditary sensory and autonomic neuropathy type 1 (HSANI) caused by a novel mutation in SPTLC2. Neurology. 80: 2106–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst D., Murphy S. M., Sathiyanadan K., Wei Y., Othman A., Laura M., Liu Y. T., Penno A., Blake J., Donaghy M., et al. . 2015. Novel HSAN1 mutation in serine palmitoyltransferase resides at a putative phosphorylation site that is involved in regulating substrate specificity. Neuromolecular Med. 17: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bode H., Bourquin F., Suriyanarayanan S., Wei Y., Alecu I., Othman A., von Eckardstein A., and Hornemann T.. 2016. HSAN1 mutations in serine palmitoyltransferase reveal a close structure-function-phenotype relationship. Hum. Mol. Genet. 25: 853–865. [DOI] [PubMed] [Google Scholar]
- 11.Laurá M., Murphy S. M., Hornemann T., Bode H., Polke J., Blake J., Houlden H., and Reilly M. M.. 2012. Hereditary sensory neuropathy type 1: correlation of severity and plasma atypical deoxy-sphyngoid bases (Abstract). Neuromuscular Disorders. 22: S18. [Google Scholar]
- 12.Eichler F. S., Hornemann T., McCampbell A., Kuljis D., Penno A., Vardeh D., Tamrazian E., Garofalo K., Lee H. J., Kini L., et al. . 2009. Overexpression of the wild-type SPT1 subunit lowers desoxysphingolipid levels and rescues the phenotype of HSAN1. J. Neurosci. 29: 14646–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garofalo K., Penno A., Schmidt B. P., Lee H. J., Frosch M. P., von Eckardstein A., Brown R. H., Hornemann T., and Eichler F. S.. 2011. Oral L-serine supplementation reduces production of neurotoxic deoxysphingolipids in mice and humans with hereditary sensory autonomic neuropathy type 1. J. Clin. Invest. 121: 4735–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oswald M. C., West R. J., Lloyd-Evans E., and Sweeney S. T.. 2015. Identification of dietary alanine toxicity and trafficking dysfunction in a Drosophila model of hereditary sensory and autonomic neuropathy type 1. Hum. Mol. Genet. 24: 6899–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Othman A., Rutti M. F., Ernst D., Saely C. H., Rein P., Drexel H., Porretta-Serapiglia C., Lauria G., Bianchi R., von Eckardstein A., et al. . 2012. Plasma deoxysphingolipids: a novel class of biomarkers for the metabolic syndrome? Diabetologia. 55: 421–431. [DOI] [PubMed] [Google Scholar]
- 16.Bertea M., Rutti M. F., Othman A., Marti-Jaun J., Hersberger M., von Eckardstein A., and Hornemann T.. 2010. Deoxysphingoid bases as plasma markers in diabetes mellitus. Lipids Health Dis. 9: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Othman A., Saely C. H., Muendlein A., Vonbank A., Drexel H., von Eckardstein A., and Hornemann T.. 2015. Plasma 1-deoxysphingolipids are predictive biomarkers for type 2 diabetes mellitus. BMJ Open Diabetes Res. Care. 3: e000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Othman A., Bianchi R., Alecu I., Wei Y., Porretta-Serapiglia C., Lombardi R., Chiorazzi A., Meregalli C., Oggioni N., Cavaletti G., et al. . 2015. Lowering plasma 1-deoxysphingolipids improves neuropathy in diabetic rats. Diabetes. 64: 1035–1045. [DOI] [PubMed] [Google Scholar]
- 19.Humpf H. U., Schmelz E. M., Meredith F. I., Vesper H., Vales T. R., Wang E., Menaldino D. S., Liotta D. C., and Merrill A. H. Jr. 1998. Acylation of naturally occurring and synthetic 1-deoxysphinganines by ceramide synthase. Formation of N-palmitoyl-aminopentol produces a toxic metabolite of hydrolyzed fumonisin, AP1, and a new category of ceramide synthase inhibitor. J. Biol. Chem. 273: 19060–19064. [DOI] [PubMed] [Google Scholar]
- 20.Abad J. L., Nieves I., Rayo P., Casas J., Fabrias G., and Delgado A.. 2013. Straightforward access to spisulosine and 4,5-dehydrospisulosine stereoisomers: probes for profiling ceramide synthase activities in intact cells. J. Org. Chem. 78: 5858–5866. [DOI] [PubMed] [Google Scholar]
- 21.Esaki K., Sayano T., Sonoda C., Akagi T., Suzuki T., Ogawa T., Okamoto M., Yoshikawa T., Hirabayashi Y., and Furuya S.. 2015. L-serine deficiency elicits intracellular accumulation of cytotoxic deoxysphingolipids and lipid body formation. J. Biol. Chem. 290: 14595–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorden D. L., Myers D. S., Ivanova P. T., Fahy E., Maurya M. R., Gupta S., Min J., Spann N. J., McDonald J. G., Kelly S. L., et al. . 2015. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J. Lipid Res. 56: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer R., Bielawski J., Kistner-Griffin E., Othman A., Alecu I., Ernst D., Kornhauser D., Hornemann T., and Spassieva S.. 2015. Neurotoxic 1-deoxysphingolipids and paclitaxel-induced peripheral neuropathy. FASEB J. 29: 4461–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steiner R., Saied E. M., Othman A., Arenz C., Maccarone A. T., Poad B. L., Blanksby S. J., von Eckardstein A., and Hornemann T.. 2016. Elucidating the chemical structure of native 1-deoxysphingosine. J. Lipid Res. 57: 1194–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakahara K., Ohkuni A., Kitamura T., Abe K., Naganuma T., Ohno Y., Zoeller R. A., and Kihara A.. 2012. The Sjogren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol. Cell. 46: 461–471. [DOI] [PubMed] [Google Scholar]
- 26.Pagano R. E., Martin O. C., Kang H. C., and Haugland R. P.. 1991. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J. Cell Biol. 113: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaebler A., Milan R., Straub L., Hoelper D., Kuerschner L., and Thiele C.. 2013. Alkyne lipids as substrates for click chemistry-based in vitro enzymatic assays. J. Lipid Res. 54: 2282–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiele C., Papan C., Hoelper D., Kusserow K., Gaebler A., Schoene M., Piotrowitz K., Lohmann D., Spandl J., Stevanovic A., et al. . 2012. Tracing fatty acid metabolism by click chemistry. ACS Chem. Biol. 7: 2004–2011. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann K., Thiele C., Schott H. F., Gaebler A., Schoene M., Kiver Y., Friedrichs S., Lutjohann D., and Kuerschner L.. 2014. A novel alkyne cholesterol to trace cellular cholesterol metabolism and localization. J. Lipid Res. 55: 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claas R. F., ter Braak M., Hegen B., Hardel V., Angioni C., Schmidt H., Jakobs K. H., Van Veldhoven P. P., and zu Heringdorf D. M.. 2010. Enhanced Ca2+ storage in sphingosine-1-phosphate lyase-deficient fibroblasts. Cell. Signal. 22: 476–483. [DOI] [PubMed] [Google Scholar]
- 31.Riley R. T., Norred W. P., Wang E., and Merrill A. H.. 1999. Alteration in sphingolipid metabolism: bioassays for fumonisin- and ISP-I-like activity in tissues, cells and other matrices. Nat. Toxins. 7: 407–414. [DOI] [PubMed] [Google Scholar]
- 32.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwakoshi N. N., Lee A. H., Vallabhajosyula P., Otipoby K. L., Rajewsky K., and Glimcher L. H.. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4: 321–329. [DOI] [PubMed] [Google Scholar]
- 34.Kolb H. C., Finn M. G., and Sharpless K. B.. 2001. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 40: 2004–2021. [DOI] [PubMed] [Google Scholar]
- 35.Haberkant P., and Holthuis J. C.. 2014. Fat & fabulous: bifunctional lipids in the spotlight. Biochim. Biophys. Acta. 1841: 1022–1030. [DOI] [PubMed] [Google Scholar]
- 36.Kuerschner L., and Thiele C.. 2014. Multiple bonds for the lipid interest. Biochim. Biophys. Acta. 1841: 1031–1037. [DOI] [PubMed] [Google Scholar]
- 37.Gaebler A., Penno A., Kuerschner L., and Thiele C.. 2016. A highly sensitive protocol for microscopy of alkyne lipids and fluorescently tagged or immunostained proteins. J. Lipid Res. 57: 1934–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jao C. Y., Roth M., Welti R., and Salic A.. 2009. Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc. Natl. Acad. Sci. USA. 106: 15332–15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pauling L. 1960. The Nature of the Chemical Bond and the Structure of Molecules and Crystals: An Introduction to Modern Structural Chemistry. Cornell University Press, Ithaca, NY. [Google Scholar]
- 40.Baird R. D., Kitzen J., Clarke P. A., Planting A., Reade S., Reid A., Welsh L., Lopez Lazaro L., de las Heras B., Judson I. R., et al. . 2009. Phase I safety, pharmacokinetic, and pharmacogenomic trial of ES-285, a novel marine cytotoxic agent, administered to adult patients with advanced solid tumors. Mol. Cancer Ther. 8: 1430–1437. [DOI] [PubMed] [Google Scholar]
- 41.Schöffski P., Dumez H., Ruijter R., Miguel-Lillo B., Soto-Matos A., Alfaro V., and Giaccone G.. 2011. Spisulosine (ES-285) given as a weekly three-hour intravenous infusion: results of a phase I dose-escalating study in patients with advanced solid malignancies. Cancer Chemother. Pharmacol. 68: 1397–1403. [DOI] [PubMed] [Google Scholar]
- 42.Vilar E., Grunwald V., Schoffski P., Singer H., Salazar R., Iglesias J. L., Casado E., Cullell-Young M., Baselga J., and Tabernero J.. 2012. A phase I dose-escalating study of ES-285, a marine sphingolipid-derived compound, with repeat dose administration in patients with advanced solid tumors. Invest. New Drugs. 30: 299–305. [DOI] [PubMed] [Google Scholar]
- 43.Massard C., Salazar R., Armand J. P., Majem M., Deutsch E., Garcia M., Oaknin A., Fernandez-Garcia E. M., Soto A., and Soria J. C.. 2012. Phase I dose-escalating study of ES-285 given as a three-hour intravenous infusion every three weeks in patients with advanced malignant solid tumors. Invest. New Drugs. 30: 2318–2326. [DOI] [PubMed] [Google Scholar]
- 44.Novgorodov S. A., Wu B. X., Gudz T. I., Bielawski J., Ovchinnikova T. V., Hannun Y. A., and Obeid L. M.. 2011. Novel pathway of ceramide production in mitochondria: thioesterase and neutral ceramidase produce ceramide from sphingosine and acyl-CoA. J. Biol. Chem. 286: 25352–25362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida H. 2007. Unconventional splicing of XBP-1 mRNA in the unfolded protein response. Antioxid. Redox Signal. 9: 2323–2333. [DOI] [PubMed] [Google Scholar]
- 46.Hoppins S. 2014. The regulation of mitochondrial dynamics. Curr. Opin. Cell Biol. 29: 46–52. [DOI] [PubMed] [Google Scholar]
- 47.Perry S. W., Norman J. P., Barbieri J., Brown E. B., and Gelbard H. A.. 2011. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques. 50: 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enes J., Langwieser N., Ruschel J., Carballosa-Gonzalez M. M., Klug A., Traut M. H., Ylera B., Tahirovic S., Hofmann F., Stein V., et al. . 2010. Electrical activity suppresses axon growth through Ca(v)1.2 channels in adult primary sensory neurons. Curr. Biol. 20: 1154–1164. [DOI] [PubMed] [Google Scholar]
- 49.Duan J., and Merrill A. H. Jr. 2015. 1-Deoxysphingolipids Encountered Exogenously and Made de Novo: Dangerous Mysteries inside an Enigma. J. Biol. Chem. 290: 15380–15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., and Zimmermann P.. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salcedo M., Cuevas C., Alonso J. L., Otero G., Faircloth G., Fernandez-Sousa J. M., Avila J., and Wandosell F.. 2007. The marine sphingolipid-derived compound ES 285 triggers an atypical cell death pathway. Apoptosis. 12: 395–409. [DOI] [PubMed] [Google Scholar]
- 52.Myers S. J., Malladi C. S., Hyland R. A., Bautista T., Boadle R., Robinson P. J., and Nicholson G. A.. 2014. Mutations in the SPTLC1 protein cause mitochondrial structural abnormalities and endoplasmic reticulum stress in lymphoblasts. DNA Cell Biol. 33: 399–407. [DOI] [PubMed] [Google Scholar]
- 53.Guntert T., Hanggi P., Othman A., Suriyanarayanan S., Sonda S., Zuellig R. A., Hornemann T., and Ogunshola O. O.. 2016. 1-Deoxysphingolipid-induced neurotoxicity involves N-methyl-D-aspartate receptor signaling. Neuropharmacology. 110: 211–222. [DOI] [PubMed] [Google Scholar]
- 54.Bennett G. J., Doyle T., and Salvemini D.. 2014. Mitotoxicity in distal symmetrical sensory peripheral neuropathies. Nat. Rev. Neurol. 10: 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pareyson D., Saveri P., Sagnelli A., and Piscosquito G.. 2015. Mitochondrial dynamics and inherited peripheral nerve diseases. Neurosci. Lett. 596: 66–77. [DOI] [PubMed] [Google Scholar]
- 56.Kijima K., Numakura C., Izumino H., Umetsu K., Nezu A., Shiiki T., Ogawa M., Ishizaki Y., Kitamura T., Shozawa Y., et al. . 2005. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum. Genet. 116: 23–27. [DOI] [PubMed] [Google Scholar]
- 57.Baloh R. H., Schmidt R. E., Pestronk A., and Milbrandt J.. 2007. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J. Neurosci. 27: 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao W. H., Zheng H., and Bennett G. J.. 2012. Characterization of oxaliplatin-induced chronic painful peripheral neuropathy in the rat and comparison with the neuropathy induced by paclitaxel. Neuroscience. 203: 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.