Abstract
High arachidonic acid (20:4n-6) and low n-3 PUFA levels impair the capacity of cultured human bone marrow mesenchymal stromal cells (hBMSCs) to modulate immune functions. The capacity of the hBMSCs to modify PUFA structures was found to be limited. Therefore, different PUFA supplements given to the cells resulted in very different glycerophospholipid (GPL) species profiles and substrate availability for phospholipases, which have preferences for polar head group and acyl chains when liberating PUFA precursors for production of lipid mediators. When supplemented with 20:4n-6, the cells increased prostaglandin E2 secretion. However, they elongated 20:4n-6 to the less active precursor, 22:4n-6, and also incorporated it into triacylglycerols, which may have limited the proinflammatory signaling. The n-3 PUFA precursor, 18:3n-3, had little potency to reduce the GPL 20:4n-6 content, while the eicosapentaenoic (20:5n-3) and docosahexaenoic (22:6n-3) acid supplements efficiently displaced the 20:4n-6 acyls, and created diverse GPL species substrate pools allowing attenuation of inflammatory signaling. The results emphasize the importance of choosing appropriate PUFA supplements for in vitro hBMSC expansion and suggests that for optimal function they require an exogenous fatty acid source providing 20:5n-3 and 22:6n-3 sufficiently, but 20:4n-6 moderately, which calls for specifically designed optimal PUFA supplements for the cultures.
Keywords: arachidonic acid, docosahexaenoic acid, eicosapentaenoic acid, glycerophospholipid, immunomodulation, lipid signaling, mass spectrometry, mesenchymal stromal/stem cell, prostaglandin E2
Mesenchymal stromal/stem cells (MSCs) are cells with multi-lineage differentiation (1) and immunomodulatory capacity (2–4). These characteristics of MSCs, especially their ability to inhibit the proliferation of antigen-specific memory T cells (5, 6), have made them excellent candidates for various cell-based therapies (7), including the treatment or prevention of graft versus host disease (8), autoimmune diseases (9), and solid organ transplantation rejections (10). In order to successfully apply these cells for therapeutic applications, substantial in vitro expansion is required. However, long-term culturing of MSCs contributes to their loss of stem cell characteristics manifested in decreased proliferation and differentiation potential, telomere shortening, and accumulation of n-6 PUFAs with signaling roles promoting inflammatory conditions (11–13). Thus, developing a safe method to preserve stem cell proliferation, differentiation, and immunomodulatory potential is of paramount importance.
PUFAs and their bioactive derivatives affect the proliferation and differentiation of various stem cells and modulate their immunological interactions with other cells (14–20). In most mammalian cell types, exogenous linoleic acid (18:2n-6) or α-linolenic acid (18:3n-3) is desaturated and elongated to arachidonic acid (20:4n-6) or to eicosapentaenoic acid (20:5n-3) and docosahexaenoic acid (22:6n-3), respectively. These resulting PUFAs are at first incorporated into membrane glycerophospholipids (GPLs), and then deliberated by phospholipases to be employed as precursors to produce various lipid mediators. Three major synthetic pathways, namely, cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450, produce lipid mediators such as prostaglandins, leukotrienes, thromboxanes, lipoxins, and resolvins, collectively termed eicosanoids and docosanoids. Prostaglandin E2 (PGE2; derivative of 20:4n-6) is known to affect the proliferation and cytokine secretion of T cells, and thus has a major role in MSC immunoregulation (21). Recently, lipoxins and resolvins have been identified as essential regulators of the resolution phase of inflammation (22) and as players in the immunoregulation of MSCs (23).
For MSC therapy, the human bone marrow-derived MSCs are the best-characterized and most widely used cells. MSCs have also been isolated from various adult and fetal tissues, including synovium, periosteum, skeletal muscle, cord blood, and adipose tissue (24–27). The fatty acid composition of bone marrow, harboring MSCs, is prone to changes due to diet, age, or clinical status of the patient (28, 29). Therefore, the variable marrow microenvironment that surrounds and nourishes human bone marrow mesenchymal stromal cells (hBMSCs) can also affect the functional properties of the cells (13). In addition, it is logical to assume that hBMSCs, which are commonly cultured with PUFA supplements very different from the PUFAs found in the human bone marrow, have altered functional properties. In order to evaluate how well the PUFA supply of hBMSCs in vitro resembles that in vivo, we first compared the fatty acid composition in human bone marrow aspirations to the fatty acid supplement of the culture media, i.e., the FBS.
The endogenous capacity of cultured hBMSCs to modify PUFA structures for their functional needs could partially compensate for suboptimal supply from the microenvironment, culture media, or the marrow in vivo. Still today, information on mammalian PUFA metabolism largely originates from studies on cultured hepatocytes, which have excellent capacity to metabolize diet-derived PUFAs (30, 31). However, some eukaryotic cell types, such as platelets and macrophages, lymphoblasts, and metastatic mammary cells, are deficient in performing desaturation steps for PUFA precursors (32). Because the PUFA metabolism of hBMSCs has not been studied before, in this work we characterized the capacity of cultured hBMSCs to modify exogenously administered 18-carbon (C18) PUFA precursors into their elongated and highly unsaturated products. The practical goal was to find out which fatty acid supplements could specifically modify the contents of the most bioactive PUFAs, i.e., 20:4-6, 20:5n-3, and 22:6n-3, of the cells and, thereby, affect their immunomodulatory properties. Because the phospholipases deliberating PUFAs for signaling act on specific GPLs, detailed analysis of the incorporation patterns of different PUFAs into different GPL classes was also carried out. It was especially important to compare the distribution of the 20:4n-6, 20:5n-3, and 22:6n-3 between the different GPL classes in the cells supplemented with either the first n-6 or n-3 family precursors or their elongated and highly unsaturated products. This work reveals the urgent need to revise the culture procedures used when expanding therapeutic MSCs and lays the foundations for future trials of manipulating MSC functionality by the means of different PUFA supplements.
MATERIALS AND METHODS
Ethics and bone marrow donors
Human bone marrow aspirates (n = 4) and hBMSC lines (n = 4) were included in the current study. The Ethical Committee of Northern Ostrobothnia Hospital District approved all the patient protocols, and the use of human material conforms to the principles outlined in the Declaration of Helsinki. Informed written consent was acquired from adult patients prior to collecting bone marrow aspirates from the iliac crest.
Materials
The hBMSCs were obtained from the clinic of Professor Petri Lehenkari and HepG2 (liver hepatocellular carcinoma) cells from BioNordika Oy (Helsinki, Finland). The α-MEM, FBS, HEPES, penicillin, streptomycin, trypsin, and L-glutamine were purchased from Gibco® Invitrogen (Paisley, UK). Eagle’s minimum essential medium (E-MEM) was purchased from American Type Culture Collection (Rockville, MD) and fatty acid-free BSA from Sigma (St. Louis, MO). FFAs 18:2n-6, 18:3n-3, 20:4n-6, 20:5n-3, and 22:6n-3 were obtained from Nu-Chek-Prep, Inc. (Elysian, MN).
Cell culture of hBMSCs and HepG2 cells
The hBMSCs that had been harvested between passages 3 and 5 were cultured in α-MEM medium and supplemented with 5% FBS, 2 mM L-glutamine, 100 u/ml penicillin, 100 μg/ml streptomycin, and 20 mM HEPES. HepG2 cells between passages 84 and 86 were cultured in E-MEM and supplemented with 10% non-heat inactivated FBS, 2 mM L-glutamine, 100 u/ml penicillin, and 100 μg/ml streptomycin. Both cell types were plated on 75 cm2 flasks at a density of 2.5 × 103 cells/cm2.
In order to compare the metabolism and incorporation pattern of different PUFAs into membrane GPLs, the cells were supplemented with specific PUFAs: 18:2n-6, 18:3n-3, 20:4n-6, 20:5n-3, or 22:6n-3 bound to BSA at 50 μM concentration (FFA/BSA 2.7:1). Ethanol was used to prepare the stock solutions of FFAs at a concentration of 100 mM. The cells were incubated in a humidified incubator at 37°C and 5% CO2, and cultured for nine consecutive days (until 70–80% confluence). The medium was renewed every 72 h and each time replenished with the same FFAs.
GC of fatty acids
Transmethylation of hBMSC and HepG2 lipids was performed according to the recommendations of Christie (33). Samples were heated in 1% H2SO4 in methanol, at a temperature of 96°C and under nitrogen atmosphere for 120 min. The fatty acid methyl esters formed were recovered with hexane and analyzed using a gas chromatograph (Shimadzu GC-2010 Plus) equipped with an auto injector (AOC-20i), flame ionization detector, and ZB-wax capillary columns (30 m, 0.25 mm ID, 0.25 μm film; Phenomenex USA). The identification of the fatty acid methyl esters was based on the retention time, the use of authentic standard mixtures of known composition, and confirmatory recordings of mass spectra (Agilent 6890N network GC with FID and 5973 MSD). Quantifications were based on FID responses corrected according to the theoretical response factors (34) and calibrations with the quantitative authentic standards. The fatty acid proportions were calculated as mole percent (later percent), and the fatty acids were marked by using the abbreviations: [carbon number]:[number of double bonds]n-[position of the first double bond calculated from the methyl end] (e.g., 22:6n-3).
MS of GPLs
For MS, total lipids were extracted from hBMSCs and HepG2 cells according to Folch, Lees, and Sloane Stanley (35). Samples dissolved in chloroform/methanol (1:2 v/v) were spiked with internal standards and supplemented with 1% NH4OH just before direct infusion of the sample solution into the ESI source of a triple quadrupole mass spectrometer (Agilent 6490 Triple Quad LC/MS with iFunnel technology; Agilent Technologies, Inc., Santa Clara, CA) at a flow rate of 10 μl/min. The instrument’s response for different GPLs is affected by the head groups and acyl chain structures. Thus, a cocktail of 13 internal standards was added to each sample solution to correct for such variations and improve the precision of quantitative analysis. In addition to the MS+ and MS− scans, MS/MS precursor ion scans were used to detect phosphatidylcholine (PC) species (precursors for the fragment ion m/z 184) and phosphatidylinositol (PI) species (precursors for m/z 241). MS/MS neutral loss scans were applied to detect phosphatidylethanolamine (PE) (neutral loss of 141 amu) and phosphatidylserine (PS) species (neutral loss of 87 amu). PE plasmalogen (PEp) species were detected according to the fragments specific for the vinyl ether chain at the sn-1 position [e.g., m/z 364, 390, and 392 for 16:0p, 18:1p, and 18:0p, respectively (36)]. Triacylglycerol (TAG) species were detected as (M+NH4)+ ions (37) and their concentrations were normalized against total PC concentration calculated from the same MS+ scan. For the MS analyses, a source temperature of 250°C and instrument collision energies of 5–45 eV (optimal settings depend on the lipid class) were used. Nitrogen was used as the nebulizing (20 psi) and the drying gas (11 μl/min at 250°C). The spectra generated by the instrument were processed by MassHunter Workstation qualitative analysis software (Agilent Technologies, Inc.) and the individual GPL species were quantified using the internal standards and free software called Lipid Mass Spectrum Analysis (LIMSA) (38). Using this software, the spectral peak intensities were converted to concentrations expressed as mole percent (later percent) for each lipid species (relative to the total amount in the lipid class). The acyl chain assemblies in each lipid species were studied by recording negative ion mode product ion scans of the anion fragments for all common fatty acids (39). For PC species, which do not ionize in negative mode as such, formate adducts served as mother ions, and yielded the anionic fragments of the acyl chains. Provided that the GC analyses had shown that only one quantitatively important double bond positional isomer was present for a certain fatty acid of the cells, these acyl chains in GPL were marked using the known accurate structure.
PGE2 production in hBMSCs
To measure PGE2 production, hBMSCs were grown in the control medium (α-MEM with 5% FBS) or in the medium supplemented with different PUFAs conjugated to BSA, as described above. After 24 h incubation, the medium was aspirated, and the cells were washed twice with PBS and once with serum-free α-MEM. The cells were then incubated in serum-free starvation α-MEM medium for 48 h. The medium was collected and centrifuged at 2,000 g for 10 min, and the supernatant was further ultracentrifuged at100,000 g for 2 h at 4°C. This latter supernatant, free of extracellular vesicles, was collected and PGE2 levels were measured by using a PGE2 ELISA kit [monoclonal item number 514010 (Cayman Chemical, Ann Arbor, MI)] according to the manufacturer’s protocol. For the absorbance (405 nm) measurements of the assay, the medium was used either undiluted or diluted 2- to 10 -fold with ELISA buffer depending on the PGE2 concentration. Confirmatory LC-MS recordings using specific multiple reaction monitoring detection for PGE2 (40) showed that potential cross-reactivity from other prostaglandins did not bias the data.
Statistical analysis
To study statistical differences between the fatty acid and lipid levels in different experimental samples, the Kruskal-Wallis nonparametric one-way ANOVA followed by a post hoc Mann-Whitney test for the means was used. The data represent four replicates of hBMSCs for each PUFA supplementation trial, four clinical bone marrow aspirates, and five FBS samples from different lots. P < 0.05 was regarded as statistically significant.
Gene expression analysis
RNA was extracted using Qiagen AllPrep DNA/RNA mini kit (Qiagen, Valencia, CA) and a Qiagen supplementary protocol (Purification of total RNA containing miRNA from animal cells using the RNAeasy Plus mini kit). A detailed description on the hybridization of labeled RNAs (onto Agilent SurePrint G3 Human GE 8 × 60 K), scanning of the slides, and processing, transforming, and modeling of the data are found in Kilpinen et al. (13). The data are available in the public data repository, Gene Expression Omnibus (GEO), and are accessible through GEO accession number GSE39035 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39035). The average expression levels for fatty acid elongases and desaturases found in MSCs and hepatocytes were retrieved from the updated version of public database, GeneSapiens, which is currently hosted by MediSapiens Ltd. under the name IST online (41).
RESULTS
Cultured hBMSCs are commonly supplemented with FBS, the PUFA composition of which differs from that of human bone marrow
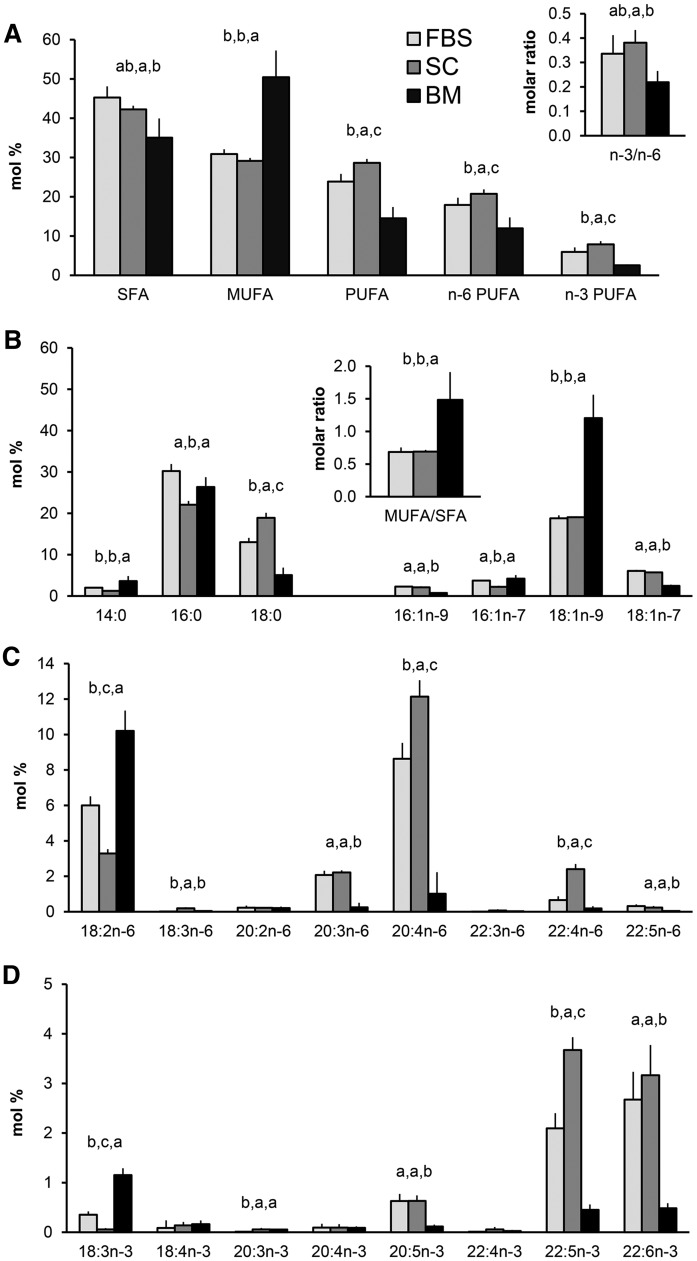
The fatty acid profiles of the hBMSCs cultured in standard medium were compared with those in the FBS and human bone marrow samples (Fig. 1). The total levels of saturated fatty acids (SFAs) in the hBMSCs were 42%, between the levels of FBS and the marrow (Fig. 1A). Among individual SFAs, the hBMSCs contained less 16:0, but more 18:0, than the FBS and marrow samples (Fig. 1B). The major MUFA, 18:1n-9, was found in hBMSCs and FBS with levels one-half of those in the marrow. Compared with hBMSC levels, both the bone marrow and the FBS were rich sources of 18:2n-6 (Fig. 1C). A striking finding was that hBMSCs, like their common FBS supplement, contained large reservoirs of 20:4n-6 (hBMSC 12%, FBS 9%), about 10 times the levels found in the bone marrow (Fig. 1C). Compared with FBS levels, the hBMSCs had a 1.4-fold increase in their relative amount of 20:4n-6. Interestingly this increase for 22:4n-6 was even 3.7-fold. The n-3 PUFA precursor, 18:3n-3, was present with 1% in the marrow, clearly exceeding the contents in the hBMSCs and FBS, but the long-chain and highly unsaturated members of the n-3 family, 20:5n-3, 22:5n-3, and 22:6n-3, were more abundant in the hBMSCs and their FBS supplement than in the marrow (Fig. 1D). When the levels of 20:5n-3 and 22:6n-3 equaled in the hBMSCs and FBS, the 22:5n-3 was enriched in the cells 1.8-fold. These findings raised the question of whether all n-3 and n-6 PUFAs provided by the culture medium or natural niche are biologically equally active and metabolized further in the hBMSCs to form even longer and more highly unsaturated fatty acids.
Fig. 1.
Fatty acid profiles (mole percent in total fatty acids, mean ± SD) in hBMSCs (SC, in control medium, n = 4) compared with those in different lots of FBS (n = 5) and in clinical samples of human bone marrow (BM, n = 4). A: Total proportions of SFAs, MUFAs, PUFAs, n-6 PUFAs, and n-3 PUFAs The ratio of n-3 to n-6 PUFAs (n-3/n-6) is shown in the insert. B: Individual SFAs and MUFAs. The ratio of MUFA total to SFA total is shown in the insert. C: Individual n-6 PUFAs. D: Individual n-3 PUFAs. As statistics, Kruskal-Wallis nonparametric one-way ANOVA followed by post hoc Mann-Whitney test for the means were used. The means with no common letter differed at the P < 0.05 level. Missing letters on the bars mean that the Kruskal-Wallis test showed no significant differences between the means.
hBMSCs elongate C18 PUFA precursors, but fail to produce highly unsaturated fatty acids
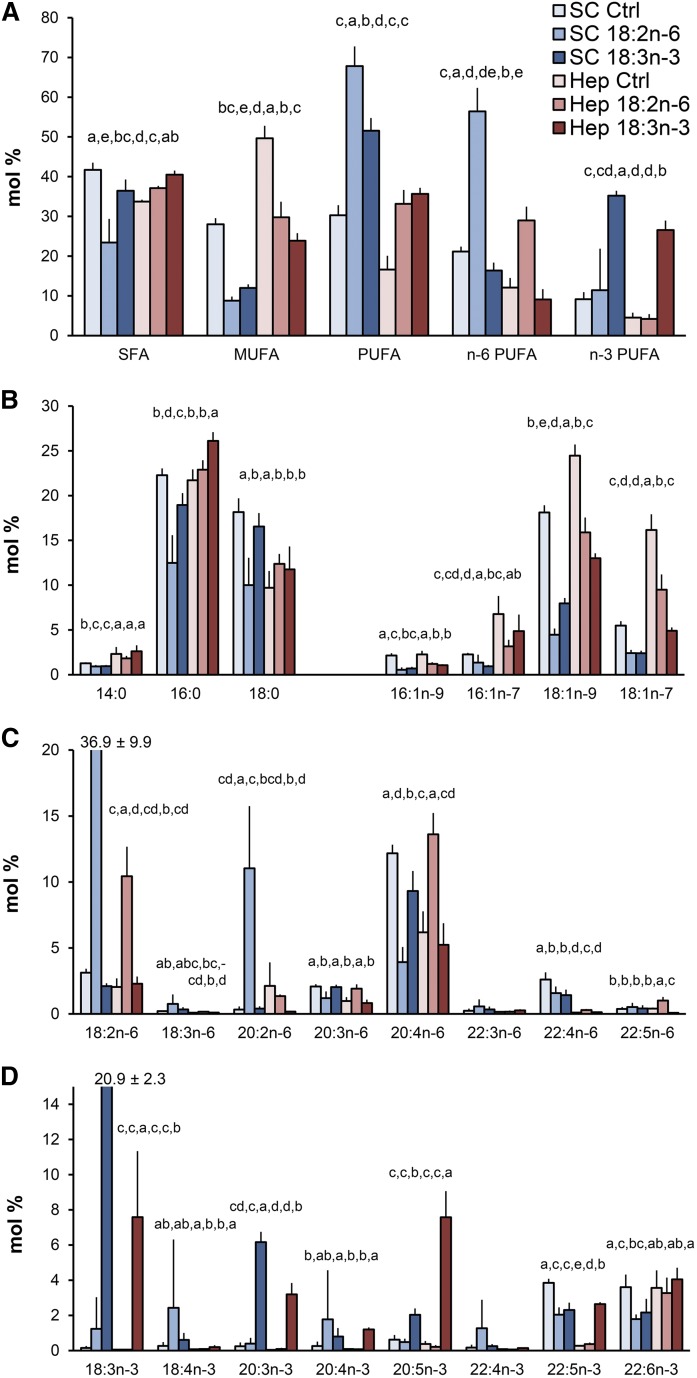
Supplementing the hBMSCs with the C18 PUFA precursors, 18:2n-6 or 18:3n-3, decreased the SFA and MUFA contents of the cells (Fig. 2A). The contents of the main individual MUFAs, 18:1n-9 and 18:1n-7, of the fatty acid-supplemented hBMSCs were halved from the levels of the cells cultured in standard medium (Fig. 2B). Incubating the hBMSCs with 18:2n-6 caused a 10-fold increase in its relative amount compared with the cells grown in standard medium, from 3% to 37% (Fig. 2C). At the same time, the level of the immediate elongation product of 18:2n-6, i.e., 20:2n-6, was increased 30-fold. In contrast, 20:4n-6 decreased by more than 60% from the level of the standard cells. Raising the 18:2n-6 concentration in the growth medium also reduced the levels of 22:5n-3 and 22:6n-3 in the hBMSCs (Fig. 2D). Similarly to the effects seen after adding 18:2n-6 to the cells, the 18:3n-3 supplementation also raised its own relative amount in the cells, from a negligible level to 21% (Fig. 2D). The immediate elongation product of 18:3n-3, i.e., 20:3n-3, showed a 25-fold increase. In addition, 20:4n-3 and 20:5n-3, which follow on the metabolic pathway, had raised their levels as a consequence of the exogenous 18:3n-3 supply. However, this trend was reversed for the longest and most highly unsaturated n-3 PUFAs, 22:5n-3 and 22:6n-3, which declined by 40% due to the supplementation. In addition, the 18:3n-3 supplement decreased the levels of 20:4n-6 and 22:4n-6 (Fig. 2C).
Fig. 2.
Comparison of the fatty acid profiles (mole percent in total fatty acids, mean ± SD) in control (Ctrl) and 18:2n-6- or 18:3n-3-supplemented hBMSCs (SC) and HepG2 (Hep) cells (n = 4). A: Total proportions of SFAs, MUFAs, PUFAs, n-6 PUFAs, and n-3 PUFAs. B: Individual SFAs and MUFAs. C: Individual n-6 PUFAs. D: Individual n-3 PUFAs. Statistics are as in Fig. 1. In the cases where a value exceeded the scale, the value was shown at the top of the bar.
Different metabolism of C18 PUFA precursors in hBMSCs and HepG2 cells
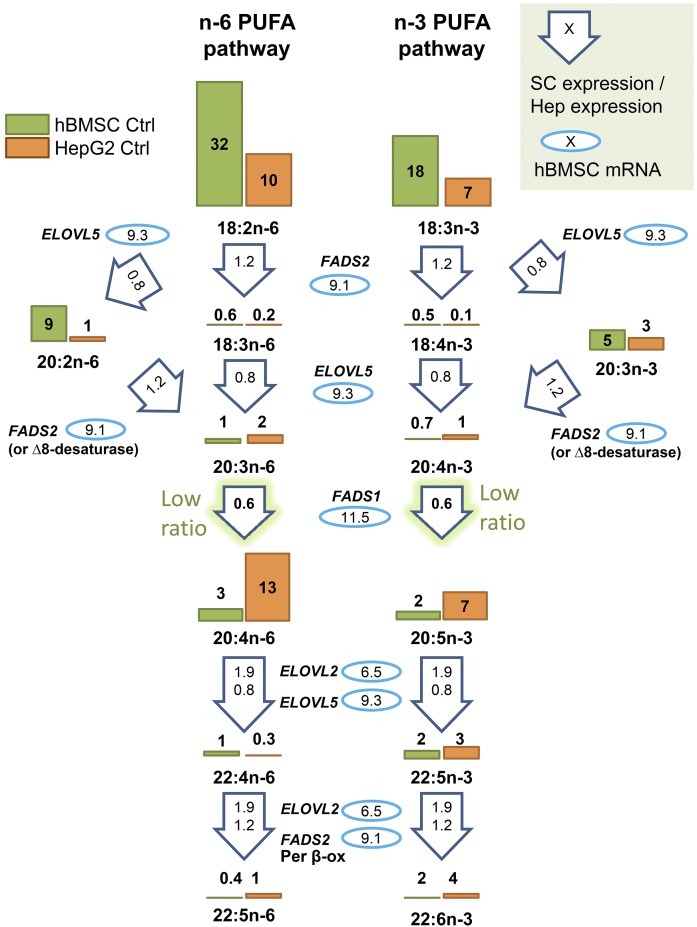
Unlike hBMSCs, the comparison HepG2 cells effectively modified exogenous 18:2n-6 and 18:3n-3 and produced longer and more unsaturated PUFAs, especially 20:4n-6 (Fig. 2C), 20:5n-3, and 22:5n-3 (Fig. 2D). Consequently, the HepG2 cells accumulated clearly less 18:2n-6 and 18:3n-3, only one-third of the relative amount that the corresponding hBMSCs accumulated. The apparent disparity in PUFA metabolism between the hBMSCs and HepG2 cells pointed to differences in the expression levels of the enzymes responsible for the acyl chain elongation or desaturation steps. Thus the mRNA levels in the hBMSCs (passage 4 cells grown in standard medium) were studied for the fatty acid elongases, ELOVL2 and ELOVL5, and for the fatty acid desaturases, FADS1 (Δ5 desaturase) and FADS2 (Δ6 desaturase) (Fig. 3). In these cells, the mRNA levels for ELOVL5, which acts in elongating C18 fatty acids (with some activity on C20 fatty acids as well), were 50% higher than the levels for ELOVL2, which acts on C20 and C22 fatty acids (on average, 9.3 versus 6.5). Expression levels for enzymes modifying fatty acid structures were also retrieved from the public online database, IST, and the values for MSCs and HepG2 cells were compared (Fig. 3). The MSC expression levels of ELOVL5 and ELOVL2 were 0.8 and 1.9 times the values of the HepG2 cells, suggesting no significant defects. The mRNA values of the hBMSCs for FADS1 and FADS2 were in the same range (11.5 and 9.1, respectively). However, the MSC expression levels for FADS1 (Δ5) were low, only 0.6 times those found in the HepG2 cells. MSC FADS2 (Δ6) expression level was 1.2 times that of HepG2 cells. In the database, the SCD (Δ9 desaturase, converting SFAs to MUFAs) expression level of MSCs was reported to be even 2.7 times higher than the levels of HepG2 cells, thus low MUFA contents of hBMSCs compared with HepG2 cells were not due to insufficient SCD expression.
Fig. 3.
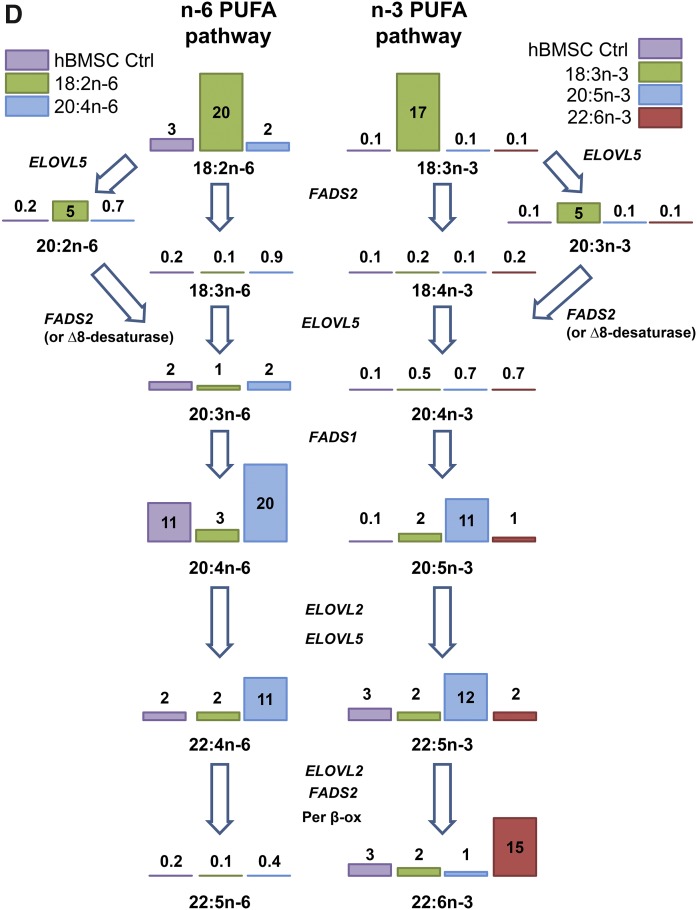
Scheme of the metabolism of n-6 and n-3 PUFAs in the hBMSCs and HepG2 cells by desaturase and elongase enzymes. The average levels of different PUFAs in hBMSCs and HepG2 cells on the pathways are visualized with bars and related values (mole percent) of the cells grown in control (Ctrl) media. The measured mRNA levels of the elongases (ELOVL5 and ELOVL2) and desaturases (FADS2, FADS1) involved are marked inside the blue ovals. The values inside the arrows are the average expression levels of the enzymes that were obtained for mesenchymal stem cells (SC) from a public online database, IST, and normalized against hepatocyte (Hep) values of the database. Per β-ox, peroxisomal partial β-oxidation.
C20 or C22 highly unsaturated fatty acid supplements efficiently and specifically modify the contents of these fatty acids in hBMSCs
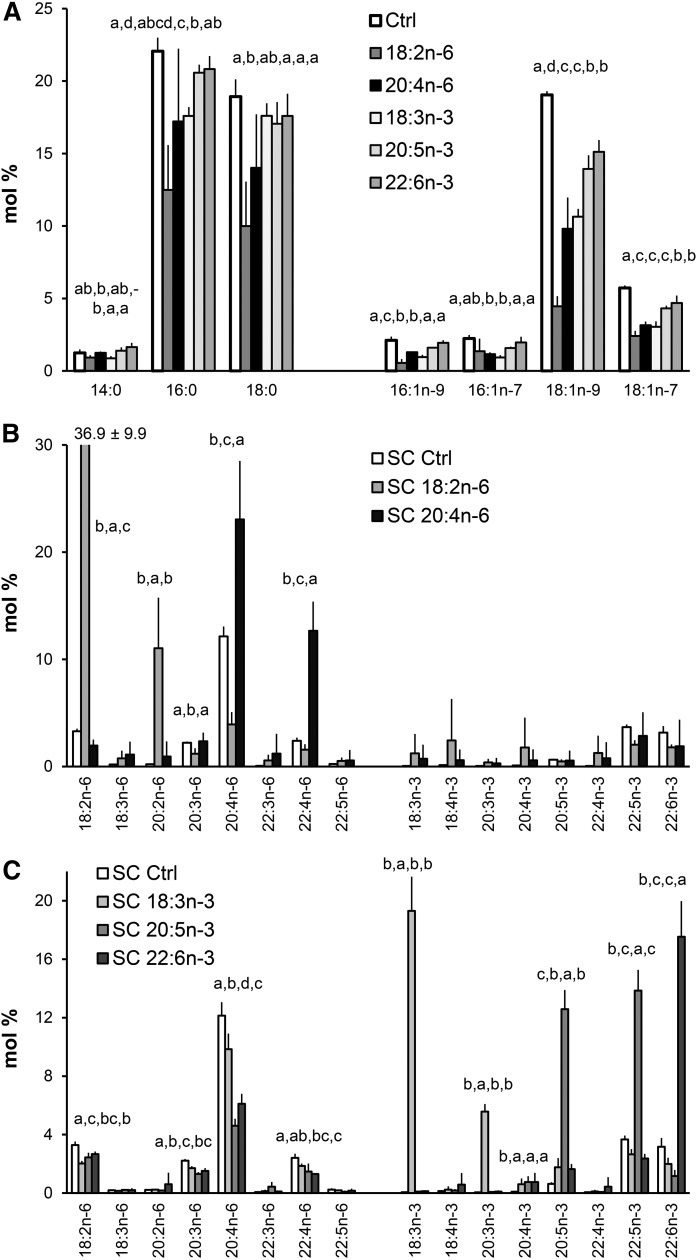
In addition to supplementing the cell cultures with C18 precursors, parallel experiments were carried out that focused on the effects of longer and highly unsaturated members of the n-6 and n-3 PUFAs on the fatty acid profiles of the hBMSCs. The levels of SFAs and especially those of MUFAs, were decreased by the C20 and C22 PUFA supplements, but in general these effects were not as strong as seen in C18 PUFA supplementation experiments (Fig. 4A).
Fig. 4.
Fatty acid profiles (mole percent in total fatty acids, mean ± SD) in hBMSCs (n = 4) cultured for 9 days in the control (Ctrl) medium or media supplemented with different n-6 and n-3 PUFAs. A: Profiles of individual SFAs and MUFAs in cells cultured in the Ctrl medium and in the cells the medium of which was supplemented with 18:2n-6, 20:4n-6, 18:3n-3, 20:5n-3, or 22:6n-3. B: Profiles of PUFAs in the cells cultured in the Ctrl medium and in the medium supplemented with 18:2n-6 or 20:4n-6. C: PUFA profiles in the cells cultured in the Ctrl medium and in the medium supplemented with 18:3n-3, 20:5n-3, or 22:6n-3. Statistics are as in Fig. 1. In the cases where the value exceeded the scale, the mean and SD are shown at the top of the bar. D: Scheme of the metabolism of n-6 and n-3 PUFAs in the hBMSCs cultured in control (Ctrl) medium or media supplemented with the different n-6 and n-3 PUFAs. The enzymes metabolizing the PUFAs are indicated (abbreviations explained in Fig. 3). The values on the bars represent mole percent of the PUFAs per total fatty acids.
As a result of 20:4n-6 supplementation, the hBMSC levels of 20:4n-6 doubled, from 12% to 23% (Fig. 4B). In addition, the supplement caused a 6-fold increase in the level of 22:4n-6, from 2% to 13%. Culturing the cells in 20:5n-3-supplemented medium resulted in a 20-fold increase in the relative amount of 20:5n-3, compared with the cells grown in standard medium, from 1% to 13%, (Fig. 4C). The added 20:5n-3 had a lowering effect on n-6 PUFA content, especially on the 20:4n-6 levels, which decreased by 60%, and a clear reduction was also found for 22:4n-6. Among n-3 PUFAs, the immediate elongation product of 20:5n-3, i.e., 22:5n-3, was raised almost 4-fold due to the 20:5n-3 supplement (Fig. 4C). However, the level of 22:6n-3 (the synthesis of which requires two chain elongations, one desaturation, and one peroxisomal chain shortening step for the precursor 22:5n-3) did not increase, but decreased by 62% in hBMSCs.
Culturing hBMSCs in 22:6n-3-supplemented medium resulted in 6-fold increase in the relative amount of 22:6n-3 compared with the cells kept in standard medium (Fig. 4C). The preceding n-3 family member, 22:5n-3, got 35% lower levels compared with the cells cultured in standard medium. The addition of 22:6n-3 lowered the hBMSC levels of 20:4n-6 and 22:4n-6 by 45% (Fig. 4C), resembling the effect of the 20:5n-3 supplement. When examining the differences in the metabolic PUFA pathways of hBMSCs due to the different PUFA supplements, it became evident that on the n-6 PUFA pathway only 20:4n-6 efficiently raised the total 20:4n-6 and 22:4n-6 contents of the cells (Fig. 4D). On the n-3 PUFA pathway, only 20:5n-3 raised the cellular levels of 20:5n-3 and 22:5n-3. The hBMSC level of 22:6n-3 was increased only by the 22:6n-3 supplement.
Pronounced TAG accumulation in hBMSCs supplemented with n-6 PUFAs
Prominent, but variable, accumulation of unsaturated TAG was detected in the hBMSCs supplemented with 18:2n-6 (supplemental Fig. S1). In addition, the TAG levels of the 20:4n-6-supplemented cells were statistically significantly higher than those of the n-3 PUFA-supplemented cells. The control cells had the lowest TAG levels.
GPL molecular species profiles of hBMSCs differ after 18:2n-6 and 20:4n-6 supplementations
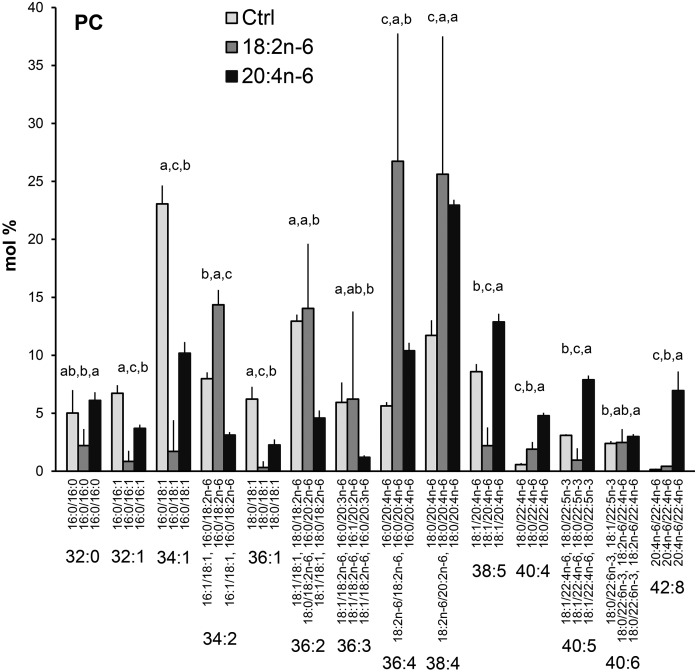
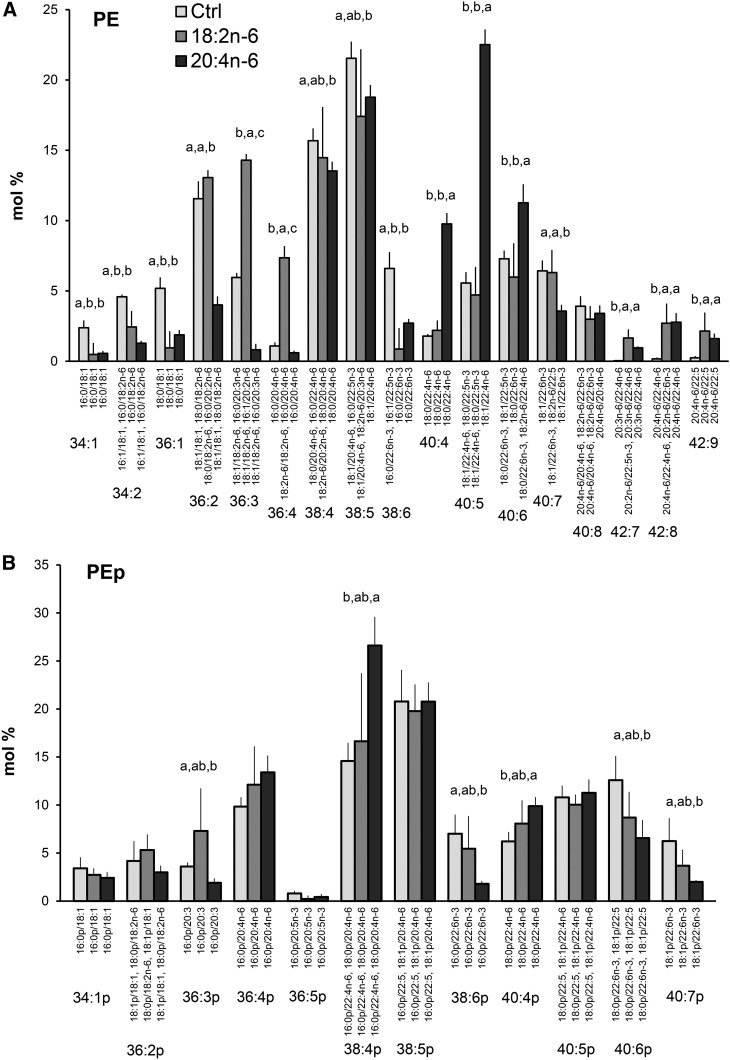
The incorporation of different PUFAs into GPL molecular species was studied in the hBMSCs that were cultured in standard growth medium or in media supplemented with either 18:2n-6 or 20:4n-6 (Figs. 5–7). Supplementing the media with the different n-6 PUFAs modified the PC molecular species profiles of the hBMSCs largely (Fig. 5). The relative amounts of monounsaturated PC species (32:1, 34:1, and 36:1) decreased due to the 18:2n-6 and 20:4n-6 supplements, in which the effect was larger for the 18:2n-6 supplemented cells. The extra 18:2n-6 was incorporated as such or after one elongation step into the PC species 34:2, 36:2, 36:3, and 36:4, which raised their percentages in the 18:2n-6 supplemented cells compared with the other cells. When the cells were supplemented with 20:4n-6, the percentages of 36:4, 38:4, and 38:5 (having 20:4n-6 as the only significant PUFA component) elevated compared with the control values (from 6, 12, and 9% to 10, 23, and 13%, respectively). In addition, highly unsaturated species with 22:4n-6 emerged, and the species 42:8 (20:4n-6/22:4n-6 with 7%) was specific for the 20:4n-6-supplemented cells. As for PC, the PE species 36:2, 36:3, and 36:4 (having 18:2n-6 or 20:2n-6 as their PUFA component) showed prominent percentages after the 18:2n-6 supplement (Fig. 6A). When 20:4n-6 was given to the cells, the percentages of the 22:4n-6-containing molecular species 40:4 and 40:5 elevated 3-fold reaching 10% and 23% levels, respectively. In addition, PE species with two PUFAs (42:7, 42:8, 42:9) emerged after both n-6 PUFA supplements. Among PEp species, the 36:4p and 38:4p served as the largest reservoirs of 20:4n-6, and only a slight increase of the 22:4n-6 containing 40:4p was found in the 20:4n-6-supplemented cells (Fig. 6B). The 20:4n-6 supplement reduced the percentages of 22:6n-3-containing PEp species 38:6p, 40:6p and 40:7p.
Fig. 5.
PC species profiles in hBMSCs cultured for 9 days in the control (Ctrl) medium or medium supplemented with 18:2n-6 or 20:4n-6 (mole percent per total PC, mean ± SD). For the isobaric species (marked horizontally) having more than one quantitatively important species (acyl chain combination marked vertically), the two most important are listed (in the reading order, the first one being the main species). When the double bond positions of the acyl chains were not marked, several isomers (revealed by GC-MS of fatty acids) were present in the cells for that particular chain. Statistics are as in Fig. 1.
Fig. 7.
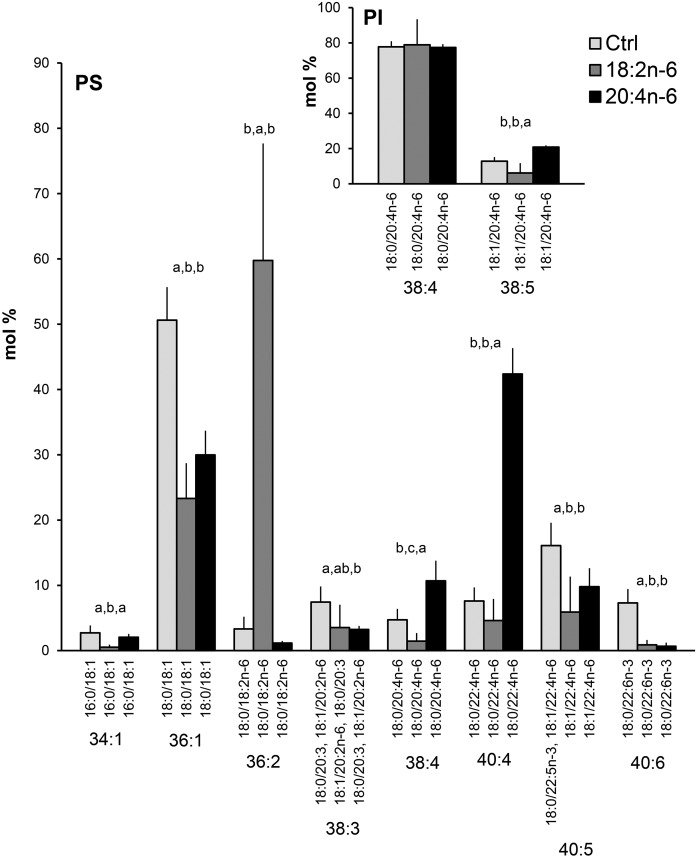
PS species profiles in hBMSCs cultured for 9 days in the control (Ctrl) medium or medium supplemented with 18:2n-6 or 20:4n-6 (mole percent per total PS, mean ± SD). For the isobaric species (marked horizontally) having more than one quantitatively important species (acyl chain combination marked vertically), the two most important were listed (in the reading order the first one being the main species). Quantitatively important PI species are shown in the insert. When the double bond positions of the acyl chains were not marked, several isomers (revealed by GC-MS of fatty acids) were present in the cells for that particular chain. Statistics are as in Fig. 1.
Fig. 6.
PE (A) and PEp (B) species profiles in hBMSCs cultured for 9 days in the control (Ctrl) medium or medium supplemented with 18:2n-6 or 20:4n-6 (mole percent per total PE or PEp, mean ± SD). For the isobaric species (marked horizontally) having more than one quantitatively important species (acyl and alkenyl chain combination marked vertically), the two most important are listed (in the reading order, the first one being the main species). When the double bond positions of the acyl chains were not marked, several isomers (revealed by GC-MS of fatty acids) were present in the cells for that particular chain. Statistics are as in Fig. 1.
Contrasting the diverse PC and PE species profiles, the hBMSC PSs consisted of only a few main species (Fig. 7). Both n-6 PUFA supplements halved the relative amounts of the monounsaturated PS species 36:1 (18:0/18:1), which was present with a roughly 50% level in the control cells and with 25‒30% in the supplemented cells (Fig. 7). When the cells received extra 18:2n-6, the species 36:2 (18:0/18:2n-6) became the dominant PS species of the cells with 60%. When 20:4n-6 was added, the proportion of 38:4 (18:0/20:4n-6) doubled compared with the control values (from 5% to 11%). The largest PS indicator species of the 20:4n-6 supplementation was 40:4 (18:0/22:4n-6), the percentages of which elevated 5-fold, from 8% in the control cells to 42% in the supplemented cells. The main 22:6n-3-containing PS species, 40:6, decreased from 7% to below 1% in the n-6 PUFA-supplemented cells. In hBMSC PIs, only two quantitatively important species were found. The main molecular species was 38:4 (18:0/20:4n-6), the percentages of which remained unaltered in the n-6 PUFA supplementations. However, the percentages of a minor component, 38:5 (18:1/20:4n-6), increased (at the expense of several trace components not shown) due to the 20:4n-6 supplement (Fig. 7 insert).
Different n-3 PUFA supplements incorporate into different GPL molecular species
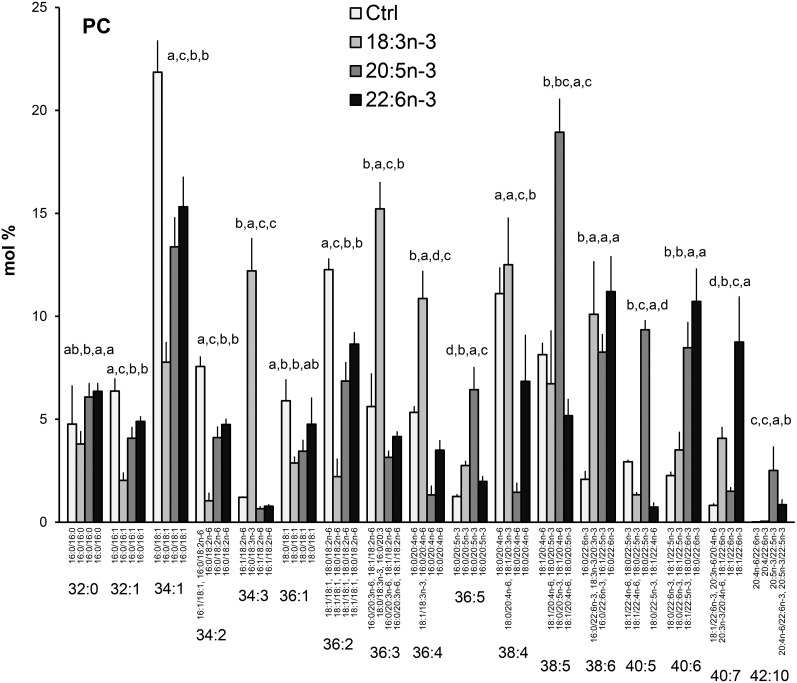
Supplementing the hBMSCs with the n-3 PUFAs, either 18:3n-3, 20:5n-3, or 22:6n-3 induced very specific GPL species profiles, different from each other and from the profiles seen in the cells grown in standard or n-6 PUFA-supplemented media (Figs. 5, 8). Among the most prominent changes seen in the PC species profile with every n-3 PUFA supplementation were the relative decreases (by more than 40%) of the SFA/MUFA species (32:1, 34:1, 36:1) and the species 34:2 and 36:2 (Fig. 8). The cells supplemented with 18:3n-3 showed the lowest percentages for these species containing mainly MUFAs. These relative reductions in MUFA-containing species were accompanied by elevations in the proportions of polyunsaturated species. For instance the 18:3n-3 supplement raised the percentage of 34:3 (16:0/18:3n-3) from the 1% level of the control cells to 12%. In addition, the percentages of 36:3, 36:4 (the main molecular species being 18:0/18:3n-3 and 18:1/18:3n-3, respectively), and 38:6 (including 18:3n-3/20:3n-3) were more than doubled compared with the control cells grown in the medium supplemented only with FBS. Compared with the control cells, the 20:5n-3 supplement caused manifold relative increases in the species 36:5 (16:0/20:5n-3), 38:5 (mainly 18:0/20:5n-3), 40:5 (18:0/22:5n-3), 38:6 (16:0/22:6n-3 and 16:1/22:5n-3), and 40:6 (18:1/22:5n-3 and 18:0/22:6n-3). Concomitantly, the 20:4n-6-containing species, 36:4 and 38:4, decreased to 1.5%, lower than with any other supplements. With extra 20:5n-3, a long-chain and highly unsaturated 42:10 emerged with almost 3%. Like the 20:5n-3 supplement, added 22:6n-3 also boosted 22:6n-3-containing PC species 38:6 and 40:6, which reached about 10% levels. In addition, the proportion of 40:7 (18:1/22:6n-3) elevated to 9% with the 22:6n-3 supplement, higher than with any other fatty acid supplementation.
Fig. 8.
PC species profiles in hBMSCs cultured for 9 days in the control (Ctrl) medium or medium supplemented with 18:3n-3, 20:5n-3, or 22:6n-3 (mole percent per total PC, mean ± SD). For the isobaric species (marked horizontally) having more than one quantitatively important species (acyl chain combination marked vertically) the two most important are listed (in the reading order, the first one being the main species). When the double bond positions of the acyl chains were not marked, several isomers (revealed by GC-MS of fatty acids) were present in the cells for that particular chain. Statistics as in Fig. 1.
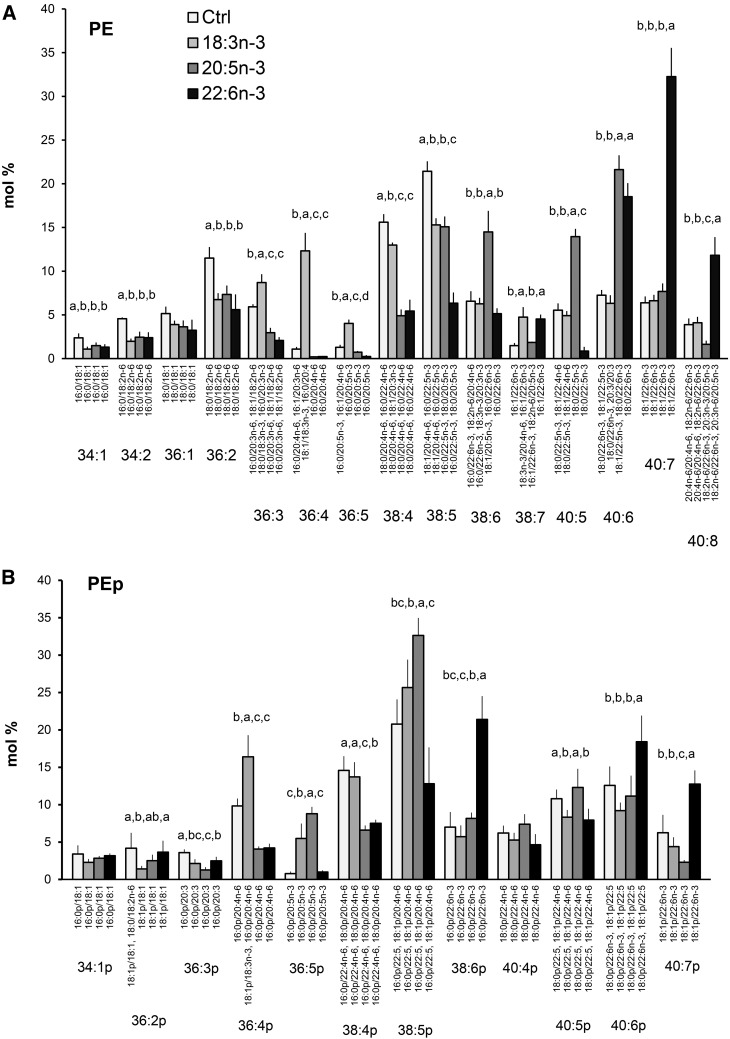
In PE profiles of hBMSCs, the species 34:1, 34:2, 36:1, and 36:2 (18:1/18:1) decreased due to all types of n-3 PUFA supplements. The n-3 PUFAs paired with SFAs and competed out MUFAs and 18:2n-6 (Fig. 9A). The added 18:3n-3 even raised the percentage of 36:4 (18:1/18:3n-3 as the main component) 10-fold (to 12%) and elevated percentages were also found for 36:3 and 36:5. Unlike the other n-3 PUFA supplements, added 18:3n-3 kept the isobaric species 38:4 high, which was due to the elevated percentage of the molecular species 18:1/20:3n-3. With the 20:5n-3 and 22:6n-3 supplements, the proportions of n-6 PUFA-containing PE species 38:4 (18:0/20:4n-6 and 16:0/22:4n-6) decreased to 5%, one-third of the values found in the cells cultured with standard medium. Addition of 20:5n-3 raised the proportions of PE 38:6 (in these cells largely 18:1/20:5n-3) and 40:5 (18:0/22:5n-3) close to 15%, 2-fold those in the other hBMSCs. When the hBMSCs were supplemented with 22:6n-3, the percentage of PE 40:7 (18:1/22:6n-3) elevated 5-fold to 32%, and the species 40:8 (in these cells largely 18:2n-6/22:6n-3) elevated 3-fold to 12%. Both the 22:5n-3 and 22:6n-3 supplements raised the percentage of PE 40:6 (18:1/22:5n-3 and 18:0/22:6n-3 as the main species, respectively). In PEp, the supplements 20:5n-3 and 22:6n-3 decreased the percentages of the 20:4n-6- and 22:4n-6-containing species 36:4p and 38:4p to one-half of the control values (Fig. 9B). In contrast, supplementing the cells with 18:3n-3 had no such effect. After the cells received extra 20:5n-3, the species 36:5p and 38:5p showed elevated percentages, and when 22:6n-3 was given, the boosted species were 38:6p (3-fold), 40:6p (1.5-fold), and 40:7p (2-fold).
Fig. 9.
PE (A) and PEp (B) species profiles in hBMSCs cultured for 9 days in the control (Ctrl) medium or medium supplemented with 18:3n-3, 20:5n-3, or 22:6n-3 (mole percent per total PE or PEp, mean ± SD). For the isobaric species (marked horizontally) having more than one quantitatively important species (acyl and alkenyl chain combination marked vertically), the two most important are listed (in the reading order, the first one being the main species). When the double bond positions of the acyl chains were not marked, several isomers (revealed by GC-MS of fatty acids) were present in the cells for that particular chain. Statistics are as in Fig. 1.
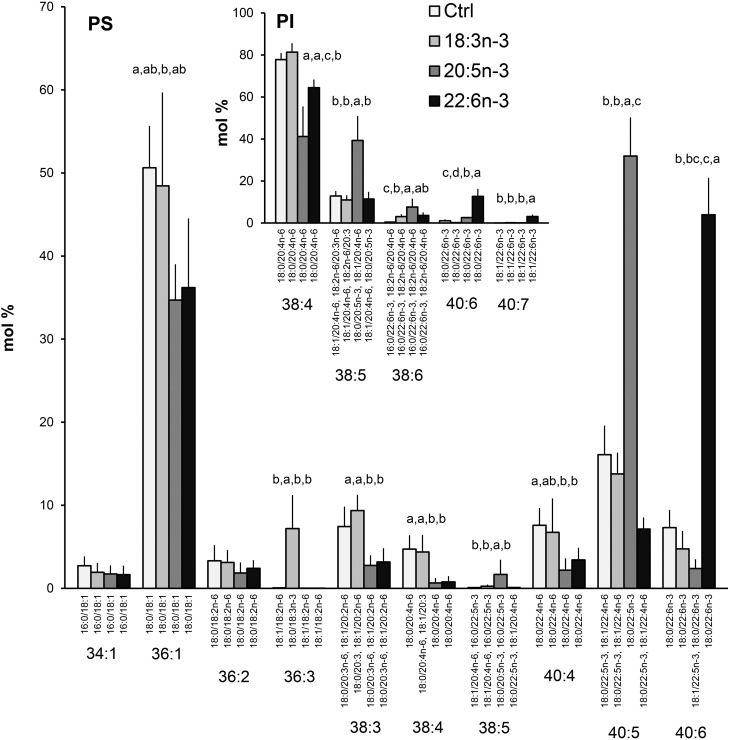
The PS species profiles of the hBMSCs supplemented with 18:3n-3 showed a specific appearance of 36:3 with a 7% level (Fig. 10). Surprisingly, 18:3n-3 was not able to significantly modify the percentages of the other five quantitatively important polyunsaturated species 38:3, 38:4, 40:4, 40:5, or 40:6. In contrast, the 20:5n-3 and 22:6n-3 supplements decreased the percentages of 38:3, 38:4, and 40:4 to one-half or more of the values found for the control and 18:3n-3-supplemented cells. Compared with the cells grown in control medium, the 20:5n-3 supplement raised the percentage of 40:5 (18:0/22:5n-3) 3-fold (to 52%), and the 22:6n-3 supplement raised the share of 40:6 (18:0/22:6n-3) 6-fold (to 45%). The PI species profiles of the hBMSCs were altered little due to the 18:3n-3 supplement (Fig. 10 insert). In contrast, exogenous 20:5n-3 and 22:6n-3 replaced part of 20:4n-6 in the main PI species, 38:4 (18:0/20:4n-6), thus causing manifold increases in the proportions of minor components with 20:5n-3 (38:5) or 22:6n-3 (40:6 and 40:7).
Fig. 10.
PS species profiles in hBMSCs cultured for 9 days in the control (Ctrl) medium or medium supplemented with 18:3n-3, 20:5n-3, or 22:6n-3 (mole percent per total PS, mean ± SD). For the isobaric species (marked horizontally) having more than one quantitatively important species (acyl chain combination marked vertically), the two most important are listed (in the reading order, the first one being the main species). Quantitatively important PI species are shown in the insert. When the double bond positions of the acyl chains were not marked, several isomers (revealed by GC-MS of fatty acids) were present in the cells for that particular chain. Statistics are as in Fig. 1.
Specific and large alterations in 20:4n-6, 20:5n-3, and 22:6n-3 distribution patterns between the GPL classes of hBMSCs supplemented with different PUFAs
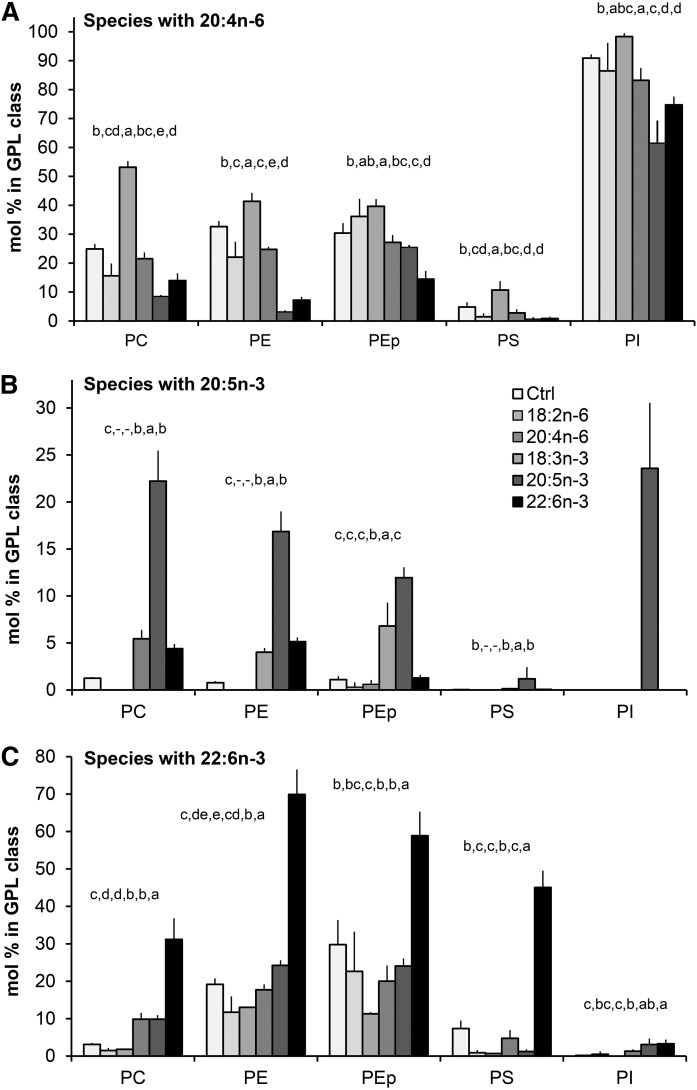
Compared with the control hBMSCs, the 18:2n-6 supplementation did not elevate the 20:4n-6 content in the GPLs of the cells, but decreased it by one-third in PC and PE (Fig. 11A). When given 18:2n-6, the PEp class kept its 20:4n-6 content unchanged. Despite the general contents of 20:4n-6 being characteristically low in PS, the percent decrease of 20:4n-6 content due to 18:2n-6 was the largest in PS. In contrast, the 20:4n-6 supplement doubled the 20:4n-6 content in PC (from 25% to 53%) and PS (from 5% to 11%), and also caused a significant increment of 20:4n-6 in PE and PEp (from 30–33% to 40–41%). Most of the PI molecules of n-6 PUFA-supplemented cells contained a 20:4n-6 residue. The n-6 PUFA supplements did not allow 20:5n-3 to be incorporated into any of the studied GPLs in any significant amount (Fig. 11B) and, in general, the n-6 PUFA supplements significantly decreased the 22:6n-3 contents of GPLs (Fig. 11C).
Fig. 11.
Distribution of lipid species carrying important PUFA precursors of signaling molecules between different GPL classes in hBMSCs cultured for 9 days in the control (Ctrl) medium or medium supplemented with 18:2n-6, 20:4n-6, 18:3n-3, 20:5n-3, or 22:6n-3 (mole percent sums of all species with the specific PUFA in each GPL class, mean ± SD). A: Distribution of species carrying 20:4n-6 between the GPL classes. B: Distribution of species carrying 20:5n-3 between the GPL classes. C: Distribution of species carrying 22:6n-3 between the GPL classes. Statistics are as in Fig. 1.
The 18:3n-3 supplement failed to replace 20:4n-6 among the PC and PS species of the hBMSCs, and compared with the control cells, small but statistically significant relative decreases of 20:4n-6 content were seen in PE (by 24%) and PI (by 8%) (Fig. 11A). The precursor 18:3n-3 was incorporated into the PC species as such or as its immediate elongation product, 20:3n-3 (Fig. 8), and thus this supplement affected the contents of 20:5n-3 and 22:6n-3 in PC to a lesser degree than when using these highly unsaturated fatty acids themselves as the supplement (Fig. 11B, C).
Contrasting the effects of the 18:3n-3 supplement, providing the cells with the long and highly unsaturated 20:5n-3 and 22:6n-3 efficiently replaced 20:4n-6 in PC, PE, and PS species, and also caused mild, but statistically significant, relative reductions of 20:4n-6 in PI (Fig. 11A). When the cells were supplemented with 20:5n-3, the 20:4n-6 content of PC, 25% in the control cells, reduced to 8%. In PE, the concomitant loss of 20:4n-6 due to the exogenous 20:5n-3 was even more pronounced, from 33% to 3% (Fig. 11A). When supplemented with 22:6n-3, the 20:4n-6 contents of PC and PE decreased from the above mentioned levels to 14% and 7%, respectively. Of note, in PEp, the reductions of 20:4n-6 due to 20:5n-3 or 22:6n-3 were much milder than the clear losses found in PE. In PS, these highly unsaturated n-3 PUFAs decreased the 20:4n-6 content from the 5% of the control cells to below 1%.
The 20:5n-3 supplement resulted in large roughly 15–20% reserves of 20:5n-3 in all studied GPL classes, except PS (Fig. 11B). Even in PI, with a strict preference for 20:4n-6, the 20:5n-3 replaced 20:4n-6 and, as the result, one-fourth of the PI species served as potential donors of 20:5n-3. With the 22:6n-3 supplement, PE contained the highest relative levels of 22:6n-3 (70%) followed by PEp (59%), PS (45%), and PC (31%) (Fig. 11C). PI adopted exogenous 22:6n-3 only with 4%. It should be noted that when the long and highly unsaturated n-3 PUFA supplements efficiently displaced 20:4n-6 in PE and PS, consequently, the PI, PEp, and PC became even more important remaining sources of 20:4n-6 in these cells (Fig. 11A).
PGE2 production in hBMSCs supplemented with 20:4n-6, 20:5n-3, or 22:6n-3
The hBMSCs initially grown in the control medium (with 5% FBS) or in the medium further supplemented with either 20:4n-6, 20:5n-3, or 22:6n-3 were studied for the PGE2 concentration they secreted into serum-free medium. The cells supplemented with 20:4n-6 released PGE2 concentrations 16-fold those released from the control cells (supplemental Fig. S2). In contrast, the n-3 PUFA-supplemented cells secreted PGE2 at concentrations close to control levels.
DISCUSSION
Bone marrow niche is a complex lipid-rich tissue where different types of stem, progenitor, and differentiated cells interact under the control of various neuronal, hormonal, and other chemical stimuli, including lipid mediators such as PGE2 derived from 20:4n-6 (42–44). Human bone marrow typically contains 50% levels of MUFAs, 30% SFAs, and 20% PUFAs (45). When hBMSCs are isolated from the marrow and expanded in culture, where the lipid and fatty acid supplements originate from bovine serum, their nourishment and activation conditions may change. Recently, physiologically relevant levels of SFAs were found to reduce hBMSC proliferation and viability, the effects of which were prevented by the major MUFA, 18:1n-9 (46). In addition, in adipocytes and circulating immune cells, SFAs have been found to activate Toll-like receptor 4 (TLR4), consequently initiating inflammatory cascades and recruiting proinflammatory M1 macrophages (47). Apparently stromal cells and other cell types of the marrow require the high MUFA/SFA ratio, 1.5 in the marrow samples of this work, for optimal functions. However, the MUFA/SFA ratio of FBS and the hBMSCs grown with FBS was only 0.7. The current work showed that compared with the bone marrow, FBS was especially rich in the PGE2 precursor, 20:4n-6 (enriched 12-fold compared with marrow), but also contained larger relative amounts of 20:5n-3 (5-fold), 22:5n-3 (7-fold), and 22:6n-3 (8-fold) than the marrow. The enzymes COX, LOX, and P450 have different specificities toward these different PUFA substrates, and the mediators formed have different functions and activities (48, 49). Because the hBMSC level of 22:4n-6 was almost 4-fold compared with the FBS level, the cells apparently had elongated part of the received bioactive 20:4n-6 to 22:4n-6. Due to its inhibitory effects on COX-1 and COX-2 activities, 22:4n-6 is regarded as biologically less active than 20:4n-6 (50, 51). In n-3 PUFAs, a similar, but less extensive, elongation of 20:5n-3 was seen, and consequently 22:5n-3, a more potent COX-1 and COX-2 inhibitor than 20:5n-3 (51, 52), was enriched in the cells almost 2-fold compared with medium FBS. Thus the common culture medium provides the hBMSCs with few MUFAs and excessively bioactive PUFA precursors, especially 20:4n-6. In these conditions, the cells may have attenuated their COX signaling by actively elongating the excess C20 PUFAs to C22 PUFAs. Notwithstanding, the data suggest that FBS with its high 20:4n-6 content, especially, is not an optimal source of fatty acids for cultured hBMSCs.
The SCD (Δ9 desaturase) expression level of MSCs is high (41), and thus the low MUFA contents of the cultured hBMSCs were not due to their inability to synthesize them, but likely that their MUFA production was downregulated by their high PUFA contents, which is regarded as a mechanism to maintain proper lipid viscosity. PUFAs have been proposed to bind to DNA regulatory areas and inhibit the production of the Δ9 desaturase enzyme responsible for the conversion of SFAs to MUFAs (53). Thus the additional PUFA supplements, when further increasing the PUFA contents of the hBMSCs, reduced the contents of MUFAs even more. In addition, we found that the ability of the hBMSCs for structural modifications of the C18 PUFA precursors was limited compared with the frequently studied HepG2 cells. In HepG2 cells, exogenous 18:2n-6 or 18:3n-3 were largely converted to their highly unsaturated products, while in hBMSCs these supplements raised the contents of their own and those of their immediate chain elongation products, but were inefficient in raising the levels of the most bioactive highly unsaturated PUFAs. The fatty acid profiles and the supporting enzyme data suggest that the desaturation steps, especially the Δ5 desaturation (FADS1) were inefficient in the hBMSCs.
On the metabolic pathway of PUFAs, the n-3 and n-6 PUFAs are known to compete for the same elongation and desaturation enzymes, and thus the n-3 PUFA supplements were expected to inhibit the metabolic modifications of the n-6 PUFAs (54–57). In hBMSCs, the ability of different n-3 PUFA supplements to inhibit the metabolism of n-6 PUFAs was very different. Supplementing the cells with the precursor, 18:3n-3, did not reduce their 20:4n-6 content as efficiently as 20:5n-3 or 22:6n-3 did. In fact, the18:3n-3 reduced the 22:5n-3 and 22:6n-3 contents of the hBMSCs, an effect not seen in the PUFA profiles of the HepG2 cells given the same supplement. Thus the manipulations intended to reduce hBMSC 20:4n-6 levels to favor anti-inflammatory instead of inflammatory signaling seem successful only with the readymade long-chain and highly unsaturated n-3 PUFA supplements. Currently, these PUFAs are not added in sufficient amounts in the standard cell culture media. Because the 18:2n-6 supplement also reduced the hBMSC levels of 20:4n-6, the biosynthetic pathways toward the long and highly unsaturated fatty acids have likely been saturated due to the excess C18 PUFA precursor (58).
The PUFA-supplemented hBMSCs partly overcame the potential harmful effects of the excess PUFAs by incorporating them into TAGs. Interestingly, the accumulation of highly unsaturated TAGs was more pronounced in the hBMSCs cultured with n-6 PUFA supplements than in the cells cultured with the same concentration of n-3 PUFAs. This may mean that producing phospholipid membranes with high n-6 PUFA contents would have been more detrimental to the cellular functions than producing membranes with high n-3 PUFA contents. In addition to finding part of the supplemented PUFAs deposited in storage TAGs, part of the PUFAs were incorporated into membrane GPLs.
Manipulating the profiles of polyunsaturated GPLs by the PUFA supplements subsequently affects the precursor pool to be used for lipid mediator production. The factors controlling PUFA availability for hydrolysis from membrane GPL and their transformation into different bioactive lipid mediators are not yet fully elucidated. However, it is widely accepted that the cytosolic phospholipase A2 type IV (cPLA2IV) is responsible for the main part of the stimulus-dependent mobilization of 20:4n-6 (59, 60). Secretory Ca2+-dependent PLA2 (sPLA2) and other lipases aid by amplifying and spreading the signal. Once 20:4n-6 or other PUFAs have been released from membrane GPLs, the excess of free PUFAs not transferred to lysoPC for re-acylation is converted to bioactive compounds by the COX, LOX, P450, and other pathways (61, 62). We measured relevant mRNA levels in the studied hBMSCs for the crucial phospholipases, PLC, PLD, cPLA2IV, and sPLA2 (supplemental Table S1), and also for COX-1 (PTGS1, 7.2), COX-2 (PTGS2, 7.1), several forms of LOX (e.g., ALOX12, 6.2; ALOX15, 6.3; ALOX5, 6.1) and numerous members of the CYP (P450) family, which suggests that the hBMSCs had the capacity to generate a wide array of lipid mediators.
The substrate pools easily accessible for the lipid mediator generating enzymes are determined by various factors. For example the structures of the polar head group, the acyl chains, and their assemblies in the GPL species affect the efficiency of cleaving the sn-2 PUFA by phospholipases. The cPLA2IV preferentially liberates 20:4n-6 from the sn-2 position and favors PC and PI as substrates, but also accepts PE with lower rates of hydrolysis (63). In human eosinophilic leukemia cells, the activity of cPLA2 toward PE species with 20:5n-3 and 22:6n-3 was found to be much lower than toward the 20:4n-6-containing PE (64). Other PLA2 forms have different GPL class preferences and also use PE and PEp species as the PUFA donor (65). The phospholipase substrate preferences partly arise from the placement of the PUFA precursors in different molecular species of membrane GPLs. In addition to the reported preferences to hydrolyze a specific sn-2 acyl chain (e.g., 20:4n-6 by cPLA2IV), the sn-1 acyl chain also affects the rate of GPL species hydrolysis due to the different degree of hydrophilicities and efflux propensities of different GPL species from the membrane bilayer (66–68). Functional consequences follow since Dong et al. (51) found that the coadministered nonessential fatty acids, 16:0, 18:1n-9, and 18:0 stimulated 20:4n-6-derived COX-2 activity, and the highest stimulation was recorded with the combination 16:0/20:4n-6, which species was kept as a minor component of the PC and PE of hBMSCs. The more prominent species of the hBMSCs, 18:0/20:4n-6, resulted in the experiment of Dong and coworkers in the lowest level of COX-2 stimulation. Thus the precise control of GPL species composition is likely crucial for the regulation of cellular functions, such as inflammation, proliferation, and apoptosis.
The emergence of diPUFA species, e.g., PC42:8 and PE42:8 and PE42:9 in the hBMSCs supplemented with 20:4n-6 (50 μM), suggests that after the primary low-capacity and high-affinity pathway for PUFA incorporation into GPLs was saturated, the cells were forced to employ the high-capacity and low-affinity de novo pathway known to produce diPUFA species (62). The rates of hydrolysis of these different diPUFA species for possible lipid mediator production are not the same. A recent study indicated that cPLA2IV hydrolyzes PC molecular species 20:4n-6/20:4n-6 with a rate superior to 22:6n-3/22:6n-3 and other PC species (68). Exogenous 20:4n-6 increased the percentages of the polyunsaturated C42 species of the hBMSCs containing 20:4n-6 coupled with 22:4n-6 or 22:5 (n-6 or n-3), which may have had significantly lower hydrolysis rates and weaker signaling potential than the 20:4n-6/20:4n-6 species. There is also other functional evidence that the elongation of 20:4n-6 to 22:4n-6 and subsequent incorporation of 22:4n-6 into membrane GPLs may serve as a mechanism to attenuate inflammatory and apoptotic signaling due to excess 20:4n-6. COX activity experiments using 22:4n-6 as the substrate resulted in activities halved or smaller than those recorded with 20:4n-6 (50, 51). Having 22:4-6 as substrate, COX-2 generates 1a,1b-dihomo PGE2, the biological activity of which is regarded as much lower than the activity of PGE2 synthesized from 20:4n-6. As far as we know, systematic studies addressing the functions and activity of the dihomo form do not exist. In line with these studies, the hBMSCs with high 20:4n-6 content and PGE2 production also had largely elevated levels of PC and PE species 18:1/22:4n-6 (40:5), perhaps to hinder the boosted proinflammatory signaling.
Adjusting the levels of 20:4n-6 in cultured therapeutic cell lines is important for controlling their immunomodulatory properties. In our previous work, cultured hBMSCs expanded in several steps had lowered capacity to suppress the proliferation of T cells and increased GPL 20:4n-6 content (13). Recently, Campos et al. (69) reported that hBMSCs stimulated by proinflammatory cytokines had enhanced levels of 20:4n-6-containing PC species 38:4. The 20:4n-6 signaling also has a pronounced role in tumor cells, which have high expression levels of PLA2s and COX-2, and enhanced production of PGE2 (70, 71). Differentiation-related increase of GPL species with 20:4n-6 was recently found in cultured neural stem cells (72). Using inhibitors for COX and PLA2 enzymes working on 20:4n-6 is one approach in the attempt to control cellular functions and fate in vivo. However, because these enzymes are part of essential pathways, and inhibitors for many PLA2 forms are unknown, an alternative holistic and relatively safe way to affect immune functions and viability of cells would be to manipulate the PUFA pool in the membranes of the therapeutic cells. As shown in this work, the PGE2 production of hBMSCs strongly reflects their supply and membrane contents of 20:4n-6, while the n-3 PUFA supplements have only a small effect on the PGE2 levels of the cells.
The 20:5n-3 or 22:6n-3 supplements lowering the 20:4n-6 contents of PC limit the 20:4n-6-related eicosanoid signaling. Despite the preference of cPLA2IV to hydrolyze PUFAs from PC and PI, the transacylases further mediate the newly acquired n-3 PUFAs to PEs and other GPLs, which can then serve as n-3 PUFA donors for various lipases (73). Increasing the ratio of 20:5n-3/20:4n-6 in membrane GPLs is known to be manifested in an increased ratio of the classic eicosanoids PGE3/PGE2, with well-known anti-inflammatory effects (51, 74). In addition, 20:5n-3 and 22:6n-3 are precursors for resolvins, protectins, and maresins, new generation mediators fortifying the resolution phase of inflammation (75). With 18:3n-3 supplement, however, the GPL levels of 20:4n-6, 20:5n-3, or 22:6n-3 were affected relatively little. This suggests that 18:3n-3, unlike 20:5n-3 (48, 49), has little potency to affect the inflammation status of the cells via eicosanoid or docosanoid signaling.
Besides being a precursor for docosanoids with anti-inflammatory properties (75), 22:6n-3 influences membrane structure and protein function by affecting membrane packing, permeability, and lateral domain structure crucial for optimal functions of various integral proteins (76). In addition, 22:6n-3 can directly bind to receptors. For example, 22:6n-3 is a ligand for macrophage GPR120 receptor initiating a signaling pathway through β-arrestin2, which then inhibits TLR4 and inflammatory cytokine receptors, and reduces the inflammatory state of cells (77). Thus, manipulating the GPL contents of 22:6n-3 may have beneficial effects on the immunomodulatory properties of the cells via different mechanisms. In addition, 22:6n-3 was the only supplement that significantly increased its relative amount in PS. All other supplements decreased PS 22:6n-3 content. The 22:6n-3 affects Ca-dependent and -independent protein kinase C (PKC)-mediated signaling cascades. In monocytic leukemia U937 cells, 22:6n-3 activated PLC, which released inositol-trisphosphate and calcium from endoplasmic reticulum-activating PKCγ. In a cell-free system, 22:6n-3 activated PKCδ by binding directly to its PS-binding site, which activation was not found for 20:4n-6, 20:5n-3, or 22:5n-3 (78). Thus, replacing 20:4n-6 by 22:6n-3 in the metabolically related PE and PS likely affects several signaling cascades via modulation of PKC activity in 22:6n-3-supplemented hBMSCs also. PC species containing 22:6n-3 were not found to promote PKC partitioning to membrane and activation (79).
The current results emphasize the importance of choosing appropriate PUFA supplements for hBMSCs and other therapeutic cells cultured and expanded with artificial PUFA sources. Because the capacity of hBMSCs to modify PUFA structures is limited, the different supplements given result in very different GPL species profiles and substrate availability for phospholipases having preferences for polar head group and acyl chains. The chromatographic and mass spectrometric data obtained suggest that the hBMSCs are able to attenuate the possible harmful effect due to excess 20:4n-6 and subsequently enhanced PGE2 production by chain elongation of 20:4n-6 to 22:4n-6. Because supplemented 20:5n-3 and 22:6n-3 replaced 20:4n-6 in membrane GPLs, they apparently influenced eicosanoid/docosanoid signaling. In addition, the n-3 PUFA supplements modified membrane structure and thus modulated integral protein function, and as ligands, they likely participated in different signaling pathways not yet elucidated for MSCs. This work lays foundations for understanding the functional consequences of different PUFA supplementations in hBMSCs, and the next question important to address in order to develop appropriate protocols for PUFA manipulations of these therapeutic cells will be to study the kinetics of the acyl chain remodeling in different GPL classes. The ultimate goal is to characterize the changes of lipid mediator profiles caused by the PUFA supplements and carry out coculture experiments with hBMSCs and other cell types to monitor changes in biological functions and cellular interactions.
Supplementary Material
Acknowledgments
The authors acknowledge the work performed by Päivi Saavalainen, Dario Greco, and Amarjit Parmar in microarray data analysis.
Footnotes
Abbreviations:
- C18
- 18-carbon
- COX
- cyclooxygenase
- cPLA2
- cytosolic phospholipase A2
- cPLA2IV
- cytosolic phospholipase A2 type IV
- GEO
- Gene Expression Omnibus
- GPL
- glycerophospholipid
- hBMSC
- human bone marrow mesenchymal stromal cell
- LOX
- lipoxygenase
- MSC
- mesenchymal stromal cell
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PEp
- phosphatidylethanolamine plasmalogen
- PGE2
- prostaglandin E2
- PI
- phosphatidylinositol
- PKC
- protein kinase C
- PS
- phosphatidylserine
- SFA
- saturated fatty acid
- TAG
- triacylglycerol
This work was partly supported by the SalWe Research Program of Intelligent Monitoring of Health and Well-being (IMO) (Tekes Grant 648/10).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., and Marshak D. R.. 1999. Multilineage potential of adult human mesenchymal stem cells. Science. 284: 143–147. [DOI] [PubMed] [Google Scholar]
- 2.Di Nicola M., Carlo-Stella C., Magni M., Milanesi M., Longoni P. D., Matteucci P., Grisanti S., and Gianni A. M.. 2002. Human bone marrow stromal cells suppress T-lymphocyte proliferation induced by cellular or nonspecific mitogenic stimuli. Blood. 99: 3838–3843. [DOI] [PubMed] [Google Scholar]
- 3.Sato K., Ozaki K., Oh I., Meguro A., Hatanaka K., Nagai T., Muroi K., and Ozawa K.. 2007. Nitric oxide plays a critical role in suppression of T-cell proliferation by mesenchymal stem cells. Blood. 109: 228–234. [DOI] [PubMed] [Google Scholar]
- 4.Ren G., Zhang L., Zhao X., Xu G., Zhang Y., Roberts A. I., Zhao R. C. C., and Shi Y.. 2008. Mesenchymal stem cell-mediated immunosuppression occurs via concerted action of chemokines and nitric oxide. Cell Stem Cell. 2: 141–150. [DOI] [PubMed] [Google Scholar]
- 5.Krampera M., Glennie S., Dyson J., Scott D., Laylor R., Simpson E., and Dazzi F.. 2003. Bone marrow mesenchymal stem cells inhibit the response of naive and memory antigen-specific T cells to their cognate peptide. Blood. 101: 3722–3729. [DOI] [PubMed] [Google Scholar]
- 6.Karlsson H., Samarasinghe S., Ball L. M., Sundberg B., Lankester A. C., Dazzi F., Uzunel M., Rao K., Veys P., Le Blanc K., et al. . 2008. Mesenchymal stem cells exert differential effects on alloantigen and virus-specific T-cell responses. Blood. 112: 532–541. [DOI] [PubMed] [Google Scholar]
- 7.Sharma R. R., Pollock K., Hubel A., and McKenna D.. 2014. Mesenchymal stem or stromal cells: a review of clinical applications and manufacturing practices. Transfusion. 54: 1418–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Blanc K., Frassoni F., Ball L., Locatelli F., Roelofs H., Lewis I., Lanino E., Sundberg B., Bernardo M. E., Remberger M., et al. . 2008. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet. 371: 1579–1586. [DOI] [PubMed] [Google Scholar]
- 9.Wang D., Zhang H., Liang J., Li X., Feng X., Wang H., Hua B., Liu B., Lu L., Gilkeson G. S., et al. . 2013. Allogeneic mesenchymal stem cell transplantation in severe and refractory systemic lupus erythematosus: 4 years of experience. Cell Transplant. 22: 2267–2277. [DOI] [PubMed] [Google Scholar]
- 10.Tan J., Wu W., Xu X., Liao L., Zheng F., Messinger S., Sun X., Chen J., Yang S., Cai J., et al. . 2012. Induction therapy with autologous mesenchymal stem cells in living-related kidney transplants: a randomized controlled trial. JAMA. 307: 1169–1177. [DOI] [PubMed] [Google Scholar]
- 11.Wagner W., Horn P., Castoldi M., Diehlmann A., Bork S., Saffrich R., Benes V., Blake J., Pfister S., Eckstein V., et al. . 2008. Replicative senescence of mesenchymal stem cells: A continuous and organized process. PLoS One. 3: e2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gharibi B., and Hughes F. J.. 2012. Effects of medium supplements on proliferation, differentiation potential, and in vitro expansion of mesenchymal stem cells. Stem Cells Transl. Med. 1: 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilpinen L., Tigistu-Sahle F., Oja S., Greco D., Parmar A., Saavalainen P., Nikkilä J., Korhonen M., Lehenkari P., Käkelä R., et al. . 2013. Aging bone marrow mesenchymal stromal cells have altered membrane glycerophospholipid composition and functionality. J. Lipid Res. 54: 622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehér I., and Gidáli J.. 1974. Prostaglandin E2 as stimulator of haemopoietic stem cell proliferation. Nature. 247: 550–551. [DOI] [PubMed] [Google Scholar]
- 15.Rajasingh J., and Bright J. J.. 2006. 15-Deoxy-Δ12,14-prostaglandin J2 regulates leukemia inhibitory factor signaling through JAK-STAT pathway in mouse embryonic stem cells. Exp. Cell Res. 312: 2538–2546. [DOI] [PubMed] [Google Scholar]
- 16.Yamazaki S., Iwama A., Takayanagi S., Morita Y., Eto K., Ema H., and Nakauchi H.. 2006. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J. 25: 3515–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Merkler K. A., Zhang X., and McLean M. P.. 2007. Prostaglandin F2alpha suppresses rat steroidogenic acute regulatory protein expression via induction of yin yang 1 protein and recruitment of histone deacetylase 1 protein. Endocrinology. 148: 5209–5219. [DOI] [PubMed] [Google Scholar]
- 18.Kim M. H. H., Kim M. O. O., Kim Y. H. H., Kim J. S. S., and Han H. J. J.. 2009. Linoleic acid induces mouse embryonic stem cell proliferation via Ca2+/PKC, PI3K/Akt, and MAPKs. Cell. Physiol. Biochem. 23: 53–64. [DOI] [PubMed] [Google Scholar]
- 19.Lee M. Y., Ryu J. M., Lee S. H., Park J. H., and Han H. J.. 2010. Lipid rafts play an important role for maintenance of embryonic stem cell self-renewal. J. Lipid Res. 51: 2082–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bieberich E. 2012. It’s a lipid’s world: bioactive lipid metabolism and signaling in neural stem cell differentiation. Neurochem. Res. 37: 1208–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aggarwal S., and Pittenger M. F.. 2005. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 105: 1815–1822. [DOI] [PubMed] [Google Scholar]
- 22.Serhan C. N., Chiang N., and Dalli J.. 2015. The resolution code of acute inflammation: Novel pro-resolving lipid mediators in resolution. Semin. Immunol. 27: 200–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang X., Abbott J., Cheng L., Colby J. K., Lee J. W., Levy B. D., and Matthay M. A.. 2015. Human mesenchymal stem (stromal) cells promote the resolution of acute lung injury in part through lipoxin A4. J. Immunol. 195: 875–881. [DOI] [PubMed] [Google Scholar]
- 24.Erices A., Conget P., and Minguell J. J.. 2000. Mesenchymal progenitor cells in human umbilical cord blood. Br. J. Haematol. 109: 235–242. [DOI] [PubMed] [Google Scholar]
- 25.De Bari C., Dell’Accio F., Tylzanowski P., and Luyten F. P.. 2001. Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum. 44: 1928–1942. [DOI] [PubMed] [Google Scholar]
- 26.Zuk P. A., Zhu M., Ashjian P., De Ugarte D. A., Huang J. I., Mizuno H., Alfonso Z. C., Fraser J. K., Benhaim P., and Hedrick M. H.. 2002. Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell. 13: 4279–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakaguchi Y., Sekiya I., Yagishita K., and Muneta T.. 2005. Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis Rheum. 52: 2521–2529. [DOI] [PubMed] [Google Scholar]
- 28.Stenderup K., Justesen J., Clausen C., and Kassem M.. 2003. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone. 33: 919–926. [DOI] [PubMed] [Google Scholar]
- 29.During A., Penel G., and Hardouin P.. 2015. Understanding the local actions of lipids in bone physiology. Prog. Lipid Res. 59: 126–146. [DOI] [PubMed] [Google Scholar]
- 30.Voss A., Reinhart M., Sankarappa S., and Sprecher H.. 1991. The metabolism of 7,10,13,16,19-docosapentaenoic acid to 4,7,10,13,16,19-docosahexaenoic acid in rat liver is independent of a 4-desaturase. J. Biol. Chem. 266: 19995–20000. [PubMed] [Google Scholar]
- 31.Sprecher H. 2000. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim. Biophys. Acta. 1486: 219–231. [DOI] [PubMed] [Google Scholar]
- 32.Grammatikos S. I., Subbaiah P. V., Victor T. A., and Miller W. M.. 1994. Diversity in the ability of cultured cells to elongate and desaturate essential (n-6 and n-3) fatty acids. Ann. N. Y. Acad. Sci. 745: 92–105. [DOI] [PubMed] [Google Scholar]
- 33.Christie W. W. 1993. Preparation of ester derivatives of fatty acids for chromatographic analysis. In Advances in Lipid Methodology–Two. W. W. Christie, editor. Oily Press, Dundee, Scotland. 69‒111. [Google Scholar]
- 34.Ackman R. G. 1992. Application of gas-liquid chromatography to lipid separation and analysis: qualitative and quantitative analysis. In Fatty Acids in Foods and Their Health Implications. C. K. Chow, editor. Marcel Dekker, New York. 47‒63. [Google Scholar]
- 35.Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 36.Zemski Berry K. A., and Murphy R. C.. 2004. Electrospray ionization tandem mass spectrometry of glycerophosphoethanolamine plasmalogen phospholipids. J. Am. Soc. Mass Spectrom. 15: 1499–1508. [DOI] [PubMed] [Google Scholar]
- 37.Duffin K. L., Henion J. D., and Shieh J. J.. 1991. Electrospray and tandem mass spectrometric characterization of acylglycerol mixtures that are dissolved in nonpolar solvents. Anal. Chem. 63: 1781–1788. [DOI] [PubMed] [Google Scholar]
- 38.Haimi P., Uphoff A., Hermansson M., and Somerharju P.. 2006. Software tools for analysis of mass spectrometric lipidome data. Anal. Chem. 78: 8324–8331. [DOI] [PubMed] [Google Scholar]
- 39.Hsu F-F., and Turk J.. 2001. Studies on phosphatidylglycerol with triple quadrupole tandem mass spectrometry with electrospray ionization: fragmentation processes and structural characterization. J. Am. Soc. Mass Spectrom. 12: 1036–1043. [Google Scholar]
- 40.Le Faouder P., Baillif V., Spreadbury I., Motta J. P., Rousset P., Chêne G., Guigné C., Tercé F., Vanner S., Vergnolle N., et al. . 2013. LC-MS/MS method for rapid and concomitant quantification of pro-inflammatory and pro-resolving polyunsaturated fatty acid metabolites. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 932: 123–133. [DOI] [PubMed] [Google Scholar]
- 41.Kilpinen S., Autio R., Ojala K., Iljin K., Bucher E., Sara H., Pisto T., Saarela M., Skotheim R. I., Björkman M., et al. . 2008. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol. 9: R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Méndez-Ferrer S., Michurina T. V., Ferraro F., Mazloom A. R., Macarthur B. D., Lira S. A., Scadden D. T., Ma’ayan A., Enikolopov G. N., and Frenette P. S.. 2010. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 466: 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikushima Y. M., Arai F., Hosokawa K., Toyama H., Takubo K., Furuyashiki T., Narumiya S., and Suda T.. 2013. Prostaglandin E2 regulates murine hematopoietic stem/progenitor cells directly via EP4 receptor and indirectly through mesenchymal progenitor cells. Blood. 121: 1995–2007. [DOI] [PubMed] [Google Scholar]
- 44.Casado-Díaz A., Ferreiro-Vera C., Priego-Capote F., Dorado G., Luque-de-Castro M. D., and Quesada-Gómez J. M.. 2014. Effects of arachidonic acid on the concentration of hydroxyeicosatetraenoic acids in culture media of mesenchymal stromal cells differentiating into adipocytes or osteoblasts. Genes Nutr. 9: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffith J. F., Yeunga D. K. W., Ahujaa A. T., Choye C. W. Y., Meia W. Y., Lamc S. S. L., Lamb T. P., Chend Z-Y., and Leunge P. C.. 2009. A study of bone marrow and subcutaneous fatty acid composition in subjects of varying bone mineral density. Bone. 44: 1092–1096. [DOI] [PubMed] [Google Scholar]
- 46.Fillmore N., Huqi A., Jaswal J. S., Mori J., Paulin R., Haromy A., Onay-Besikci A., Ionescu L., Thébaud B., Michelakis E., et al. . 2015. Effect of fatty acids on human bone marrow mesenchymal stem cell energy metabolism and survival. PLoS One. 10: e0120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi H., Kokoeva M. V., Inouye K., Tzameli I., Yin H., and Flier J. S.. 2006. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Invest. 116: 3015–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouzer C. A., and Marnett L. J.. 2011. Endocannabinoid oxygenation by cyclooxygenases, lipoxygenases, and cytochromes P450: cross-talk between the eicosanoid and endocannabinoid signaling pathways. Chem. Rev. 111: 5899–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stables M. J., and Gilroy D. W.. 2011. Old and new generation lipid mediators in acute inflammation and resolution. Prog. Lipid Res. 50: 35–51. [DOI] [PubMed] [Google Scholar]
- 50.Zou H., Yuan C., Dong L., Sidhu R. S., Hong Y. H., Kuklev D. V., and Smith W. L.. 2012. Human cyclooxygenase-1 activity and its responses to COX inhibitors are allosterically regulated by nonsubstrate fatty acids. J. Lipid Res. 53: 1336–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong L., Zou H., Yuan C., Hong Y. H., Kuklev D. V., and Smith W. L.. 2016. Different fatty acids compete with arachidonic acid for binding to the allosteric or catalytic subunits of cyclooxygenases to regulate prostanoid synthesis. J. Biol. Chem. 291: 4069–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akiba S., Murata T., Kitatani K., and Sato T.. 2000. Involvement of lipoxygenase pathway in docosapentaenoic acid-induced inhibition of platelet aggregation. Biol. Pharm. Bull. 23: 1293–1297. [DOI] [PubMed] [Google Scholar]
- 53.Ntambi J. M. 1999. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J. Lipid Res. 40: 1549–1558. [PubMed] [Google Scholar]
- 54.Holman R. T., and Mohrhauer H.. 1963. A hypothesis involving competitive inhibitions in the metabolism of polyunsaturated fatty acids. Acta Chem. Scand. 17: S84–S90. [Google Scholar]
- 55.Jakobsson A., Westerberg R., and Jacobsson A.. 2006. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 45: 237–249. [DOI] [PubMed] [Google Scholar]
- 56.Schmitz G., and Ecker J.. 2008. The opposing effects of n-3 and n-6 fatty acids. Prog. Lipid Res. 47: 147–155. [DOI] [PubMed] [Google Scholar]
- 57.Brenna J. T., Kothapalli K. S., and Park W. J.. 2010. Alternative transcripts of fatty acid desaturase (FADS) genes. Prostaglandins Leukot. Essent. Fatty Acids. 82: 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mohrhauer H., and Holman R. T.. 1963. The effect of dose level of essential fatty acids upon fatty acid composition of the rat liver. J. Lipid Res. 4: 151–159. [PubMed] [Google Scholar]
- 59.Fujishima H., Sanchez Mejia R. O., Bingham C. O. 3rd, Lam B. K., Sapirstein A., Bonventre J. V., Austen K. F., and Arm J. P.. 1999. Cytosolic phospholipase A2 is essential for both the immediate and the delayed phases of eicosanoid generation in mouse bone marrow-derived mast cells. Proc. Natl. Acad. Sci. USA. 96: 4803–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Astudillo A. M., Balgoma D., Balboa M. A., and Balsinde J.. 2012. Dynamics of arachidonic acid mobilization by inflammatory cells. Biochim. Biophys. Acta. 1821: 249–256. [DOI] [PubMed] [Google Scholar]
- 61.Chilton F. H., Fonteh A. N., Surette M. E., Triggiani M., and Winkler J. D.. 1996. Control of arachidonate levels within inflammatory cells. Biochim. Biophys. Acta. 1299: 1–15. [DOI] [PubMed] [Google Scholar]
- 62.Pérez-Chacón G., Astudillo A. M., Balgoma D., Balboa M. A., and Balsinde J.. 2009. Control of free arachidonic acid levels by phospholipases A2 and lysophospholipid acyltransferases. Biochim. Biophys. Acta. 1791: 1103–1113. [DOI] [PubMed] [Google Scholar]
- 63.Hanel A. M., Schüttel S., and Gelb M. H.. 1993. Processive interfacial catalysis by mammalian 85-kilodalton phospholipase A2 enzymes on product-containing vesicles: application to the determination of substrate preferences. Biochemistry. 32: 5949–5958. [DOI] [PubMed] [Google Scholar]
- 64.Shikano M., Masuzawa Y., Yazawa K., Takayama K., Kudo I., and Inoue K.. 1994. Complete discrimination of docosahexaenoate from arachidonate by 85 kDa cytosolic phospholipase A2 during the hydrolysis of diacyl- and alkenylacylglycerophosphoethanolamine. Biochim. Biophys. Acta. 1212: 211–216. [DOI] [PubMed] [Google Scholar]
- 65.Diez E., Chilton F. H., Stroup G., Mayer R. J., Winkler J. D., and Fonteh A. N.. 1994. Fatty acid and phospholipid selectivity of different phospholipase A2 enzymes studied by using a mammalian membrane as substrate. Biochem. J. 301: 721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haimi P., Hermansson M., Batchu K. C., Virtanen J. A., and Somerharju P.. 2010. Substrate efflux propensity plays a key role in the specificity of secretory A-type phospholipases. J. Biol. Chem. 285: 751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Batchu K. C., Hokynar K., Jeltsch M., Mattonet K., and Somerharju P.. 2015. Substrate efflux propensity is the key determinant of Ca2+-independent phospholipase A-β (iPLAβ)-mediated glycerophospholipid hydrolysis. J. Biol. Chem. 290: 10093–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Batchu K. C. 2016. Factors Regulating the Substrate Specificity of A-type Phospholipases - A Mass-Spectrometric Study. PhD Thesis. University of Helsinki, Helsinki, Finland.
- 69.Campos A. M., Maciel E., Moreira A. S., Sousa B., Melo T., Domingues P., Curado L., Antunes B., Domingues M. R., and Santos F.. 2016. Lipidomics of mesenchymal stromal cells: understanding the adaptation of phospholipid profile in response to pro-inflammatory cytokines. J. Cell. Physiol. 231: 1024–1032. [DOI] [PubMed] [Google Scholar]
- 70.Bae S. H., Jung E. S., Park Y. M., Kim B. S., Kim B. K., Kim D. G., and Ryu W. S.. 2001. Expression of cyclooxygenase-2 (COX-2) in hepatocellular carcinoma and growth inhibition of hepatoma cell lines by a COX-2 inhibitor, NS-398. Clin. Cancer Res. 7: 1410–1418. [PubMed] [Google Scholar]
- 71.Park J. B., Lee C. S., Jang J. H., Ghim J., Kim Y. J., You S., Hwang D., Suh P. G., and Ryu S. H.. 2012. Phospholipase signalling networks in cancer. Nat. Rev. Cancer. 12: 782–792. [DOI] [PubMed] [Google Scholar]
- 72.Itokazu Y., Tajima N., Kerosuo L., Somerharju P., Sariola H., Yu R. K., and Käkelä R.. 2016. A2B5+/GFAP+ cells of rat spinal cord share a similar lipid profile with progenitor cells: A comparative lipidomic study. Neurochem. Res. 41: 1527–1544. [DOI] [PubMed] [Google Scholar]
- 73.Hishikawa D., Hashidate T., Shimizu T., and Shindou H.. 2014. Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J. Lipid Res. 55: 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miles E. A., and Calder P. C.. 1998. Modulation of immune function by dietary fatty acids. Proc. Nutr. Soc. 57: 277–292. [DOI] [PubMed] [Google Scholar]
- 75.Serhan C. N., and Petasis N. A.. 2011. Resolvins and protectins in inflammation resolution. Chem. Rev. 111: 5922–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stillwell W., Shaikh S. R., Zerouga M., Siddiqui R., and Wassall S. R.. 2005. Docosahexaenoic acid affects cell signaling by altering lipid rafts. Reprod. Nutr. Dev. 45: 559–579. [DOI] [PubMed] [Google Scholar]
- 77.Oh D. Y., Talukdar S., Bae E. J., Imamura T., Morinaga H., Fan W., Li P., Lu W. J., Watkins S. M., and Olefsky J. M.. 2010. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 142: 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aires V., Hichami A., Filomenko R., Plé A., Rébé C., Bettaieb A., and Khan N. A.. 2007. Docosahexaenoic acid induces increases in [Ca2+]i via inositol 1,4,5-triphosphate production and activates protein kinase Cγ and -δ via phosphatidylserine binding site: Implication in apoptosis in U937 Cells. Mol. Pharmacol. 72: 1545–1556. [DOI] [PubMed] [Google Scholar]
- 79.Giorgione J., Epand R. M., Buda C., and Farkas T.. 1995. Role of phospholipids containing docosahexaenoyl chains in modulating the activity of protein kinase C. Proc. Natl. Acad. Sci. USA. 92: 9767–9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.