Abstract
Variations in the gene LDAH (C2ORF43), which encodes lipid droplet-associated hydrolase (LDAH), are among few loci associated with human prostate cancer. Homologs of LDAH have been identified as proteins of lipid droplets (LDs). LDs are cellular organelles that store neutral lipids, such as triacylglycerols and sterol esters, as precursors for membrane components and as reservoirs of metabolic energy. LDAH is reported to hydrolyze cholesterol esters and to be important in macrophage cholesterol ester metabolism. Here, we confirm that LDAH is localized to LDs in several model systems. We generated a murine model in which Ldah is disrupted but found no evidence for a major function of LDAH in cholesterol ester or triacylglycerol metabolism in vivo, nor a role in energy or glucose metabolism. Our data suggest that LDAH is not a major cholesterol ester hydrolase, and an alternative metabolic function may be responsible for its possible effect on development of prostate cancer.
Keywords: lipase, lipoprotein metabolism, cholesterol efflux, triglycerides, animal models
Lipid droplets (LDs) are cellular organelles that are important for energy and lipid metabolism (1, 2). LD accumulation is a hallmark of obesity and is linked to the metabolic syndrome and type II diabetes. LD accumulation is central to atherosclerosis development, in which macrophages in arterial walls accumulate cholesterol esters (CEs) in LDs to become foam cells. Finally, LDs accumulate in many carcinomas (3), and LDs and lipid metabolism are connected to renal clear cell carcinoma and prostate cancer (4–8).
Among genes that encode LD proteins, LDAH is associated with prostate cancer. The SNP rs13385191 in intron 6 of LDAH is associated with increased prostate cancer risk (9–12). A rare A>G variant is associated with a difference in LDAH mRNA abundance, and prostate cancer risk is inversely correlated with its expression (13, 14). In addition, rs13385191 is associated with nonfatal outcome of prostate cancer (9, 12). Linkage of the LDAH locus with prostate cancer suggests that loss of the lipid droplet-associated hydrolase (LDAH) function has a role in prostate tumorigenesis.
The single polypeptide encoded by LDAH is predicted to be a serine hydrolase of the α/β-fold type (15, 16). Homologs in multiple species, including yeast (YPR147C), suggest a conserved function at LDs; however, LDAH’s molecular function remains uncertain but has recently been investigated. Goo et al. reported that LDAH is a CE hydrolase (17). This finding is intriguing inasmuch as recent studies have linked accumulation of CEs in LDs to prostate and breast cancer aggressiveness (18, 19). Supporting a function in CE metabolism, two other SNPs in LDAH are associated with changes in LDL cholesterol (20, 21). However, LDAH is near APOB on chromosome 2, and these SNPs were originally linked to APOB, a confounding factor because APOB is involved in cholesterol metabolism.
Besides LDAH, other lipases have been implicated in CE hydrolysis. NCEH1 has been reported to hydrolyze CEs (22), but at least in mice, it has also been reported to hydrolyze ether lipid 2-acetyl monoalkylglycerol (23). The LD-localized hormone-sensitive lipase (HSL) contributes to CE hydrolysis (23–25). Whether HSL has a major role in CE hydrolysis in macrophages is debated because CE hydrolysis still occurs in its absence (26). Lysosomal acid lipase also contributes to cellular CE metabolism and regulates macrophage cholesterol efflux, potentially through lipophagy (27–29), but whether it has access to LDs under normal conditions is not clear. Thus, the enzymes that hydrolyze CEs at LDs are uncertain.
In this study, we tested the reported role for LDAH in CE hydrolysis and the metabolism of other neutral lipids by generating and analyzing a knockout mouse model lacking this enzyme.
MATERIALS AND METHODS
Amino acid sequence analysis
CG9186 (dLDAH) secondary structure was predicted with JPred4 (30) and PSIPRED (31, 32).
Cell culture and transfection
HeLa cells were cultured in DMEM with 10% FBS and PenStrep. S2 cell culture was performed as described (33). HeLa and S2 cells were transfected using FuGENE HD (Promega, Madison, WI) and Effectene (Qiagen, Germantown, MD) transfection reagents, respectively, according to the manufacturer’s instructions. LDs were induced and stained as described (33–35), S2 cells were induced with 1 mM oleic acid-BSA, and HeLa cells were induced with 0.5 mM oleic acid coupled to BSA. For colocalization experiments, a C-terminally tagged ADRP-YFP fusion construct or dsRed2-ER (Clontech, Mountain View, CA) was cotransfected into HeLa cells or GFP-Sec61β for S2 cells. For localization studies in mammalian cells we generated expression plasmids containing human full-length LDAH with N-terminal mCherry- or GFP-tag under the CMV promoter. For LD-targeting studies, we expressed Drosophila full-length CG9186 with C-terminal mCherry-tag, the LD domain alone (amino acids (aa) 152–201 of Drosophila CG9186 (CG9186aa152-201)), or Drosophila CG9186 with aa157–200 replaced by a AAAGGGGSGGGGS-linker (Δ aa157–200) under the actin promoter.
Fluorescence microscopy and image analysis
Immunofluorescence and spinning-disk confocal microscopy (100 × 1.4 NA oil immersion objective [Olympus], iMIC [Till], CSU22 [Yokugawa], iXonEM 897 [Andor]) were as described (33). LD area per cell was quantified as described (35).
Mouse experiments
All animal studies followed guidelines issued by Yale and Harvard universities’ institutional animal care and use committees. Mice were housed at 12-h light/12-h dark cycle with ad libitum access to food and water unless indicated otherwise.
The mouse strain used for this research project was created from embryonic stem cell clone 14003A-H3 (C57BL/6Ntac background, injected into B6(Cg)-Tyrc-2J/J blastocysts), obtained from the Knockout Mouse Project Repository (www.komp.org) and generated by Regeneron Pharmaceuticals. Forward primers S30636 (wild-type; 5′- CATCTCACCTCCTCTCCGTC-3′) or NeoF (knockout; 5′- TCATTCTCAGTATTGTTTTGCC-3′) and reverse primer SD (5′-CAGAGTCCTTCCCATGTCAC-3′) were used for genotyping.
Velocigene targeted alleles were created as described (36). Mice with germ-line transmission of the knockout allele were backcrossed from a C57BL/6Ntac onto a C57BL/6J background for a minimum of three generations. Mice were, therefore, a mixture of these C57BL/6 strains. All animals were generated through breeding heterozygous animals.
Major determinants of whole-body energy balance were evaluated by the Yale School of Medicine Mouse Metabolic Phenotyping Center using the Comprehensive Lab Animal Monitoring System with Oxymax (Columbus Labs), including VO2, VCO2, activity, feeding, and drinking behavior. Body composition was determined by proton-NMR with the Bruker Minispec. Glucose homeostasis was evaluated by glucose tolerance test, according to recommendations of the NIH-funded Mouse Metabolic Phenotyping Consortium (37). Briefly, mice were fasted overnight before we collected a basal plasma sample for glucose and insulin measure. Mice were dosed intraperitoneally with 1 mg/g dextrose, and plasma was collected at set intervals for glucose and insulin measures. Glucose was measured by the glucose oxidase method using YSI, and insulin measured by radioimmunoassay.
Histology slides were prepared by the Yale School of Medicine Research Histology core or the Harvard Medical School Rodent Histology core.
For the cold-exposure experiments, animals were fasted overnight and placed at 4°C for indicated times. Temperatures were determined using a rectal thermometer. Blood glucose was measured as described below. A 60% high-fat diet (D12492) was obtained from Research Diets (New Brunswick, NJ).
Thin-layer chromatography
Lipids were extracted from tissue lysates (38), separated on silica TLC plates (Merck, Kenilworth, NJ) with n-heptane/isopropyl ether/acetic acid (60/40/4), and detected by cerium molybdate staining. Bands were identified by comparing with standards.
Isolation of bone marrow-derived macrophages
Femurs and tibia were dissected, and muscle was removed. Bone marrow was flushed with 5 ml of DMEM/F12 (Thermo Fisher Scientific, Waltham, MA) with a 5 ml syringe and a 25 gauge needle. Cells were centrifuge at 500 rpm for 10 min, resuspended in medium (DMEM/F12 + 20% HI-FBS + 20% L929 conditioned medium), and plated on petri dishes. On day 4, fresh medium was added to the plates. Experiments were performed on day 7.
Cholesterol ester quantification and turnover measurement
Cells were seeded in 6-well plates, and medium was changed to contain 1% FBS and loaded with 50 µg/ml of acetylated low-density lipoproteins (acLDLs) (Alfa Aesar, Ward Hill, MA) for 18 h. For quantification of total CEs, lipids were extracted. Cells were washed in PBS, and 750 µl of hexane:isopropanol (2:3) were added to each well and incubated rocking for 10 min at room temperature. The organic solvent phase was collected and dried. Lipids were resuspended in 40 µl of chloroform, spotted on a TLC plate, and developed in hexanes:ethylether:acetic acid (80:20:1). Cholesterol esters were quantified by charring with CuSO4 and densitometry. Values were normalized to protein content determined by BCA assay.
To measure cholesterol ester turnover, cells were loaded with 50 µg/ml acLDL for 12 h and subsequently labeled with 0.25 µCi/ml 14C-oleate (Perkin-Elmer, Waltham, MA) for 6 h. The medium was changed, and 10 µg/ml Sandoz 58-035 ACAT inhibitor and 2 µg/ml methyl-β-cyclodextrin (both from Sigma Aldrich, St. Louis, MO) were added. Lipids were extracted after 0, 8, and 24 h and separated by TLC as described above. The CE band was scraped and quantified by scintillation counting.
Metabolite and hormone measurements
Blood glucose levels were measured using a FreeStyle Lite glucosemeter (Abbott Diabetes Care, Alameda, CA). Serum leptin levels and liver glycogen levels were determined by the Yale Mouse Metabolic Phenotyping Center Analytical Core. Testosterone and corticosterone assays were performed by the Vanderbilt University Medical Center Hormone Assay and Analytical Services Core.
Lipidomics
Lipidomics analysis of white adipose tissue (WAT), brown adipose tissue (BAT), and liver of 4-week ad libitum-fed animals fed a high-fat diet (HFD) was performed as described (39). For liquid chromatography-mass spectrometer analysis of lipids from livers of 22-week HFD ad libitum-fed animals, lipids were extracted from liver corresponding to 75 μg of protein by chloroform/methanol extraction (38). Detected lipids were identified using LipidSearch (MKI, Tokyo, Japan). Peaks were defined through raw files, product ion, and precursor ion accurate masses. Lipid species were identified by database (>1,000,000 entries) search of positive and negative ion adducts. The accurate mass-extracted ion chromatograms were integrated for each identified lipid species and peak areas obtained for quantitation. An internal standard for phosphatidylinositol (PI 17:0–20:4; Avanti Polar Lipids, Alabaster, AL), which spiked prior to extraction, was used for normalization.
Proteomics
WATs and livers from wild-type and Ldah knockout mice were collected and processed by filter-aided sample preparation as described (40). Eluted peptides were analyzed by HPLC (EASY-nLC 1000, Thermo Scientific), combined with an Orbitrap mass spectrometer (Q Exactive HF, Thermo Scientific). Raw mass spectrometry data were processed by the MaxQuant software version 1.5.1.2, and statistical analyses were performed with the Perseus software (Max Planck Institute of Biochemistry, Munich, Germany (41)).
Lipid hydrolase activity assays
Mouse tissues were homogenized in buffer A (0.25 M sucrose, 1 mM EDTA, 1 mM DTT, 20 μg/ml leupeptine, 2 μg/ml antipain, 1 μg/ml pepstatin) followed by centrifugation at 20,000 g for 30 min at 4°C. The protein content of the 20,000 g infranatant was then determined by the Bio-Rad Protein Assay Kit with BSA as a standard.
Measurement of in vitro triacylglycerol (TG) hydrolase activity was as described (42). Briefly, 10 μg of WAT protein or 100 μg of liver protein in a total volume of 100 μl buffer A were incubated with 100 μl of a phospholipid-emulsified triolein substrate solution. The substrate for the measurement of TG-hydrolytic activity in WAT contained 1.67 mM triolein, 190 μM phosphatidylcholine/phosphatidylinositol (ratio 3:1), and 10 μCi/ml 3H-triolein and was prepared by sonication in 100 mM potassium phosphate buffer, pH 7.0 with 2% fatty acid-free BSA. For measurement of TG hydrolase activity in the liver, the substrate solution consisted of 0.32 mM triolein, 45 μM phosphatidylcholine/phosphatidylinositol (ratio 3/1), and 10 μCi/ml [9,10-3H] triolein and was prepared as described above. After 1 h at 37°C, released free fatty acids (FFAs) were extracted and quantified by liquid scintillation counting.
The measurement of in vitro CE hydrolase activities in WAT and liver was performed according to the measurement of TG-hydrolase activity using a phospholipid-emulsified cholesteryl oleate substrate solution, which consisted of 0.45 mM cholesteryl oleate, 0.45 mM PC/PI (ratio 3:1), and 1 μCi / ml 14C-cholesteryl oleate.
To measure hydrolase activity of LDAH protein in vitro, we used lysates of cells overexpressing LDAH or CG9186 for the lipid hydrolase activity assays.
Western blots
Tissues were lyzed in RIPA buffer with a dounce homogenizer and sonicated. For Western blot analysis, 50 µg protein of lysates were loaded. LDAH was detected using a polyclonal antibody directed against the C terminus of murine LDAH at a dilution 1:500 (17). The tubulin antibody was purchased from Sigma-Aldrich (Cat. no. T5168) and used at a dilution of 1:2000.
qRT-PCR analysis
The following primers were used in qRT-PCR analysis. Expression was normalized to the average of β-actin and cyclophilin levels.
mLDAH: 5′-CTTCACGTGATGAAGCGAGT-3′ (forward primer), 5′-AGTTGGGAAGAGCAGAAAGG-3′ (reverse primer); mHSL: 5′-ACGAGACAGGCCTCAGTGTGA-3′ (forward primer), 5′-CCACGCAACTCTGGGTCTATG-3′ (reverse primer); mATGL: 5′-GAGCCCCGGGGTGGAACAAGAT-3′ (forward primer), 5′-AAAAGGTGGTGGGCAGGAGTAAGG-3′ (reverse primer); mβ-Actin: 5′-CATCGTGGGCCGCTCTA-3′ (forward primer), 5′-CACCCACATAGGAGTCCTTCTG-3′ (reverse primer); mCyclophilin: 5′-TGGAAGAGCACCAAGACAACA-3′ (forward primer), 5′-TGCCGGAGTCGACAATGAT-3′ (reverse primer).
Statistics
Statistical significance was tested using Student t-test. For experiments with multiple time points, a two-way ANOVA was used (GraphPad Prism Software).
Value less than 0.05 would have been considered significant in all statistical analyses.
RESULTS
LDAH/C2Orf43 orthologs localize to lipid droplets via a hydrophobic domain
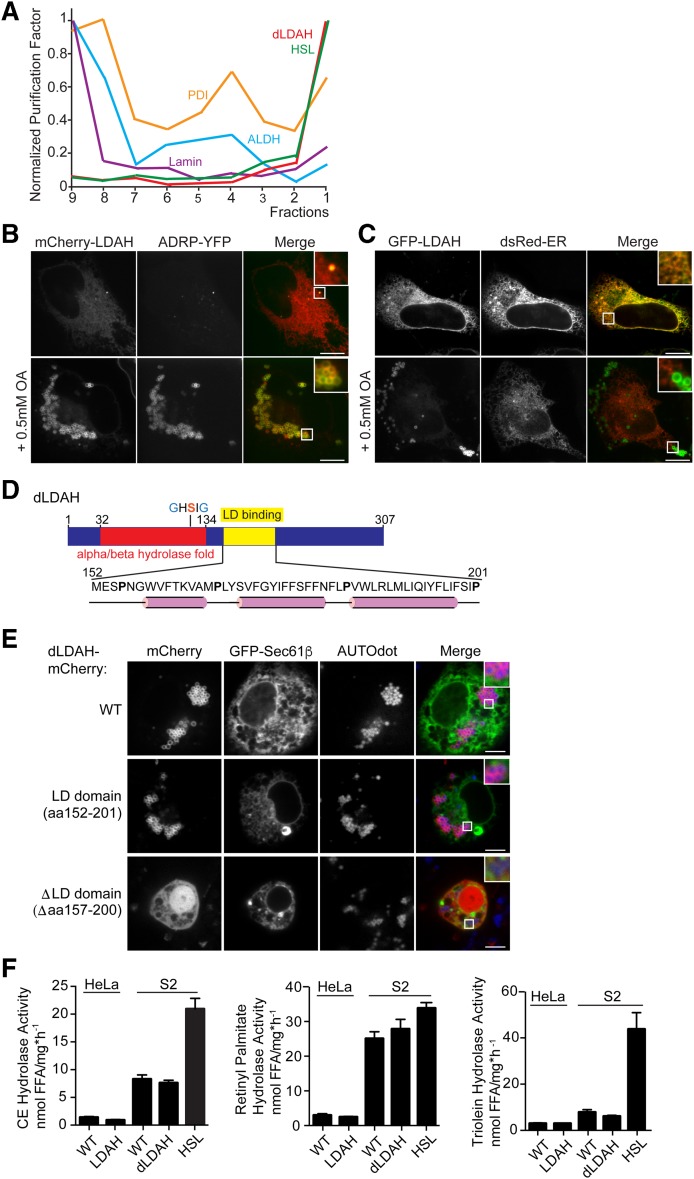
In Drosophila S2 cells and Saccharomyces cerevisiae, LDAH homologs copurify with LD proteins (43, 44). Drosophila CG9186 (referred to as dLDAH hereinafter) was highly enriched in the top fraction of a sucrose gradient used to separate cellular proteins, a purification profile typical of bona fide LD proteins, such as HSL (Fig. 1A). Consistent with this result, Drosophila and human LDAH localize to LDs (17, 45).
Fig. 1.
LDAH homologs localize to LDs with a hydrophobic hairpin. A: The Drosophila homolog of LDAH has the purification profile of a LD protein. Normalized purification factors of different organelle markers across a cellular fractionation are plotted. HSL is the LD marker; protein disulfide isomerase is the ER marker; alcohol dehydrogenase is the cytosolic marker; lamin is the nuclear marker. B, C: LDAH localizes to LDs or to the ER in the absence of LDs. B: mCherry-tagged LDAH colocalizes with ADRP in HeLa cells in the presence of LDs. Cells were transfected with constructs and treated with 0.5 mM oleic acid overnight. Representative images are shown. Scale bar, 10 μm. C: GFP-tagged LDAH localized to the ER in the absence of LDs. Cells were transfected with constructs and imaged the next day. Representative images are shown. Scale bar, 10 μm. D, E: A hydrophobic hairpin motif localizes LDAH to LDs. D: A hydrophobic segment comprising amino acids 160–195 of Drosophila CG9186/LDAH is predicted to have a hairpin structure and is responsible for LD binding. The α/β-hydrolase fold and the catalytic GxSxG-motif are indicated. α-Helices predicted by PSIPRED and JPred 4 are shown in pink. E: Full-length mCherry-tagged dLDAH and amino acids 152–201 of dLDAH localize to LDs after oleic acid treatment, while deletion of amino acids 157–200 results in ablation of LD binding. GFP-Sec61β was used to visualize the ER. Cells were transfected with constructs and treated with 1 mM oleic acid overnight. LDs were stained with AUTOdot (blue). Representative images are shown. Scale bar, 5 μm. F: LDAH overexpression does not affect cholesterol esterase, retinol esterase, or triacylglycerol hydrolysis activity. Nanomoles of free fatty acids (FFA) per (hour per milligram protein) ± SD. Values are means (n = 4). Activities were determined in lysates of WT HeLa or S2 cells and cells overexpressing the LDAH homologs using phospholipid-emulsified 3H-labeled lipids at neutral pH. S2 cells overexpressing HSL were used as a positive control. ALDH, alcohol dehydrogenase; PDI, protein disulfide isomerase
To confirm the cellular localization of human LDAH, also known as C2ORF43 (17), we expressed the protein fused to an mCherry or GFP-tag and examined its localization by confocal microscopy. mCherry-tagged LDAH colocalized with the LD marker ADRP in oleate-loaded HeLa cells (Fig. 1B). In the absence of LDs, LDAH localized to the endoplasmic reticulum (ER) (Fig. 1C).
Many LD proteins bind to LDs via a hydrophobic, membrane-embedded sequence (class I binding proteins) or an amphipathic helix (class II binding proteins) (46, 47). dLDAH contains a short hydrophobic motif (amino acids 152–201), predicted to be mostly α-helical but containing prolines (Fig. 1D), conserved among species, suggesting a hydrophobic hairpin, class I LD-binding motif. To examine this, we expressed a fusion construct of amino acids 152–201 with an N-terminal mCherry-tag in S2 cells. This protein localized to LDs in cells treated with oleic acid (Fig. 1E). In contrast, dLDAH in which amino acids 157–200 were replaced by a generic linker sequence (N-AAAGGGGSGGGGS-C) did not target LDs.
Since LDs store neutral lipids, such as CE, TG, and retinol esters, we measured hydrolase activity toward these substrates in lysates from cells overexpressing LDAH versus control cells (Fig. 1F). Consistent with a previous report (45), overexpression of mammalian or Drosophila LDAH did not increase hydrolase activity, whereas overexpression of HSL increased CE and TG hydrolysis activities
LDAH protein is absent in mice with a targeted gene-disruption allele
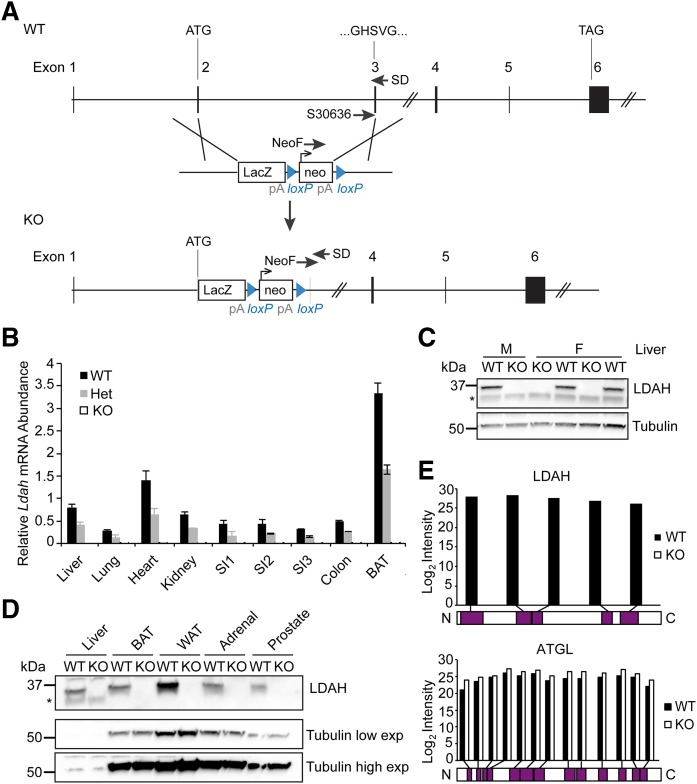
To investigate the physiological function of LDAH/C2ORF43, we generated an Ldah knockout mouse and evaluated it for metabolic phenotypes. Sequencing of the genomic locus of these animals confirmed replacement of a region from exon 2, downstream of the start codon, to base 169 of exon 3, including the predicted catalytic GxSxG motif (schematic shown in Fig. 2A), with a LacZ-Neomycin targeting cassette. Ldah mRNA was absent in tissues of Ldah knockout mice and ∼50% reduced in Ldah heterozygous mice (Fig. 2B).
Fig. 2.
LDAH is absent in mice with a targeted gene-disruption allele. A: A gene knockout cassette disrupts exons 2 and 3 of the Ldah gene. B–E: Ldah mRNA and protein are absent in Ldah KO animals. Schematic of the Ldah gene locus and targeting cassette (Knockout Mouse Project). Genotype of animals was confirmed with SD30636, NeoF, and SD primers. B: Ldah mRNA is reduced to 50% of the gene product in heterozygous mice and absent in Ldah KO animals. Relative Ldah mRNA abundance ± SD in different tissues of Ldah WT (black bars), heterozygous (gray bars), and KO mice (white bars) determined by qPCR. Ldah values were normalized to the average of β-actin and cyclophilin. Values are means (n = 3–4). C: Western blots against LDAH in liver tissue from male and female Ldah WT and KO animals confirmed loss of LDAH protein. Tubulin was used as a loading control. D: LDAH protein is undetectable in tissues of male Ldah KO animals by Western blot. Low and high exposures are shown for tubulin. E: Mass spectrometry (MS) analysis confirmed absence of LDAH in Ldah KO animals. Peptides that were identified by MS for LDAH (top) or ATGL (bottom) in WT or Ldah KO animals are mapped to the protein sequence. For ATGL, peptides were identified in both WT and KO tissue across the length of the protein. For LDAH, no peptides were identified in KO animals, and peptides from various parts of the protein were detected in lysates from WT tissue. Data from WAT and livers of two animals per genotype were combined for the graph.
Western blot analysis with an antibody binding an LDAH C-terminal antigen showed that the LDAH protein was present in many tissues of wild-type animals, including prostate, with highest levels found in WAT (Fig. 2D, supplemental Fig. S1A). In contrast, LDAH protein was undetectable in liver, BAT and WAT, adrenal gland, prostate, brain, spleen, kidney, testis, and bone marrow–derived macrophages (BMDM) of Ldah knockout mice (Fig. 2C, D, supplemental Fig. S1A). To further confirm the absence of the LDAH protein in knockout mice, we performed mass-spectrometry analysis of lysates from WAT and livers of wild-type and Ldah knockout mice. No LDAH peptides were detected in tissues of knockout mice, whereas five different, unique peptides covering different parts of the protein were detected in the wild-type samples (Fig. 2E). In contrast, the abundance of peptides derived from the TG lipase adipose triglyceride lipase (ATGL) were found at similar levels in each genotype. Consistent with these observations, we found that LDAH deletion did not result in a compensatory up-regulation of the major neutral lipid lipases ATGL or HSL, as determined by qPCR (supplemental Fig. S1B, C).
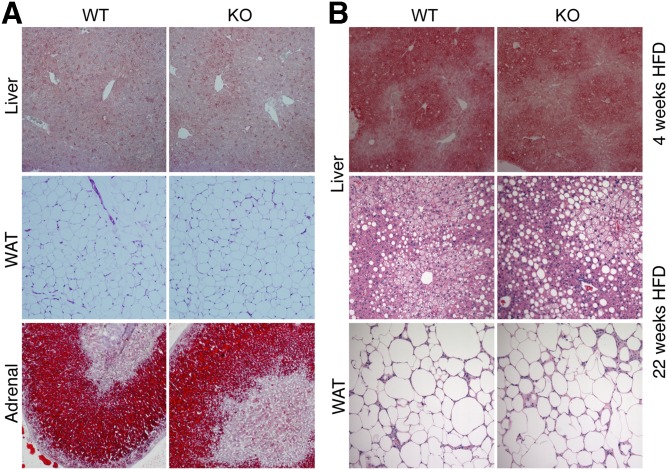
Ldah knockout animals were born at the expected Mendelian ratio (data not shown) and displayed no gross phenotypic changes or alterations in tissue morphology at 8–12 weeks of age (Fig. 3, supplemental Fig. S2). The size of adipocytes and amount of oil red O-staining of livers and adrenal glands were similar in Ldah wild-type and knockout animals (Fig. 3A, B, supplemental Fig. S2).
Fig. 3.
Histological analysis revealed no abnormalities in Ldah KO animals. A, B: Histology of tissues from animals fed a chow diet (A) or a HFD (B). A: Oil red O staining of liver and adrenal glands, and hematoxylin and eosin (H and E) staining of white adipose tissue (WAT) from 10- to 12-week-old Ldah wild-type or knockout animals fed a chow diet. B: Oil red O staining of livers after 4-week HFD feeding (top panel), and H and E staining of livers and WAT after 22 weeks on a HFD (bottom panels). Tissues are shown at a magnification of ×10.
Loss of LDAH does not affect body mass, body composition, glucose tolerance, or tissue lipid composition
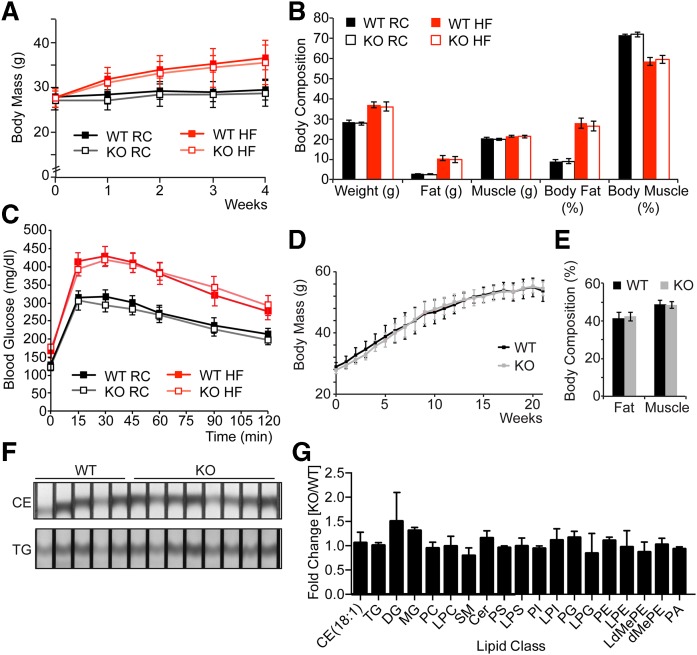
To analyze and challenge the metabolism of Ldah knockout animals, we placed male mice on rodent chow or lard-based 60% kcal% fat-containing diet. While all mice gained body weight more rapidly when fed a high-fat diet (HFD), knockout mice gained weight at a rate similar to their wild-type littermates (Fig. 4A). After 4 weeks on an HFD, we evaluated their energy metabolism. There were no differences in body composition (Fig. 4B), energy expenditure, locomotor activity, water or food intake, respiratory exchange ratio, oxygen consumption, or carbon dioxide production (supplemental Figs. S3, S4). Glucose homeostasis, as determined by oral glucose tolerance test, was also not affected on chow or HFD (Fig. 4C).
Fig. 4.
Loss of LDAH does not affect body mass gain, body composition, glucose tolerance, or tissue lipid composition. Loss of LDAH does not affect body mass gain, glucose tolerance, or body composition on chow or HFD. A: Ldah KO mice gain body mass as WT animals on chow or HFD. Body mass (g) ± SD of Ldah WT (closed squares) and KO (open squares) animals on chow (black squares) or 4-week HFD (red squares). Values are means (n = 6–8). B: Body composition (g or %) ± SEM of Ldah WT (closed bars) and KO (open bars) animals on chow (black bars) or 4-week HFD (red bars). Values are means (n = 6–8). C: Blood glucose (mg/dl) ± SEM of Ldah WT (closed squares) and KO (open squares) animals on chow (black squares) or 4-week HFD (red squares) after oral glucose tolerance test. Values are means (n = 6–8). D: Body mass (g) ± SD of Ldah WT (black squares) and KO (gray squares) animals on 22-week HFD. Values are means (n = 6–9). E: Body composition (g or %) ± SEM of Ldah WT (black bars) and KO (gray bars) animals on 22-week HFD. Values are means (n = 6–9). F, G: Lipid composition of livers after 22 weeks on HFD is not affected by loss of LDAH. F: CEs and TGs in liver lysates separated by TLC and stained by cerium molybdate as described [Krahmer et al., 2011 (34)]. G: Fold-change of lipid classes ± SD in Ldah KO versus WT animals determined by MS. Values are means (n = 4).
We assayed for a role of LDAH in lipid metabolism, but found no changes between genotypes on a HFD in serum lipids or levels of major lipid species in liver, WAT or BAT (Table 1, supplemental Fig. S5A, B). Neither did we detect differences in the accumulation of neutral lipids in the livers of Ldah knockout and control mice on chow or HFD as determined by oil red O staining (Fig. 3B). Major metabolic parameters and lipid classes were not changed in Ldah knockout mice on HFD (Fig. 3, supplemental Figs. S3–S5).
TABLE 1.
Serum parameters of Ldah WT and KO mice
| WT (n) | KO (n) | P | |
| Corticosterone (ng/ml) | 97.26 ± 75.65 | 114.02 ± 80.13 | 0.35 |
| (6) | (8) | ||
| Testosterone (ng/ml) | 2.03 ± 1.97 | 1.1 ± 1.34 | 0.18 |
| (7) | (6) | Chow diet, ad lib | |
| Cholesterol (mg/dl) | 132.62 ± 30.05 | 137.87 ± 8.21 | 0.34 |
| (6) | (6) | ||
| HDL (mg/dl) | 97.85 ± 21.02 | 99.91 ± 5.04 | 0.41 |
| (6) | (6) | ||
| LDL (mg/dl) | 17.36 ± 3.17 | 17.62 ± 2.04 | 0.43 |
| (6) | (6) | ||
| TG (mg/dl) | 52.45 ± 25.24 | 64.34 ± 30.58 | 0.24 |
| (6) | (6) | ||
| NEFA (mmol/L) | 0.92 ± 0.23 | 0.99 ± 0.21 | 0.29 |
| (6) | (6) | ||
| B-Hb (mmol/L) | 0.18 ± 0.06 | 0.14 ± 0.03 | 0.08 |
| (6) | (6) | 4 weeks 60% high-fat diet, ad lib | |
| TG (mg/dl) | 84.02 ± 7.97 | 75.09 ± 9.21 | 0.11 |
| (3) | (4) | ||
| B-Hb (mmol/L) | 0.51 ± 0.17 | 0.49 ± 0.11 | 0.44 |
| (3) | (4) | ||
| Glucose (mg/dl) | 166.00 ± 12.31 | 171.88 ± 20.25 | 0.26 |
| (6) | (9) | ||
| Leptin (ng/ml) | 44.19 ± 7.29 | 44.54 ± 6.02 | 0.45 |
| (6) | (9) | 21 weeks 60% high-fat diet, fasted | |
| Cholesterol (mg/dl) | 68.49 ± 8.14 | 67.46 ± 3.78 | 0.38 |
| (6) | (7) | ||
| HDL (mg/dl) | 51.60 ± 3.86 | 51.20 ± 4.36 | 0.43 |
| (6) | (7) | ||
| LDL (mg/dl) | 5.50 ± 0.80 | 5.73 ± 0.31 | 0.25 |
| (6) | (7) | ||
| TG (mg/dl) | 65.38 ± 9.05 | 62.01 ± 10.84 | 0.28 |
| (6) | (7) | ||
| NEFA (mmol/L) | 1.93 ± 0.28 | 1.81 ± 0.41 | 0.28 |
| (6) | (7) | ||
| B-Hb (mmol/L) | 1.96 ± 0.41 | 2.19 ± 0.58 | 0.22 |
| (6) | (7) | Fasted, cold-exposed |
Serum parameters measured in different experiments for WT and Ldah KO animals are reported. The number of animals of a certain genotype (n) used for each study is shown in parentheses. Measurements belonging to one experiment are grouped together, with different experiments being separated by double rules. The feeding state and, where applicable, additional treatments of animals in a given experiment are indicated in the last column. Student’s t-test was performed for statistical analysis. B-Hb, β-hydroxybutyrate; NEFA, nonesterified fatty acid.
Because phenotypes related to lipid metabolism were not observed on chow diet or with short-term HFD, we further challenged the metabolism of the Ldah knockout mice by feeding them a 60% HFD for 22 weeks. Despite the longer dietary challenge, body weight gain was not affected by LDAH loss (Fig. 4D). Neither was the percentage of body fat (Fig. 4E), fasted serum glucose, leptin, TG, or ketone bodies (Table 1). There were no apparent changes in total TG, CEs, or other major lipid species as determined by thin layer chromatography and lipidomics (Fig. 4F, G). Liver glycogen content was comparable between genotypes (supplemental Fig. S5C). The histology of liver, WAT and BAT, and heart tissues were not affected in Ldah knockout mice after 22 weeks on HFD (Fig. 3B, supplemental Fig. S2).
In an overnight fasting experiment, with subsequent cold exposure without access to food, knockout animals tolerated this stress slightly better than wild-type animals by maintaining their body temperature in the cold. However, the differences were minor and only significant at some time points, and overall both genotypes maintained blood glucose levels and body weight (supplemental Fig. S6).
Loss of LDAH does not affect cholesterol ester turnover or hydrolysis
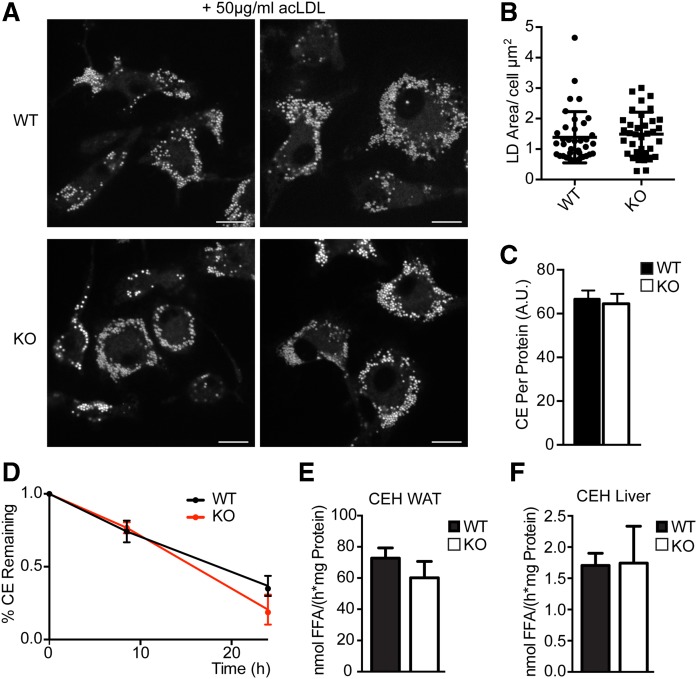
Since LDAH had been implicated in CE hydrolysis and efflux from macrophages (17), we assayed CE metabolism in Ldah knockout mice. We isolated macrophages from bone marrow of wild-type and knockout mice and tested CE accumulation and turnover after acetylated LDL (acLDL) treatment. After overnight incubation with 50 µg/ml acLDL, Ldah wild-type and knockout cells had similar amounts of LDs and CEs (Fig. 5A–C). Using radioactive oleic acid to label lipids, we determined the turnover of CEs with ACAT1 inhibitor and β-cyclodextrin as a cholesterol acceptor in the medium and found no differences in macrophages lacking LDAH (Fig. 5D). Consistent with these results, we observed no changes in CE hydrolysis activity in WAT or liver lysates from Ldah knockout animals (Fig. 5E, F). TG hydrolysis activity was also similar (supplemental Fig. S7). Corticosterone and testosterone levels were similar in wild-type and knockout mice, suggesting it is unlikely that LDAH affects murine steroid hormone metabolism (Table 1).
Fig. 5.
Loss of LDAH does not affect cholesterol ester (CE) turnover or hydrolysis. CE storage in bone marrow-derived macrophages was not affected by LDAH loss. Cells were treated with 50 µg/ml AcLDL for 18 h before imaging or lipid extraction. A: Representative images are shown. LDs were stained with BODIPY. Scale bar, 10 μm. B: LD area per cell (µm2). Values are means, and individual data points are plotted (n > 30). C: CEs per protein determined by thin-layer chromatography. D: LDAH does not play a role in cholesterol ester turnover in bone marrow-derived macrophages. Percentage CEs ± SD remaining. Values are means (n = 3). E, F: LDAH deficiency does not affect cholesterol esterase activity. CE hydrolase activity in white adipose tissue (WAT) and liver of Ldah WT (black bars) and KO mice (white bars). Activities were determined in the 20,000 g infranatant using phospholipid-emulsified 14C-labeled cholesterol oleate at neutral pH. E: Nanomoles of free fatty acids (FFA) per (hours per milligram protein) ± SD in WAT of Ldah WT (black bars) or KO mice (white bars). Values are means (n = 5). F: Nanomoles of FFA per (hours per milligrams protein) ± SD in livers of Ldah WT (black bars) or KO mice (white bars). Values are means (n = 7–8). CEH, cholesterol ester hydrolase.
DISCUSSION
We show here that the LD-localized putative lipase LDAH does not have a major, physiologically detectable role in murine CE metabolism. These findings contrast with previous in vitro studies that reported LDAH functions as a CE hydrolase (17). In our animal studies, LDAH deletion did not change CE levels, CE hydrolysis activity, or the metabolism of TGs or other major lipid classes. Moreover, we found no evidence for CE hydrolysis activity in in vitro assays of lysates with the overexpressed enzyme. We also found no evidence for a role of LDAH in whole-body energy metabolism.
Because we found no changes in CE metabolism in LDAH-deficient mice, it is unlikely that LDAH has CE hydrolysis activity and any role in macrophage cholesterol efflux. The previous report (17) had limitations. For example, the CE hydrolysis activity reported was minimal in comparison with the activity of the known CE hydrolase HSL, and the differences shown for overexpression of LDAH in comparison with a catalytically dead enzyme were negligible. Our results are consistent with another study that detected no activity toward any major lipid species, including CEs in in vitro assays (45). However, at present, it remains possible that under specific physiological or pathological conditions, LDAH might play a role in CE or TG hydrolysis, or both, in macrophages. We also cannot exclude possible redundancy of LDAH activity with other murine cholesterol esterases, which might compensate for LDAH loss in vivo.
LDAH will need to be tested for alternative activities. We ruled out lipase activity toward major species of CEs, TGs, and other lipid classes. However, LDAH might hydrolyze a structurally similar, potentially lowly abundant or difficult-to-detect molecule, such as a modified sterol ester, oxysterol ester, or ether lipid. We found no differences in total sterols or oxysterols in WAT or liver from LDAH knockout mice and no activity toward ether TG in overexpression experiments (data not shown). Our efforts to reveal the substrate of LDAH by untargeted mass spectrometry-based lipidomics have been, thus far, unsuccessful.
LDAH homologs localize to LDs via a hydrophobic hairpin targeting motif, in agreement with previous reports (45). Accordingly, LDAH is a class I LD protein (i.e., targeting LDs from the ER). The hydrophobicity of LDAHs’ LD targeting motif and low propensity to be displaced from shrinking LDs during lipolysis (35) suggest that LDAH evolved for optimal substrate access to the hydrophobic LD core. However, whether LDAH has an enzymatic function on LDs is not known.
With no known function, the mechanism linking LDAH to human disease remains enigmatic. A SNP in the LDAH gene associated with reduced mRNA expression is also associated with an increased risk for prostate cancer (9, 10, 13, 14). Prostate cancer is one of the cancer types known to upregulate lipid metabolism, and CE accumulation in LDs is associated with increased prostate cancer aggressiveness (6, 18). An intriguing possibility is that cancer development could be linked to changes in LDAH enzymatic activity. Lower expression of LDAH in the prostate itself due to a sequence variation could result in remodeling of lipid metabolism to promote cancer growth. However, we did not find a role for LDAH in lipid metabolism or in prostate cancer development, because we have found no signs of prostate neoplasia or cancer in the knockout mice, albeit we analyzed only a small number of animals at 1 year of age (data not shown). One confounding factor in interpreting the human genetic studies and a potential connection among LDAH, lipid metabolism, and cancer is the proximity of the LDAH and APOB loci. The modest linkage disequilibrium found between the LDAH SNP with SNPs near APOB makes the effect of genetic variants of LDAH on lipid metabolism and cancer risk difficult to dissect from those which might be driven by apoB. SNPs can also exert regulatory effects over hundreds of kilobases (48). Thus, the LDAH SNP (or linked polymorphisms close by) could have an effect on APOB expression, which in turn might be responsible for the genetic associations observed for this locus.
The expression pattern of LDAH in humans suggests an additional link to prostate cancer. Although LDAH levels appear to be high in WAT in mice, LDAH levels are high in adrenal glands in humans (http://www.humanproteomemap.org; www.proteomicsdb.org; www.gtexportal.org), the primary production site of steroid hormones such as androgens from cholesterol (49). Many prostate cancer cells depend on androgenic signals from the adrenal gland (50, 51), and therefore, LDAH might affect risk for prostate cancer by affecting androgenic metabolism in adrenal gland LDs.
In summary, our data suggest that SNPs in LDAH affect prostate cancer risk through a mechanism other than CE hydrolysis activity and that LDAH has an alternative LD-associated metabolic function. Whether LDAH functions in the metabolism of lipids, other metabolites, or perhaps xenobiotics remains to be elucidated.
Supplementary Material
Acknowledgments
The authors thank members of the Farese and Walther laboratory and Drs. Lorelei Mucci, Ericka Abot, and Stefan Stender for advice and helpful discussions. We also thank Grisell Diaz-Ramirez and Drs. Huajin Wang and Carrie Grueter for experimental help. The authors thank Dr. Michael Jurczak and the Yale Mouse Metabolic Phenotyping Center for mouse in vivo metabolism analyses and Dr. Roderick Bronson (at the rodent pathology core at Harvard Medical School) for pathology expertise. We also thank Drs. Jeffrey McDonald and Sarah Martin for sterol and additional lipid analysis, Dr. Christoph Heier for ether lipid activity assays (data not shown), and Dr. Alan Saghatelian for advice on untargeted lipidomics. The authors thank Gary Howard for editorial assistance and Dr. Young-Hwa Goo for the generous gift of the LDAH antibody.
Footnotes
Abbreviations:
- ATGL
- adipose triglyceride lipase
- BAT
- brown adipose tissue
- CE
- cholesterol ester
- ER
- endoplasmic reticulum
- HFD
- high-fat diet
- HSL
- hormone sensitive lipase
- LD
- lipid droplet
- LDAH
- lipid droplet-associated hydrolase
- TG
- triacylglycerol
- WAT
- white adipose tissue
This work was supported by the National Institues of Health Grant 1R01GM097194 (to T.C.W.), Grant R01DK101579 (to R.V.F.), the G. Harold and Leila Y. Mathers Foundation (to T.C.W.), and J. David Gladstone Institute (to R.V.F.). N.K. was supported by an American Heart Association predoctoral fellowship. National Institutes of Health grants to Velocigene at Regeneron Inc. (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted embryonic stem cells for 8,500 genes in the Knockout Mouse Project (KOMP) and archived and distributed by the KOMP Repository at the University of California, Davis, and Children’s Hospital Oakland Research Institute (U42RR024244). Testosterone and corticosterone assays were performed by the Vanderbilt University Medical Center Hormone Assay and Analytical Services Core, which is supported by National Institutes of Health Grants DK059637 and DK020593. T.C.W is an investigator of the Howard Hughes Medical Institute. Dr. Michael Jurczak and the Yale Mouse Metabolic Phenotyping Center were funded by National Institutes of Health Grant DK059635. The content is solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Hashemi H. F., and Goodman J. M.. 2015. The life cycle of lipid droplets. Curr. Opin. Cell Biol. 33: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krahmer N., Farese R. V. Jr., and Walther T. C.. 2013. Balancing the fat: lipid droplets and human disease. EMBO Mol. Med. 5: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Straub B. K., Herpel E., Singer S., Zimbelmann R., Breuhahn K., Macher-Goeppinger S., Warth A., Lehmann-Koch J., Longerich T., Heid H., et al. . 2010. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Mod. Pathol. 23: 480–492. [DOI] [PubMed] [Google Scholar]
- 4.Hager M. H., Solomon K. R., and Freeman M. R.. 2006. The role of cholesterol in prostate cancer. Curr. Opin. Clin. Nutr. Metab. Care. 9: 379–385. [DOI] [PubMed] [Google Scholar]
- 5.Schlaepfer I. R., Rider L., Rodrigues L. U., Gijón M. A., Pac C. T., Romero L., Cimic A., Sirintrapun S. J., Glodé L. M., Eckel R. H., et al. . 2014. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Mol. Cancer Ther. 13: 2361–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu X., Daniels G., Lee P., and Monaco M. E.. 2014. Lipid metabolism in prostate cancer. Am. J. Clin. Exp. Urol. 2: 111–120. [PMC free article] [PubMed] [Google Scholar]
- 7.Tamura K., Makino A., Hullin-Matsuda F., Kobayashi T., Furihata M., Chung S., Ashida S., Miki T., Fujioka T., Shuin T., et al. . 2009. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 69: 8133–8140. [DOI] [PubMed] [Google Scholar]
- 8.Drabkin H. A., and Gemmill R. M.. 2012. Cholesterol and the development of clear-cell renal carcinoma. Curr. Opin. Pharmacol. 12: 742–750. [DOI] [PubMed] [Google Scholar]
- 9.Shui I. M., Lindström S., Kibel A. S., Berndt S. I., Campa D., Gerke T., Penney K. L., Albanes D., Berg C., Bueno-de-Mesquita H. B., et al. . 2014. Prostate cancer (PCa) risk variants and risk of fatal PCa in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Eur. Urol. 65: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takata R., Akamatsu S., Kubo M., Takahashi A., Hosono N., Kawaguchi T., Tsunoda T., Inazawa J., Kamatani N., Ogawa O., et al. . 2010. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat. Genet. 42: 751–754. [DOI] [PubMed] [Google Scholar]
- 11.Long Q. Z., Du Y. F., Ding X. Y., Li X., Song W. B., Yang Y., Zhang P., Zhou J. P., and Liu X. G.. 2012. Replication and fine mapping for association of the C2orf43, FOXP4, GPRC6A and RFX6 genes with prostate cancer in the Chinese population. PLoS One. 7: e37866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindström S., Schumacher F. R., Campa D., Albanes D., Andriole G., Berndt S. I., Bueno-de-Mesquita H. B., Chanock S. J., Diver W. R., Ganziano J. M., et al. . 2012. Replication of five prostate cancer loci identified in an Asian population—results from the NCI Breast and Prostate Cancer Cohort Consortium (BPC3). Cancer Epidemiol. Biomarkers Prev. 21: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penney K. L., Sinnott J. A., Tyekucheva S., Gerke T., Shui I. M., Kraft P., Sesso H. D., Freedman M. L., Loda M., Mucci L. A., et al. . 2015. Association of prostate cancer risk variants with gene expression in normal and tumor tissue. Cancer Epidemiol. Biomarkers Prev. 24: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Innocenti F., Cooper G. M., Stanaway I. B., Gamazon E. R., Smith J. D., Mirkov S., Ramirez J., Liu W., Lin Y. S., Moloney C., et al. . 2011. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 7: e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon G. M., and Cravatt B. F.. 2010. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J. Biol. Chem. 285: 11051–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenfant N., Hotelier T., Velluet E., Bourne Y., Marchot P., and Chatonnet A.. 2013. ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 41: D423–D429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goo Y. H., Son S. H., Kreienberg P. B., and Paul A.. 2014. Novel lipid droplet-associated serine hydrolase regulates macrophage cholesterol mobilization. Arterioscler. Thromb. Vasc. Biol. 34: 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue S., Li J., Lee S. Y., Lee H. J., Shao T., Song B., Cheng L., Masterson T. A., Liu X., Ratliff T. L., et al. . 2014. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 19: 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Gonzalo-Calvo D., López-Vilaró L., Nasarre L., Perez-Olabarria M., Vázquez T., Escuin D., Badimon L., Barnadas A., Lerma E., and Llorente-Cortés V.. 2015. Intratumor cholesteryl ester accumulation is associated with human breast cancer proliferation and aggressive potential: a molecular and clinicopathological study. BMC Cancer. 15: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lettre G., Palmer C. D., Young T., Ejebe K. G., Allayee H., Benjamin E. J., Bennett F., Bowden D. W., Chakravarti A., Dreisbach A., et al. . 2011. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 7: e1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen H., Damcott C. M., Rampersaud E., Pollin T. I., Horenstein R. B., McArdle P. F., Peyser P. A., Bielak L. F., Post W. S., Chang Y. P., et al. . 2010. Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the old order amish. Arch. Intern. Med. 170: 1850–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Igarashi M., Osuga J., Uozaki H., Sekiya M., Nagashima S., Takahashi M., Takase S., Takanashi M., Li Y., Ohta K., et al. . 2010. The critical role of neutral cholesterol ester hydrolase 1 in cholesterol removal from human macrophages. Circ. Res. 107: 1387–1395. [DOI] [PubMed] [Google Scholar]
- 23.Buchebner M., Pfeifer T., Rathke N., Chandak P. G., Lass A., Schreiber R., Kratzer A., Zimmermann R., Sattler W., Koefeler H., et al. . 2010. Cholesteryl ester hydrolase activity is abolished in HSL−/− macrophages but unchanged in macrophages lacking KIAA1363. J. Lipid Res. 51: 2896–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraemer F. B., and Shen W. J.. 2002. Hormone-sensitive lipase: control of intracellular tri-(di-)acylglycerol and cholesteryl ester hydrolysis. J. Lipid Res. 43: 1585–1594. [DOI] [PubMed] [Google Scholar]
- 25.Sekiya M., Osuga J., Yahagi N., Okazaki H., Tamura Y., Igarashi M., Takase S., Harada K., Okazaki S., Iizuka Y., et al. . 2008. Hormone-sensitive lipase is involved in hepatic cholesteryl ester hydrolysis. J. Lipid Res. 49: 1829–1838. [DOI] [PubMed] [Google Scholar]
- 26.Contreras J. A. 2002. Hormone-sensitive lipase is not required for cholesteryl ester hydrolysis in macrophages. Biochem. Biophys. Res. Commun. 292: 900–903. [DOI] [PubMed] [Google Scholar]
- 27.Lohse P., Lohse P., Chahrokh-Zadeh S., and Seidel D.. 1997. Human lysosomal acid lipase/ cholesteryl ester hydrolase and human gastric lipase: site-directed mutagenesis of Cys227 and Cys236 results in substrate-dependent reduction of enzymatic activity. J. Lipid Res. 38: 1896–1905. [PubMed] [Google Scholar]
- 28.Ouimet M., and Marcel Y. L.. 2012. Regulation of lipid droplet cholesterol efflux from macrophage foam cells. Arterioscler. Thromb. Vasc. Biol. 32: 575–581. [DOI] [PubMed] [Google Scholar]
- 29.Du H., Duanmu M., Witte D., and Grabowski G. A.. 1998. Targeted disruption of the mouse lysosomal acid lipase gene: long-term survival with massive cholesteryl ester and triglyceride storage. Hum. Mol. Genet. 7: 1347–1354. [DOI] [PubMed] [Google Scholar]
- 30.Drozdetskiy A., Cole C., Procter J., and Barton G. J.. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43: W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchan D. W., Minneci F., Nugent T. C., Bryson K., and Jones D. T.. 2013. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41: W349–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195–202. [DOI] [PubMed] [Google Scholar]
- 33.Wilfling F., Wang H., Haas J. T., Krahmer N., Gould T. J., Uchida A., Cheng J. X., Graham M., Christiano R., Frohlich F., et al. . 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24: 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H. W., Schmidt-Supprian M., Vance D. E., Mann M., et al. . 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14: 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kory N., Thiam A. R., Farese R. V. J., and Walther T. C.. 2015. Protein crowding is a determinant of lipid droplet composition. Dev. Cell. 34: 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valenzuela D. M., Murphy A. J., Frendewey D., Gale N. W., Economides A. N., Auerbach W., Poueymirou W. T., Adams N. C., Rojas J., Yasenchak J., et al. . 2003. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol. 21: 652–659. [DOI] [PubMed] [Google Scholar]
- 37.Ayala J. E., Samuel V. T., Morton G. J., Obici S., Croniger C. M., Shulman G. I., Wasserman D. H., and McGuinness O. P., and NIH Mouse Metabolic Phenotyping Center Consortium . 2010. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis. Model. Mech. 3: 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 39.Saghatelian A., Trauger S. A., Want E. J., Hawkins E. G., Siuzdak G., and Cravatt B. F.. 2004. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 43: 14332–14339. [DOI] [PubMed] [Google Scholar]
- 40.Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M.. 2009. Universal sample preparation method for proteome analysis. Nat. Methods. 6: 359–362. [DOI] [PubMed] [Google Scholar]
- 41.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J.. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13: 731–740. [DOI] [PubMed] [Google Scholar]
- 42.Schweiger M., Eichmann T. O., Taschler U., Zimmermann R., Zechner R., and Lass A.. 2014. Measurement of lipolysis. Methods Enzymol. 538: 171–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krahmer N., Hilger M., Kory N., Wilfling F., Stoehr G., Mann M., Farese R. V. Jr., and Walther T. C.. 2013. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell. Proteomics. 12: 1115–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Currie E., Guo X., Christiano R., Chitraju C., Kory N., Harrison K., Haas J., Walther T. C., and Farese R. V. Jr. 2014. High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J. Lipid Res. 55: 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiel K., Heier C., Haberl V., Thul P. J., Oberer M., Lass A., Jäckle H., and Beller M.. 2013. The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J. Cell Sci. 126: 2198–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiam A. R., Farese R. V. Jr., and Walther T. C.. 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14: 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kory N., Farese R. V. Jr., and Walther T. C.. 2016. Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol. 26: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Heyningen V., and Bickmore W.. 2013. Regulation from a distance: long-range control of gene expression in development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20120372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker L. N. 1991. Control of adrenal androgen secretion. Endocrinol. Metab. Clin. North Am. 20: 401–421. [PubMed] [Google Scholar]
- 50.Chatterjee B. 2003. The role of the androgen receptor in the development of prostatic hyperplasia and prostate cancer. Mol. Cell. Biochem. 253: 89–101. [DOI] [PubMed] [Google Scholar]
- 51.Huggins C. 1942. Effect of orchiectomy and irradiation on cancer of the prostate. Ann. Surg. 115: 1192–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.