Abstract
GPIHBP1, an endothelial cell protein, binds LPL in the interstitial spaces and shuttles it to its site of action inside blood vessels. For years, studies of human GPIHBP1 have been hampered by an absence of useful antibodies. We reasoned that monoclonal antibodies (mAbs) against human GPIHBP1 would be useful for 1) defining the functional relevance of GPIHBP1’s Ly6 and acidic domains to the binding of LPL; 2) ascertaining whether human GPIHBP1 is expressed exclusively in capillary endothelial cells; and 3) testing whether GPIHBP1 is detectable in human plasma. Here, we report the development of a panel of human GPIHBP1-specific mAbs. Two mAbs against GPIHBP1’s Ly6 domain, RE3 and RG3, abolished LPL binding, whereas an antibody against the acidic domain, RF4, did not. Also, mAbs RE3 and RG3 bound with reduced affinity to a mutant GPIHBP1 containing an Ly6 domain mutation (W109S) that abolishes LPL binding. Immunohistochemistry studies with the GPIHBP1 mAbs revealed that human GPIHBP1 is expressed only in capillary endothelial cells. Finally, we created an ELISA that detects GPIHBP1 in human plasma. That ELISA should make it possible for clinical lipidologists to determine whether plasma GPIHBP1 levels are a useful biomarker of metabolic or vascular disease
Keywords: chylomicrons, dyslipidemias, endothelial cells, lipoprotein lipase, triglycerides
LPL, a triglyceride hydrolase secreted by myocytes and adipocytes, is crucial for the lipolytic processing of triglyceride-rich lipoproteins inside blood vessels (1–3). For decades, the mechanism by which LPL reaches its site of action inside blood vessels was mysterious. However, we now know that GPIHBP1, a GPI-anchored protein of endothelial cells, binds LPL in the subendothelial spaces and transports it across endothelial cells to the capillary lumen (4, 5). GPIHBP1 is a member of the Ly6/uPAR protein family. The hallmark of this family is an ∼80–amino acid “Ly6 domain” containing 8 or 10 cysteines—all in a characteristic spacing pattern and all disulfide bonded so as to create a three-fingered fold (6). Unlike other Ly6 family members, GPIHBP1 contains an acidic domain at its amino terminus, with 21 of 26 consecutive residues in human GPIHBP1 being aspartate or glutamate (7). Surface plasmon resonance (SPR) studies with purified proteins have strongly suggested that GPIHBP1’s Ly6 domain is largely responsible for high-affinity interactions with LPL, whereas the acidic domain simply facilitates the initial binding event and stabilizes LPL activity (8). It would be desirable to confirm that finding by testing the capacity of monoclonal antibodies (mAbs) against GPIHBP1’s Ly6 and acidic domains to block the binding of LPL to GPIHBP1. Unfortunately, mAb tools for studying GPIHBP1 have been lacking.
Thus far, most of our understanding of GPIHBP1/LPL physiology has come from studies of mice, and the majority of those studies have relied on a rat mAb against mouse GPIHBP1, 11A12, that binds downstream from GPIHBP1’s Ly6 domain (9). Experiments with mAb 11A12 were essential for proving that GPIHBP1 transports LPL to the capillary lumen (4, 5, 10). Also, immunohistochemistry studies with mAb 11A12 showed that GPIHBP1 was expressed in endothelial cells of capillaries but not in endothelial cells of larger blood vessels (e.g., venules) (5). The majority of the LPL in mouse tissues was located on capillaries, mirroring the expression of GPIHBP1. Currently, it is unclear whether this peculiar pattern of GPIHBP1 expression (i.e., specificity for capillary endothelial cells) is unique to the mouse or is also found in humans. Unfortunately, monoclonal antibody 11A12 was not helpful for resolving this issue because it binds exclusively to mouse GPIHBP1.
In this study, our goal was to create high-affinity mAbs against human GPIHBP1—for three reasons. First, we wanted to generate mAbs against both the acidic and Ly6 domains of GPIHBP1 and then use the mAbs to elucidate the relevance of those domains for LPL binding. Second, we wanted to determine whether GPIHBP1 is expressed only in capillary endothelial cells in humans or whether it might be expressed more broadly in all endothelial cells. Studies of the domestic pig (11) and guinea pig (12) found LPL along endothelial cells of large blood vessels, raising the possibility that GPIHBP1 might be expressed in all endothelial cells in some mammalian species. Third, we wanted to determine whether GPIHBP1 is present in human plasma. We were not successful in detecting GPIHBP1 in mouse plasma by Western blotting, but we were intrigued that another GPI-anchored Ly6 protein, urokinase-type plasminogen activator receptor (uPAR), is easily detectable in human plasma with solid-phase immunoassays (ELISAs) (13). For that reason, we wanted to test whether an ELISA would be capable of detecting GPIHBP1 in human plasma. We reasoned that the development of a GPIHBP1 ELISA might allow clinical investigators to test whether GPIHBP1 levels are perturbed in the setting of metabolic or vascular diseases.
MATERIAL AND METHODS
GPIHBP1 expression vectors
The production of human GPIHBP1 in insect cells was described previously (14). Briefly, a secreted version of human GPIHBP1 with an N-terminal uPAR epitope tag (detectable by mAb R24) (15) and a carboxyl-terminal 11A12 epitope tag from the mouse GPIHBP1 sequence (9) was expressed in Drosophila S2 cells. The medium was concentrated 20-fold with Amicon Ultra 10k MWCO centrifugal filters (Millipore), and the concentration of human GPIHBP1 in the medium was determined by Western blotting using a highly purified GPIHBP1 standard (8). Point mutations in GPIHBP1 were introduced in expression vectors by PCR with the QuickChange Lightning kit (Agilent Technologies). Deletions were introduced by linearizing the wild-type expression vector by PCR (using 5′-phosphorylated primers), followed by ligation. Expression vectors for S-protein–tagged CD59 and a GPIHBP1–CD59 chimeric protein were described previously (16). The integrity of all vectors was confirmed by DNA sequencing.
Monoclonal antibodies
Mice were immunized intraperitoneally with purified full-length human GPIHBP1 (8). Antibody titers in the plasma of immunized mice were monitored by ELISA, and splenocytes were fused with Sp2/0-Ag14 myeloma cells. Hybridomas were grown under azaserine hypoxanthine selection, and ∼20,000 hybridoma supernatants were screened for high-affinity antibodies with a high-throughput antigen microarray and an ELISA. The top 24 clones were expanded and subcloned by serial dilution. Monoclonal antibodies were isotyped by commercially available assay kits (IsoStrip, Roche) and adapted to serum-free medium. Antibodies were purified from cell culture medium with a protein G-agarose column. All monoclonal antibodies are available upon request.
Western blots
Purified GPIHBP1 proteins or conditioned medium from GPIHBP1-expressing Drosophila S2 cells were size-fractioned on 12% Bis-Tris SDS-PAGE gels in MES buffer (Thermo Fisher Scientific). After transferring the proteins to a nitrocellulose membrane, the membrane was incubated with GPIHBP1-specific mAbs (4 μg/ml) in blocking buffer (LI-COR). After washing, binding of primary antibodies was detected with an IRDye800-labeled donkey anti–mouse IgG (1:2,000; LI-COR). In other Western blots, we used an IRDye680-labeled antibody 11A12 (1:500); an IRDye680-labeled antibody R24 (1:500); or an IRDye800-labeled V5 antibody (1:500). Western blots were scanned—and band intensities quantified—with an Odyssey infrared scanner (LI-COR).
Immunocytochemistry studies
CHO pgsA-745 cells (1 × 106 cells) were electroporated with 2 μg of plasmid DNA and then plated on coverslips in 24-well plates. The next day, the cells were fixed in 100% methanol, permeabilized with 0.2% Triton X-100, and blocked in 10% donkey serum. The cells were then incubated overnight at 4°C with GPIHBP1-specific mAbs (diluted to 10 μg/ml in blocking buffer), followed by an Alexa488-conjugated donkey anti–mouse IgG (Thermo Fisher Scientific; 1:800), a goat polyclonal antibody against the S-protein tag (Abcam; 1:800), and an Alexa555-conjugated donkey anti–goat IgG (Thermo Fisher Scientific; 1:800). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were recorded with an Axiovert 200M confocal fluorescence microscope and processed with the Zen 2010 software (all from Zeiss).
Kinetics for the interaction between mAbs and GPIHBP1 by SPR
Purified mAbs RG3 and RE3 in 10 mM of sodium acetate (pH 5.0) were covalently immobilized on a CM5 sensor chip that had been preactivated with NHS/EDC (N-ethyl-N′-[3-dimethylaminopropyl] carbodiimide), with the goal of achieving a surface density of 1,500 resonance units. mAb RF4 could be immobilized by this procedure, but the immobilized RF4 did not bind GPIHBP1. In hindsight, this was probably due to the fact that this mAb binds the disordered acidic domain of GPIHBP1 containing a high density of carboxylates. We suspect that mAb RF4 bound noncovalently to the carboxymethylated dextran matrix on the sensor chip and that this binding event inactivated the mAb. To circumvent this problem, we captured mAb RF4 on the sensor chip via a high-affinity interaction with covalently immobilized rabbit anti-mouse IgG (GE Healthcare Life Science, Uppsala, Sweden). Binding was recorded at 20°C, and the buffer flow rate was 50 μl/min (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, pH 7.4, containing 0.05% [v/v] surfactant P20). For multicycle kinetics, three-fold dilution series of GPIHBP1 (spanning a concentration from 1 to 90 nM) were injected for 200 s, followed by a 1,200-s dissociation step. For single-cycle kinetic titration of the RF4 × GPIHBP1 interaction, five consecutive injections of 20 μl of purified GPIHBP1 (two-fold dilutions ranging from 12 to 200 nM) were recorded. In the between cycles, the sensor chip was regenerated with two consecutive 10-μl injections of 0.1 M acetic acid/HCl (pH 2.5) in 0.5 M NaCl and 20 mM H3PO4. For multicycle analyses, the kinetic rate constants (kon and koff) for the mAb × GPIHBP1 interactions were derived by local nonlinear regression fitting of the data after double-buffer referencing to a simple bimolecular interaction model assuming pseudo first-order reaction conditions with BIA evaluation 4.1 software (Biacore, Uppsala, Sweden). For single-cycle kinetic analyses of the interaction between captured mAb RF4 and GPIHBP1, the rate constants were fitted to a simple bimolecular interaction model with global fitting (T200 Evaluation Software 2.0, GE Healthcare Life Science).
Epitope binning of GPIHBP1 mAbs was performed with a Biacore3000 (GE Healthcare Life Science), as described (17).
Testing the ability of GPIHBP1-specific mAbs to block LPL binding in a cell-free LPL–GPIHBP1 binding assay
Human GPIHBP1 containing carboxyl-terminal sequences encoding the mAb 11A12 epitope (from Drosophila S2 cells) was first incubated for 1 h at 4°C with mAb 11A12–coated agarose beads and then incubated for 30 min at 4°C with or without GPIHBP1-specific mAbs (final concentration, 5 μg/ml) and V5-tagged human LPL (18). After washing the beads, the GPIHBP1 (and any GPIHBP1-bound LPL) were eluted from the agarose beads by heating in SDS sample buffer for 5 min at 90°C. The amounts of GPIHBP1 and LPL in the flow-through, washes, and elution fractions were assessed by Western blotting with an IRDye680-labeled mAb 11A12 and an IRdye800-labeled V5 antibody, respectively.
Testing the ability of GPIHBP1-specific antibodies to block LPL binding to GPIHBP1-expressing cells
CHO pgsA-745 cells (2 × 106 cells and 2 μg plasmid DNA) were electroporated with expression vectors for S-protein–tagged human wild-type GPIHBP1 (wt) or a mutant GPIHBP1 that cannot bind LPL (GPIHBP1-W109S) (14). After 1 day, cells were washed and subsequently incubated for 1 h at 4°C with mAbs RG3, RF4, or RE3 (20 μg/ml). After washing, the cells were incubated for 1 h at 4°C with V5-tagged human LPL (200 ng/well). The cells were then washed six times in PBS and fixed in 100% methanol, and we performed immunocytochemistry studies on nonpermeabilized cells. After blocking with 10% donkey serum in PBS/Ca/Mg, the cells were incubated with an Alexa488-conjugated donkey anti–mouse IgG (Thermo Fisher Scientific; 50 ng/ml), a rabbit anti-S–protein tag (Abcam; 1:1,000), an Alexa647-conjugated donkey anti–rabbit IgG (Thermo Fisher Scientific; 2.5 μg/ml), and an Alexa555-conjugated mouse anti–V5 antibody (1:50). DNA was stained with DAPI. Images were recorded with an Axiovert 200M confocal fluorescence microscope.
Immunohistochemistry studies on human adipose tissue
Frozen sections (20 μm) of human cardiac adipose tissue (from the Duke Human Heart Repository) were placed on glass slides, fixed with 3% paraformaldehyde, permeabilized in 0.2% Triton X-100, blocked in 10% donkey serum, and then incubated overnight at 4°C with a mixture of mAbs RE3 and RF4 (10 μg/ml each), a rabbit polyclonal antibody against human von Willebrand factor (vWF) (Dako; 1:200), and a goat polyclonal antibody against human collagen IV (Novus Biologicals; 1:200), followed by 1-h incubations with an Alexa647-conjugated donkey anti–mouse IgG (Thermo Fisher Scientific; 1:500), an Alexa488-conjugated donkey anti–rabbit IgG (Thermo Fisher Scientific; 1:500), and an Alexa555-conjugated donkey anti–goat IgG (Thermo Fisher Scientific; 1:500). DNA was stained with DAPI. Images were recorded with an Axiovert 200M microscope with a ×20 objective and processed with the Zen 2010 software (Zeiss).
Immunoperoxidase studies were performed with the ImmPRESS Excel Staining Kit (Vector Laboratories). The sections were quenched of endogenous peroxidase activity with the BLOXALL buffer (Vector Laboratories) and blocked in 10% normal horse serum. Next, the sections were incubated for 1 h with mAb RF4 (1 μg/ml) or a rabbit polyclonal antibody against vWF (Dako; 1:200), followed by a 15-min incubation with a goat anti-mouse IgG (Vector Laboratories) or a goat anti-rabbit IgG (Vector Laboratories), and a 30-min incubation with a horse anti-goat IgG (ImmPRESS Excel Reagent, Vector Laboratories). Sections were then stained with ImmPACT DAB EqV (Vector Laboratories) until color change was apparent (∼10 s). Finally, sections were counterstained with hematoxylin and mounted with 90% glycerol in PBS. Images were taken with a Nikon Eclipse E600 microscope (Plan Fluor ×20/0.50 NA or ×60/0.75 NA objectives) equipped with a DS-Fi2 camera (Nikon).
An ELISA to detect the binding of LPL to human GPIHBP1
We coated 96-well plates overnight at 4°C with mAb R24 (0.5 μg/well). On the next day, the plates were blocked for 4 h at room temperature in Starting Block buffer (Pierce) and then incubated overnight at 4°C with 0.5 μg/well of human GPIHBP1–uPAR fusion protein from Drosophila S2 cells in the presence or absence of mAbs RG3, RF4, and RE3. Serial dilutions of the mAbs were tested in triplicate (from 20 to 0.1 μg/ml of mAb). After washing, the wells were incubated with 200 ng/well of V5-tagged human LPL for 1 h at 4°C, then washed and subsequently incubated for 1 h at 4°C with an HRP-labeled V5 antibody (Thermo Fisher Scientific; 1:5,000) to detect bound LPL. The presence of GPIHBP1 on the plates was verified in duplicate wells incubated with HRP-labeled 11A12 (1:50,000). To document mAb binding to GPIHBP1, we incubated replicate wells with HRP-labeled versions of RG3, RF4, and RE3. After washing the plates, 50 μl of one-step ultra 3,3’,5,5’-tetramethylbenzidine (TMB) ELISA substrate (Pierce) was added to each well. The plate was incubated at room temperature for 5 min before stopping the reaction with 50 μl of 2 M sulfuric acid. The optical density was read immediately at 450 nm on a Spectra Max 190 plate reader (Molecular Devices).
An ELISA to detect GPIHBP1 in human plasma
We coated 96-well plates with 1 μg/well of mAb RF4 overnight at 4°C. After blocking overnight at 4°C with PBS containing 1% BSA and 0.05% NaN3, the wells were incubated overnight at 4°C with the plasma samples. Serial dilution of the plasma samples (1:2 to 1:256) was performed in 1% BSA, 0.05% Tween 20, 0.05% ProClin 300, 50 μg/ml normal mouse IgG, and 5g/L polyoxyethylene alkyl ether in PBS. On the next day, plates were washed, and the captured GPIHBP1 was detected with 0.5 μg/well of HRP-labeled mAb RE3 Fab′ (diluted in PBS containing 1% BSA, 0.05% Tween 20, and 0.05% ProClin 300). After incubating for 30 min at 4°C, plates were washed, and 50 μl of TMB substrate (Kem-en-Tec) was added per well. After 30 min, the reaction was stopped by adding 50 μl of 2 M sulfuric acid. The optical density was read at 450 nm.
RESULTS
Defining the binding properties of the monoclonal antibodies
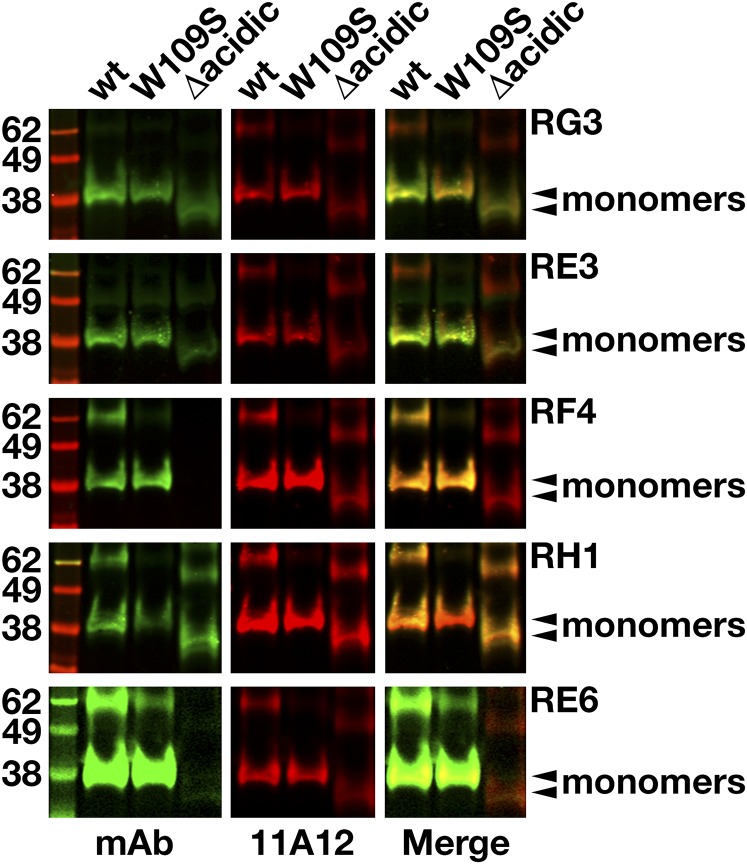
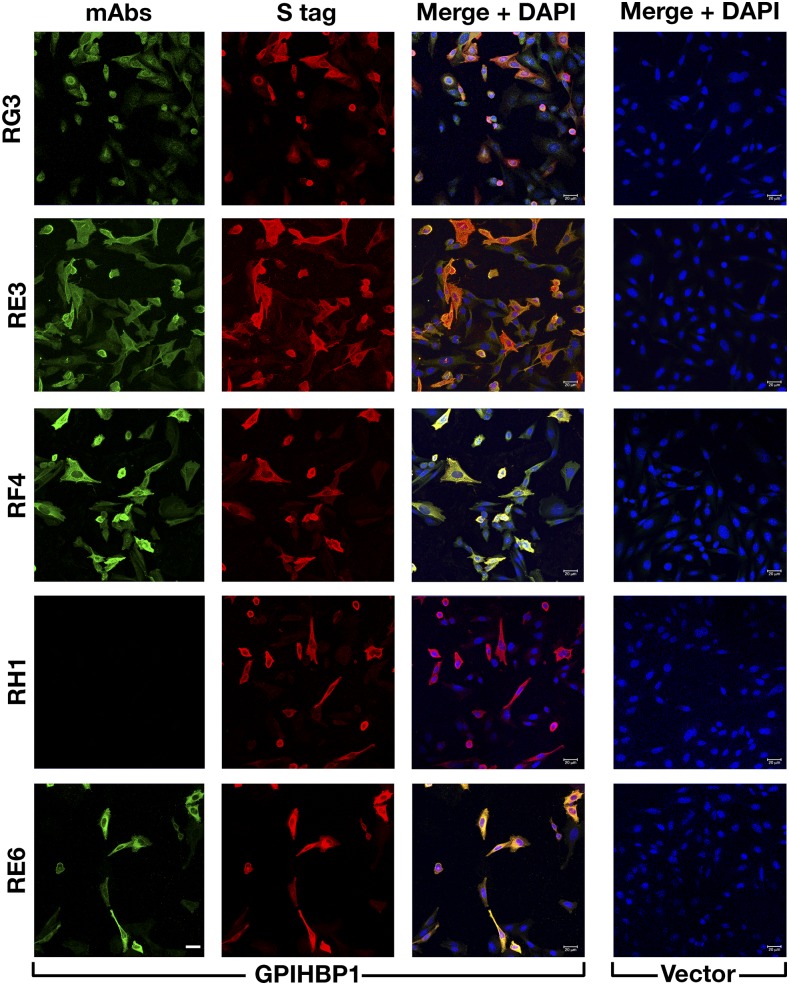
By screening hybridoma culture supernatants with Western blots, we identified 23 clones that secreted mAbs that bound to GPIHBP1 (supplemental Fig. S1). Some of the mAbs bound exclusively to nonreduced GPIHBP1; some bound exclusively to GPIHBP1 monomers; others bound both monomers and multimers. Several mAbs did not bind to a mutant GPIHBP1 lacking the acidic domain (GPIHBP1-Δacidic) (supplemental Fig. S2). We chose five mAbs for further study: RG3 (IgG2bκ), RE3 (IgG2aκ), RH1 (IgG1κ), RE6 (IgG1κ), and RF4 (IgG2bκ). RG3, RE3, and RH1 bound to wild-type GPIHBP1 and GPIHBP1-Δacidic. RE6 bound to wild-type GPIHBP1 and weakly to GPIHBP1-Δacidic. RG3 and RE3 bound preferentially to GPIHBP1 monomers, whereas RE6, RH1, and RF4 bound to both monomers and dimers. RF4 did not bind to GPIHBP1-Δacidic (Fig. 1) and bound avidly to a GPIHBP1–CD59 chimera containing GPIHBP1’s acidic domain but CD59’s Ly6 domain (supplemental Fig. S3). All mAbs except RH1 detected GPIHBP1 on the surface of GPIHBP1-transfected CHO cells (Fig. 2).
Fig. 1.
Western blots with GPIHBP1-specific monoclonal antibodies. Soluble versions of wild-type (wt) human GPIHBP1, GPIHBP1-W109S, and GPIHBP1-Δacidic (in which GPIHBP1’s acidic domain had been deleted) were expressed in Drosophila S2 cells; all constructs had a carboxyl-terminal mAb 11A12 epitope tag (9). Western blot analysis was performed on the conditioned medium of the Drosophila S2 cell cultures under nonreducing conditions using IRDye800-labeled GPIHBP1-specific mAbs RG3, RE3, RF4, RH1, and RE6 (green) and an IRDye680-labeled mAb 11A12 (red). Monomers are indicated by arrowheads.
Fig. 2.
Testing the ability of GPIHBP1-specific mAbs to bind to GPIHBP1 on the surface of GPIHBP1-transfected cells. CHO pgsA-745 cells were transiently transfected with empty vector or with an expression vector for S-protein–tagged human GPIHBP1. After 1 day, the cells were fixed with methanol, and immunocytochemistry studies were performed on permeabilized cells with mAbs RG3, RE3, RF4, RH1, and RE6 (10 μg/ml; green) and a rabbit antibody against the S-protein tag (red). DNA was stained with DAPI (blue). Scale bar (lower left panel), 20 μm.
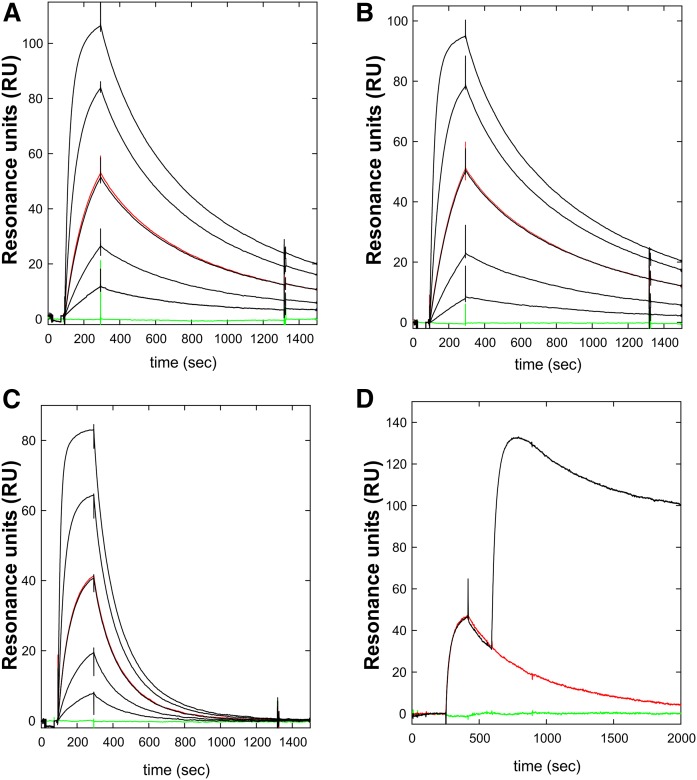
The binding of three mAbs (RE3, RG3, RF4) was evaluated by real-time surface plasmon resonance studies. All three mAbs bound GPIHBP1 with high affinity (KD ≤ 5 nM) Table 1, Fig. 3). The kinetics of RE3 and RG3 binding to full-length GPIHBP1 and GPIHBP1-Δacidic were virtually identical, indicating that their epitopes are not dependent on GPIHBP1’s acidic domain. Both RG3 and RE3 bound to GPIHBP1-W109S with reduced affinity—a consequence of an increase in koff (Table 1, Fig. 3). The W109S mutation eliminates LPL binding and does so without disrupting the formation of disulfide bonds in the Ly6 domain (19).
TABLE 1.
Kinetic rate constants for GPIHBPl-specific mAbs by SPR
| mAb | Analyte | kon (105 M–1s–1) | koff (10−3s–1) | KD (nM) | n | Nonoverlapping Epitopes |
| RE3 | GPIHBP1 | 4.2 ± 1.8 | 1.64 ± 0.27 | 4.4 ± 1.8 | 4 | RF4 |
| RE3 | GPIHBP1-W109S | 8.2 ± 1.9 | 7.82 ± 1.80 | 10.0 ± 3.3 | 3 | |
| RE3 | GPIHBP1-Δacidic | 7.6 ± 2.7 | 1.34 ± 0.11 | 2.0 ± 0.6 | 3 | |
| RG3 | GPIHBP1 | 0.64 ± 0.22 | 0.17 ± 0.04 | 3.1 ± 1.8 | 4 | RF4 |
| RG3 | GPIHBP1-W109S | 0.93 ± 0.15 | 1.23 ± 0.06 | 13.7 ± 2.4 | 3 | |
| RG3 | GPIHBP1-Δacidic | 1.26 ± 0.21 | 0.27 ± 0.11 | 2.3 ± 1.3 | 2 | |
| RF4 | GPIHBP1 | 5.4 ± 1.9 | 0.74 ± 0.06 | 1.4 ± 0.2 | 3 | RE3, RG3 |
Kinetics rate constants were derived from data recorded with a BiacoreT200 for three-fold dilutions of various purified GPIHBP1 proteins (8) and were fitted to a 1:1 binding model. GPIHBP1 is full-length GPIHBP1; GPIHBP1-W109S contains a serine for tryptophan mutation in a highly conserved region of the Ly6 domain; GPIHBP1-Δacidic contains a deletion of the acidic domain (the first 31 amino acids of the mature protein). Data for mAbs RE3 and RG3 were processed by a multicycle protocol with mAbs that had been directly immobilized on the chip (1,000 RU), whereas the data for mAb RF4 were processed by a single-cycle protocol in which mAb RF4 was captured by an immobilized rabbit anti-mouse IgG. Epitope mapping was performed with sequential injections, as is illustrated by Fig. 3.
Fig. 3.
Kinetics for the interaction between mAb RE3 and GPIHBP1 by surface plasmon resonance. The real-time kinetic interactions between immobilized mAb RE3 and different GPIHBP1 proteins were measured with a BiacoreT200 system. A three-fold dilution series of wild-type GPIHBP1 (A); GPIHBP1-Δacidic (B); and GPIHBP1-W109S (C) were injected between 100 and 300 s, followed by a dissociation phase from 300 to 1,500 s. The concentrations analyzed were 90, 30, 10, 3, and 1 nM GPIHBP1 (black curves). One repeat measurement of 10 nM GPIHBP1 was performed at the end of each analysis (red); a buffer control curve is also shown (green). (D) Example of epitope binning. RE3 was immobilized on the sensor chip and 100 nM GPIHBP1 was captured by injection at 300 s, followed by a second injection of either buffer (red curve) or 100 nM mAb RF4 (black curve). The sensorgrams show that RE3 and RF4 belong to separate epitope bins and that their binding to GPIHBP1 was not mutually exclusive.
Epitope binning by pair-wise comparison of antibody binding to GPIHBP1 by SPR (Table 1, Fig. 3, supplemental Fig. S4) revealed that the epitopes for RE3 and RG3 are overlapping and distinct from that of RF4. This finding suggested a strategy for developing a sandwich ELISA for detecting GPIHBP1 (using mAb RF4 to capture GPIHBP1 and either RE3 or RG3 to detect GPIHBP1).
Testing the ability of GPIHBP1-specific mAbs to block LPL binding to GPIHBP1
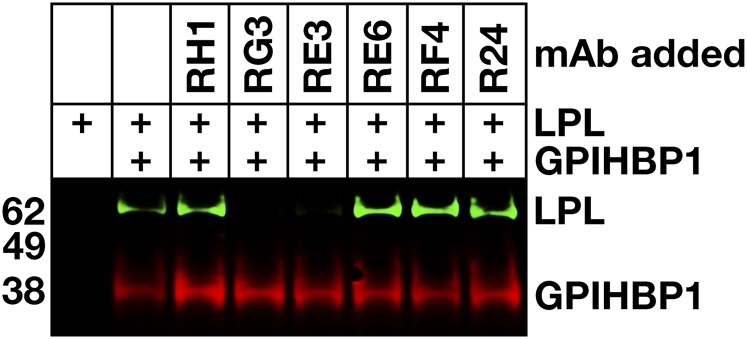
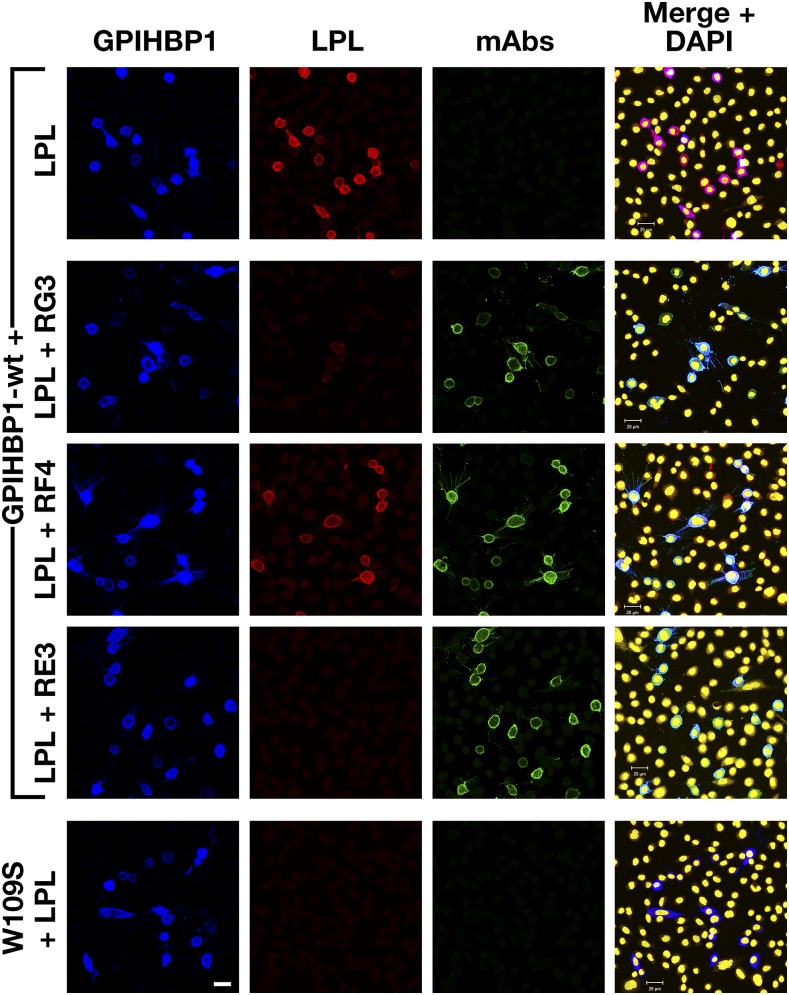
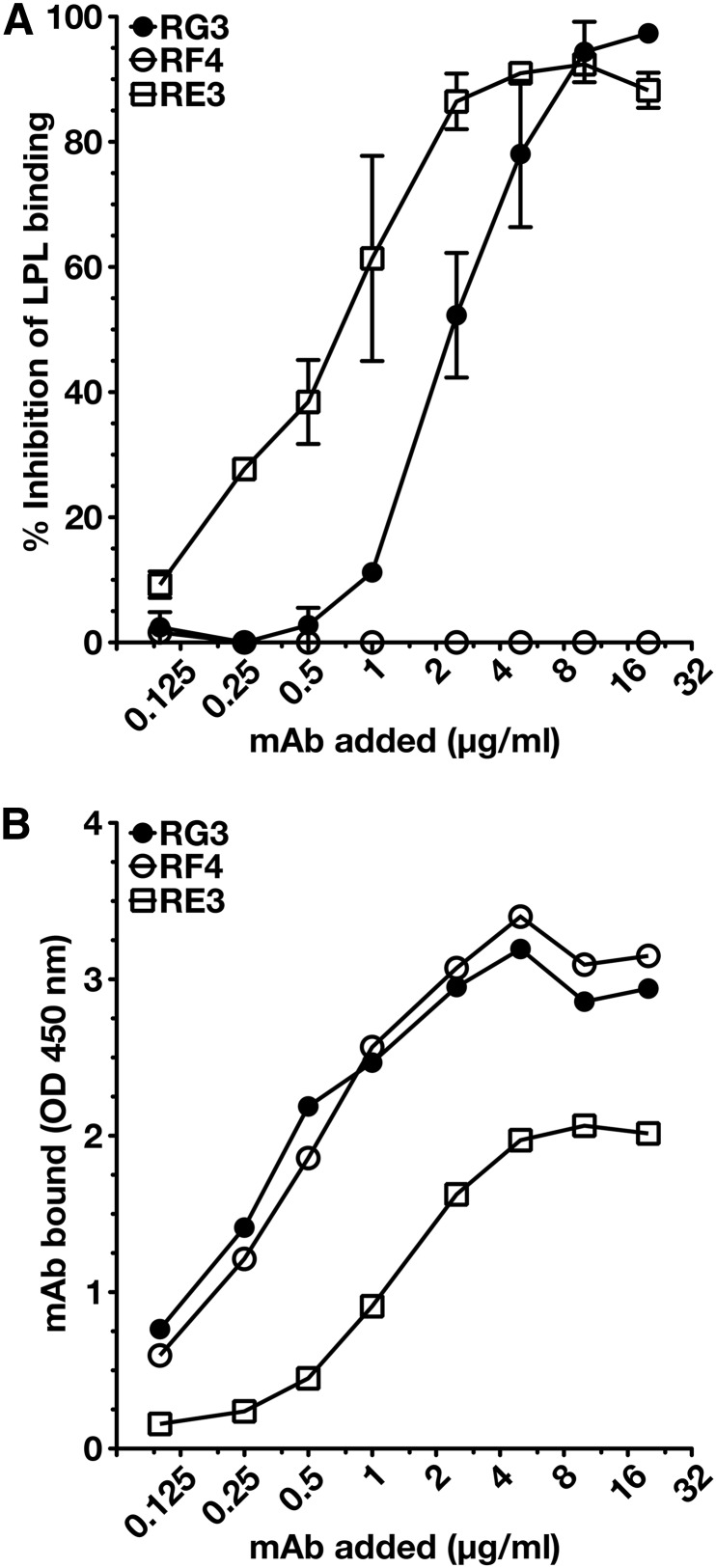
To determine whether any of the mAbs blocked the binding of LPL to GPIHBP1, we initially utilized a cell-free LPL–GPIHBP1 binding assay (9). Equimolar amounts of LPL, recombinant human GPIHBP1 harboring the mAb 11A12 epitope, and GPIHBP1-specific mAbs were incubated with agarose beads coated with mAb 11A12. After 1 h, the beads were washed, and the amount of GPIHBP1 (and GPIHBP1-bound LPL) captured by the beads was assessed by Western blotting. RG3 and RE3 abolished the binding of LPL to GPIHBP1, whereas RH1, RE6, and RF4 did not (Fig. 4). The ability of GPIHBP1-specific mAbs to block LPL binding was further tested with a cell-based LPL–GPIHBP1 binding assay and an ELISA. Again, these studies revealed that the binding of mAbs RG3 and RE3 to GPIHBP1 abolished LPL binding (Figs. 5, 6). Of note, mAb RF4 bound avidly to GPIHBP1 but did not block LPL binding (Figs. 5, 6).
Fig. 4.
Monoclonal antibodies RG3 and RE3 block LPL binding to GPIHBP1 in a cell-free LPL–GPIHBP1 binding assay. Soluble wild-type human GPIHBP1 with a carboxyl-terminal 11A12 epitope tag was incubated with 11A12-coated agarose beads and V5-tagged human LPL, in the presence or absence of mAbs RH1, RG3, RE3, RE6, RF4, or R24. After washing the beads, GPIHBP1 and any GPIHBP1-bound LPL was eluted from the beads by heating in SDS loading buffer. Shown is a Western blot on the eluted proteins with IRDye680-labeled mAb 11A12 (red) and an IRDye800-labeled V5 antibody (green).
Fig. 5.
RG3 and RE3, but not RF4, block the binding of LPL to GPIHBP1 on the surface of cultured cells. CHO pgsA-745 cells were transfected with vectors for S-protein–tagged versions of wild-type human GPIHBP1 (wt) or GPIHBP1-W109S. After 1 day, the cells were washed and preincubated with mAbs RG3, RF4, or RE3 (20 μg/ml) or PBS alone. After washing, the cells were incubated with V5-tagged human LPL (200 ng/well). The cells were washed and fixed with methanol, and immunocytochemistry was performed on nonpermeabilized cells with an Alexa488-conjugated donkey anti–mouse IgG (green), a rabbit anti-S–protein tag (blue), and an Alexa555-conjugated mouse anti–V5 antibody (red). DNA was stained with DAPI (yellow). Scale bar (lower left panel), 20 μm.
Fig. 6.
Dose-dependent inhibition of LPL binding to GPIHBP1 by mAbs RE3 and RG3. We coated 96-well ELISA plates with the uPAR-specific mAb R24, blocked, and then incubated them with uPAR-tagged human GPIHBP1 (0.5 μg/well) in the presence or absence of serial dilutions of mAbs RG3, RE3, or RF4. After washing, the plates were incubated with V5-tagged LPL (200 ng/well). GPIHBP1-bound LPL was detected with an HRP-labeled V5 antibody. The binding of the GPIHBP1-specific mAbs to GPIHBP1 was documented in an independent assay with HRP-labeled mAbs RG3, RE3, or RF4. (A) Inhibition of LPL binding to GPIHBP1 by mAbs RE3 and RG3. Antibody RF4 had no effect on LPL binding. (B) Amount of mAb bound to GPIHBP1 for each amount of the GPIHBP1-specific mAb added.
Detecting GPIHBP1 in capillaries of human adipose tissue
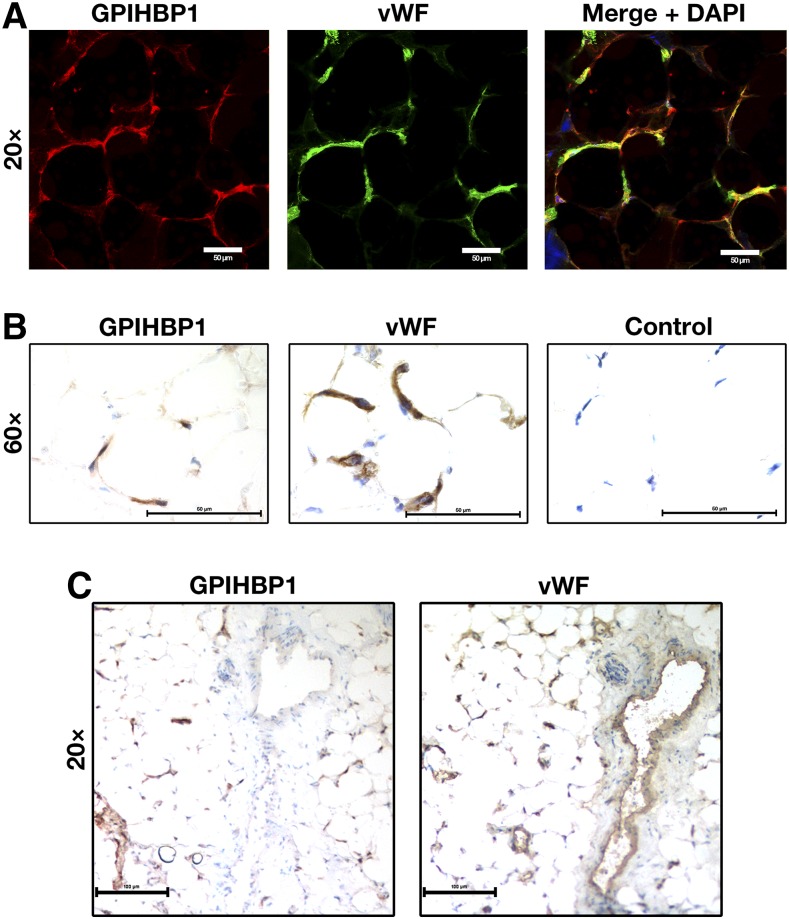
We tested the ability of RE3 and RF4 to detect human GPIHBP1 in human adipose tissue by confocal immunofluorescence microscopy and by immunoperoxidase staining. RE3 and RF4 detected GPIHBP1 in capillaries of epicardial adipose tissue, colocalizing with vWF (Fig. 7, supplemental Fig. S5). GPIHBP1 was found only in capillaries and not in endothelial cells of venules, whereas vWF was found in endothelial cells of both capillaries and venules (Fig. 7, supplemental Fig. S5). Despite considerable effort, we were unsuccessful in detecting GPIHBP1 in capillaries of postmortem human heart tissue by confocal immunofluorescence microscopy or immunoperoxidase staining. This was not entirely surprising because the expression of GPIHBP1 in human heart is much lower than in human adipose tissue, as judged by RNA seq data (Human Protein Atlas database; www.proteinatlas.org) (20). A caveat to our immunohistochemistry studies on human heart is that we examined heart tissue that was harvested from patients with devastating and irreversible disease after the heart had stopped beating (“donation after cardiac death”), and we simply do not know how these circumstances affected GPIHBP1 expression in capillaries.
Fig. 7.
Detection of GPIHBP1 in human tissues with GPIHBP1-specific monoclonal antibodies. Sections of human cardiac adipose tissue (20 μm) were fixed with 3% paraformaldehyde and processed for confocal immunofluorescence microscopy (A) or light microscopy (B, C). (A) Confocal microscopy images showing GPIHBP1 (in this case, detected by a combination of mAbs RE3 and RF4, 10 μg/ml each; red) and von Willebrand Factor (vWF, a marker for endothelial cells; green) in the capillaries of human cardiac adipose tissue. (B) Consecutive HRP-stained sections showing GPIHBP1 (left panel, mAb RF4, 1 μg/ml) and vWF (middle panel) in capillaries of human cardiac adipose tissue. No primary antibody was added in the control panel (right). (C) Consecutive HRP-stained sections showing expression of vWF in both capillaries and a large venule, whereas GPIHBP1 was expressed in endothelial cells of capillaries but not in endothelial cells of the venule. In B and C, sections were counterstained with hematoxylin. Scale bar for A, 20 μm; B, 50 μm; C, 100 μm.
Detecting GPIHBP1 in human plasma
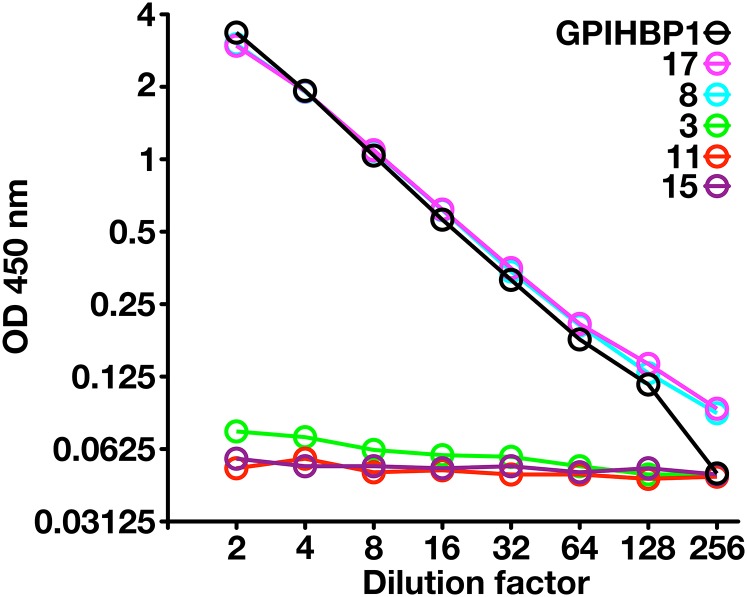
Because another GPI-anchored Ly6 protein, uPAR, can be detected in human plasma by ELISA (13), we suspected that it might be possible to detect GPIHBP1 in plasma. To explore that possibility, we created a sandwich ELISA in which mAb RF4 was used to capture the GPIHBP1 in plasma, and HRP-labeled mAb RE3 was used to detect the captured GPIHBP1. This ELISA readily detected a recombinant GPIHBP1 standard and was able to detect GPIHBP1 in normal human plasma. In two normal subjects, the plasma GPIHBP1 levels were ∼1 ng/ml (Fig. 8). As negative controls, we included three subjects who were homozygous for GPIHBP1 mutations [a deletion of the entire GPIHBP1 gene in subject 3 (21), and a C89X nonsense mutation in subjects 11 and 15 (supplemental Fig. S6)]. As was expected, the plasma levels of GPIHBP1 in those subjects were essentially undetectable (Fig. 8).
Fig. 8.
An ELISA to detect GPIHBP1 in human plasma. We coated 96-well plates with mAb RF4 (1 μg/well), blocked them with BSA, and incubated them overnight at 4°C with dilutions of human plasma (ranging from 1:2 to 1:256) or dilutions of purified human GPIHBP1. In the case of the purified GPIHBP1, the “1:2 dilution” corresponds to 500 pg/ml of recombinant GPIHBP1, and the “1:256 dilution” corresponds to buffer alone (no recombinant GPIHBP1). After washing the plates, GPIHBP1 captured by mAb RF4 was detected with HRP-labeled RE3 Fab′. Samples 8 and 17 were normal control plasma samples; sample 3 was from a subject homozygous for a deletion of GPIHBP1 (21); samples 11 and 15 were from subjects homozygous for a GPIHBP1 nonsense mutation (C89×). The plasma GPIHBP1 levels in samples 8 and 17 were 1,043 and 1,051 pg/ml, respectively. As expected, the GPIHBP1 levels in the GPIHBP1-deficient subjects were essentially absent. (Note that GPIHBP1-C89X is not bound by mAb RE3 [see supplemental Fig. S6] and therefore cannot be detected by this ELISA.) The plot represents a log-transformation of the data.
DISCUSSION
We identified 23 mAbs against human GPIHBP1 in our initial screening efforts and selected five for further study—four against GPIHBP1’s cysteine-rich Ly6 domain (RG3, RE3, RH1, RE6) and one against the acidic domain (RF4). These mAbs proved to be useful for three lines of investigation. First, we found that two mAbs against the Ly6 domain, RG3 and RE3, blocked the binding of LPL to GPIHBP1, whereas a mAb against the acidic domain (RF4) did not. Of note, both RG3 and RE3 bound preferentially to nonreduced GPIHBP1, implying that the epitopes for those mAbs depend on intact disulfide bonds in the Ly6 domain. Also, by SPR, mAbs RG3 and RE3 bound with reduced affinity to GPIHBP1-W109S, an “Ly6 domain mutant” that lacks the capacity to bind LPL (19). Those findings provide strong support for recent SPR studies (8) that concluded that GPIHBP1’s Ly6 domain is largely responsible for high-affinity interactions with LPL. Second, immunohistochemistry studies of human adipose tissue with mAbs RE3 and RF4 revealed that GPIHBP1 is expressed only in capillary endothelial cells and not in venules—the same pattern observed previously in mice (5). From the standpoint of lipoprotein physiology, this pattern of expression makes sense. LPL is secreted by adipocytes and is subject to local regulation by ANGPTL4 (22). The fact that GPIHBP1 is expressed specifically in capillaries—the blood vessels that are immediately adjacent to adipocytes—facilitates the capture of locally produced LPL and serves to focus lipolytic activity, according to the requirements of nearby parenchymal cells. Third, we found, using a mAb-based ELISA, that GPIHBP1 can be detected in the plasma of normal subjects but not subjects with GPIHBP1 deficiency.
In our immunohistochemistry studies, we were able to detect GPIHBP1 on capillary endothelial cells of human adipose tissue by confocal immunofluorescence microscopy and by immunoperoxidase staining, but we could not detect GPIHBP1 in capillaries of the heart. Our inability to detect GPIHBP1 in heart capillaries was initially perplexing because we invariably observed robust GPIHBP1 expression in mouse heart capillaries (4, 5). Follow-up studies revealed that our inability to detect GPIHBP1 in heart capillaries was probably related to lower amounts of GPIHBP1 expression in human heart.
One of our principal goals in creating GPIHBP1-specific monoclonal antibodies was to develop an ELISA for the detection of GPIHBP1 in human plasma, just as others had done for uPAR, another GPI-anchored Ly6 protein (13). The combination of mAb RF4 (to capture GPIHBP1 in the plasma) and HRP-labeled mAb RE3 (to detect captured GPIHBP1) proved successful. Our ELISA detected GPIHBP1 in serial dilutions of plasma from normal subjects but not from subjects with GPIHBP1 deficiency. At this point, we do not understand why GPIHBP1 circulates in the plasma, but there are several possibilities. One is that exosomes containing GPIHBP1 are shed from capillary endothelial cells. Another is that GPIHBP1 is released into the plasma by an enzyme that cleaves the GPI anchor or cleaves GPIHBP1 downstream from the Ly6 domain. Another possibility is that soluble GPIHBP1 (GPIHBP1 lacking a GPI anchor) is secreted from capillary endothelial cells as a consequence of inefficiencies in the transamidase reaction that ordinarily replaces GPIHBP1’s carboxyl-terminal hydrophobic signal peptide with a GPI anchor (23). We favor the latter possibility because cultured cells that overexpress the GPI-anchored form of GPIHBP1 invariably secrete large amounts of soluble GPIHBP1 (9). Regardless of the mechanism, our discovery that GPIHBP1 can be detected in human plasma is exciting. With the experimental approaches described here and our new GPIHBP1-specific mAbs, clinical lipidologists will now be able to test the utility of plasma GPIHBP1 levels as a biomarker for metabolic disease, vascular disease, or both.
Supplementary Material
Acknowledgments
We thank personnel at Finsen Laboratory for the purified GPIHBP1 immunogen; the Monash Antibody Technologies Facility for creating the monoclonal antibodies; and Helen H. Hobbs for plasma samples from subjects with GPIHBP1 deficiency.
Footnotes
Abbreviations:
- DAPI
- 4′,6-diamidino-2-phenylindole
- mAb
- monoclonal antibody
- SPR
- surface plasmon resonance
- uPAR
- urokinase-type plasminogen activator receptor
- vWF
- von Willebrand factor.
This work was supported by grants from the National Heart, Lung, and Blood Institute (HL090553, HL087228, and HL125335) and a Transatlantic Network Grant from the Leducq Foundation (12CVD04). Xuchen Hu was supported by a Ruth L. Kirschstein National Research Service Award (T32HL69766); the National Institutes of Health National Institute of General Medical Science Training Grant (T32GM08042); and the University of California, Los Angeles, Medical Scientist Training Program. The authors have no financial conflicts of interest to declare.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Korn E. D. 1955. Clearing factor, a heparin-activated lipoprotein lipase. I. Isolation and characterization of the enzyme from normal rat heart. J. Biol. Chem. 215: 1–14. [PubMed] [Google Scholar]
- 2.Korn E. D. 1955. Clearing factor, a heparin-activated lipoprotein lipase. II. Substrate specificity and activation of coconut oil. J. Biol. Chem. 215: 15–26. [PubMed] [Google Scholar]
- 3.Havel R. J., and Gordon R. S. Jr. 1960. Idiopathic hyperlipemia: metabolic studies in an affected family. J. Clin. Invest. 39: 1777–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beigneux A. P., Davies B., Gin P., Weinstein M. M., Farber E., Qiao X., Peale P., Bunting S., Walzem R. L., Wong J. S., et al. . 2007. Glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5: 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies B. S., Beigneux A. P., Barnes R. H. 2nd, Tu Y., Gin P., Weinstein M. M., Nobumori C., Nyren R., Goldberg I., Olivecrona G., et al. . 2010. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fry B. G., Wuster W., Kini R. M., Brusic V., Khan A., Venkataraman D., and Rooney A. P.. 2003. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 57: 110–129. [DOI] [PubMed] [Google Scholar]
- 7.Ioka R. X., Kang M-J., Kamiyama S., Kim D-H., Magoori K., Kamataki A., Ito Y., Takei Y. A., Sasaki M., Suzuki T., et al. . 2003. Expression cloning and characterization of a novel glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein, GPI-HBP1. J. Biol. Chem. 278: 7344–7349. [DOI] [PubMed] [Google Scholar]
- 8.Mysling S., Kristensen K. K., Larsson M., Beigneux A. P., Gardsvoll H., Fong L. G., Bensadouen A., Jorgensen T. J., Young S. G., and Ploug M.. 2016. The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. eLife. 5: e12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beigneux A. P., Gin P., Davies B. S. J., Weinstein M. M., Bensadoun A., Fong L. G., and Young S. G.. 2009. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J. Biol. Chem. 284: 30240–30247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies B. S., Goulbourne C. N., Barnes R. H. 2nd, Turlo K. A., Gin P., Vaughan S., Vaux D. J., Bensadoun A., Beigneux A. P., Fong L. G., et al. . 2012. Assessing mechanisms of GPIHBP1 and lipoprotein lipase movement across endothelial cells. J. Lipid Res. 53: 2690–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vodenicharov A., Atanassova P., Yonkova P., Kostadinov G., and Hristov H.. 2007. Expression of lipoprotein lipase in the renal artery and vein of the domestic pig–an enzyme-histochemical study. Bulg. J. Vet. Med. 10: 155–160. [Google Scholar]
- 12.Camps L., Reina M., Llobera M., Vilaro S., and Olivecrona T.. 1990. Lipoprotein lipase: cellular origin and functional distribution. Am. J. Physiol. 258: C673–C681. [DOI] [PubMed] [Google Scholar]
- 13.Piironen T., Laursen B., Pass J., List K., Gardsvoll H., Ploug M., Dano K., and Hoyer-Hansen G.. 2004. Specific immunoassays for detection of intact and cleaved forms of the urokinase receptor. Clin. Chem. 50: 2059–2068. [DOI] [PubMed] [Google Scholar]
- 14.Beigneux A. P., Davies B. S. J., Tat S., Chen J., Gin P., Voss C. V., Weinstein M. M., Bensadoun A., Pullinger C. R., Fong L. G., et al. . 2011. Assessing the role of the glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J. Biol. Chem. 286: 19735–19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gårdsvoll H., Hansen L. V., Jorgensen T. J., and Ploug M.. 2007. A new tagging system for production of recombinant proteins in Drosophila S2 cells using the third domain of the urokinase receptor. Protein Expr. Purif. 52: 384–394. [DOI] [PubMed] [Google Scholar]
- 16.Gin P., Yin L., Davies B. S. J., Weinstein M. M., Ryan R. O., Bensadoun A., Fong L. G., Young S. G., and Beigneux A. P.. 2008. The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J. Biol. Chem. 283: 29554–29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B., Gandhi S., Yuan C., Luo Z., Li R., Gardsvoll H., de Lorenzi V., Sidenius N., Huang M., and Ploug M.. 2015. Stabilizing a flexible interdomain hinge region harboring the SMB binding site drives uPAR into its closed conformation. J. Mol. Biol. 427: 1389–1403. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Zeev O., Mao H. Z., and Doolittle M. H.. 2002. Maturation of lipoprotein lipase in the endoplasmic reticulum. Concurrent formation of functional dimers and inactive aggregates. J. Biol. Chem. 277: 10727–10738. [DOI] [PubMed] [Google Scholar]
- 19.Beigneux A. P., Fong L. G., Bensadoun A., Davies B. S., Oberer M., Gardsvoll H., Ploug M., and Young S. G.. 2015. GPIHBP1 missense mutations often cause multimerization of GPIHBP1 and thereby prevent lipoprotein lipase binding. Circ. Res. 116: 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., et al. . 2015. Proteomics. Tissue-based map of the human proteome. Science. 347: 1260419. [DOI] [PubMed] [Google Scholar]
- 21.Rios J. J., Shastry S., Jasso J., Hauser N., Garg A., Bensadoun A., Cohen J. C., and Hobbs H. H.. 2012. Deletion of GPIHBP1 causing severe chylomicronemia. J. Inherit. Metab. Dis. 35: 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida K., Shimizugawa T., Ono M., and Furukawa H.. 2002. Angiopoietin-like protein 4 is a potent hyperlipidemia-inducing factor in mice and inhibitor of lipoprotein lipase. J. Lipid Res. 43: 1770–1772. [DOI] [PubMed] [Google Scholar]
- 23.Kinoshita T., and Fujita M.. 2016. Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J. Lipid Res. 57: 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.