Abstract
The 1-deoxysphingolipids (1-deoxySLs) are atypical sphingolipids (SLs) that are formed when serine palmitoyltransferase condenses palmitoyl-CoA with alanine instead of serine during SL synthesis. The 1-deoxySLs are toxic to neurons and pancreatic β-cells. Pathologically elevated 1-deoxySLs cause the inherited neuropathy, hereditary sensory autonomic neuropathy type 1 (HSAN1), and are also found in T2D. Diabetic sensory polyneuropathy (DSN) and HSAN1 are clinically very similar, suggesting that 1-deoxySLs may be implicated in both pathologies. The 1-deoxySLs are considered to be dead-end metabolites, as they lack the C1-hydroxyl group, which is essential for the canonical degradation of SLs. Here, we report a previously unknown metabolic pathway, which is capable of degrading 1-deoxySLs. Using a variety of metabolic labeling approaches and high-resolution high-accuracy MS, we identified eight 1-deoxySL downstream metabolites, which appear to be formed by cytochrome P450 (CYP)4F enzymes. Comprehensive inhibition and induction of CYP4F enzymes blocked and stimulated, respectively, the formation of the downstream metabolites. Consequently, CYP4F enzymes might be novel therapeutic targets for the treatment of HSAN1 and DSN, as well as for the prevention of T2D.
Keywords: sphingolipids, lipids/chemistry, mass spectrometry, obesity, diabetes, neurotoxicity, metabolic pathway
Sphingolipid (SL) de novo synthesis typically starts with the condensation of serine and palmitoyl-CoA, a reaction catalyzed by serine palmitoyltransferase (SPT) (1–3). SPT can also use alanine in certain conditions (4), which results in the formation of the atypical cytotoxic 1-deoxysphingolipids (1-deoxySLs). These lack the C1-hydroxyl group of regular SLs, which is essential for further metabolic steps and degradation (4, 5). As such, 1-deoxySLs are commonly believed to be “dead-end” metabolites that accumulate within cells, tissues, and body fluids.
Pathologically elevated 1-deoxySLs have been found in a number of neurological and metabolic disorders. They are a hallmark of hereditary sensory autonomic neuropathy type 1 (HSAN1), a rare autosomal dominant inherited peripheral neuropathy that is caused by various mutations in SPT. Plasma 1-deoxySLs are also elevated in patients with nondiabetic metabolic syndrome (MetS) and T2D (6) and may contribute to the development of diabetic sensory polyneuropathy (DSN), which is clinically very similar to HSAN1 (7, 8). Both conditions start in the lower extremities with a loss of sensation often accompanied by positive sensory symptoms such as hyperpathia and neuropathic pain, as well as the development of painless wounds leading to ulcers (9, 10). L-serine supplementation was shown to effectively lower 1-deoxySL levels while improving neuropathic symptoms in a HSAN1 mouse model as well as in a diabetic rat model (11, 12). In addition, 1-deoxySLs were shown to be elevated under Taxol treatment and might contribute to Taxol-induced peripheral neuropathy (13). Furthermore, 1-deoxySLs were found to be increased in patients with nonalcoholic steatohepatitis (14) and to interfere with the function and survival of pancreatic β-cells (15). Elevated 1-deoxySL plasma levels are also predictive for the risk to develop T2D (6).
The 1-deoxysphinganine (1-deoxySA), which is formed by SPT through the condensation of palmitoyl-CoA and alanine, is N-acylated to 1-deoxy-dihydroceramides (5, 16, 17). However, while canonical dihydroceramides are converted to ceramides by the introduction of a Δ4,5 trans double bond (DB) (18, 19), a Δ14,15 cis DB is most commonly introduced in 1-deoxy-dihydroceramides (20), indicating that 1-deoxySLs follow a different metabolic route than canonical SLs. Upon degradation, ceramides are typically hydrolyzed to sphingosine (SO), and then converted to the catabolic intermediate, sphingosine-1-phosphate (S1P) (21), which is irreversibly degraded by S1P lyase (22). This is not possible for 1-deoxysphingosine (1-deoxySO), as the C1-hydroxyl group is missing.
In this work, we addressed the issue of 1-deoxySL metabolism and degradation in more detail. We demonstrated for the first time that 1-deoxySO is metabolized to a variety of downstream metabolites through reactions that are at least partly mediated by enzymes of the cytochrome P450 (CYP)4F subfamily.
MATERIALS AND METHODS
Cell culture
Mouse embryonic fibroblasts (MEFs) were grown and maintained in DMEM (Sigma) with 10% FBS (Fisher Scientific; FSA15-043), and 1% penicillin/streptomycin (P/S; 100 units per milliliter/0.1 mg per milliliter; Sigma) at 37°C and 5% CO2.
Transfection of Cyp4a10, Cyp4f13 in HEK cells
HEK293 cells were grown in DMEM + 10% FBS + 1% P/S. Four micrograms of the Cyp4a10 and Cyp4f13 plasmids (Origene, Rockville, MD) were diluted in 400 μl of Opti-MEM I (Gibco; 51985) and then gently vortexed. Eight microliters of TurboFect transfection reagent (ThermoFisher) were added to the diluted DNA, mixed by gently pipetting, and incubated for 20 min at room temperature. The TurboFect/DNA mixture was then added drop-wise to the cells. Forty-eight hours after transfection, the cells were split at a 1:10 dilution into 10 ml of growth medium plus 400 μg/μl Geneticin (Gibco) for selection and passaged three times before being used for experiments. Expression of the CYP4A10 and CYP4F13 proteins was confirmed by Western blot using a rabbit anti-FLAG-tag antibody (Serotec).
Metabolic labeling assays
Deuterated and unlabeled 1-deoxySL 1 mM stock solutions were prepared by dissolving the lipids in 100% ethanol (Sigma).
For the pulse-chase experiments, MEF cells were treated with 1 μM deuterium-labeled (d3) 1-deoxySA or d7-SA (Avanti Polar Lipids) for 2 h, after which they were chased for 0, 1, 4, 8, 24, and 48 h with unsupplemented growth medium (DMEM), or for 1, 2, 3, 4, 6, and 8 days for the long-term chase experiments. For the experiments with HET0016, the inhibitor was present in the medium throughout the entire duration of both the pulse and chase periods. Cells were harvested at the time points specified above by first washing with PBS, followed by trypsinization in 500 μl for 3 min. An equal volume of growth medium was added to stop trypsinization and cells were counted (Z2 Coulter Counter; Beckman Coulter, Brea, CA) and pelleted (800 g, 5 min at 4°C). The cell pellet was subsequently resuspended in 1 ml cold PBS and centrifuged, and PBS was removed by aspiration.
The pulse medium was collected upon exchanging with unsupplemented growth medium. Before harvesting the cells, the chase medium from all the time points was also collected, lyophilized overnight (Christ Alpha 2-4 LSC Freeze Dryer; Kuhne, Birsfelden, Switzerland), and stored at −20°C until extraction of the SLs.
For the metabolic order assay, MEF cells were treated with 1 μM of either d3-deoxySA-OH, d3-deoxySO-OH, d3-deoxySA, or three synthetic 1-deoxySO analogs with different DB positions (4,5-trans; 14,15-trans; and 14,15-cis). Cells were harvested after 48 h.
For the CYPA/CYP4F inhibition and induction assays, MEF cells treated with 1 μM d3-deoxySA alone or in combination with HET0016 (5 μM; Santa Cruz Biotechnology), fenofibrate (20 μM; Sigma), or retinoic acid (20 μM, Sigma), respectively, and harvested after 24 h. For the downstream metabolite inhibition curves with HET0016, MEF cells were treated with 1 μM d3-deoxySA and the following concentrations of the inhibitor for 24 h: 50 nM, 100 nM, 500 nM, 1 μM, 2.5 μM, 5 μM, and10 μM.
HEK-WT, HEK-Cyp4a10, and HEK-Cyp4f13 cells were treated with 1 μM d3-deoxySA. After 48 h, cells were washed with 2 ml PBS and then harvested in 1 ml PBS, counted, and pelleted (800 g, 5 min at 4°C).
Please note that for the pulse-chase experiments, units are in total picomoles to account for the dilution of the labeled lipids taken up in the pulse period by the multiplication of cells, while for the steady state experiments, units are in picomoles per 1e6 cells.
Extraction and analysis of sphingoid and deoxysphingoid bases
The frozen cell pellets were resuspended in 100 μl PBS and the lyophilized medium was also dissolved in 100 μl of PBS prior to extraction. Five hundred microliters of methanol (Honeywell), including 200 pmol of internal standard [d7-sphinganine (d7-SA) and d7-sphingosine (d7-SO); Avanti Polar Lipids] was added to each sample. Lipids were extracted by shaking in a Thermomixer for 1 h at 1,000 rpm and 37°C (Thermomixer Comfort; Eppendorf, Hamburg, Germany). To pellet precipitated protein, samples were centrifuged at 16,000 g for 5 min and the supernatant was transferred to a new tube. Lipids were acid/base hydrolyzed to remove the N-acyl chain, as described earlier (4, 23).
Before analysis by LC/MS, lipids were redissolved in 75 μl of 56.7% methanol, 33.3% ethanol, 10% water, and derivatized with 5 μl OPA working solution [990 μl boric acid (3%) + 10 μl o-phthalaldehyde (50 mg/ml in ethanol) + 0.5 μl 2-mercaptoethanol]. OPA was used for derivatization in order to improve the chromatographic separation of peaks and also to aid with the specificity of the metabolite search later on, as only SL metabolites with an amino group would be derivatized.
The hydrolyzed sphingoid bases were separated on a C18 column (Uptispere 120 Å, 5 μm, 125 × 2 mm; Interchim, Montluçon, France) using a Transcend UPLC pump (Thermo, Reinach, BL, Switzerland) at a flow rate of 0.3 ml/min with a binary solvent system, where solvent A was 1:1 methanol/ammonium acetate (5 mM) in water, and solvent B was methanol. The column was first equilibrated using a mixture of 50% mobile phase A and 50% mobile phase B, and then 25 μl of sample was injected, which was eluted by the same mixture for 1 min. This was followed by a ramp gradient over 25 min to 100% phase B, which remained until 32 min, followed by a ramp gradient back down to 50% solvent A and 50% solvent B for 1 min, and column equilibration with the same mixture for 2 min. The sphingoid and deoxysphingoid bases were ionized using an atmospheric pressure chemical ionization source and detected on a Q-Exactive hybrid quadrupole Orbitrap mass spectrometer (Thermo). The detection was performed in full scan mode, with a scan range of m/z 120–1,200, mass resolution of 140,000, automatic gain control target 3.00E+06, and maximum injection time of 512 ms.
Differential analysis
The software Sieve 2.0 (ThermoFisher) was used to perform differential analysis of the lipidomics dataset obtained for the different time points of the pulse-chase experiments. The basic workflow used to perform the differential analysis was described by Snyder et al. (24). The software aligns the retention times of the chromatographic peaks between the two conditions being compared, after which it defines frames based on retention time, mass, and intensity (i.e., the area under the peak), where each frame represents an individual metabolite, its isotopes, or any fragment measured. A list of 486 frames was populated from the comparison of time points 0 and 48 h of the chase, translating into approximately 312 components after filtering out natural isotopologues. The software calculated a ratio of the change in levels between the two conditions compared, and also the statistical significance of the change, as triplicate measurements were used. All frames were normalized to the spiked internal standard d7-SA.
The components that were classified as changing between the time points were then filtered based on the following criteria: 400 < m/z < 510, as that is the mass range of OPA-derivatized sphingoid bases; a retention time <26 min, as that is when the washing of the column begins; a ratio >1, as this would indicate a change between the two time points; and a P value <0.05. We further filtered the components by selecting for lipid metabolites with odd-numbered masses because these would be expected to contain the d3 label. Because samples were derivatized with o-phthalaldehyde (OPA) before running on the MS, we applied the added criteria that the molecular formula should contain one sulfur, which comes from the OPA working solution. We then generated the molecular formula from the accurate mass (within 5 ppm) of the remaining compounds after all restrictions were applied.
Visual analysis of the total ion chromatogram
To ensure that no metabolites were excluded by the Sieve software due to low levels or incomplete peak separation at baseline, the total ion current plot was analyzed scan by scan over the duration of each run (35 min), using the same restrictions as above (i.e., odd-numbered mass, the accurate mass of the compound should be able to generate a formula containing one sulfur). We analyzed the mass spectra of each scan in the total ion current plot for the most intense m/z present in order to identify new metabolites containing the d3 label.
Deuterium exchange assay with deuterium-labeled substrates
MEF cells overexpressing the SPT enzyme with the mutation SPTLC1-C133W, which were obtained from Vincent Timmerman (University of Antwerp, Belgium), were grown in DMEM with 10% FBS, 1% P/S, and 4 μg/ml Geneticin. The cells were treated with methyl-d3 palmitic acid (Cambridge Isotopes) as a 1:1 molar complex with fatty acid-free BSA, as well as with d4-alanine (Cambridge Isotopes), and harvested after 48 h. SLs were extracted and acid/base hydrolyzed as described and measured by LC/MS. The 1-deoxySLs with mass offsets of +6, +5, +4, +3, +2, +1, as well as the unlabeled mass M were quantified in order to account for the natural isotopologues, as well as the +3 labeling arising from the d3-palmitic acid or the d4-alanine (one deuterium is lost from the alanine upon condensation with the palmitoyl-CoA) and the +6 labeling arising from the condensation of both labeled substrates.
LC fraction separation
Pure amounts of some of the newly identified 1-deoxySL metabolites were obtained by fractionating the LC eluate at the specific retention times of the metabolites. Each fraction was collected by using a divert valve after the LC column. The fractions were eluted in 50% methanol and 50% 1:1 methanol/ammonium acetate. The mobile phase was evaporated under a steady stream of nitrogen (Techne Sample Concentrator; Bibby Scientific Ltd., Staffordshire, UK). In order to remove the ammonium acetate from the separated fractions, the lipids were extracted by redissolving in 100 μl PBS + 666 μl methanol + 333 μl chloroform. The samples were then agitated at 1,000 rpm 37°C for 1 h (Thermomixer Comfort; Eppendorf), followed by the addition of 500 μl chloroform and 200 μl of alkaline water. The upper (water) phase was removed by aspiration, and the lower (organic) phase was washed three times with 1 ml of alkaline water. Lipids were dried under a steady stream of nitrogen and resuspended in ethanol. Fraction purity and concentration were measured by spiking with internal standards (d7-SA and d7-SO) and running on the LC/MS as described above. There was less than 1% contamination with any of the other d3-deoxySL metabolites in the purified d3-deoxySO-OH fraction and 5% contamination in the d3-deoxySA-OH fraction with d3-deoxySO-OH.
Statistical analysis
Statistical significance was calculated by a two-tailed Student’s t-test and ANOVA using GraphPad Prism 6.
RESULTS
The decrease in intracellular d3-deoxySA over time is not compensated for by the increase in d3-deoxySO
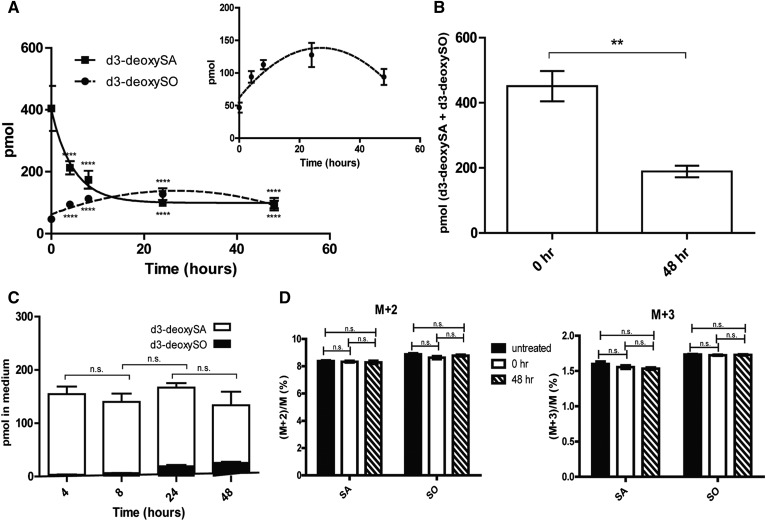
To examine whether 1-deoxySO is the end product of 1-deoxySL metabolism, we supplemented cells with a pulse of the isotope-labeled precursor d3-deoxySA (2 h) followed by a chase with unsupplemented growth medium. Lipids were extracted and acid/base hydrolyzed to remove the N-acyl chain, and the d3-deoxysphingoid bases were quantified by LC/MS. We observed a time-dependent decrease of d3-deoxySA within 48 h to approximately half of the initial concentration (Fig. 1A). However, this decrease was not compensated for by an equivalent increase in the downstream product d3-deoxySO. In fact, d3-deoxySO increased only for the first 24 h, and then started to decline again (Fig. 1A). At 48 h, the sum of d3-deoxySA and d3-deoxySO was less than half of what was measured directly after the pulse (Fig. 1B). The total levels of d3-labeled deoxysphingoid bases (d3-deoxySA + d3-deoxySO) in the medium were constant over time (Fig. 1C).
Fig. 1.
Total levels of d3-labeled 1-deoxySLs decrease over a 48 h chase period. A: MEFs were treated with a 2 h pulse of d3-deoxySA followed by a chase with unsupplemented growth medium; time point 0 represents the start of the chase period. The levels of d3-deoxySA decreased over the duration of the chase, while d3-deoxySO increased up to 24 h and then also decreased (after acid/base hydrolysis). Inlay is a magnified version of the d3-deoxySO levels over time. Statistical significance for the difference in the levels of d3-deoxySA and d3-deoxySO at each time point when compared with time point 0 was calculated using a one-way ANOVA with a Bonferroni correction for comparing selected pairs. B: Total d3-deoxySA + d3-deoxySO levels decreased by a factor of 2 between the 0 and 48 h time points of the chase. C: The growth medium was collected at all time points of the chase and the lipids were extracted. The total levels of d3-labeled deoxysphingoid bases (d3-deoxySA + d3-deoxySO) in the medium did not change over time. D: The ratio of (M+2)/M and (M+3)/M SA and SO, where M is the mass of the canonical unlabeled sphingoid bases, were unchanged between the 0 and 48 h time points of the chase, as well as when compared with untreated control cells. Points and bars represent averages ± SEM. Statistical significance was calculated using a one-way ANOVA followed by Bonferroni correction. **P < 0.01, ****P < 0.0001; n.s., not significant.
One explanation for this loss in labeled d3-deoxySLs could be the conversion to canonical C18-SLs by a hydroxylation/oxygenation at C1. The addition of a hydroxyl group at C1 would result in the replacement of one of the deuteriums, which would therefore result in a d2-labeled C18 sphingoid base. Alternatively, an oxygen could also be added to C1 by nucleophilic attack, as occurs through the action of heme-dependent mono-oxygenases (see supplemental Fig. S1 for a depiction of the two reactions). In this case there would be no deuterium loss, and the resulting SA and SO bases would have a d3 label (see supplemental Fig. S1 for a depiction of the two different reactions). As most elements occur as a mixture of isotopes, this results in compounds having different isotopologues whose relative amounts are based on the occurrence of the atomic isotopes in nature. Here, we refer to the mass of the canonical SA/SO as M, where M+2 and M+3 are usually the natural isotopologues having two or three 13C atoms, respectively. However, in this case, we have also used them to refer to the potential d2- and d3-labeled SA and SO, as these labels would also result in a mass of +2 and +3, respectively, when compared with the canonical unlabeled lipids. The incorporation of the label into canonical SA or SO would result in an altered (M+2)/M or (M+3)/M ratio. However, as there was no change in the relative amounts of M+2 and M+3 between the 0 and 48 h time points, nor when compared with untreated cells (Fig. 1D), this indicates that there was no conversion of the 1-deoxySLs to canonical SLs.
Eight novel 1-deoxySLs identified increase over time
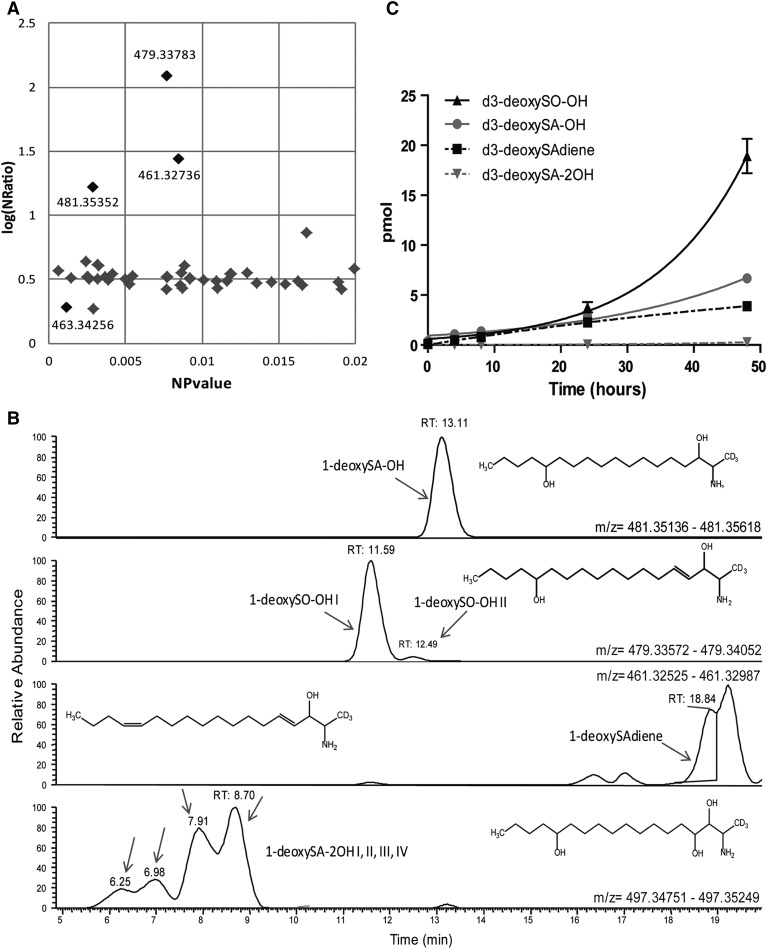
Next, we systemically screened for unknown labeled 1-deoxySL downstream metabolites that changed between the 0 and 48 h time points. We used the metabolic profiling software “Sieve” to perform differential analysis of the mass spectra. This identified 312 components that showed significant differences. Of them, 56 remained valid after applying further filtering criteria, as described in the Materials and Methods section. The three components that showed the greatest change over time (NRatio with NPvalue < 0.05) had odd-numbered masses, as would be expected for a d3-labeled deoxysphingoid base (Fig. 2A).
Fig. 2.
Identification of eight novel 1-deoxySL metabolites that increase over time. A: Differential analysis using Sieve was performed for all metabolites between the 0 and 48 h time points of the chase in unsupplemented growth medium, after a 2 h pulse with d3-deoxySA. The volcano plot indicates all components identified with an m/z between 400 and 510, ratio >1, and P value <0.02. The P value and ratio shown are after normalization to the spiked internal standard d7-SA. The three black dots in the upper left indicate the metabolites with the greatest change in levels (these also had odd masses that could have a d3 label). B: Extracted ion chromatograms from the 48 h time point of the masses that passed the applied filtering criteria, as well as an additional mass (497.34838) identified by visual scanning of the total ion chromatogram. The structures were deduced after generating the chemical formula from the accurate mass (the conjugated OPA group is not shown for clarity). Note that the exact positions of the OH groups (except at C3) and the DBs along the carbon chain are unknown. The retention time of each metabolite on a C18 reverse phase column is indicated. The labeling of the individual 1-deoxySO-OH (I and II) and 1-deoxySA-2OH (I, II, III, and IV) peaks refers to the different retention times. C: Levels of all of the newly identified d3-labeled metabolites increased over the 48 h chase period. Data are represented as mean ± SEM.
In addition, the total-ion chromatogram was visually screened, leading to the identification of five additional d3-labeled 1-deoxySL metabolites (Fig. 2B). From the accurate mass (5 ppm) and the constraint that the molecule should contain a sulfur group originating from the o-phthalaldehyde/β-mercaptoethanol derivatization, a specific chemical composition could be assigned to each of the eight metabolites. The identified compounds were hydroxyl-deoxySA (1-deoxySA-OH), hydroxyl-deoxySO I and II (1-deoxySO-OH; same exact mass, but different retention times), deoxysphingadiene (1-deoxySAdiene), and four isobaric peaks corresponding to dihydroxyl-deoxySA (1-deoxySA-2OH) (see Fig. 2B for putative structures).
The putative molecular formulas were further validated by comparing the retention times of the identified metabolites. The addition of hydroxyl groups and DBs decrease hydrophobicity, meaning that these compounds would elute earlier from the column in reverse-phase LC (25, 26). Aligning the identified metabolites by their putative number of hydroxyl groups, one would predict an elution in the following order: 1-deoxySA-2OH (3 OHs), 1-deoxySO-OH (2OHs, 1 DB), 1-deoxySA-OH (2 OHs), 1-deoxySAdiene (1 OH, 2 DBs), 1-deoxySO (1 OH, 1 DB), 1-deoxySA (1 OH). This exactly reflects the observed elution order of the identified molecules (Fig. 2B).
All the newly identified downstream metabolites increased over the 48 h of the chase (Fig. 2C). To assess whether the same downstream metabolites are also formed when 1-deoxySA is synthesized de novo, instead of being supplemented to the cells, we treated cells expressing the SPTLC1p.C133W HSAN1 mutant with ω-d3-labeled palmitic acid and/or d4-alanine. SPTLC1p.C133W is the most common mutation in HSAN1 and cells expressing this mutant show a significant increase in 1-deoxySL formation. The downstream metabolites were identified and quantified after acid/base hydrolysis according to the +3 mass differences arising either from the ω-d3-palmitic acid or the d4-alanine, as well as the +6 mass differences resulting from the conjugation of both labeled substrates. All identified 1-deoxySL downstream metabolites were formed from the labeled palmitic acid and/or labeled alanine (supplemental Fig. S2).
The metabolic conversion of 1-deoxySLs is slow compared with canonical SLs
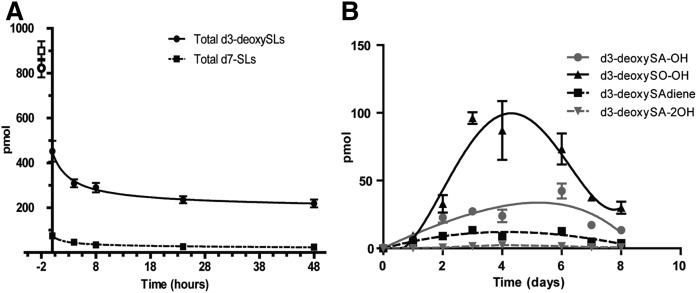
To compare the degradation of 1-deoxySLs and canonical SLs, we performed a 2 h pulse with either 1 μM d3-deoxySA or 1 μM d7-SA, followed by a 48 h chase. Labeled sphingoid and 1-deoxysphingoid bases were quantified by LC/MS after acid/base hydrolysis. The d7-labeled sphingoid bases disappeared almost completely during the 48 h chase, whereas the decrease in d3-deoxySLs (including all of the newly identified metabolites) was much less pronounced (Fig. 3A). Also, while the same amount of d7-SA and d3-deoxySA was added, the concentration of d7-labeled sphingoid bases compared with the d3-labeled deoxysphingoid bases was about 80% lower at the beginning of the chase (t = 0 h) (Fig. 3A). This indicates that a great proportion of the supplemented d7-SA was already metabolized within the 2 h of the pulse.
Fig. 3.
The metabolic conversion of 1-deoxySLs is slower than for canonical SLs. A: MEFs were treated with either 1 μM d7-SA or d3-deoxySA for a 2 h pulse, followed by a chase period in unsupplemented growth medium up to 48 h. The pulse medium was also collected and the d7-SA and d3-deoxySA content was measured. The total d3-labeled 1-deoxySLs (including the new metabolites) decreased over the chase period, although less than the d7-labeled canonical SLs did. While the total d7-labeled SLs were already much lower at time point 0, this was not a function of availability of the labeled lipids, as can be seen by the points at −2 h, which represent the total amount of d3-deoxySA and d7-SA in the pulse medium. B: MEFs were treated with a 2 h pulse of d3-deoxySA, followed by a chase period up to 8 days in unsupplemented growth medium. All of the d3-deoxySL metabolites increased until 4–6 days, and then drastically decreased by the final time point at 8 days. Data are represented as mean ± SEM.
To determine whether the newly identified 1-deoxySL downstream metabolites are converted further, we performed an extended pulse-chase experiment over a period of 8 days. During this long-term chase, the newly identified downstream metabolites increased within the first 4–6 days and then declined again (Fig. 3B).
The novel metabolites are all downstream of 1-deoxySO and form a branched pathway
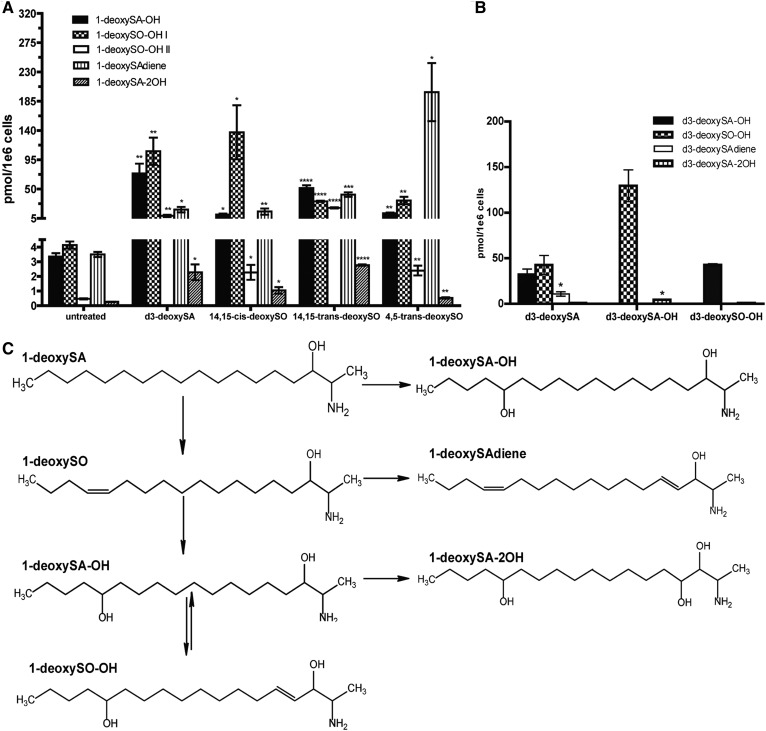
As demonstrated recently, the 1-deoxySO that is the most abundant in a variety of cell types as well as in human plasma bears a Δ14,15-cis DB in contrast to the Δ4,5-trans DB of canonical sphingosine (20). To elucidate the order in which the newly identified metabolites are formed and to determine where they integrate into the 1-deoxySL metabolic pathway, we treated MEF cells for 48 h with either d3-deoxySA, native Δ14,15-cis-deoxySO, or two synthetic 1-deoxySO isomers (Δ4,5-trans and Δ14,15-trans). For d3-deoxySA-treated cells, the d3-labeled metabolites were quantified, while in the 1-deoxySO-treated cells, the unlabeled metabolites were quantified (after acid/base hydrolysis). The levels of 1-deoxySAdiene, 1-deoxySA-OH, 1-deoxySO-OH, and 1-deoxySA-2OH all significantly increased after adding Δ14,15-cis-deoxySO, indicating that all of these metabolites are formed downstream of 1-deoxySO (Fig. 4A). Also, treatment with the synthetic isomers Δ4,5-trans-deoxySO and Δ14,15-trans-deoxySO significantly increased the levels of all downstream metabolites, suggesting that the responsible enzymes are not very substrate specific (Fig. 4A). However, the relative proportion of the formed downstream metabolites differed between treatments. Cells treated with Δ4,5-trans-deoxySO formed approximately 10-fold more 1-deoxySAdiene compared with cells treated with d3-deoxySA, Δ14,15-trans-deoxySO, or Δ14,15-cis-deoxySO (Fig. 4A).
Fig. 4.
All identified 1-deoxySL metabolites are produced downstream of 1-deoxySO and form a branched pathway. A: MEF cells were treated with d3-deoxySA, 14,15-cis-1-deoxySO, 14,15-trans-1-deoxySO, and 4,5-trans-1-deoxySO for 48 h (indicated along the x axis) and the resulting 1-deoxySL downstream metabolites were measured by LC/MS (d3-labeled for the d3-deoxySA treated condition and unlabeled for the other conditions; indicated by the different patterns). All metabolites were significantly increased compared with untreated controls upon treatment with any of the 1-deoxySO isomers. Statistical significance was calculated for individual metabolites arising from each treatment versus untreated controls using the Student’s t-test. B: MEF cells were treated with isolated d3-deoxySA, d3-deoxySA-OH, and d3-deoxySO-OH (a mix of both I and II; indicated along the x axis) for 48 h, and the resulting downstream d3-labeled 1-deoxySL metabolites were measured by LC/MS (indicated by the different patterns). The amounts of each downstream metabolite produced from the different treatments were compared in pairs and significance was calculated using the Student’s t-test. C: Proposed metabolic pathway order. All bars represent averages ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
In addition, we supplemented cells with native d3-deoxySA-OH and d3-deoxySO-OH (I + II), isolated by LC fractionation after a prolonged treatment with d3-deoxySA. The addition of d3-deoxySA-OH led to an increase in d3-deoxySO-OH (I + II), and conversely, adding d3-deoxySO-OH increased the levels of d3-deoxySA-OH, indicating that this reaction is reversible (Fig. 4B). In contrast, d3-deoxySA-2OH (sum of all four) was formed in the largest quantities after treatment with d3-deoxySA-OH, denoting that the di-hydroxylated form is likely the direct product of d3-deoxySA-OH (Fig. 4B). However, 1-deoxySAdiene was only formed when supplementing the cells with d3-deoxySA or the different 1-deoxySO standards, but not with the fractionated d3-deoxySA-OH or d3-deoxySO-OH (I + II). This indicates that 1-deoxySAdiene is formed directly downstream of 1-deoxySO (Fig. 4A, B).
In summary, all eight metabolites were formed downstream of 1-deoxySO, constituting three metabolic branches (Fig. 4C). It appears that 1-deoxySA-OH can also be formed directly from 1-deoxySA, as levels were higher when cells were supplemented with d3-deoxySA than with 1-deoxySO.
The downstream metabolism of 1-deoxySO is mediated by CYP4F enzymes
The observation that 1-deoxySLs are converted into several mono- or poly-hydroxylated metabolites indicates a role of hydroxylases in this process. We hypothesized that CYP enzymes could be involved in the downstream metabolism of 1-deoxySLs, as these enzymes are hydroxylases of both endogenous and exogenous substrates. In particular, we considered the CYP4A and CYP4F sub-families, which are involved in fatty acid hydroxylation, as their activity is enhanced by peroxisome proliferators like fibrates (27, 28), and fenofibrate has been shown to lower 1-deoxySA and Δ14,15-cis-deoxySO levels in the plasma of dyslipidemic patients (29). Thus, we inferred that members of the CYP4A or CYP4F subfamilies could be involved in the downstream metabolism of 1-deoxySLs.
To test whether PPARα activation stimulates the downstream metabolism of 1-deoxySLs, we treated MEF cells with d3-deoxySA and fenofibrate (20 μM) for 24 h and measured the levels of all d3-labeled 1-deoxySL metabolites (after acid/base hydrolysis). Fenofibrate increased the formation of all labeled downstream metabolites, although only the increase in d3-deoxySA-OH, d3-deoxySO-OH II, and d3-deoxySAdiene was statistically significant (supplemental Fig. S3).
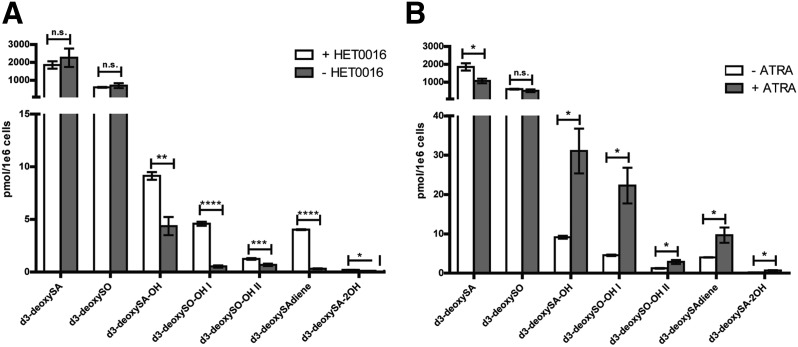
To further test the hypothesis that CYP4A or CYP4F enzymes are involved in 1-deoxySL metabolism, we supplemented cells for 24 h with d3-deoxySA and HET0016 (5 μM), a pan inhibitor of the CYP4A/CYP4F subfamilies (30, 31). The presence of HET0016 significantly decreased the levels of all downstream metabolites in a dose-dependent manner (Fig. 5A, supplemental Fig. S4A). Furthermore, in pulse-chase experiments, d3-deoxySO no longer decreased after 24 h in the presence of the inhibitor, but instead continued to increase (supplemental Fig. S4B).
Fig. 5.
Inhibition of CYP4F enzymes decreases the formation of the downstream d3-deoxySL metabolites, while induction of these enzymes increases their levels. Cells were treated with d3-deoxySA (1 μM) and either the inhibitor HET0016 (5 μM) or ATRA (20 μM) for 24 h, after which the d3-deoxySLs were extracted and quantified by LC/MS. A: While the differences in d3-deoxySA and d3-deoxySO were not significant between controls and HET0016 treatment, the downstream d3-deoxySL metabolites were significantly decreased in the presence of the inhibitor. B: The d3-deoxySA was significantly lower in the ATRA-treated cells, while the downstream metabolites were all significantly and drastically increased. Statistical significance of the differences between untreated and treated cells was calculated using the Student’s t-test. Bars represent averages ± SEM. *P < 0.05, **P < 0.01; n.s., not significant. See also supplemental Figs. S3, S4.
As HET0016 inhibits both CYP4A and CYP4F enzymes, we treated cells with all-trans retinoic acid (ATRA; 20 μM), which preferentially induces the expression of CYP4F genes (32–34). ATRA treatment significantly increased all the d3-deoxySL downstream metabolites. In parallel, levels of d3-deoxySA were significantly lower with ATRA treatment, likely because of the increased downstream conversion (Fig. 5B).
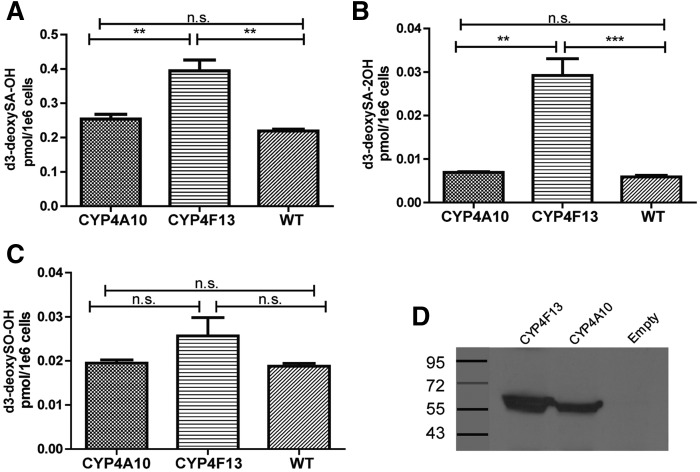
Finally, to further analyze the role of specific CYP4A and CYP4F enzymes, we overexpressed mouse Cyp4a10 or Cyp4f13 in HEK cells, and then treated cells with d3-deoxySA. Overexpression of Cyp4a10 did not affect levels of the downstream metabolites, whereas overexpression of Cyp4f13 increased d3-deoxySA-OH and d3-deoxySA-2OH more than 2-fold as compared with wild-type cells (Fig. 6).
Fig. 6.
Levels of d3-deoxySA-OH and d3-deoxySA-2OH significantly increase upon overexpression of Cyp4f13. HEK WT cells or HEK cells overexpressing either mouse Cyp4a10 or Cyp4f`13 were treated with d3-deoxySA for 48 h, and the d3-labeled 1-deoxySL downstream metabolites were measured by LC/MS. Levels of d3-deoxySA-OH (A) and d3-deoxySA-2OH (B) were significantly higher in Cyp4f13-overexpressing cells. Levels of d3-deoxySO-OH (C) did not differ between the three cell lines. The sum of all four d3-deoxySA-2OH peaks and both d3-deoxySO-OH peaks is shown. D: Western blot of CYP4F13 and CYP4A10 expression. Bars represent averages ± SEM. Statistical significance was calculated using one-way ANOVA followed by Bonferroni correction. **P < 0.01, ***P < 0.001; n.s., not significant.
DISCUSSION
Current belief holds that 1-deoxySLs are metabolic “dead-end” products that cannot be degraded by the canonical SL catabolic pathway due to their missing C1 hydroxyl group. Here, we showed that, instead, 1-deoxySLs are metabolized by CYP4F enzymes.
We identified eight novel 1-deoxySL downstream metabolites. The enzymes involved in the downstream metabolism of 1-deoxySLs appear not to have a high specificity, as nonnatural 1-deoxySO analogs were also metabolized (Fig. 3). In contrast to canonical SO, which bears a Δ4-5-trans DB, the most abundant native 1-deoxySO contains a Δ14,15-cis DB (20), suggesting that the DB in 1-deoxySO is introduced by a desaturase different to DES1. The identity of this enzyme has not yet been elucidated, but interestingly, treating cells with synthetic Δ4,5-trans-1-deoxySO strongly stimulated the formation of 1-deoxySAdiene in comparison to treatment with the Δ14,15-cis-deoxySO isomer. This indicates that the same enzyme that introduces the Δ14,15-cis DB in 1-deoxySA is also responsible for forming 1-deoxySAdiene from Δ4,5-trans-deoxySO. Of note, sphingadiene, a fairly abundant canonical C18-SL, also contains both a Δ4,5-trans and a Δ14,15-cis DB (35, 36).
The pathways downstream of 1-deoxySO appear to be highly branched and could be part of a cellular detoxification/elimination pathway that typically involves CYP enzymes (37). Furthermore, compared with canonical SL catabolism, which occurs within minutes to hours, the conversion of 1-deoxySLs is slow and takes place over several days, which is also true of the detoxification process. Detoxification usually occurs by two sequential steps referred to as phase I and phase II. In phase I, CYP enzymes typically hydroxylate compounds to increase polarity; whereas in phase II, hydrophilic moieties like glucuronic acid or sulfate esters are conjugated to further increase water solubility (37). The 1-deoxySLs have been shown to be cytotoxic in a variety of cell types (5, 38–40), including cultured primary chicken dorsal root ganglia neurons (4) and pancreatic β-cells (15), as well as to produce a neurotoxic phenotype in vivo (41, 42). Therefore, it is feasible that hydroxylation is a physiological way to prevent the toxic accumulation of these lipids. Initial attempts to identify glucuronidated or sulfonated 1-deoxySL downstream products were not successful, which may be due to low abundance and/or the quick elimination of these metabolites. However, it is likely that this metabolic conversion is the reason we see the decline in all of the metabolites after 4 days, and that these glucuronidated or sulfonated products make up the remaining missing d3 label not accounted for by the newly identified metabolites. In the future, identification of glucuronidated or sulfonated metabolites might be better possible in urine of HSAN1 patients, which would be expected to contain a significant concentration of these 1-deoxySL metabolites. Unfortunately, urine from HSAN1 patients was not available at this time.
With regard to the potential role of CYP enzymes in this process, we specifically focused on the CYP4A and CYP4F subfamilies, which are known to be fatty acid hydroxylases and are moderately-to-highly expressed in MEF cells (43). The activity of CYP4A and CYP4F enzymes can be enhanced by peroxisome proliferators like fibrates (27, 28), and fenofibrate has been shown to lower 1-deoxySA and Δ14,15-cis-deoxySO levels in the plasma of dyslipidemic patients (29). Thus, we inferred that CYP4A or CYP4F could potentially be involved in the downstream metabolism of 1-deoxySLs. However, while fibrates strongly induce CYP4A expression, they regulate CYP4F enzymes in an isoform-, tissue-, and species-specific manner, which could explain why fenofibrate had a weaker effect in MEFs compared with ATRA (supplemental Fig. S3) (44–46). On the other hand, ATRA has been shown to be a strong inducer of CYP4Fs (32, 33), while it suppressed expression of CYP4A11 (34). Upon treatment with ATRA, we observed a more than 2-fold increase in the downstream metabolites, suggesting that CYP4F rather than CYP4A enzymes are involved. This is further supported by our finding that overexpression of Cyp4f13, but not of Cyp4a10, increased the production of two of the downstream metabolites in HEK293 cells. Pan inhibition of CYP4A/CYP4F enzymes prevented the downstream metabolism of 1-deoxySO.
Interestingly, CYP4A and CYP4F genes are differentially regulated by conditions that increase the flux of free fatty acids, such as fasting (47), high-fat diet (48), or T2D (49). CYP4A expression is increased under these conditions, whereas CYP4F expression is decreased. A 60- to 700-fold increase in mouse Cyp4a mRNA was observed during fasting (50), while Cyp4f14 was found to be highly and significantly downregulated under these conditions (51). In mice on a high-fat diet, Cyp4a mRNA was increased 2.5- to 100-fold, while Cyp4f genes were downregulated by 20–80% (28). Preliminary data from Hardwick et al. (52) suggest that Cyp4f genes are also downregulated in an ob/ob mouse model of fatty liver disease. The downregulation of CYP4F enzymes in metabolic conditions has interesting implications, as it suggests that reduced degradation rather than increased formation is the reason for the elevated 1-deoxySL levels seen in MetS and T2D.
CYP4F enzymes have been shown to mediate the degradation of fingolimod (FTY720), an S1P analog used in treating relapsing multiple sclerosis (53). Its elimination occurs primarily by hydroxylation mediated by CYP4F2 and possibly other enzymes of the CYP4F subfamily. Although ω-hydroxylated fingolimod was the major biotransformation product observed, six other metabolites with internal hydroxylations were identified (53). This goes along with our identification of multiple isomers of both 1-deoxySO-OH and 1-deoxySA-2OH. Furthermore, although the main function of CYP enzymes is hydroxylation, they can also act as desaturases (54), which might explain the formation of the 1-deoxySAdiene metabolites.
Considering the neuro- and β-cell toxicity of 1-deoxySLs, it will be important to elucidate ways to lower these lipids in the context of neuropathies and prediabetes. Oral L-serine supplementation was demonstrated to efficiently suppress 1-deoxySL formation and is currently being tested as a treatment in HSAN1 (https://clinicaltrials.gov/ct2/show/NCT01733407). The positive effects of lowering 1-deoxySLs on peripheral nerve function was demonstrated in transgenic HSAN1 mice and HSAN1 patients (11), as well as in a diabetic rat model (12). In all three instances, 1-deoxySL levels decreased while mechanical sensitivity and nerve function improved. The induction of CYP4F enzymes could therefore present a novel therapeutic approach for lowering 1-deoxySLs. This would be synergistic to serine supplementation, which blocks 1-deoxySL de novo synthesis.
In conclusion, neurotoxic and β-cell toxic 1-deoxySLs are not catabolized by the canonical SL pathway, but by several hydroxylation and desaturation reactions that appear to be primarily mediated by CYP4F enzymes. Increased 1-deoxySA and 1-deoxySO levels in patients with MetS and T2D might therefore be explained by a downregulation of these enzymes. Consequently, induction of these enzymes could be a new therapeutic strategy in T2D, DSN, and HSAN1.
Supplementary Material
Footnotes
Abbreviations:
- ATRA
- all-trans retinoic acid
- CYP
- cytochrome P450
- DB
- double bond
- 1-deoxySA
- 1-deoxysphinganine
- 1-deoxySAdiene
- deoxysphingadiene
- 1-deoxySA-OH
- hydroxyl-deoxysphinganine
- 1-deoxySA-2OH
- dihydroxyl-deoxysphinganine
- 1-deoxySL
- 1-deoxysphingolipid
- 1-deoxySO
- 1-deoxysphingosine
- 1-deoxySO-OH
- hydroxyl-deoxysphingosine
- DSN
- diabetic sensory polyneuropathy
- HSAN1
- hereditary sensory autonomic neuropathy type 1
- MEF
- mouse embryonic fibroblast
- MetS
- metabolic syndrome
- P/S
- penicillin/streptomycin
- SA
- sphinganine
- SL
- sphingolipid
- SO
- sphingosine
- S1P
- sphingosine-1-phosphate
- SPT
- serine palmitoyltransferase
This work was supported by the European Commission Seventh Framework Programme (Project 305707), the Swiss National Science Foundation (Project 31003A_153390/1), the Hurka Foundation, the Novartis Foundation, and the Rare Disease Initiative Zurich (“radiz”, Clinical Research Priority Program for Rare Diseases, University of Zurich) (I.A., A.O., T.H., and A.v.E.). Other support was provided by the Deutsche Forschungsgemeinschaft (SFB645 and TRR83) (A.O.).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Weiss B., and Stoffel W.. 1997. Human and murine serine-palmitoyl-CoA transferase–cloning, expression and characterization of the key enzyme in sphingolipid synthesis. Eur. J. Biochem. 249: 239–247. [DOI] [PubMed] [Google Scholar]
- 2.Hanada K., Hara T., Nishijima M., Kuge O., Dickson R. C., and Nagiec M. M.. 1997. A mammalian homolog of the yeast LCB1 encodes a component of serine palmitoyltransferase, the enzyme catalyzing the first step in sphingolipid synthesis. J. Biol. Chem. 272: 32108–32114. [DOI] [PubMed] [Google Scholar]
- 3.Buede R., Rinkerschaffer C., Pinto W. J., Lester R. L., and Dickson R. C.. 1991. Cloning and characterization of Lcb1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J. Bacteriol. 173: 4325–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penno A., Reilly M. M., Houlden H., Laura M., Rentsch K., Niederkofler V., Stoeckli E. T., Nicholson G., Eichler F., Brown R. H. Jr., et al. . 2010. Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J. Biol. Chem. 285: 11178–11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zitomer N. C., Mitchell T., Voss K. A., Bondy G. S., Pruett S. T., Garnier-Amblard E. C., Liebeskind L. S., Park H., Wang E., Sullards M. C., et al. . 2009. Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxysphinganine: a novel category of bioactive 1-deoxysphingoid bases and 1-deoxydihydroceramides biosynthesized by mammalian cell lines and animals. J. Biol. Chem. 284: 4786–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Othman A., Saely C. H., Muendlein A., Vonbank A., Drexel H., von Eckardstein A., and Hornemann T.. 2015. Plasma 1-deoxysphingolipids are predictive biomarkers for type 2 diabetes mellitus. BMJ Open Diabetes Res. Care. 3: e000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callaghan B. C., Cheng H. L. T., Stables C. L., Smith A. L., and Feldman E. L.. 2012. Diabetic neuropathy: clinical manifestations and current treatments. Lancet Neurol. 11: 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dohrn M. F., Othman A., Hirshman S. K., Bode H., Alecu I., Fahndrich E., Karges W., Weis J., Schulz J. B., Hornemann T., et al. 2015. Elevation of plasma 1-deoxy-sphingolipids in type 2 diabetes mellitus: a susceptibility to neuropathy? Eur. J. Neurol. 22: 806–814. [DOI] [PubMed] [Google Scholar]
- 9.Bejaoui K., Uchida Y., Yasuda S., Ho M., Nishijima M., Brown R. H., Holleran W. M., and Hanada K.. 2002. Hereditary sensory neuropathy type 1 mutations confer dominant negative effects on serine palmitoyltransferase, critical for sphingolipid synthesis. J. Clin. Invest. 110: 1301–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawkins J. L., Hulme D. J., Brahmbhatt S. B., Auer-Grumbach M., and Nicholson G. A.. 2001. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat. Genet. 27: 309–312. [DOI] [PubMed] [Google Scholar]
- 11.Garofalo K., Penno A., Schmidt B. P., Lee H. J., Frosch M. P., von Eckardstein A., Brown R. H., Hornemann T., and Eichler F. S.. 2011. Oral L-serine supplementation reduces production of neurotoxic deoxysphingolipids in mice and humans with hereditary sensory autonomic neuropathy type 1. J. Clin. Invest. 121: 4735–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Othman A., Bianchi R., Alecu I., Wei Y., Porretta-Serapiglia C., Lombardi R., Chiorazzi A., Meregalli C., Oggioni N., Cavaletti G., et al. . 2015. Lowering plasma 1-deoxysphingolipids improves neuropathy in diabetic rats. Diabetes. 64: 1035–1045. [DOI] [PubMed] [Google Scholar]
- 13.Kramer R., Bielawski J., Kistner-Griffin E., Othman A., Alecu I., Ernst D., Kornhauser D., Hornemann T., and Spassieva S.. 2015. Neurotoxic 1-deoxysphingolipids and paclitaxel-induced peripheral neuropathy. FASEB J. 29: 4461–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorden D. L., Myers D. S., Ivanova P. T., Fahy E., Maurya M. R., Gupta S., Min J., Spann N. J., McDonald J. G., Kelly S. L., et al. . 2015. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J. Lipid Res. 56: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuellig R. A., Hornemann T., Othman A., Hehl A. B., Bode H., Guntert T., Ogunshola O. O., Saponara E., Grabliauskaite K., Jang J. H., et al. . 2014. Deoxysphingolipids, novel biomarkers for type 2 diabetes, are cytotoxic for insulin-producing cells. Diabetes. 63: 1326–1339. [DOI] [PubMed] [Google Scholar]
- 16.Humpf H. U., Schmelz E. M., Meredith F. I., Vesper H., Vales T. R., Wang E., Menaldino D. S., Liotta D. C., and Merrill A. H. Jr. 1998. Acylation of naturally occurring and synthetic 1-deoxysphinganines by ceramide synthase. Formation of N-palmitoyl-aminopentol produces a toxic metabolite of hydrolyzed fumonisin, AP1, and a new category of ceramide synthase inhibitor. J. Biol. Chem. 273: 19060–19064. [DOI] [PubMed] [Google Scholar]
- 17.Abad J. L., Nieves I., Rayo P., Casas J., Fabrias G., and Delgado A.. 2013. Straightforward access to spisulosine and 4,5-dehydrospisulosine stereoisomers: probes for profiling ceramide synthase activities in intact cells. J. Org. Chem. 78: 5858–5866. [DOI] [PubMed] [Google Scholar]
- 18.Ternes P., Franke S., Zahringer U., Sperling P., and Heinz E.. 2002. Identification and characterization of a sphingolipid Delta 4-desaturase family. J. Biol. Chem. 277: 25512–25518. [DOI] [PubMed] [Google Scholar]
- 19.Michel C., G. van Echten-Deckert, J. Rother, K. Sandhoff, E. Wang, and A. H. Merrill. 1997. Characterization of ceramide synthesis. A dihydroceramide desaturase introduces the 4,5-trans-double bond of sphingosine at the level of dihydroceramide. J. Biol. Chem. 272: 22432–22437. [DOI] [PubMed] [Google Scholar]
- 20.Steiner R., Saied E. M., Othman A., Arenz C., Maccarone A. T., Poad B. L., Blanksby S. J., von Eckardstein A., and Hornemann T.. 2016. Elucidating the chemical structure of native 1-deoxysphingosine. J. Lipid Res. 57: 1194–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang E., Norred W. P., Bacon C. W., Riley R. T., and Merrill A. H.. 1991. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 266: 14486–14490. [PubMed] [Google Scholar]
- 22.Van Veldhoven P. P., Gijsbers S., Mannaerts G. P., Vermeesch J. R., and Brys V.. 2000. Human sphingosine-1-phosphate lyase: cDNA cloning, functional expression studies and mapping to chromosome 10q22. Biochim. Biophys. Acta. 1487: 128–134. [DOI] [PubMed] [Google Scholar]
- 23.Riley R. T., Norred W. P., Wang E., and Merrill A. H.. 1999. Alteration in sphingolipid metabolism: bioassays for fumonisin- and ISP-I-like activity in tissues, cells and other matrices. Nat. Toxins. 7: 407–414. [DOI] [PubMed] [Google Scholar]
- 24.Snyder N. W., Khezam M., Mesaros C. A., Worth A., and Blair I. A.. 2013. Untargeted metabolomics from biological sources using ultraperformance liquid chromatography-high resolution mass spectrometry (UPLC-HRMS). J. Vis. Exp. 75: e50433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jungalwala F. B., Hayssen V., Pasquini J. M., and McCluer R. H.. 1979. Separation of molecular species of sphingomyelin by reversed-phase high-performance liquid chromatography. J. Lipid Res. 20: 579–587. [PubMed] [Google Scholar]
- 26.Vollhardt K. P. C., and Schore N. E.. 2007. Organic Chemistry: Structure and Function. W.H. Freeman, New York. [Google Scholar]
- 27.Park E. C., Kim S. I., Hong Y., Hwang J. W., Cho G. S., Cha H. N., Han J. K., Yun C. H., Park S. Y., Jang I. S., et al. . 2014. Inhibition of CYP4A reduces hepatic endoplasmic reticulum stress and features of diabetes in mice. Gastroenterology. 147: 860–869. [DOI] [PubMed] [Google Scholar]
- 28.Hardwick J. P. 2008. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem. Pharmacol. 75: 2263–2275. [DOI] [PubMed] [Google Scholar]
- 29.Othman A., Benghozi R., Alecu I., Wei Y., Niesor E., von Eckardstein A., and Hornemann T.. 2015. Fenofibrate lowers atypical sphingolipids in plasma of dyslipidemic patients: A novel approach for treating diabetic neuropathy? J. Clin. Lipidol. 9: 568–575. [DOI] [PubMed] [Google Scholar]
- 30.Sato M., Ishii T., Kobayashi-Matsunaga Y., Amada H., Taniguchi K., Miyata N., and Kameo K.. 2001. Discovery of a N′-hydroxyphenylformamidine derivative HET0016 as a potent and selective 20-HETE synthase inhibitor. Bioorg. Med. Chem. Lett. 11: 2993–2995. [DOI] [PubMed] [Google Scholar]
- 31.Miyata N., Taniguchi K., Seki T., Ishimoto T., Sato-Watanabe M., Yasuda Y., Doi M., Kametani S., Tomishima Y., Ueki T., et al. . 2001. HET0016, a potent and selective inhibitor of 20-HETE synthesizing enzyme. Br. J. Pharmacol. 133: 325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., and Hardwick J. P.. 2000. Regulation of CYP4F2 leukotriene B4 omega-hydroxylase by retinoic acids in HepG2 cells. Biochem. Biophys. Res. Commun. 279: 864–871. [DOI] [PubMed] [Google Scholar]
- 33.Kikuta Y., Yamashita Y., Kashiwagi S., Tani K., Okada K., and Nakata K.. 2004. Expression and induction of CYP4F subfamily in human leukocytes and HL60 cells. Biochim. Biophys. Acta. 1683: 7–15. [DOI] [PubMed] [Google Scholar]
- 34.Antoun J., Amet Y., Simon B., Dreano Y., Corlu A., Corcos L., Salaun J. P., and Plee-Gautier E.. 2006. CYP4A11 is repressed by retinoic acid in human liver cells. FEBS Lett. 580: 3361–3367. [DOI] [PubMed] [Google Scholar]
- 35.Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., et al. . 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renkonen O., and Hirvisalo E. L.. 1969. Structure of plasma sphingadienine. J. Lipid Res. 10: 687–693. [PubMed] [Google Scholar]
- 37.Foye W. O., Lemke T. L., and Williams D. A.. 2013. Foye’s Principles of Medicinal Chemistry. 7th edition. Chapter 4. T. L. Lemke and D. A. Williams, editors. Lippincott Williams and Wilkins/Wolters Kluwer Health, Philadelphia, PA. [Google Scholar]
- 38.Sánchez A. M., Malagarie-Cazenave S., Olea N., Vara D., Cuevas C., and Díaz-Laviada I.. 2008. Spisulosine (ES-285) induces prostate tumor PC-3 and LNCaP cell death by de novo synthesis of ceramide and PKCzeta activation. Eur. J. Pharmacol. 584: 237–245. [DOI] [PubMed] [Google Scholar]
- 39.Salcedo M., Cuevas C., Alonso J. L., Otero G., Faircloth G., Fernandez-Sousa J. M., Avila J., and Wandosell F.. 2007. The marine sphingolipid-derived compound ES 285 triggers an atypical cell death pathway. Apoptosis. 12: 395–409. [DOI] [PubMed] [Google Scholar]
- 40.Cuadros R., Montejo de Garcini E., Wandosell F., Faircloth G., Fernandez-Sousa J. M., and Avila J.. 2000. The marine compound spisulosine, an inhibitor of cell proliferation, promotes the disassembly of actin stress fibers. Cancer Lett. 152: 23–29. [DOI] [PubMed] [Google Scholar]
- 41.Baird R. D., Kitzen J., Clarke P. A., Planting A., Reade S., Reid A., Welsh L., Lopez Lazaro L., de las Heras B., Judson I. R., et al. . 2009. Phase I safety, pharmacokinetic, and pharmacogenomic trial of ES-285, a novel marine cytotoxic agent, administered to adult patients with advanced solid tumors. Mol. Cancer Ther. 8: 1430–1437. [DOI] [PubMed] [Google Scholar]
- 42.Schöffski P., Dumez H., Ruijter R., Miguel-Lillo B., Soto-Matos A., Alfaro V., and Giaccone G.. 2011. Spisulosine (ES-285) given as a weekly three-hour intravenous infusion: results of a phase I dose-escalating study in patients with advanced solid malignancies. Cancer Chemother. Pharmacol. 68: 1397–1403. [DOI] [PubMed] [Google Scholar]
- 43.Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., and Zimmermann P.. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawashima H., Kusunose E., Thompson C. M., and Strobel H. W.. 1997. Protein expression, characterization, and regulation of CYP4F4 and CYP4F5 cloned from rat brain. Arch. Biochem. Biophys. 347: 148–154. [DOI] [PubMed] [Google Scholar]
- 45.Sehgal N., Agarwal V., Valli R. K., Joshi S. D., Antonovic L., Strobel H. W., and Ravindranath V.. 2011. Cytochrome P4504f, a potential therapeutic target limiting neuroinflammation. Biochem. Pharmacol. 82: 53–64. [DOI] [PubMed] [Google Scholar]
- 46.Cui X., Kawashima H., Barclay T. B., Peters J. M., Gonzalez F. J., Morgan E. T., and Strobel H. W.. 2001. Molecular cloning and regulation of expression of two novel mouse CYP4F genes: expression in peroxisome proliferator-activated receptor alpha-deficient mice upon lipopolysaccharide and clofibrate challenges. J. Pharmacol. Exp. Ther. 296: 542–550. [PubMed] [Google Scholar]
- 47.Corton J. C., Apte U., Anderson S. P., Limaye P., Yoon L., Latendresse J., Dunn C., Everitt J. I., Voss K. A., Swanson C., et al. . 2004. Mimetics of caloric restriction include agonists of lipid-activated nuclear receptors. J. Biol. Chem. 279: 46204–46212. [DOI] [PubMed] [Google Scholar]
- 48.Neat C. E., Thomassen M. S., and Osmundsen H.. 1981. Effects of high-fat diets on hepatic fatty acid oxidation in the rat. Isolation of rat liver peroxisomes by vertical-rotor centrifugation by using a self-generated, iso-osmotic, Percoll gradient. Biochem. J. 196: 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnett C. R., Gibson G. G., Wolf C. R., Flatt P. R., and Ioannides C.. 1990. Induction of cytochrome P450III and P450IV family proteins in streptozotocin-induced diabetes. Biochem. J. 268: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patsouris D., Reddy J. K., Muller M., and Kersten S.. 2006. Peroxisome proliferator-activated receptor alpha mediates the effects of high-fat diet on hepatic gene expression. Endocrinology. 147: 1508–1516. [DOI] [PubMed] [Google Scholar]
- 51.Bauer M., Hamm A. C., Bonaus M., Jacob A., Jaekel J., Schorle H., Pankratz M. J., and Katzenberger J. D.. 2004. Starvation response in mouse liver shows strong correlation with life-span-prolonging processes. Physiol. Genomics. 17: 230–244. [DOI] [PubMed] [Google Scholar]
- 52.Hardwick J. P., Osei-Hyiaman D., Wiland H., Abdelmegeed M. A., and Song B. J.. 2009. PPAR/RXR regulation of fatty acid metabolism and fatty acid omega-hydroxylase (CYP4) isozymes: implications for prevention of lipotoxicity in fatty liver disease. PPAR Res. 2009: 952734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin Y., Zollinger M., Borell H., Zimmerlin A., and Patten C. J.. 2011. CYP4F enzymes are responsible for the elimination of fingolimod (FTY720), a novel treatment of relapsing multiple sclerosis. Drug Metab. Dispos. 39: 191–198. [DOI] [PubMed] [Google Scholar]
- 54.Rettie A. E., Sheffels P. R., Korzekwa K. R., Gonzalez F. J., Philpot R. M., and Baillie T. A.. 1995. CYP4 isozyme specificity and the relationship between omega-hydroxylation and terminal desaturation of valproic acid. Biochemistry. 34: 7889–7895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.