Abstract
Mutations in the lamin A/C gene encoding nuclear lamins A and C (lamin A/C) cause familial partial lipodystrophy type 2 (FPLD2) and related lipodystrophy syndromes. These are mainly characterized by redistribution of adipose tissue associated with insulin resistance. Several reports suggest that alterations in the extracellular matrix of adipose tissue leading to fibrosis play a role in the pathophysiology of lipodystrophy syndromes. However, the extent of extracellular matrix alterations in FPLD2 remains unknown. We show significantly increased fibrosis and altered expression of genes encoding extracellular matrix proteins in cervical subcutaneous adipose tissue from a human subject with FLPD2. Similar extracellular matrix alterations occur in adipose tissue of transgenic mice expressing an FPLD2-causing human lamin A variant and in cultured fibroblasts from human subjects with FPLD2 and related lipodystrophies. These abnormalities are associated with increased transforming growth factor-β signaling and defects in matrix metalloproteinase 9 activity. Our data demonstrate that lamin A/C gene mutations responsible for FPLD2 and related lipodystrophies are associated with transforming growth factor-β activation and an extracellular matrix imbalance in adipose tissue, suggesting that targeting these alterations could be the basis of novel therapies
Keywords: adipocytes, adipose tissue, fibrosis, nuclear envelope
Mutations in the lamin A/C gene (LMNA) encoding the nuclear intermediate filament lamins A and C (lamin A/C) cause multiple diseases often referred to as laminopathies (1). These include cardiomyopathy and muscular dystrophy (2–5), Hutchinson-Gilford progeria syndrome (6, 7), mandibuloacral dysplasia type A (8), peripheral neuropathy (9), and familial partial lipodystrophy type 2 (FPLD2), and related lipodystrophies (10–13). FPLD2, also known as Dunnigan-type familial partial lipodystrophy, is an autosomal dominant inherited disease characterized by abnormal body fat distribution occurring around the onset of puberty. Major features are loss of subcutaneous adipose tissue from trunk, limbs, and gluteal region contrasting with its accumulation in face, neck, axilla, and visceral region (14, 15). Metabolic defects resulting from these adipose tissue alterations include dyslipidemia, especially hypertriglyceridemia, hepatic steatosis, premature atherosclerosis, and insulin resistance leading to type 2 diabetes (15–18).
Most LMNA mutations causing FPLD2 generate amino acid substitutions that change the surface charge of an immunoglobulin-like fold in lamin A and lamin C (19, 20). In 90% of patients, this occurs as a result of the substitution of a basic arginine with a neutral tryptophan, leucine or glutamine (R482W/L/Q) (10–12). LMNA mutations causing amino acid substitutions at other codons may cause related atypical lipodystrophy syndromes (13). The disease-causing mutations likely generate “gain of function” or “dominant negative” variants of lamin A and lamin C, because the disease occurs in heterozygous patients and Lmna null mice do not develop lipodystrophy (21). Only one published study has examined transgenic mice overexpressing an FPLD2-causing lamin A variant (R482Q) in adipose tissue (22). When fed a high-fat diet, these mice develop subcutaneous lipoatrophy, insulin resistance, and hepatic steatosis and demonstrate an inability of adipose tissue self-renewal with preadipocytes unable to differentiate into adipocytes (22). This is consistent with a report showing that overexpression of lamin A and FPLD2-causing lamin A variants blocks in vitro differentiation of 3T3-L1 preadipocytes into adipocytes (23).
Some studies have suggested that alterations in adipose tissue extracellular matrix (ECM) play a role in the pathophysiology of lipodystrophy syndromes. Adipose tissue fibrosis has been reported in subcutaneous lipoatrophic areas of patients with partial lipodystrophy caused by perilipin deficiency (24) and in patients with lipodystrophy receiving antiretroviral therapy for HIV-1 infection (25). Subcutaneous lipoatrophic abdominal adipose tissue from a patient with lipodystrophy caused by mutation in the gene encoding DNA polymerase δ also has been reported to have increased fibrosis and increased expression of extracellular matrix genes, transforming growth factor-β (TGF-β), matrix metalloproteinase (MMP) 14, and fibronectin (26). Considering the scarcity of subcutaneous adipose tissue from patients diagnosed with FPLD2, only a few studies have focused on the histological alterations in affected human tissue. Béréziat et al. described a significant increase in fibrosis with accumulation of collagen fibrils in hypertrophic cervical adipose tissue from patients with LMNA-related lipodystrophies (27). In contrast, one study of perilipomatous adipose tissue from lipoatrophic fat of patients with FPLD2 reported a near-normal histological phenotype (28). We therefore hypothesized that ECM remodeling, fibrosis, and related cell signaling alterations occur in adipose tissue and cells from human subjects and transgenic mice expressing lamin A/C variants that cause FPLD2 and related lipodystrophies.
MATERIALS AND METHODS
Human subjects
Hypertrophic cervical subcutaneous adipose tissue was collected during plastic surgery from a female patient with FPLD2 (LMNA p.R482W mutation) and from five nonobese, nondiabetic control subjects during surgery to treat benign thyroid nodules or parotid tumors. Primary dermal fibroblast cultures were established after punch biopsy from three female subjects with LMNA mutations (p.R482W, p.R399H, and p.L387V) and from two nonobese, nondiabetic women. All subjects provided written informed consent to participate in the research protocol, which was approved by the Comité de Protection des Personnes, Hôpital Saint-Louis (Paris, France).
Mice
Transgenic mice were generated at the Herbert Irving Comprehensive Cancer Center Transgenic Mouse Facility at Columbia University Medical Center. We generated plasmids containing from 5′ to 3′: a 5.4-kb fatty acid binding protein 4 (Fabp4) promoter, cDNA encoding a FLAG epitope tag fused in frame to either full-length wild-type human prelamin A or R482Q prelamin A, an SV40 splice site, and a polyadenylation site. Minigenes were excised from plasmids by restriction endonuclease digestion and microinjected separately into superovulated B6/CBA F1 fertilized oocytes in vitro. Oocytes were then transferred to pseudopregnant foster mothers to produce transgenic founders. Founder transgenic mice were identified by PCR analysis of DNA from tail biopsies using two primer pairs corresponding to sequences in FLAG and human lamin A. The first pair (forward: 5′-ATGGACTACAAGGACGACGATGACA-3′ reverse: 5′-AGTTCAGCAGAGCCTCCAGGTCCTT-3′) produces a PCR product, which includes FLAG and a sequence within human lamin A/C. The second primer pair (forward: 5′-AGGACCTGCAGGAGCTCAATGATCG-3′ reverse: 5′-AGTTCAGCAGAGCCTCCAGGTCCTT-3′) corresponds to a sequence in human lamin A. Transgenic mice were backcrossed to wild-type Friend Virus B (FVB) mice (Jackson Laboratory) at least eight generations to obtain stable transgenic offspring and adequate numbers of individuals for further experiments. Mice were maintained on a 12-h light/dark cycle. Starting at 12 weeks of age, they were fed a high-fat diet containing 45% of calories from fat (D12451, Research Diets). At 40 weeks of age after overnight fasting, blood was obtained and, after sacrifice, adipose tissue collected from three different fat depots. Nontransgenic control mice were used in all experiments. For some experiments, we also used a line of transgenic mice (R482Q “Wojtanik”) overexpressing R482Q prelamin A under control of a Fabp4 promoter that has been described previously (22); the late Dr. Constantine Londos (National Institute of Diabetes and Digestive and Kidney Diseases Laboratory of Cellular and Developmental Biology) provided these mice. The Columbia University Medical Center Institutional Animal Care and Use Committee approved all protocols involving mice.
Histology
Human subcutaneous adipose tissue samples were collected and prepared as described previously (27). Mouse subcutaneous adipose tissue samples were obtained after sacrifice. Samples were placed in 10% neutral buffered formalin for 48 h, embedded in paraffin, and sectioned at 5 μm. Adipose tissue sections were deparaffinized and stained with Sirius red for 1 h to detect collagen fibrils. The surface area stained with Sirius red was quantified with ImageJ software on three randomly chosen regions for each section, and all values were normalized to an arbitrary control section.
Glucose tolerance tests
After 4–5 h of fasting, blood glucose levels were measured by using OneTouch Ultra Blood Glucose Monitoring System (Johnson & Johnson), and mice then received an intraperitoneal injection of 1 mg/kg glucose. Blood glucose levels were measured immediately after glucose administration and at 5, 15, 30, 60, and 120 min afterward.
Plasma lipid concentrations
Blood from overnight-fasted mice was collected from the retro-orbital plexus. Plasma total cholesterol and triglycerides were measured using Infinity Kit (Thermo Fisher Scientific). Free fatty acids were measured using NEFA C kit (Wako).
Cell culture
Fibroblasts were grown in DMEM (Thermo Fisher Scientific) containing 1 g/l glucose, 20 mM L-glutamine, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 110 mg/ml sodium pyruvate, 10% FBS, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (all from Thermo Fischer Scientific) at 37°C in 5% CO2 per 95% air. Fibroblasts were used between culture passages 4 and 7.
RT-quantitative PCR
Total RNA was extracted from human and mouse tissue using the RNeasy Lipid Tissue Isolation Kit (Qiagen) following the manufacturer’s instructions. Mouse adipose tissue was collected at 40 weeks of age. For primary fibroblast cultures, RNA was extracted using RNeasy Isolation Kit (Qiagen). Quality and concentrations of RNA were measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific). The cDNA was synthesized using Superscript First Strand Synthesis System (Thermo Fisher Scientific), according to the manufacturer’s instructions. Quantitative real-time PCR was performed on an ABI 7300 Real-Time PCR System (Applied Biosystems) using HotStart-IT SYBR green qPCR Master Mix (Affymetrix). Relative levels of mRNA expression were calculated using the ΔΔCT method (29). Individual expression values were normalized and compared with mRNA encoding hypoxanthine phosphoribosyltransferase 1. Values are shown graphically as mRNA relative expression with one arbitrary control subject in the control group of each experiment. Primers used for RT-qPCR of human mRNAs are listed in supplemental Table S1. Primers used for RT-qPCR of murine mRNAs are listed in supplemental Table S2.
Protein extraction and immunoblotting
Frozen adipose tissues were homogenized in 300 µl of 3× Laemmli buffer (30), heated 6 min at 95°C, and centrifuged at 12,000 g for 10 min. The upper phase containing all the lipids was removed, and samples were subjected to SDS-PAGE and blotted onto nitrocellulose membranes. For primary fibroblast cultures, cells were isolated and homogenized in Cell Extraction Buffer (Thermo Fisher Scientific) with Protease Inhibitor Cocktail (Roche), plus 1.0 mM of phenylmethylsulfonyl fluoride (Sigma-Aldrich). Proteins in samples were denatured in Laemmli sample buffer containing β-mercaptoethanol by boiling for 5 min, separated by SDS-PAGE, and transferred to nitrocellulose membranes. For immunoblotting, nitrocellulose membranes were washed with blocking buffer (4% BSA in Tris-buffered saline with 0.1% polysorbate 20) and probed with primary antibodies diluted in blocking buffer overnight at 4°C. The primary antibodies used for immunoblotting were M5 anti-FLAG (Sigma-Aldrich), rabbit anti-lamin B1 (31), anti-connective tissue growth factor (CTGF) (Abcam, #AB-6992), anti-TGF-β1/2/3, anti-phosphorylated Smad2/3, anti-total Smad2/3, anti-Smad4, anti-MMP 9, anti-TIMP1, and anti-β-actin (Santa Cruz Biotechnologies; #SC-7892, #SC-11769, #SC-6032, #SC-7966, #SC-10737, #SC-5538, #SC-47778, respectively). Blots were washed with Tris-buffered saline containing 0.1% polysorbate 20 and then incubated in blocking buffer with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare) at room temperature. Recognized proteins were visualized by enhanced chemiluminescence (Thermo Fisher Scientific) and detected by exposure on X-ray films (Labscientific). To quantify signals, we scanned immunoblots and densities of the bands quantified by using ImageJ software. The TGF-β, Smad4, and CTGF band densities were normalized to β-actin signal density of each sample. For Smad2/3, band intensity of phosphorylated proteins was normalized to the band intensity signal for total proteins.
Immunofluorescence microscopy
Frozen sections of adipose tissue embedded in optimal cutting temperature compound (Tissue-Tek) were fixed in cold methanol for 10 min at −20°C. Sections were permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature, washed three times with 0.1% polysorbate 20 in PBS (Solution A), and incubated with the primary antibodies diluted in PBS containing 0.1% polysorbate 20 and 2% BSA (Solution B) for 2 h at room temperature. Primary antibodies used were mouse anti-FLAG M5 monoclonal antibody and rabbit anti-lamin B1 polyclonal antibody. After washing four times with Solution A, sections were incubated with secondary antibodies diluted in Solution B for 1 h at room temperature. Secondary antibodies used were fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G and rhodamine-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Labs). Fibroblasts were grown on glass coverslips at subconfluency and were fixed in cold methanol for 10 min at −20°C. Rabbit antibody directed against type I collagen (Abcam, #ab-292) was revealed by anti-rabbit immunoglobulin G coupled to Texas Red (Thermo Fisher Scientific). Cell nuclei were visualized by diamidino-2-phenylindole hydrochloride staining. Intensity of collagen I staining per unit area was quantified using ImageJ software, and all values normalized to an arbitrary measurement from a control human fibroblast.
Gelatin zymography
Zymography was performed on supernatants from cultured fibroblasts to assess MMP activity. Fibroblasts were cultured in serum-free medium for 24 h. Gelatin-degrading activity was examined by SDS-PAGE on a 7.5%-gel containing gelatin (1.0 mg/ml) without prior heating or reduction. Volumes of 10–15 µl of supernatants were run in parallel. Volumes were determined after cellular density quantification. After electrophoresis, gels were washed in 2.5% Triton-X 100 and then incubated in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM CaCl2, and 0.02% NaN3. Gels were stained with Coomassie Brilliant Blue R250 (Thermo Fisher Scientific) and destained with a solution of 50% methanol, 40% acetic acid, 10% water. Gelatinolytic activity of each gelatinase is evident as a clear band against the blue background of stained gelatin.
Statistical analysis
All results were from triplicate experiments and are expressed as means ± SEs. Statistical analyses were performed using Prism (GraphPad Software). For analysis of mouse adipose tissue and human fibroblasts, one-way ANOVAs with post hoc Tukey analysis were performed on data at a minimum P < 0.05 threshold.
RESULTS
ECM is altered in cervical subcutaneous adipose tissue from a patient with FPLD2
We examined anterior cervical subcutaneous adipose tissue from one patient with FPLD2 caused by the LMNA p.R482W mutation and from unaffected controls. Features of the control subjects and patient have been described previously (13, 27, 32). These features, and the subjects from whom adipose tissue sections or adipose tissue RNA were available, are summarized in Table 1.
TABLE 1.
Features of unaffected controls and patient with FPLD2 caused by the LMNA p.R482W mutation from whom cervical adipose tissue sections or RNA from adipose tissue was available
| Control A | Control B | Control C | Control D | Control E | R482W | |
| Sex | Female | Female | Female | Male | Male | Female |
| Age (years) | 48 | 54 | 87 | 62 | 82 | 46 |
| Body mass index (kg/m2) | 24.5 | 22.1 | 23 | 23 | 24.8 | 22.1 |
| Peripheral lipoatrophy | No | No | No | No | No | Yes |
| Cervical fat accumulation | No | No | No | No | No | Yes |
| Glucose tolerance | Normal | Normal | Normal | Normal | Normal | Diabetes |
| Triglycerides (mM) | Normal | Normal | Normal | Normal | Normal | 2.5 |
| Tissue section available | Yes | No | No | No | No | Yes |
| RNA sample available | Yes | Yes | Yes | Yes | Yes | Yes |
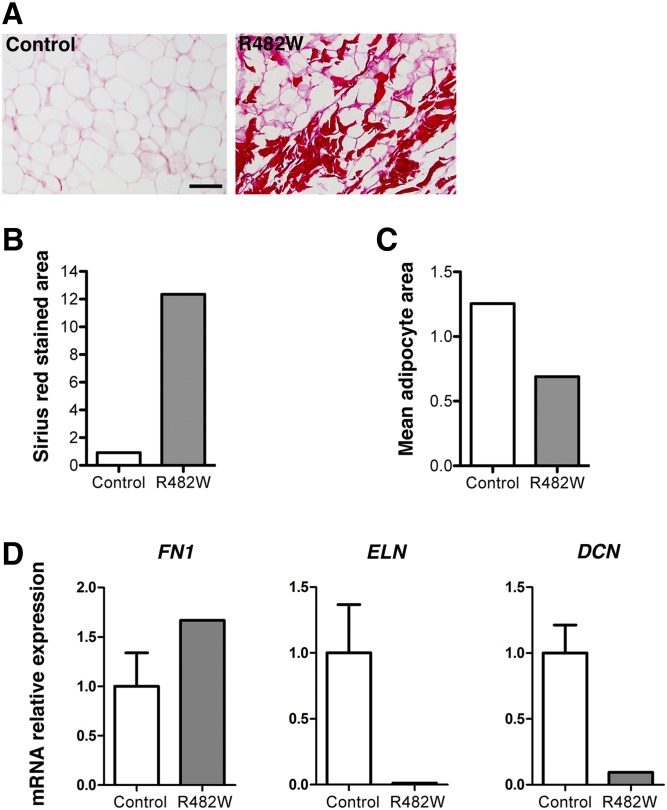
We first confirmed the presence of fibrosis in adipose tissue of the patient with the LMNA p.R482W mutation with tissue available from one unaffected control as assessed by Sirius red staining of sections (Fig. 1A). The surface area of cervical subcutaneous fat stained with Sirius red was 12-fold greater in the affected patient than in the control (Fig. 1B). Adipocyte surface area was also less in tissue sections from the affected patient than in the control (Fig. 1C). These results confirmed similar findings reported by Béréziat et al. (27). We then examined in cervical adipose tissue of the patient and five unaffected controls the expression of mRNAs for three different ECM proteins: fibronectin, which binds type I collagen and is involved in the maintenance of adipocyte shape (33); elastin, a major component of elastic fibers providing strength and flexibility to connective tissue (34); and decorin, which also binds to type I collagen and functions in matrix assembly (35). Expression of FN1, encoding fibronectin, was increased, whereas expression of the ELN and DCN genes, encoding elastin, and decorin respectively, were decreased in the patient compared with the control subjects (Fig. 1D). These data showed increased fibrosis and altered expression of genes encoding ECM proteins in cervical subcutaneous adipose tissue from a human subject with FLPD2.
Fig. 1.
Analysis of fibrosis in subcutaneous adipose tissue from a patient with FLPD2 caused by LMNA p.R482W mutation. A: Sirius red staining of subcutaneous adipose tissue sections from one unaffected human subject (Control) and one patient with LMNA p.R482W mutation (R482W); scale bar = 100 µm. B: Bar graph showing surface area of subcutaneous adipose tissue sections stained with Sirius red from one unaffected human subject (Control) and one patient with LMNA p.R482W mutation (R482W). C: Bar graph showing mean cellular area of adipocytes from one unaffected human subject (Control) and one patient with LMNA p.R482W mutation (R482W) adipose tissue sections. In A–C, the control is Control A from Table 1. D: Relative expression of mRNAs encoded by FN1, ELN, and DCN in subcutaneous adipose tissue from unaffected human subjects (Control) and a patient with LMNA p.R482W mutation (R482W). Values are means ± SEs for n = 5 controls and n = 1 patient.
ECM alterations in adipose tissue of transgenic mice expressing R482Q lamin A
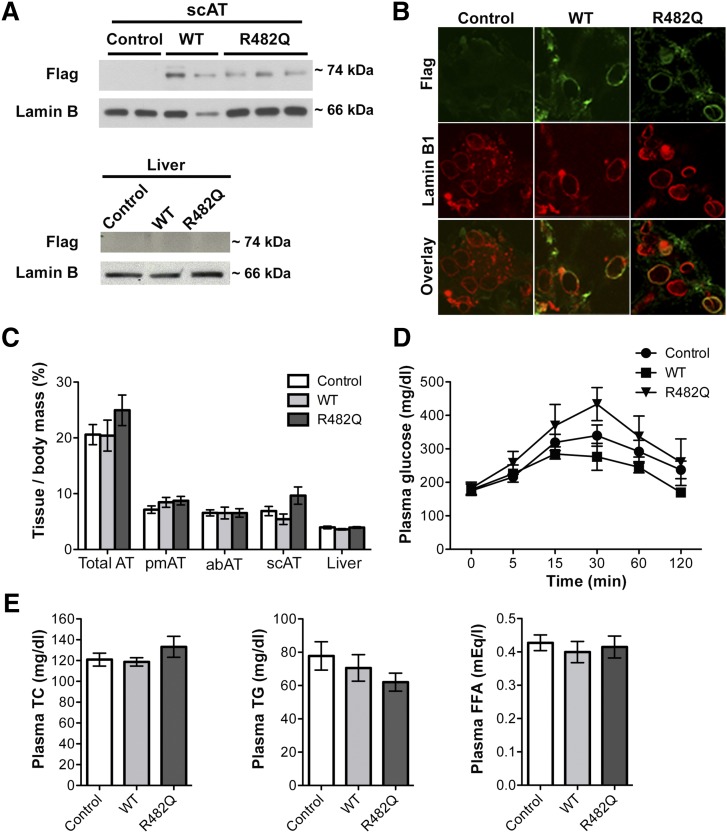
To further investigate the effects of FPLD2-causing LMNA mutations, we generated transgenic mice overexpressing human R482Q lamin A and wild-type lamin A selectively in adipose tissue. As in the patient with the LMNA p.R482W mutation, this amino acid substitution similarly changes the surface charge of the immunoglobulin-like fold in lamin A/C and is commonly found in patients with FPLD2 (10–12, 19, 20, 32). In the mice, cDNAs encoding wild-type or R482Q human prelamin A with FLAG epitope tags at their amino termini were expressed under the control of the Fabp4 promoter. To confirm selective expression of the transgene in adipose tissue, we compared the expression of the FLAG-tagged proteins in subcutaneous adipose tissue and liver from the transgenic mice and nontransgenic controls. The FLAG-tagged proteins were selectively expressed in extracts of adipose tissue but not liver from the transgenic mice (Fig. 2A). Immunofluorescence microscopic analysis of adipose tissue sections confirmed that FLAG-tagged wild-type and R482Q lamin A were correctly localized to the nuclear envelope, where they colocalized with lamin B1 in cells expressing the Fab4 promoter-containing transgene (Fig. 2B). However, R482Q lamin A transgenic mice did not develop gross anatomic or metabolic signs of partial lipodystrophy. In female mice fed a high-fat diet for 28 weeks, the ratios of adipose tissue mass to total body mass did not show significant differences compared with either transgenic mice expressing wild-type human lamin A or nontransgenic controls (Fig. 2C). Female R482Q lamin A transgenic mice demonstrated a trend toward impaired glucose tolerance; however, the results did not reach statistical significance compared with transgenic mice expressing wild-type human lamin A or nontransgenic controls (Fig. 2D). There were also no significant differences in plasma cholesterol, triglycerides, or free fatty acid concentrations among the three groups (Fig. 2E). Male mice demonstrated the same phenotypes (supplemental Fig. S1).
Fig. 2.
Characterization of transgenic mice overexpressing FLAG-tagged human wild-type or R482Q lamin A. A: Immunoblots of protein extracts of subcutaneous adipose tissue (scAT; upper panel) and liver (lower panel) from nontransgenic control mice (Control), transgenic mice overexpressing FLAG-tagged human wild-type lamin A (WT), or FLAG-tagged human R482Q lamin A (R482Q). Blots were probed with antibody against FLAG (Flag) or antibody against lamin B1 (Lamin B) as a loading control; each lane shows an extract from a different mouse. B: Photomicrographs showing immunofluorescence labeling of FLAG (green) and lamin B1 (red) in adipose tissue sections from Control, WT, and R482Q mice. C: Bar graphs showing ratios of total adipose tissue (AT), parametrial depots (pmAT), abdominal depots (abAT), subcutaneous depots (scAT), and liver masses to total body mass of Control, WT, and R482Q female mice at 40 weeks of age. Values are means ± SEs (Control n = 14; WT n = 7; R482Q n = 6). D: Plasma glucose concentrations immediately prior to (0 min) and after administration of 1 mg/kg of glucose in Control (circle), WT (square), and R482Q (triangle) female mice at 39 weeks of age. Values are means ± standard deviations (control n = 14; WT n = 5; R482Q n = 6). E: Bar graphs showing concentrations of plasma total cholesterol (TC), triglyceride (TG), and free fatty acids (FFA) in Control, WT, and R482Q female mice at 40 weeks of age. Values are means ± SEs (Control n = 14; WT n = 7; R482Q n = 6).
Whereas our R482Q lamin A transgenic mice did not develop lipoatrophy or metabolic abnormalities, Wojtanik et al. generated a similar line of transgenic mice (R482Q “Wojtanik”) that were reportedly unable to accumulate fat and had decreased insulin sensitivity and hepatic steatosis (22). We therefore examined the relative expression of human and mouse lamin A/C mRNA by RT-qPCR using primers that recognized both human and mouse transcripts. There was approximately a 5-fold increase in lamin A/C mRNA in the transgenic mice that we generated that expressed either wild-type or R482Q lamin A compared with nontransgenic mice, whereas the transgenic mice generated by Wojtanik et al. had an approximately 8.5-fold increase in expression of R482Q lamin A (Table 2). R482Q lamin A variant expression was significantly increased in transgenic mice generated by Wojtanik et al. compared with the transgenic mice that we generated expressing this variant, suggesting that the severity of the lipodystrophy phenotype depends on the expression level of the pathogenic lamin A variant when two normal Lmna alleles are present.
TABLE 2.
Expression levels of mouse plus human lamin A/C mRNA in adipose tissue from transgenic mice overexpressing human wild-type lamin A (WT) or human R482Q lamin A (R482Q) generated in this study and in adipose tissue from R482Q lamin A transgenic mice reported by Wojtanik et al. (22)
| Mouse | N | Lamin A/C relative mRNA expression | P compared with nontransgenic mice | P compared with mice expressing WT lamin A | P compared with mice expressing R482Q lamin A |
| Control | 6 | 1.00 ± 0.58 | — | <0.001 | — |
| WT | 6 | 4.91 ± 0.44 | <0.001 | — | — |
| R482Q | 6 | 5.16 ± 0.36 | <0.001 | ns | — |
| R482Q “Wojtanik” | 6 | 8.49 ± 0.87 | <0.001 | <0.001 | <0.001 |
We used primers recognizing sequences present in both human and mouse lamin A/C cDNA (forward: 5′-GGACCTGCAGGAGCTCAATG-3′ reverse: 5′-TCCTCACGCACTTTGCTCAG-3′). Nontransgenic mice (Control) are shown as a reference, in which lamin A/C mRNA is normalized to 1.00. Values are means ± SEs (control n = 6; WT n = 6; R482Q n = 6; R482Q “Wojtanik” n = 6) and P values given for various comparisons; ns, not significant.
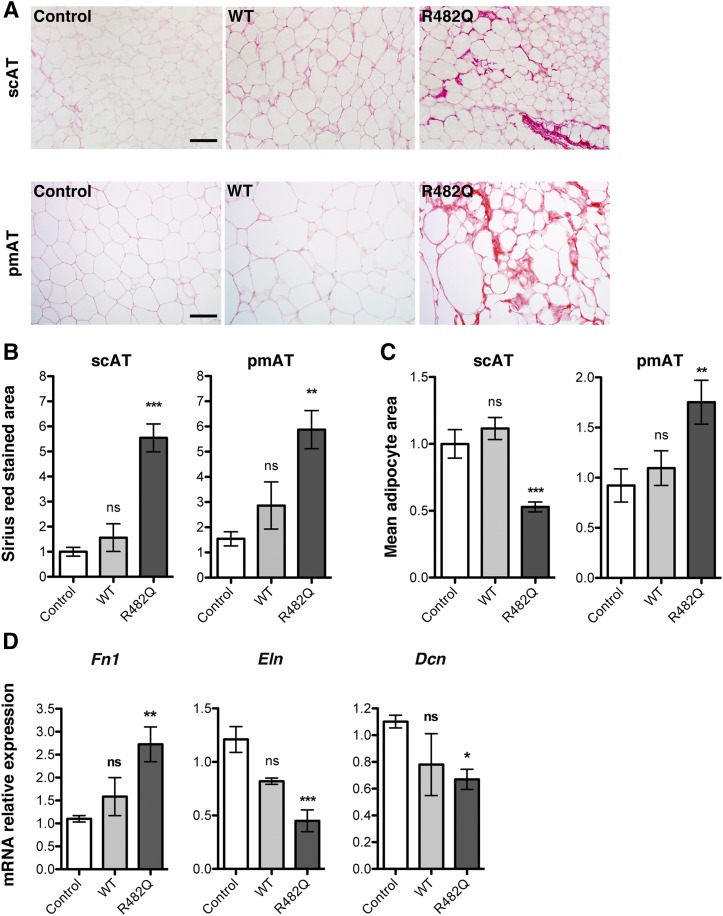
Although the R482Q lamin A transgenic mice we generated did not develop regional lipoatrophy or metabolic abnormalities, histological examination of adipose tissue revealed changes similar to those seen in the human subject with the LMNA p.R482W mutation. Compared with nontransgenic mice and transgenic mice expressing wild-type lamin A, subcutaneous and parametrial adipose tissue from the R482Q lamin A transgenic mice displayed an increase in Sirius red staining of collagen fibrils (Fig. 3A). The surface area of subcutaneous and parametrial fat stained with Sirius red was a statistically significant 5-fold to 6-fold greater amount in the R482Q lamin A transgenic mice than in the nontransgenic controls (Fig. 3B). Hence, although the mass of these fat depots in relation to body mass was not decreased in these transgenic mice, there was more collagen in relation to fat. Adipocyte surface area was also significantly smaller in subcutaneous adipose tissue from the transgenic mice than that from the nontransgenic controls, but surface area was larger in parametrial fat of the transgenic mice than in controls (Fig. 3C). Expression of mRNA from Fn1 encoding fibronectin was significantly increased, and expressions of Dcn-encoding decorin and Eln-encoding elastin were significantly reduced in subcutaneous adipose tissue from R482Q lamin A transgenic mice compared with nontransgenic controls (Fig. 3D). Overall, there was a very similar phenotype of altered ECM and fibrosis in subcutaneous and visceral adipose tissue from R482Q lamin A transgenic mice compared with cervical subcutaneous adipose tissue from a human subject with the LMNA mutation p.R482W. These ECM alterations occurred in mice that did not have overt lipoatrophy or its metabolic consequences, suggesting that they may be early defects in the pathogenesis of FPLD2.
Fig. 3.
Analysis of adipose tissue from transgenic mice overexpressing FLAG-tagged human wild-type or R482Q lamin A. A: Photomicrographs showing Sirius red staining of subcutaneous adipose tissue (scAT) and parametrial adipose tissue (pmAT) sections from nontransgenic mice (Control), transgenic mice expressing FLAG-tagged wild-type human lamin A (WT), or transgenic mice expressing FLAG-tagged human lamin A R482Q (R482Q); scale bar = 100 µm. B: Bar graphs showing surface area of subcutaneous adipose tissue (scAT) and parametrial adipose tissue (pmAT) sections stained with Sirius red from nontransgenic Control, WT, and R482Q mice. For each section, Sirius red-stained area was calculated on three randomly chosen regions using ImageJ software. Values are means ± SEs (Control n = 3 mice; WT n = 3 mice; R482Q n = 4 mice). **P < 0.01; ***P < 0.001; ns, not significant compared with nontransgenic mice (Control). C: Bar graphs showing mean cellular area of adipocytes from subcutaneous adipose tissue (scAT) and parametrial adipose tissue (pmAT) sections from nontransgenic Control, WT, and R482Q mice. For each section, mean adipocyte areas were calculated with ImageJ software on three randomly chosen regions. Values are means ± SEs (Control n = 3 mice; WT n = 3 mice; R482Q n = 4 mice). **P < 0.01; ***P < 0.001; ns, not significant compared with nontransgenic mice (Control). D: Means ± SEs of relative expression of mRNAs encoded by Fn1, Eln, and Dcn in subcutaneous adipose tissue from Control, WT, and R482Q mice. Values are means ± SEs (Control n = 3 mice; WT n = 3 mice; R482Q n = 4 mice). *P < 0.05; **P < 0.01; ns, not significant compared with nontransgenic mice (Control).
ECM is altered in dermal fibroblasts from patients with LMNA mutations
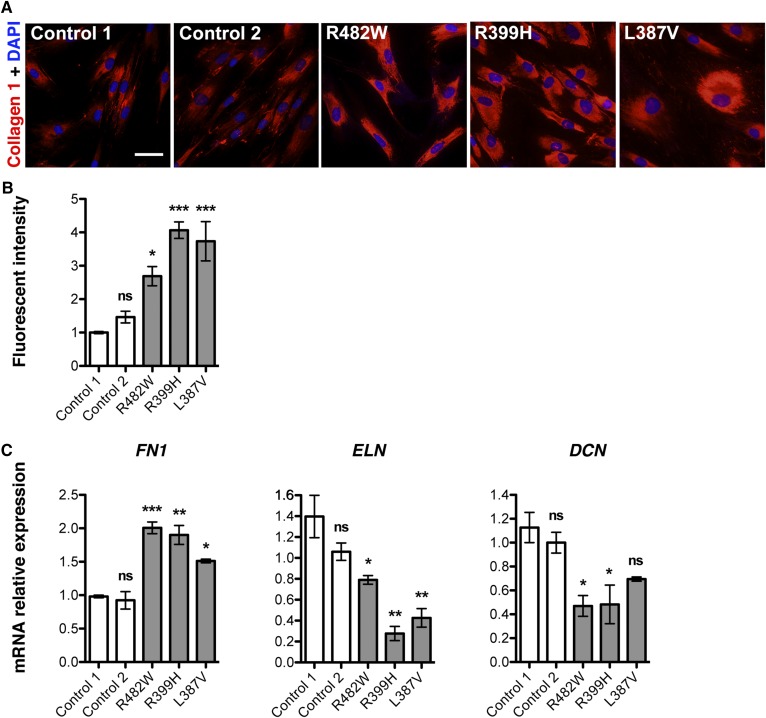
Because adipose tissue from both a patient with the LMNA p.R482W mutation and R482Q lamin A transgenic mice similarly displayed fibrosis and altered expression of ECM proteins, we decided to investigate ECM alterations in human cells bearing lipodystrophy-causing LMNA mutations. We studied primary cultures of skin dermal fibroblasts obtained from patients with FPLD2 or atypical lipodystrophies due to three different LMNA mutations (p.R482W, p.R399H, and p.L387V) and from two unaffected controls. Clinical features of these patients and controls have previously been described (13, 32) and are summarized in Table 3. There was significantly increased expression of type I collagen in fibroblasts from patients with LMNA mutations compared with controls when examined by immunofluorescence microscopy (Fig. 4A, B). RT-qPCR showed significantly increased expression of FN1-encoding fibronectin and significantly decreased expression of ELN and DCN respectively encoding elastin and decorin in fibroblasts with LMNA mutations compared with controls (Fig. 4C). These ECM alterations in fibroblasts from patients with lipodystrophy-causing LMNA mutations were similar to those in subcutaneous adipose tissue from the patient with FPLD2 due to LMNA p.R482W mutation and from R482Q lamin A transgenic mice.
TABLE 3.
Clinical features of control subjects and patients with the LMNA mutations indicated from whom fibroblasts were obtained
| Control 1 | Control 2 | R482W | R399H | L387V | |
| Sex | Female | Female | Female | Female | Female |
| Age (years) | 43 | 33 | 60 | 54 | 61 |
| Body mass index (kg/m2) | 24.6 | 21.2 | 24.5 | 23.3 | 27 |
| Clinical peripheral lipoatrophy | No | No | Four limbs and trunk | Lower limbs | No |
| Cervical fat accumulation | No | No | Yes | No | No |
| Glucose tolerance | Normal | Normal | Diabetes | Diabetes | Diabetes |
| Triglycerides (mM) | NA | NA | 2.3 | 17.1 | 1.5 |
NA, not applicable.
Fig. 4.
Analysis of ECM in dermal fibroblasts from patients with FPLD2 and different LMNA mutations. A: Photomicrographs showing immunofluorescence labeling of type I collagen (collagen 1; red) in cultured dermal fibroblasts from control human subjects and from patients with FPLD2 and LMNA p.R482W (R482W), p.R399H (R399H), and p.L387V (L387V) mutations. Nuclei were stained with diamidino-2-phenylindole hydrochloride (DAPI) (blue). Scale bar = 50 µm. B: Bar graph showing quantification of fluorescence intensity of type I collagen labeling using ImageJ software. Each bar represents means ± SEs from immunofluorescence staining performed on three consecutive passages of dermal fibroblast cultures from control individuals (Control 1, Control 2) and patients with FPLD2 and the indicated LMNA mutations. *P < 0.05; ***P < 0.001; ns, not significant compared with Control 1. C: Means ± SEs of relative expression of mRNAs encoded by FN1, ELN, and DCN in three consecutive passages of dermal fibroblast cultures from control individuals (Control 1, Control 2) and patients with FPLD2 and the indicated LMNA mutations. *P < 0.05; **P < 0.01; ***P < 0.001, ns, not significant compared with Control 1.
ECM alterations caused by LMNA mutations are associated with TGF-β signaling activation and matrix metalloproteinase 9 alterations
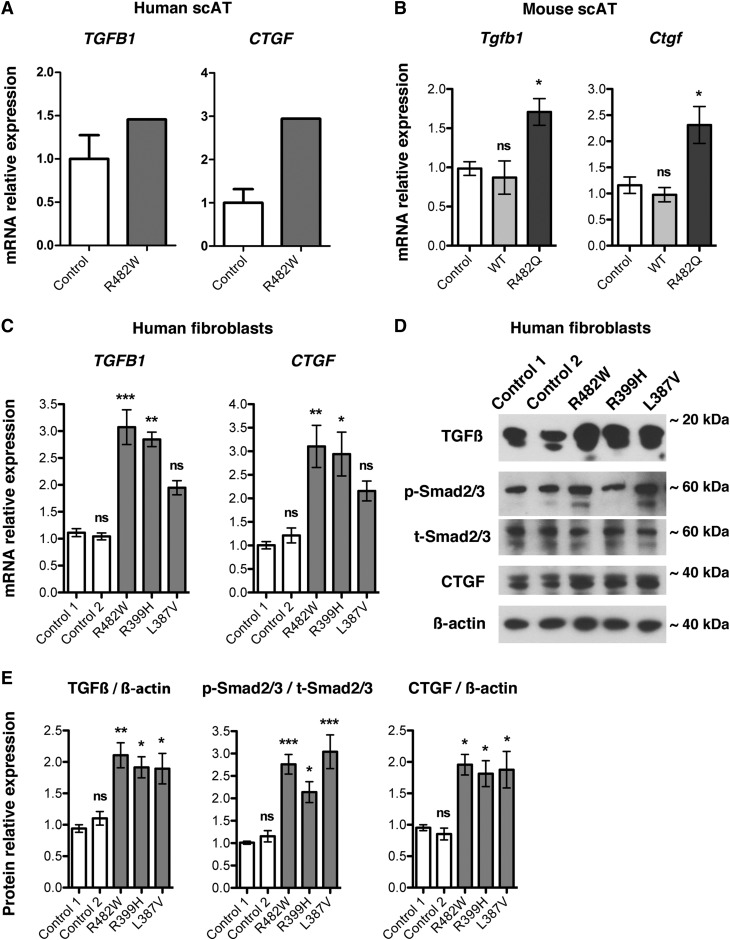
We found increased fibrosis in cervical adipose tissue from a patient with a FPLD2-causing LMNA mutation, in subcutaneous and visceral fat from mice overexpressing lamin A R482Q in adipose tissue, and in dermal fibroblasts from patients with lipodystrophies caused by LMNA mutations. Activation of TGF-β signaling causes matrix deposition by promoting expression of ECM genes (36, 37). We therefore investigated TGF-β signaling in adipose tissue and fibroblasts with lipodystrophy-causing lamin A/C alterations. We first assessed expression of TGFB1 encoding TGF-β1 and CTGF encoding connective tissue growth factor (CTGF) in RNA obtained from human subcutaneous adipose tissue using RT-qPCR. CTGF expression is induced by TGF-β (36). TGFB1 and CTGF expression was increased in subcutaneous adipose tissue from the patient with p.R482W LMNA mutation compared with that in controls (Fig. 5A). Tgfb1 and Ctgf mRNAs were similarly significantly increased in subcutaneous adipose tissue from R482Q lamin A transgenic mice compared with nontransgenic controls and transgenic mice expressing wild-type lamin A (Fig. 5B). We then investigated the TGF-β signaling pathway in human fibroblasts with lipodystrophy-causing LMNA mutations. There was a significant increase in both TGFB1 and CTGF expression in fibroblasts with LMNA p.R482W and p.R399H mutations compared with control cells and a nonsignificant trend toward increased expression in cells with the LMNA p.L387V mutation (Fig. 5C). We then examined the expression of TGF-β1 and CTGF proteins as well as phosphorylation of Smad2/3, mediators of TGF-β signaling that are phosphorylated by its activated receptor (38). Expression of TGF-β1, phosphorylation of Smad2/3, and expression of CTGF were all significantly increased in fibroblasts with p.R482W, p.R399H, and p.L387V LMNA mutations compared with that of controls (Fig. 5D, E). These results indicate that the TGF-β signaling pathway is activated in fibrotic tissue and cells expressing lipodystrophy-causing LMNA mutations.
Fig. 5.
Effects of FPLD2-causing LMNA mutations on TGF-β signaling. A: Relative expression of mRNAs encoded by TGFB1 and CTGF in cervical subcutaneous human adipose tissue (Human scAT) from control subjects (Control) and subject with LMNA p.R482W mutation (R482W). Values are means ± SEs for n = 5 controls and n = 1 patient. B: Means ± SEs of relative expression of mRNAs encoded by Tgfb1 and Ctgf in scAT from nontransgenic mice (Control), transgenic mice overexpressing wild-type lamin A (WT), or transgenic mice overexpressing lamin A R482Q (R482Q) in adipose tissue; n = 3 per group. *P < 0.05; ns, not significant compared with nontransgenic mice (Control). C: Means ± SEs of relative expression of mRNAs encoded by TGFB1 and CTGF in three consecutive passages of dermal fibroblast cultures control individuals (Control 1, Control 2) and patients with FPLD2 and LMNA mutations p.R482W (R482W), p.R399H (R399H), and p.L387V (L387V). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant compared with Control 1. D: Immunoblots using antibodies against TGF-β1/2/3 (TGF-β), phosphorylated Smad2/3 (p-Smad2/3), total Smad2/3 (t-Smad2/3), and CTGF of protein extracts from fibroblasts of control individuals (Control 1, Control 2) and patients with the indicated LMNA mutations. Immunoblots using anti-β-actin are shown as a loading control; migrations of molecular mass stands are indicated at the right of each blot. E: Bar graphs showing quantification of the ratio of the protein signals to β-actin signals for TGF-β and CTGF and ratio of the phosphorylated Smad2/3 signals to respective total Smad2/3 signals in protein extracts from three consecutive passages of dermal fibroblast cultures of control individuals (Control 1, Control 2) and patients with FPLD2 and the indicated LMNA mutations. Values are means ± SEs (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant compared with Control 1.
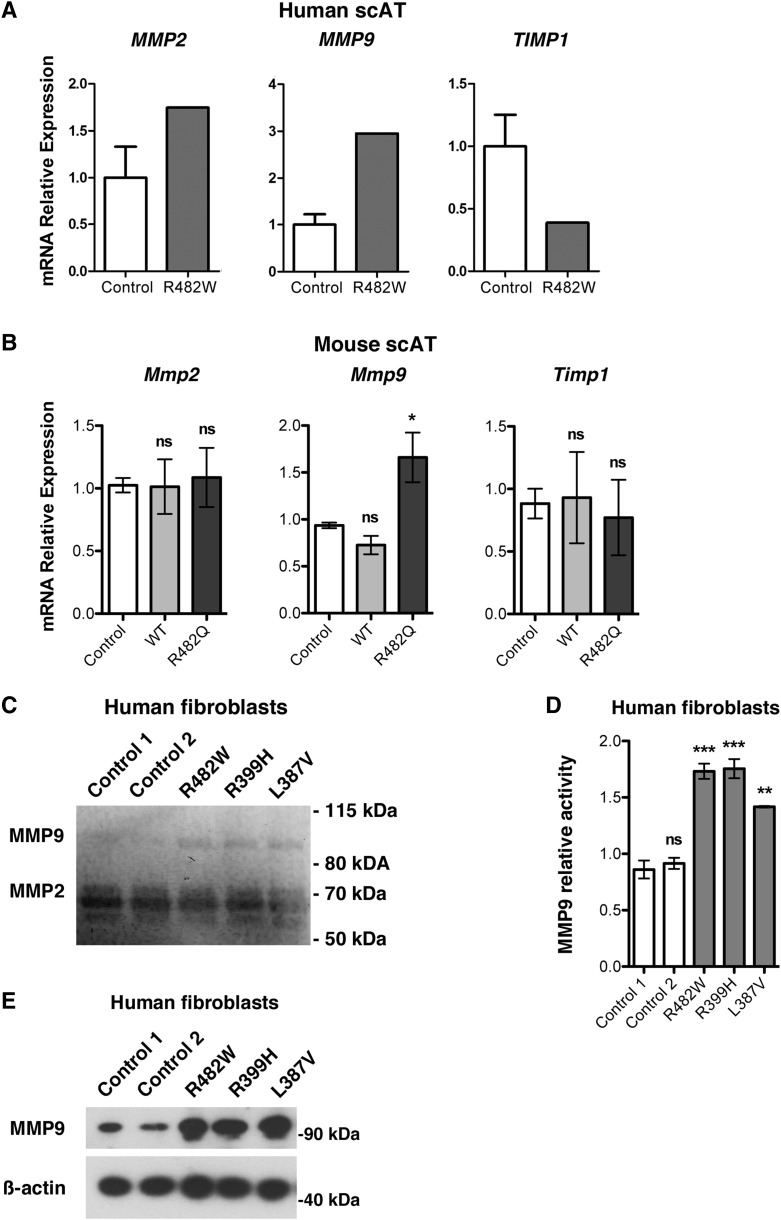
We determined whether ECM degradation was altered in adipose tissue and cultured cells expressing lipodystrophy-causing lamin A/C variants. Matrix metalloproteinases (MMPs) are endopeptidases that degrade ECM proteins such as collagen, fibronectin, and laminin, and their tissue inhibitors regulate their activity (39, 40). We first examined expression of MMP2, MMP9, and TIMP1, which encodes TIMP metallopeptidase inhibitor 1. Whereas MMP2 expression was unaffected, MMP9 expression was increased approximately 3-fold and TIMP1 expression was decreased approximately 2.5-fold in cervical subcutaneous adipose tissue of the patient with LMNA p.R482W compared with controls (Fig. 6A). In subcutaneous adipose tissue from R482Q lamin A transgenic mice, we found a similar result with unchanged expression of Mmp2, an approximate 1.5-fold increase of Mmp9 expression, but no significant change in Timp1 expression compared with nontransgenic controls and transgenic mice expressing wild-type lamin A (Fig. 6B). To assess whether or not LMNA mutations affect MMP activity, we performed gelatin zymography with supernatants of cultured fibroblasts, showing gelatinolytic activity of MMP2 and MMP9 (Fig. 6C). Quantification showed a significant increase in MMP9 activity in all supernatants of cultured fibroblasts from patients with LMNA mutations compared with that in controls (Fig. 6D). We also found elevated expression of MMP9 in fibroblasts from patients compared with that from controls (Fig. 6E). These data demonstrated increased MMP9 expression and activity in cells expressing lipodystrophy-causing lamin A/C variants.
Fig. 6.
MMP expression and activity in LMNA-associated lipodystrophies. A: Relative expression of mRNAs encoded by MMP2, MMP9, and TIMP1 in human cervical subcutaneous adipose tissue (Human scAT) from control human subjects (Control) and patient with LMNA p.R482W mutation (R482W). Values are means ± SEs for n = 5 controls and n = 1 patient. B: Means ± SEs of relative expression of mRNAs encoded by Mmp2, Mmp9, and Timp1 in mouse subcutaneous adipose tissue (Mouse scAT) from nontransgenic mice (Control), transgenic mice expressing wild-type lamin A (WT) or transgenic mice expressing lamin A R482Q (R482Q); n = 3 per group. *P < 0.05; ns, not significant compared with nontransgenic mice (Control). C: Gelatin zymography using supernatants from serum-starved fibroblasts from control individuals (Control 1, Control 2) and patients with FPLD2 and the indicated LMNA mutations was performed to assess the enzymatic activity of MMP2 and MMP9. Equal amounts of supernatants of protein extracts collected after 24 h were loaded onto a gelatin-containing gel and subjected to electrophoresis under nonreducing/nondenaturing conditions. Negative image of the gel is shown so that MMP2 and MMP9 activity appears as dark bands against a light background. Migrations of molecular mass standards are indicated at the right and migration of MMP9 and MMP2 at the left. D: Bar graphs showing quantification of MMP9 activity assessed by gelatin zymography in protein supernatants from three consecutive passages of dermal fibroblast cultures of control individuals (Control 1, Control 2) and patients with FPLD2 and the indicated LMNA mutations. Values were calculated using ImageJ software and are means ± SEs (n = 3). **P < 0.01; ***P < 0.001; ns, not significant compared with Control 1. Results are presented as fold change over Control 1. E: Immunoblots using antibody against MMP9 on protein extracts from fibroblasts of control individuals (Control 1, Control 2) and patients FPLD2 and the indicated LMNA mutations. Immunoblots using anti-β-actin are shown as a loading control; migrations of molecular mass stands are shown at the right of each blot.
DISCUSSION
Our results support the hypothesis that ECM remodeling, fibrosis, and abnormal TGF-β signaling occurs early in the pathogenesis of FPLD2 and related lipodystrophies caused by LMNA mutations. We have shown these alterations in cervical subcutaneous adipose tissue from a patient with the p.R482W LMNA mutation that causes FPLD2, in subcutaneous adipose tissue of transgenic mice expressing R482Q lamin A in adipose tissue, and in fibroblasts from patients with lipodystrophy-causing LMNA mutations. All three of these model systems showed hyperactivation of TGF-β signaling and accumulation of collagen, as assessed by staining with Sirius red, an anionic dye stains collagen by reacting, via its sulphonic acid groups, with basic groups present in the protein (41). Collagen accumulation was associated with alterations in the expression of genes encoding ECM proteins. Antagonists of TGF-β, if administered early enough, may therefore have potential therapeutic benefits in FPLD2.
The transgenic mice we generated overexpressing FLAG-tagged human R482Q lamin A selectively in adipose tissue did not have significant fat loss or metabolic abnormalities associated with lipodystrophy. However, they had significantly increased fibrosis in adipose tissue, indicating that expression of an FPLD2-causing lamin A variant can directly induce adipose tissue fibrosis. Adipose tissue from these mice also demonstrated increased expression of Tgfb1, Ctgf, and Mmp9. Wojtanik et al. (22) generated a similar transgenic mouse line that had decreased body adipose tissue as well as decreased insulin sensitivity and hepatic steatosis. Although Wojtanik et al. (22) mentioned leukocyte infiltration and adipocyte size heterogeneity, a hematoxylin and eosin-stained section of white adipose tissue shown in their article demonstrated increase stromal staining, which could have been fibrosis. Differences between the transgenic mice generated by Wojtanik et al (22) and by us selectively expressing R482Q lamin A in adipose tissue are that (1) we included a FLAG epitope TAG at the amino-terminus of the lamin A variant, and (2) our line had relatively lower expression of the lamin A variant in adipose tissue. Although both lines were on the FVB background, there may also be other subtle genetic differences between them.
Increased TGF-β signaling and fibrosis have been described in laminopathies affecting tissues other than adipose. Hearts from mice with an Lmna mutation that causes cardiomyopathy have marked fibrosis and evidence of enhanced TGF-β and CTGF activity (42, 43). Hearts of human subjects with cardiomyopathy caused by LMNA mutations similarly have prominent fibrosis (44, 45). Adventitial fibrosis in coronary arteries has been reported in children with Hutchinson-Gilford progeria syndrome, which is caused by LMNA mutation (46). Expression of a lamin A variant that causes mandibuloacral dysplasia type A and expression of unprocessed prelamin A in osteoblast-like cells lead to increased secretion of TGF-β (47). MMP9 activity has also been reported to be elevated in serum from patients with mandibuloacral dysplasia type A (48). Even in normal tissues, there appears to be a general relationship between tissue stiffness, collagen expression, and lamin A expression (49). Hence, various genetic alterations in lamin A may lead to pathology in different tissues by inducing fibrosis.
ECM remodeling is a key factor in adipose tissue dysfunction with the level of fibrosis inversely associated with overall metabolic fitness of fat and the ability to adapt to changing nutrient conditions (50). Composition of the ECM also influences cell differentiation. Differentiation of 3T3-F442A preadipocytes into adipocytes is inhibited by fibronectin (33). Culturing adult adipose-derived stem cells on gels that mimic the native stiffness of adipose tissue upregulates adipogenic markers in the absence of exogenous growth factors, whereas increasing the stiffness blocks expression of these markers (51). Depletion of collagen VI, a major component of the ECM of adipose tissue, results in expansion of adipocytes and improvements of energy homeostasis in ob/ob mice (52). These findings further support the hypothesis that ECM remodeling and increased adipose tissue fibrosis induced by lipodystrophy-causing lamin A/C variants underlie pathogenesis.
Lipodystrophies caused by LMNA mutations are rare genetic disorders that share features with metabolic syndrome and central obesity, which are common in the developed world. FPLD2 is characterized by altered distribution of fat, with loss from the periphery and excess in other body regions such as the face and neck. More commonly in the general population, excess body fat alone can lead to insulin resistance, dyslipidemia, hepatic steatosis, and an increased risk for diabetes mellitus and atherosclerosis. As in the models of FPLD2 we examined, white adipose tissue from obese human subjects has increased fibrosis and expression of ECM components (53–55). ECM remodeling and fibrosis may therefore be a common defect in FPLD2 and obesity, reducing adipose tissue expandability and the ability to store an excess of energy, ultimately leading to lipotoxicity and metabolic dysfunction (50, 56). Although this is a parsimonious explanation for the metabolic abnormalities that occur in FPDL2, pleiotropic effects of disease-causing lamin A/C variants in tissues other than adipose could also be a contributing factor.
Supplementary Material
Acknowledgments
The authors thank the late Dr. Constantine “Dean” Londos for providing transgenic mice and Dr. Yi-Hao Yu and Dr. Henry N. Ginsberg for a vector containing the Fabp4 promoter.
Footnotes
Abbreviations:
- CTGF
- connective tissue growth factor
- ECM
- extracellular matrix
- Fabp4
- fatty acid binding protein 4
- FPLD2
- familial partial lipodystrophy type 2
- MMP
- matrix metalloproteinase
- RT-qPCR
- RT-quantitative PCR
- TGF-β
- transforming growth factor-β
This work was supported by National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR048997 (to H.J.W). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. C. LD. was a recipient of a “Young French Speaking Researcher” postdoctoral fellowship from Société Francophone du Diabète (SFD pfe/cc 11-014).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Worman H. J., and Bonne G.. 2007. “Laminopathies”: a wide spectrum of human diseases. Exp. Cell Res. 313: 2121–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonne G., Di Barletta M. R., Varnous S., Bécane H. M., Hammouda E. H., Merlini L., Muntoni F., Greenberg C. R., Gary F., Urtizberea J. A., et al. 1999. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 21: 285–288. [DOI] [PubMed] [Google Scholar]
- 3.Fatkin D., MacRae C., Sasaki T., Wolff M. R., Porcu M., Frenneaux M., Atherton J., Jr Vidaillet H. J., Spudich S., De Girolami U., et al. 1999. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 341: 1715–1724. [DOI] [PubMed] [Google Scholar]
- 4.Brodsky G. L., Muntoni F., Miocic S., Sinagra G., Sewry C., and Mestroni L.. 2000. Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement. Circulation. 101: 473–476. [DOI] [PubMed] [Google Scholar]
- 5.Muchir A., Bonne G., van der Kooi A. J., van Meegen M., Baas F., Bolhuis P. A., de Visser M., and Schwartz K.. 2000. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum. Mol. Genet. 9: 1453–1459. [DOI] [PubMed] [Google Scholar]
- 6.De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C. L., Munnich A., Le Merrer M., et al. 2003. Lamin a truncation in Hutchinson-Gilford progeria. Science. 300: 2055. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson M., Brown W. T., Gordon L. B., Glynn M. W., Singer J., Scott L., Erdos M. R., Robbins C. M., Moses T. Y., Berglund P., et al. 2003. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 423: 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novelli G., Muchir A., Sangiuolo F., Helbling-Leclerc A., D’Apice M. R., Massart C., Capon F., Sbraccia P., Federici M., Lauro R., et al. 2002. Mandibuloacral dysplasia is caused by a mutation in LMNA-encoding lamin A/C. Am. J. Hum. Genet. 71: 426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Sandre-Giovannoli A., Chaouch M., Kozlov S., Vallat J. M., Tazir M., Kassouri N., Szepetowski P., Hammadouche T., Vandenberghe A., Stewart C. L., et al. 2002. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. Am. J. Hum. Genet. 70: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao H., and Hegele R. A.. 2000. Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 9: 109–112. [DOI] [PubMed] [Google Scholar]
- 11.Shackleton S., Lloyd D. J., Jackson S. N., Evans R., Niermeijer M. F., Singh B. M., Schmidt H., Brabant G., Kumar S., Durrington P. N., et al. 2000. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat. Genet. 24: 153–156. [DOI] [PubMed] [Google Scholar]
- 12.Speckman R. A., Garg A., Du F., Bennett L., Veile R., Arioglu E., Taylor S. I., Lovett M., and Bowcock A. M.. 2000. Mutational and haplotype analyses of families with familial partial lipodystrophy (Dunnigan variety) reveal recurrent missense mutations in the globular C-terminal domain of lamin A/C. Am. J. Hum. Genet. 66: 1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decaudain A., Vantyghem M. C., Guerci B., Hécart A. C., Auclair M., Reznik Y., Narbonne H., Ducluzeau P. H., Donadille B., Lebbé C., et al. 2007. New metabolic phenotypes in laminopathies: LMNA mutations in patients with severe metabolic syndrome. J. Clin. Endocrinol. Metab. 92: 4835–4844. [DOI] [PubMed] [Google Scholar]
- 14.Dunnigan M. G., Cochrane M. A., Kelly A., and Scott J. W.. 1974. Familial lipoatrophic diabetes with dominant transmission. A new syndrome. Q. J. Med. 43: 33–48. [PubMed] [Google Scholar]
- 15.Guénantin A. C., Briand N., Bidault G., Afonso P., Béréziat V., Vatier C., Lascols O., Caron-Debarle M., Capeau J., and Vigouroux C.. 2014. Nuclear envelope-related lipodystrophies. Semin. Cell Dev. Biol. 29: 148–157. [DOI] [PubMed] [Google Scholar]
- 16.Garg A. 2004. Acquired and inherited lipodystrophies. N. Engl. J. Med. 350: 1220–1234. [DOI] [PubMed] [Google Scholar]
- 17.Lüdtke A., Genschel J., Brabant G., Bauditz J., Taupitz M., Koch M., Wermke W., Worman H. J., and Schmidt H. H.. 2005. Hepatic steatosis in Dunnigan-type familial partial lipodystrophy. Am. J. Gastroenterol. 100: 2218–2224. [DOI] [PubMed] [Google Scholar]
- 18.Robbins A. L., and Savage D. B.. 2015. The genetics of lipid storage and human lipodystrophies. Trends Mol. Med. 21: 433–438. [DOI] [PubMed] [Google Scholar]
- 19.Dhe-Paganon S., Werner E. D., Chi Y. I., and Shoelson S. E.. 2002. Structure of the globular tail of nuclear lamin. J. Biol. Chem. 277: 17381–17384. [DOI] [PubMed] [Google Scholar]
- 20.Krimm I., Östlund C., Gilquin B., Couprie J., Hossenlopp P., Mornon J. P., Bonne G., Courvalin J. C., Worman H. J., and Zinn-Justin S.. 2002. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 10: 811–823. [DOI] [PubMed] [Google Scholar]
- 21.Cutler D. A., Sullivan T., Marcus-Samuels B., Stewart C. L., and Reitman M. L.. 2002. Characterization of adiposity and metabolism in Lmna-deficient mice. Biochem. Biophys. Res. Commun. 291: 522–527. [DOI] [PubMed] [Google Scholar]
- 22.Wojtanik K. M., Edgemon K., Viswanadha S., Lindsey B., Haluzik M., Chen W., Poy G., Reitman M., and Londos C.. 2009. The role of LMNA in adipose: a novel mouse model of lipodystrophy based on the Dunnigan-type familial partial lipodystrophy mutation. J. Lipid Res. 50: 1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boguslavsky R. L., Stewart C. L., and Worman H. J.. 2006. Nuclear lamin A inhibits adipocyte differentiation: implications for Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 15: 653–663. [DOI] [PubMed] [Google Scholar]
- 24.Gandotra S., Le Dour C., Bottomley W., Cervera P., Giral P., Reznik Y., Charpentier G., Auclair M., Delépine M., Barroso I., et al. 2011. Perilipin deficiency and autosomal dominant partial lipodystrophy. N. Engl. J. Med. 364: 740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jan V., Cervera P., Maachi M., Baudrimont M., Kim M., Vidal H., Girard P. M., Levan P., Rozenbaum W., Lombès A., et al. 2004. Altered fat differentiation and adipocytokine expression are inter-related and linked to morphological changes and insulin resistance in HIV-1-infected lipodystrophic patients. Antivir. Ther. 9: 555–564. [PubMed] [Google Scholar]
- 26.Weedon M. N., Ellard S., Prindle M. J., Caswell R., Lango Allen H., Oram R., Godbole K., Yajnik C. S., Sbraccia P., Novelli G., et al. 2013. An in-frame deletion at the polymerase active site of POLD1 causes a multisystem disorder with lipodystrophy. Nat. Genet. 45: 947–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Béréziat V., Cervera P., Le Dour C., Verpont M. C., Dumont S., Vantyghem M. C., Capeau J., and Vigouroux C. , and . Lipodystrophy Study Group. 2011. LMNA mutations induce a non-inflammatory fibrosis and a brown fat-like dystrophy of enlarged cervical adipose tissue. Am. J. Pathol. 179: 2443–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araújo-Vilar D., Victoria B., González-Méndez B., Barreiro F., Fernández-Rodríguez B., Cereijo R., Gallego-Escuredo J. M., Villarroya F., and Pañeda-Menéndez A.. 2012. Histological and molecular features of lipomatous and nonlipomatous adipose tissue in familial partial lipodystrophy caused by LMNA mutations. Clin. Endocrinol. (Oxf.). 76: 816–824. [DOI] [PubMed] [Google Scholar]
- 29.Ponchel F., Toomes C., Bransfield K., Leong F. T., Douglas S. H., Field S. L., Bell S. M., Combaret V., Puisieux A., Mighell A. J., et al. 2003. Real-time PCR based on SYBR-Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 31.Cance W. G., Chaudhary N., Worman H. J., Blobel G., and Cordoncardo C.. 1992. Expression of the nuclear lamins in normal and neoplastic human tissues. J. Exp. Clin. Cancer Res. 11: 233–246. [Google Scholar]
- 32.Vigouroux C., Magré J., Vantyghem M. C., Bourut C., Lascols O., Shackleton S., Lloyd D. J., Guerci B., Padova G., Valensi P., et al. 2000. Lamin A/C gene: sex-determined expression of mutations in Dunnigan-type familial partial lipodystrophy and absence of coding mutations in congenital and acquired generalized lipoatrophy. Diabetes. 49: 1958–1962. [DOI] [PubMed] [Google Scholar]
- 33.Spiegelman B. M., and Ginty C. A.. 1983. Fibronectin modulation of cell shape and lipogenic gene expression in 3T3-adipocytes. Cell. 35: 657–666. [DOI] [PubMed] [Google Scholar]
- 34.Alkhouli N., Mansfield J., Green E., Bell J., Knight B., Liversedge N., Tham J. C., Welbourn R., Shore A. C., Kos K., et al. 2013. The mechanical properties of human adipose tissues and their relationships to the structure and composition of the extracellular matrix. Am. J. Physiol. Endocrinol. Metab. 305: E1427–E1435. [DOI] [PubMed] [Google Scholar]
- 35.Svensson L., Heinegård D., and Oldberg A.. 1995. Decorin-binding sites for collagen type I are mainly located in leucine-rich repeats 4–5. J. Biol. Chem. 270: 20712–20716. [DOI] [PubMed] [Google Scholar]
- 36.Leask A., and Abraham D. J.. 2004. TGF-beta signaling and the fibrotic response. FASEB J. 18: 816–827. [DOI] [PubMed] [Google Scholar]
- 37.Rosenbloom J., Castro S. V., and Jimenez S. A.. 2010. Narrative review: fibrotic diseases: cellular and molecular mechanisms and novel therapies. Ann. Intern. Med. 152: 159–166. [DOI] [PubMed] [Google Scholar]
- 38.Kamato D., Burch M. L., Piva T. J., Rezaei H. B., Rostam M. A., Xu S., Zheng W., Little P. J., and Osman N.. 2013. Transforming growth factor-β signalling: role and consequences of Smad linker region phosphorylation. Cell. Signal. 25: 2017–2024. [DOI] [PubMed] [Google Scholar]
- 39.Page-McCaw A., Ewald A. J., and Werb Z.. 2007. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hopps E., and Caimi G.. 2012. Matrix metalloproteinases in metabolic syndrome. Eur. J. Intern. Med. 23: 99–104. [DOI] [PubMed] [Google Scholar]
- 41.Junqueira L. C., Bignolas G., and Brentani R. R.. 1979. Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. Histochem. J. 11: 447–455. [DOI] [PubMed] [Google Scholar]
- 42.Arimura T., Helbling-Leclerc A., Massart C., Varnous S., Niel F., Lacène E., Fromes Y., Toussaint M., Mura A. M., Keller D. I., et al. 2005. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet. 14: 155–169. [DOI] [PubMed] [Google Scholar]
- 43.Chatzifrangkeskou M., Le Dour C., Wu W., Morrow J. P., Joseph L. C., Beuvin M., Sera F., Homma S., Vignier N., Mougenot N., et al. 2016. ERK1/2 directly acts on CTGF/CCN2 expression to mediate myocardial fibrosis in cardiomyopathy caused by mutations in the lamin A/C gene. Hum. Mol. Genet. 25: 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raman S. V., Sparks E. A., Baker P. M., McCarthy B., and Wooley C. F.. 2007. Mid-myocardial fibrosis by cardiac magnetic resonance in patients with lamin A/C cardiomyopathy: possible substrate for diastolic dysfunction. J. Cardiovasc. Magn. Reson. 9: 907–913. [DOI] [PubMed] [Google Scholar]
- 45.Holmström M., Kivistö S., Heliö T., Jurkko R., Kaartinen M., Antila M., Reissell E., Kuusisto J., Kärkkäinen S., Peuhkurinen K., et al. 2011. Late gadolinium enhanced cardiovascular magnetic resonance of lamin A/C gene mutation related dilated cardiomyopathy. J. Cardiovasc. Magn. Reson. 13: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olive M., Harten I., Mitchell R., Beers J. K., Djabali K., Cao K., Erdos M. R., Blair C., Funke B., Smoot L., et al. 2010. Cardiovascular pathology in Hutchinson-Gilford progeria: correlation with the vascular pathology of aging. Arterioscler. Thromb. Vasc. Biol. 30: 2301–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evangelisti C., Bernasconi P., Cavalcante P., Cappelletti C., D’Apice M. R., Sbraccia P., Novelli G., Prencipe S., Lemma S., Baldini N., et al. 2015. Modulation of TGFbeta 2 levels by lamin A in U2-OS osteoblast-like cells: understanding the osteolytic process triggered by altered lamins. Oncotarget. 6: 7424–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lombardi F., Fasciglione G. F., D’Apice M. R., Vielle A., D’Adamo M., Sbraccia P., Marini S., Borgiani P., Coletta M., and Novelli G.. 2008. Increased release and activity of matrix metalloproteinase-9 in patients with mandibuloacral dysplasia type A, a rare premature ageing syndrome. Clin. Genet. 74: 374–383. [DOI] [PubMed] [Google Scholar]
- 49.Swift J., Ivanovska I. L., Buxboim A., Harada T., Dingal P. C., Pinter J., Pajerowski J. D., Spinler K. R., Shin J. W., Tewari M., et al. 2013. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 341: 1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun K., Tordjman J., Clément K., and Scherer P. E.. 2013. Fibrosis and adipose tissue dysfunction. Cell Metab. 18: 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young D. A., Choi Y. S., Engler A. J., and Christman K. L.. 2013. Stimulation of adipogenesis of adult adipose-derived stem cells using substrates that mimic the stiffness of adipose tissue. Biomaterials. 34: 8581–8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan T., Muise E. S., Iyengar P., Wang Z. V., Chandalia M., Abate N., Zhang B. B., Bonaldo P., Chua S., and Scherer P. E.. 2009. Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Mol. Cell. Biol. 29: 1575–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Divoux A., Tordjman J., Lacasa D., Veyrie N., Hugol D., Aissat A., Basdevant A., Guerre-Millo M., Poitou C., Zucker J. D., et al. 2010. Fibrosis in human adipose tissue: composition, distribution, and link with lipid metabolism and fat mass loss. Diabetes. 59: 2817–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henegar C., Tordjman J., Achard V., Lacasa D., Cremer I., Guerre-Millo M., Poitou C., Basdevant A., Stich V., Viguerie N., et al. 2008. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol. 9: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mutch D. M., Tordjman J., Pelloux V., Hanczar B., Henegar C., Poitou C., Veyrie N., Zucker J. D., and Clément K.. 2009. Needle and surgical biopsy techniques differentially affect adipose tissue gene expression profiles. Am. J. Clin. Nutr. 89: 51–57. [DOI] [PubMed] [Google Scholar]
- 56.Virtue S., and Vidal-Puig A.. 2010. Adipose tissue expandability, lipotoxicity and the metabolic syndrome—an allostatic perspective. Biochim. Biophys. Acta. 1801: 338–349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.