Abstract
Objective
Early detection of Heart Failure (HF) could mitigate the enormous individual and societal burden from this disease. Clinical detection is based, in part, on recognition of the multiple signs and symptoms comprising the Framingham HF diagnostic criteria that are typically documented, but not necessarily synthesized, by primary care physicians well before more specific diagnostic studies are done. We developed a natural language processing (NLP) procedure to identify Framingham HF signs and symptoms among primary care patients, using electronic health record (EHR) clinical notes, as a prelude to pattern analysis and clinical decision support for early detection of HF.
Design
We developed a hybrid NLP pipeline that performs two levels of analysis: (1) At the criteria mention level, a rule-based NLP system is constructed to annotate all affirmative and negative mentions of Framingham criteria. (2) At the encounter level, we construct a system to label encounters according to whether any Framingham criterion is asserted, denied, or unknown.
Measurements
Precision, recall, and F-score are used as performance metrics for criteria mention extraction and for encounter labeling.
Results
Our criteria mention extractions achieve a precision of 0.925, a recall of 0.896, and an F-score of 0.910. Encounter labeling achieves an F-score of 0.932.
Conclusion
Our system accurately identifies and labels affirmations and denials of Framingham diagnostic criteria in primary care clinical notes and may help in the attempt to improve the early detection of HF. With adaptation and tooling, our development methodology can be repeated in new problem settings.
Keywords: Natural language processing, Text mining, Heart failure, Electronic health records, Diagnostic criteria
1. Introduction and objective
The individual and societal impact of heart failure (HF) is staggering. One in five US citizens over age 40 is expected to develop HF during their lifetimes. It is currently the leading cause of hospitalization among Medicare beneficiaries and, with an aging U.S. population, HF prevalence and related costs will only increase, as prevalence of HF is expected to double by 2030 [1]. Individual and societal burdens may be mitigated through early detection of HF and intervention with lifestyle changes and proven preventive therapies.
Identifying the early manifestations of HF in the primary care setting is not straightforward. HF is a complex pathophysiologically heterogeneous syndrome, with substantial individual variability in expression. Moreover, because the signs and symptoms are also expressed for multiple causal factors unrelated to HF (e.g., chronic obstructive pulmonary disease, venous insufficiency, kidney disease), both false positive and false negative rates of diagnosis are relatively high in primary care [2,3].
The Framingham heart failure criteria published in 1971 [4] are based on clinical data acquired in the 1950s and 1960s but are still the most common HF signs and symptoms documented by primary care physicians (PCPs) today, usually well before more specific diagnostic studies are considered. But, relatively little is known about how these criteria are documented by PCPs or the extent to which these criteria vary in their sensitivity and specificity to HF diagnosis. In fact, when originally developed, the Framingham criteria only identified approximately half of the patients who had previously been diagnosed clinically with HF [4]. While other clinical criteria for HF have been developed, the agreement among different criteria is poor to moderate at best [5].
Ambulatory care is rapidly changing, especially with regard to adoption of electronic health records (EHR). Despite the structured information in EHRs – such as diagnosis codes, medications, and lab results – large portions of EHR data are still in narrative text format, principally in clinical encounter notes and imaging notes. There are widely recognized barriers to the application of NLP tools to such data [6–8].
This paper presents results of using NLP to extract Framingham criteria from clinical notes of primary care patients with and without HF. This work is part of a larger project, called PredMED, which is focused on the early detection and management of HF [9,10]. In PredMED, the extracted criteria serve as features for various downstream statistical and machine-learning applications. To the best of our knowledge, there are no published studies of text extraction for the Framingham HF criteria as they are documented in primary care. Lin et al. [11] reported some success in using the MedLEE parser [12] on discharge summaries and radiology reports to predict ICD-9 codes for HF diagnosis. More recent work [13,14] is based on the Unstructured Information Management Architecture (UIMA) framework, as is ours. More generally, the NLP extraction work done within the i2b2 competition [15–20] is similar to work we describe herein, with the crucial difference that our EHR dataset does not have pre-existing reference standard annotations. We describe iterative annotation methods similar to those found in other NLP work on EHR entity extraction [21,23–26] that were essential to developing our reference standards.
2. Materials and methods
An NLP application was developed and validated for identifying affirmations and denials of fifteen of the seventeen Framingham criteria for HF shown in Table 1.
Table 1.
Framingham diagnostic criteria for heart failure. Circulation time and change in vital capacity are not routinely evaluated in current clinical practice.
| Extracted criteria code names | |
|---|---|
| Major criteria | |
| Paroxysmal nocturnal dyspnea or orthopnea | PNDyspnea (PND) |
| Neck vein distention | JVDistension (JVD) |
| Rales | Rales (RALE) |
| Radiographic cardiomegaly | RCardiomegaly (RC) |
| Acute pulmonary edema | APEdema (APED) |
| S3 gallop | S3Gallop (S3G) |
| Central venous pressure > 16 cm H2O | ICV Pressure (ICV) |
| Circulation time of 25 s | (not extracted) |
| Hepatojugular reflux | HJReflux (HJR) |
| Weight loss of 4.5 kg in 5 days, in response to HF treatment | WeightLoss (WTL) |
| Minor criteria | |
| Bilateral ankle edema | AnkleEdema (ANKED) |
| Nocturnal cough | NightCough (NC) |
| Dyspnea on ordinary exertion | DOExertion (DOE) |
| Hepatomegaly | Hepatomegaly (HEP) |
| Pleural effusion | PleuralEffusion (PLE) |
| A decrease in vital capacity by 1/3 of max | (not extracted) |
| Tachycardia (rate of ≥120 min−1) | Tachycardia (TACH) |
2.1. Source of data
Data for this study were obtained from the Geisinger Clinic (GC) primary care practice EHRs. The dataset consisted of the full encounter records for 6355 incident primary care HF patients diagnosed between 2003 and 2010, as previously described [27], and up to ten clinic-, sex-, and age-matched control patients for each HF case. There were 26,052 controls. In total, there were over 3.3 million clinical notes, comprising over 4 gigabytes of text. While there were 56 different encounter types, “Office Visit” accounted for 81% of all encounters, followed by “Case Manager” (8%) and “Radiology” (7%).
2.2. Tools
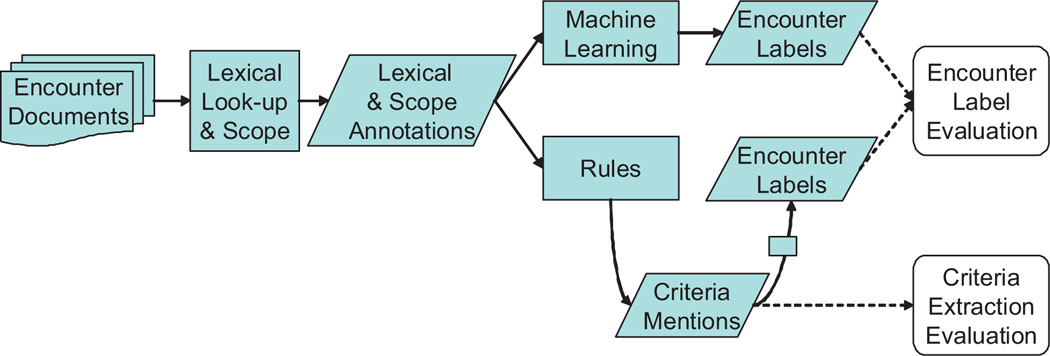
We built a text analysis pipeline (Fig. 1) to extract Framingham criteria, using Language Ware [28] for basic text processing and the IBM LanguageWare Resource Workbench (LRW) to develop dictionaries and grammars. The resulting analytics were then inserted into a UIMA [29,30] pipeline, which provides for acquisition of the clinical note texts and the other steps in Fig. 1. We also used a concordance program [31] to avoid overtraining to the development encounters, by letting us understand the behavior of our analytics on the entire encounter corpus.
Fig. 1.
High-level PredMED text analysis pipeline.
2.3. Methods
Our development and evaluation process comprised the following steps:
A cardiologist and a linguist analyzed a development dataset of 65 encounter documents rich in Framingham criteria, to learn the linguistics of criteria mentions.
The linguist used the NLP tools to build initial extractors for assertions and denials of Framingham criteria (this section).
The clinical expert and linguist incrementally measured and improved the performance of the extractors on the development documents (Section 2.4).
The clinical expert used annotation guidelines he developed to train coders, who manually created gold standard annotations on an evaluation dataset of 400 randomly selected encounter documents (Section 3).
The linguist used the gold standard to measure the performance of the final extractors on two tasks, criteria extraction and encounter labeling (Section 4).
Text analysis involved the following tasks:
Basic text processing encompassed standard Language-Ware analytics, including paragraph and sentence boundary detection, tokenization, dictionary look-up, morphological analysis, and part-of-speech tagging.
Dictionaries and grammars served to recognize words and phrases used to express Framingham HF criteria and other possible indicators of HF, segment beginnings, and various syntactic structures.
Text Analysis Engines (TAEs) were built for disambiguating and for applying constraints to candidate criteria mentions produced by the LanguageWare components. TAEs were also used to decide when criteria were negated or occurred in counterfactual contexts.
2.3.1. Dictionaries
There are 10 dictionaries in PredMED. The principal dictionary, FramSymptomVocab, contains entries for most of the Framingham diagnostic criteria. Each entry contains a main spelling (the “lemma”) and variant forms; for example, the entry for AnkleEdema has ankle edema as its lemma, along with variants for edema, leg edema, pedal edema, oedema, etc. There are around 75 such entries, comprising hundreds of variants.
In addition to FramSymptomVocab, there are dictionaries for:
negating words and counterfactual triggers, such as denies and if.
segment header words.
weight loss phrases, time value words, weight unit words, and diuretic words, used in the WeightLoss criterion extractor.
2.3.2. Grammars
Grammars are implemented as LanguageWare “parsers” and consist of cascaded finite-state automata that build UIMA annotations over recognized spans of text, using the shallow parser technique described by Boguraev [32]. Since diagnostic criteria occur mostly in noun phrases, the most important PredMED grammar recognizes compound noun phrases. An example, containing two Framingham criteria (underlined), is chest pain, DOE, or night cough. As a by-product of noun phrase recognition, we also find “positive noun phrases,” such as some moderate DOE, which play a role in computing negated scopes.
The grammar that creates “negated scope” annotations uses various syntactic combinations of negating word annotations and [compound] noun phrases to decide how far the negated scope extends around a negating word. Similarly, the “counterfactual scope” grammar makes the same decisions for counterfactual trigger annotations, using conditional and subjunctive constructions, among others. An example, with the counterfactual scope underlined, is: Patient should call if she experiences shortness of breath. Popular non-grammatical approaches to finding negated and counter-factual scopes are NegEx and ConText [33,34].
While most criteria extractors rely mainly on finding appropriate FramSymptomVocab items in the text, a few need further syntactic analysis. For the WeightLoss criterion, a grammar creates an annotation containing phrases – from the same or adjacent sentences – that denote the amount of weight, length of time, and diuretic treatment. For the Tachycardia criterion, the beats-per-minute value is stored.
2.3.3. Text analysis engines
Once the dictionaries and grammars have been applied, the UIMA data store contains “candidate criteria mentions,” among other annotations. TAEs filter those candidates based on all the information in the data store, yielding the final set of affirmed and denied Framingham criteria as output. Filtering is performed using the following devices.
Co-occurrence constraints
For each Framingham criterion, two word lists help constrain its textual occurrences. The lists contain words which must (or must not) co-occur in the same sentence with the candidate criterion. For example, the criterion Tachycardia may not co-occur with any of the words bruce, exercise, treadmill, stress, ekg, echo, predicted, maximal, etc.
Disambiguation
Several of the FramSymptomVocab forms are ambiguous. An example is edema, which can signal either AnkleEdema or APEdema. Our disambiguation heuristic begins by choosing a “default” meaning for the ambiguous term. For edema, the default is AnkleEdema. Next, we look for evidence in the UIMA data store that can “prove” the non-default meaning. We disambiguate edema to APEdema if (a) we see nearby terms such as x-ray, or cxr, (b) the term occurs within a Radiology encounter note, or (c) the containing sentence mentions anatomy remote from the legs.
Negation
In general, any FramSymptomVocab item that occurs within a negated scope annotation is marked as negated. An exception is sometimes given for “positive noun phrases” even when they are within a negated scope. Further special negation handling concerns superordinate terms. If a term that is semantically superordinate to a criterion is denied, then the criterion itself is also denied. For example, There is no evidence of organomegaly supports the negation of the Hepatomegaly criterion, because the concept organomegaly is superordinate to hepatomegaly. By contrast, occurrence of organomegaly in a non-negated scope would not be sufficient for PredMED to affirm Hepatomegaly.
Counterfactuals
Similar to negation, if a candidate criterion mention occurs within a counterfactual scope annotation, it receives special treatment. In this case, it is marked as invalid, to prevent the affirmation (or denial) of criteria that may not have actually occurred. For example, PredMED does not assert DOExertion for the sentence Patient should call if she experiences shortness of breath.
Segment constraints
Certain criteria have segment type restrictions. For example, edema can be disambiguated to APEdema, if it occurs in a “Chest X-ray” segment. Other segment types can prevent affirmation or denial of all criteria. In the text Monitor Symptoms: call the clinic for signs of SOB, the DOExertion will not be affirmed because “Monitor Symptoms:” is an “instruction” segment type, where we may not assume the condition has occurred.
Numeric constraints
For criteria that have numeric constraints, such as WeightLoss (“4.5 kg in 5 days”) or Tachycardia (“rate of >120min−1”), candidate criteria are invalidated if the associated values are not within bounds.
2.3.4. Encounter labeling
PredMED applications need to know the dates on which criteria are documented for patients. We obtain this information by first labeling each encounter with the names of criteria it mentions and then using the encounter dates. We use two approaches to assign criteria labels to encounters. The first, similar to the one used by Garla et al. [25], relies on a machine-learning (CHAID decision-tree) classifier [35] that uses, as features, the lexical annotations and the [negated and counterfactual] scope annotations, but without constraint checking or disambiguation. The classifier is trained using the development reference annotations. The second method is rule-based and simply labels each encounter with criteria that are extracted by the NLP pipeline.
2.4. Iterative annotation refinement for the development reference set
For extractor development, we used a procedure called “iterative annotation refinement” (IAR) to create the reference Framingham criteria annotations on our development dataset. To start, a cardiologist and a computational linguist discussed the meanings of the Framingham criteria and the diverse ways they are expressed in text. Initial criteria extractors were built and run against the 65 development documents. The extracted criteria mentions were then manually evaluated by the clinical expert and judged correct or incorrect, thus providing a first assessment of the extractors’ precision. To expand the set of expert annotations so that recall could be measured, while at the same time improving the extractors, we iterated on the following steps:
Automatically annotate the development documents with the currently implemented extractors.
Automatically compare the new annotations with the current reference annotations, producing a new performance measurement along with a tabulation of the disagreements.
- Manually review the error tabulation, resulting in:
- the clinical expert updating the reference annotations,
- the clinical expert updating the annotation guidelines, and
- the linguist changing the criteria extractors, in part guided by the updated guidelines.
IAR is similar to other methods used to annotate clinical notes text and to create corresponding annotation guidelines and extractors, although none of the earlier methods produces all three items (i.e., guidelines, annotations, and extractors) simultaneously. Those methods include the Annotation Induction method reported by Chapman and others [21,22] and the CDKRM (“cancer disease knowledge representation model”) annotation method described by Coden et al. [23]. In addition, Carrell et al. [24] report on a model, called TALLAL (“tag a little, learn a little”), for iteratively training a machine-learning annotator without however creating annotation guidelines. Finally, Garla et al. [25] use iterative refinement to tune document labels and a YTEX classifier for abdominal imaging reports. Table 2 compares IAR to those methods, along several dimensions.
Table 2.
Comparison of iterative methods for creating annotations, guidelines, and extractors.
| Extraction target | Result of using the method |
Sources of annotations compared in each iteration |
Arbiter for disagreements at each iteration |
Objective (and metric) for each iteration |
|
|---|---|---|---|---|---|
| IAR | Framingham HF criteria |
|
Expert and extractor | Expert | Improve extractor performance (F-score) |
| Annotation Induction | Clinical conditions |
|
Expert and linguist | Consensus | Improve inter-annotator agreement (F-score) |
| CDKRM | Classes in the cancer disease model |
|
2 experts | Consensus | Improve inter-annotator agreement (agreement %) |
| TALLAL | PHI (protected health information) classes |
|
Expert and extractor | Expert | Annotate full dataset (to the expert’s satisfaction) |
| YTEX | Document Classifications |
|
Expert and classifier | Expert | Resolve misclassification errors |
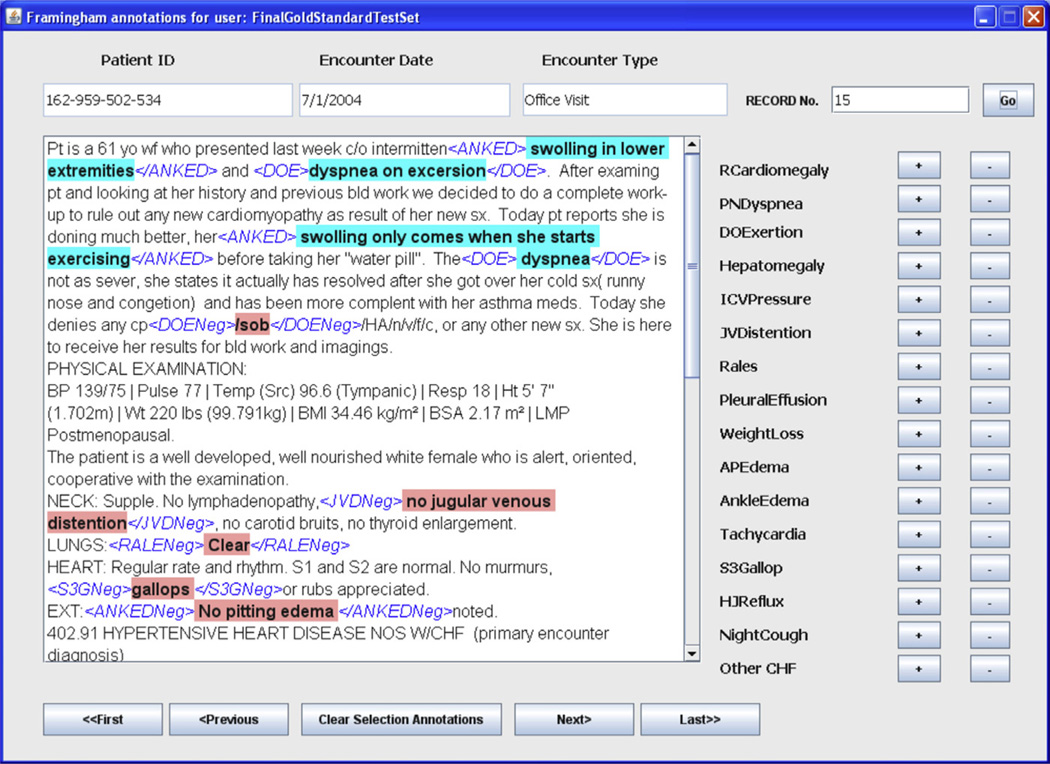
We developed a user interface (Fig. 2) for managing expert annotations on text files.
Fig. 2.
User interface for the annotation tool, which was used to manage expert annotations during IAR. Criteria abbreviations are as given in Table 1. Note the misspellings and the contradictions in the annotations for DOExertion and AnkleEdema.
3. Evaluation setup
Our evaluation dataset consisted of clinical notes from 400 randomly selected encounters: 200 from HF patients and 200 from control patients. To build the evaluation gold standard, the clinical expert trained three additional coders, using the guidelines developed during iterative refinement of the development annotations. Each coder annotated 200 encounter notes and each note was annotated by two coders. Disagreements were adjudicated by consensus between the two coders, with the clinical expert breaking ties. The coders were not told which encounters were for cases and which were for controls. Since the gold standard reference annotations were the result of consensus among the 4 coders, calculation of a Kappa score for inter-annotator agreement was not meaningful.
After initial coding, we did a single iteration of IAR steps 1, 2, and 3a which yielded the final evaluation gold standard. An objective was to inspire improvement in the coders’ recall. This is similar to the “pre-annotation” intervention studied during generation of the i2b2 reference standard [36]. Assisted annotation has long been used by the information retrieval community to annotate multimedia document repositories [37,38], with the precise purpose of improving human recall. To confirm PredMED’s need for this procedure, we measured the initial recall of one of our coders to be only 0.943, when measured against the final (assisted) consensus gold standard.
We evaluated PredMED criteria extractors in two ways. In the first, we assessed their ability to correctly label encounter documents with the criteria they contain. This is “Encounter Label Evaluation,” shown in Fig. 3. We measured the performance of both the machine-learning and rule-based labelers.
Fig. 3.
Evaluation flow.
For the second evaluation – “Criteria Extraction Evaluation,” at the bottom of Fig. 3 – we measured the performance of the extractors directly, by comparing criteria extracted from the evaluation dataset against the gold standard annotations.
We used the following metrics (where “TP” is true positives, “FP” is false positives, and “FN” is false negatives):
Precision [which is the same as Positive Predictive Value] is: TP/(TP + FP).
Recall [which is the same as Sensitivity] is: TP/(TP + FN).
F-score is: (2 × Precision × Recall)/(Precision + Recall).
For evaluating criteria extraction, any extracted annotation whose span overlaps a gold standard annotation of the same type is treated as a true positive. (Most cases of non-exact overlap are caused by coder annotations that include adjacent punctuation marks.) The labeling gold standard is derived from the criteria gold standard by simply asserting any criterion to be an encounter label for the encounters in which it occurs.
4. Results
4.1. Iterative annotation refinement
Our development dataset contained 65 encounter notes from heart failure cases. IAR produced a development reference set containing 1225 criteria mentions. With respect to that reference set, the initial set of clinical expert annotations on the development dataset achieved [Precision: 0.946; Recall: 0.812; F-score: 0.874]. As expected, initial coder recall is much poorer than initial precision. Over the development period, IAR improved the PredMED extractors’ performance from [Precision: 0.638; Recall: 0.506; F-score: 0.564] to [Precision: 0.931; Recall: 0.939; F-score: 0.935] (on the development dataset).
4.2. Encounter Label Evaluation
Results of the encounter labeling evaluation (on the evaluation dataset) are shown in Table 3.
Table 3.
Machine-learning encounter labeling vs. rule-based encounter labeling.
| Machine-learning method |
Rule-based method |
|||||
|---|---|---|---|---|---|---|
| Recall | Precision | F-score | Recall | Precision | F-score | |
| Affirmed | 0.675000 | 0.754190 | 0.712401 | 0.738532 | 0.899441 | 0.811083 |
| Denied | 0.945556 | 0.905319 | 0.925000 | 0.987599 | 0.931915 | 0.958949 |
| Overall | 0.896364 | 0.881144 | 0.888689 | 0.938462 | 0.926720 | 0.932554 |
Bold values represent the overall performance of the machine-learning method and the rule-based method respectively.
The rule-based method (F-score 0.932) outperformed the machine-learning method (F-score 0.888). However the 99% confidence intervals ((0.900–0.964) and (0.848–0.929), respectively) indicate that the methods are not significantly different in performance.
4.3. Criteria mention evaluation
Table 4 shows the performance of the criteria extractors measured against the evaluation gold standard. The overall performance has an F-score of 0.910. To ensure that sample criteria extracted from case and control encounters belong to the same population, we also measured performance separately for cases and controls. Although extraction seems to perform slightly better for cases, there is no significant difference, as is shown by the large overlap in the 99% confidence intervals.
Table 4.
Performance of Framingham diagnostic criteria extraction.
| Precision | Recall | F-score | 99% confidence interval (F-score) |
|
|---|---|---|---|---|
| Overall (exact) | 0.925234 | 0.896864 | 0.910828 | 0.891–0.929 |
| Cases (N = 993) | 0.930107 | 0.900104 | 0.914859 | 0.892–0.937 |
| Controls (N = 499) | 0.915401 | 0.890295 | 0.902673 | 0.868–0.936 |
| Overall (relaxed) | 0.948239 | 0.919164 | 0.933475 | 0.916–0.950 |
| Affirmed (N = 367) | 0.747801 | 0.789474 | 0.768072 | 0.711–0.824 |
| Denied (N = 1125) | 0.982857 | 0.928058 | 0.954672 | 0.938–0.970 |
| Overall (exact) with unassisted GoldStd | 0.904385 | 0.890934 | 0.897609 | 0.877–0.917 |
Bold value represents the overall performance of criteria extraction.
We also measured the performance of “relaxed” criterion extraction, which ignores negation and reflects the underlying ability to recognize Framingham criteria independent of context. That performance has an F-score of 0.933. The performance on exact criterion extraction is slightly worse, because of errors made in negative scope detection. Measured in this way, the penalty we pay for those errors is an F-score decline of 0.023.
The relatively poorer performance on affirmed criteria (N = 367), when compared to denied criteria (N = 1125), is likely due to the greater syntactic diversity of affirmations. For example, PredMED missed the affirmation of PleuralEffusion in the text Blunting of the right costophrenic angle.
Finally, we also measured extractor performance against the initial gold standard, before “pre-annotation” assistance. As expected, the measured precision is lower (0.904 vs. 0.925), because of the poorer initial recall of the coders. In other words, the extractors found valid criteria mentions which the coders had initially missed.
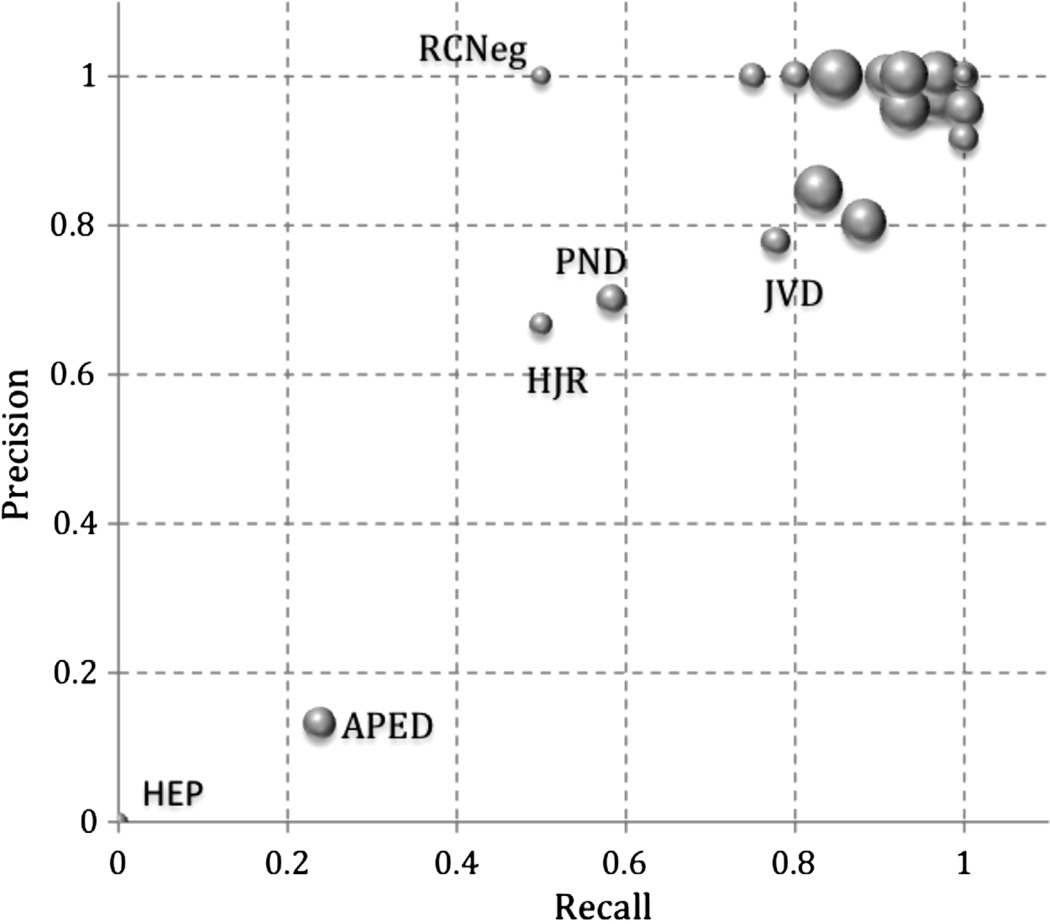
Recall and precision were calculated for individual criteria, as shown in Fig. 4. For most criteria, performance is good, as shown by the cluster in the upper-right corner. The “HEP” with the vanishingly small circle at (0.0, 0.0) reflects the fact that there was a single affirmation of Hepatomegaly in the evaluation dataset and PredMED failed to find it. Similarly, “RCNeg,” the denial of RCardiomegaly, occurred twice but PredMED found it only once, yielding a precision of 1.0 with a recall of 0.5. “APED” in the lower-left quadrant results from our disambiguation often incorrectly assigning edema to APEdema.
Fig. 4.
Precision and recall for individual criteria. Criteria abbreviations are as given in Table 1. Each circle represents a criterion and its size reflects the criterion’s occurrence frequency in the extracted results.
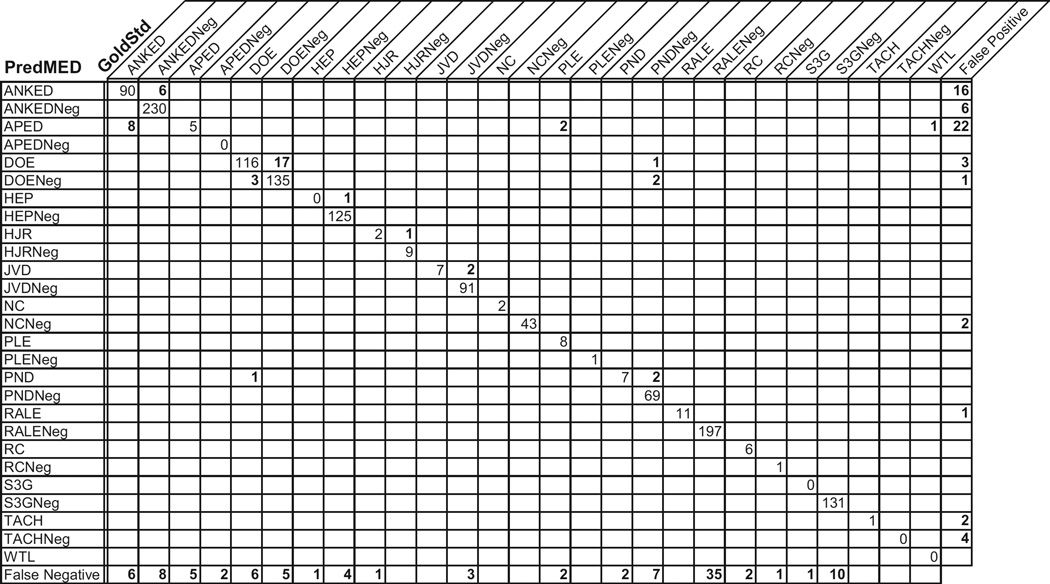
Fig. 5 shows a confusion matrix for 1492 extracted criteria in our evaluation run. Each value in the matrix gives the number of times a criterion was extracted with a certain category (i.e., its row) when it should have had the category assigned by the gold standard (i.e., its column). For example, the “6” in the second column of the first row indicates that on 6 occasions, PredMED called a mention “ANKED” when the gold standard says that it should have been “ANKEDNeg.”
Fig. 5.
PredMED extractions vs. gold standard annotations – a detailed performance analysis, presented as a confusion matrix over assertions and denials of Framingham criteria. Criteria abbreviations are as given in Table 1. Denials are marked with a “-Neg” suffix. Zero values off the diagonal have been blanked, for readability.
As expected, the largest numbers appear along the diagonal, reflecting the large overall agreement between PredMED and the gold standard. The values just off the diagonal represent the degree to which the extractors mistakenly recognize an affirmation as a denial, and vice versa. Further away from the diagonal, the values show us cases where ambiguities result in mistakes in PredMED’s extractions. For example, “APED” was confused with “ANKED” 8 times and with “PLE” twice (for example, in the text CXR suggests fluid at bases).
Finally, the rightmost column shows extractions for which there were no corresponding mentions in the gold standard (i.e., the false positives). An example here is that “APED” was extracted 22 times more that it should have been. (Upon analysis, we realized that the extractor often incorrectly recognized fluid as a mention of APEdema.) Similarly, the bottom row counts the occasions where gold standard annotations were missed by the extractors (i.e., the false negatives).
5. Discussion
Evaluation results reveal a few general error types, involving data quality, human anatomy, and syntactic complexity.
One manifestation of the data quality issue was that 26 of the 237 errors encountered in the final evaluation run - over 10% - were due to spelling errors. These include simple typographical errors, as in Fig. 2. Further errors occurred when text was dumped from the EHR system and reassembled in the PredMED analysis environment, a process that introduced sentence boundary errors that misled the extractors’ algorithms.
Assessing a potential mention of a candidate Framing-ham criterion often requires accurate knowledge of human anatomy. For example, the word swollen could be a mention of AnkleEdema if the word calf occurs in the same phrase, but not if the word nose does. A better approach than our co-occurrence-constraint wordlists would be to have a general facility for assessing when and which anatomical regions are being discussed.
Our strategy for recognizing Framingham criteria and, especially, their denials relies on the fact that most of their mentions can be parsed as noun phrases. Unfortunately, not all can be. For example, the following is an example of an expert-annotated denial of RCardiomegaly that PredMED missed: The cardiac silhouette is at the upper limits of normal. Dealing with such language will require use of more powerful parsing machinery than we are currently using.
Much research in medical NLP requires the existence of well-annotated reference standard document datasets [15–20]. For specific applications, however, it is often the case that (1) there is no locally available annotated dataset for development and evaluation in the new domain, (2) obtaining suitable annotated datasets from elsewhere is often impossible, because of institutional and EHR system differences and HIPAA constraints, and (3) creating such a dataset locally often taxes the skill and resources of the institution. In such a situation, IAR has the following advantages:
The error analysis step provides for excellent communication between the expert and the computational linguist who develops the extractor. This is much broader bandwidth communication than separately written annotation guidelines would provide.
The process produces a (rule-based) extractor that is consistent with the annotation guidelines.
The extractor can be used for pre-annotation to assist coders in creating higher-quality reference standards, by improving their recall.
The resulting annotations may be used to train machine-learning systems, if desired.
One use case for our development approach is in the creation of criteria extraction systems for medical criteria beyond the Framingham HF set. The NYHA [39] and the ESC [40], as well as the MedicalCriteria.com website [41], present hundreds of sets of medical criteria, many of which could be addressed with systems like PredMED and would benefit from IAR. Furthermore, many specific criteria reappear in multiple criteria sets, pointing to further opportunities for re-use. With appropriate tooling for reusing extractors, for managing expert annotations, and for IAR error analysis, our approach can be an attractive alternative to current development methods.
6. Conclusion
The Framingham criteria extractors are effective at finding criteria mentions with high precision and recall. Furthermore, those criteria can serve as the basis for accurately labeling clinical notes with respect to the criteria that they document, a prerequisite for downstream clinical applications of EHRs. Iterative annotation refinement is an effective tool for creating criteria extraction systems for applications where there is no preexisting dataset of suitably annotated text.
Summary points.
What was already known on the topic
Framingham criteria are known to correlate with heart failure diagnosis.
Iterative development methods have been used to separately create annotation guidelines and machine-learning-based extractors for entity mentions in clinical notes.
What this study added to our knowledge
We now know that Framingham criteria can be reliably detected in clinical notes taken by primary care physicians.
IAR is the first method that explicitly creates rule-based entity extractors, annotation guidelines, and annotations all at the same time.
We successfully addressed the tendency of human coders to exhibit low initial recall when annotating criteria mentions, using IAR (during development) and pre-annotation (during creation of the evaluation gold standard).
Acknowledgments
We are grateful to Zahra Daar, Craig Wood, Harry Stavropou-los, Parikshit Sondhi, Benjamin Taymore, and Rajakrishnan Vijayadrishnan for a variety of contributions to this work, including project coordination, data preparation, tool building, and manual annotation.
Footnotes
Author contributions
Following are the contributions of the authors to the work described in this paper.
Roy Byrd created the criteria extractors, participated in the IAR (iterative annotation refinement) process, as the computational linguist, performed the performance evaluation and analysis for criteria extraction, and wrote the bulk of the paper text.
Steven Steinhubl participated in the IAR process, as the clinical expert (in cardiology), created the annotation guidelines, and managed the human coding process that created the evaluation gold standard annotations.
Jimeng Sun did the random selection for the 400 evaluation documents and created the machine-learning classifier for encounter labeling and did its performance evaluation. Shahram Ebadolahi is the project co-leader and researched the use of annotation assistance for reference dataset creation.
Walter “Buzz” Stewart is the principal investigator, lead the project, and suggested the evaluation methodology.
Competing interests
There are no competing interests.
References
- 1.Heidenreich PA, Trogdon JG, Khavjou OA, et al. Forecasting the future of cardiovascular disease in the United States. Circulation. 2011;123(8):933–944. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 2.Remes J, Miettinen H, Reunanen A, Pyorala K. Validity of clinical diagnosis of heart failure in primary health care. Eur. Heart J. 1991;12:315–321. doi: 10.1093/oxfordjournals.eurheartj.a059896. [DOI] [PubMed] [Google Scholar]
- 3.Rutten FH, Moons KGM, Cramer M-JM, Grobbee DE, Zuithoff NPA, Lammers J-WJ, Hoes AW. Recognizing heart failure in elderly patients with stable chronic obstructive pulmonary disease in primary care: cross sectional diagnostic study. BMJ. 2005;331:1379. doi: 10.1136/bmj.38664.661181.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKee PA, Castelli WP, McNamara PM, Kannel WB. The natural history of congestive heart failure: the Framingham study. N. Engl. J. Med. 1971;285(26):1441–1446. doi: 10.1056/NEJM197112232852601. [DOI] [PubMed] [Google Scholar]
- 5.Di Bari M, Pozzi C, Cavallini MC, Innocenti F, Baldereschi G, De Alfieri W, et al. The diagnosis of heart failure in the community. Comparative validation of four sets of criteria in unselected older adults: the ICARe Dicomano Study. J. Am. Coll. Cardiol. 2004;44:1601–1608. doi: 10.1016/j.jacc.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 6.Chapman WW, Nadkarni PM, Hirschman L, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J. Am. Med. Inform. Assoc. 2011;18:540–543. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier N, Nazarenko A, Baud R, Ruch P.Recent advances in natural language processing for biomedical applications Int. J. Med. Inform 200675413–41.7 [DOI] [PubMed] [Google Scholar]
- 8.Suominen H, Lehtikunnas T, Back B, et al. Applying language technology to nursing documents: pros and cons with a focus on ethics. Int. J. Med. Inform. 2007;76S:S293–S301. doi: 10.1016/j.ijmedinf.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Vijayakrishnan R, Steinhubl SR, Sun J, et al. Poster at the American College of Cardiology Scientific Session & Expo. Chicago: 2012. Mar, Potential impact of predictive models for early detection of heart failure on the initiation of evidence-based therapies. [Google Scholar]
- 10.Steinhubl SR, Williams B, Sun J, Byrd RJ, Daar Z, Ebadollahi S, Stewart WF. Text and data mining of longitudinal electronic health records in a primary care population can identify heart failure patients months to years prior to formal diagnosis using the Framingham criteria, Poster at the American Heart Association Scientific Sessions, November 2011. Orlando (also available as Circulation. 2011;124:A12035. [Google Scholar]
- 11.Lin M, Chuang J, Liuo D, Chen C. Application of MedLEE to Process Medical Text Reports in Taiwan. 2012 Apr; libir.tmu.edu.tw/bitstream/987654321/21442/1/MISTT055_full.doc. [Google Scholar]
- 12.Friedman C, Hripcsak G, Shagina L, et al. Representing information in patient reports using natural language processing and the extensible markup language. J. Am. Med. Inform. Assoc. 1999;6(1):76–87. doi: 10.1136/jamia.1999.0060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garvin JH, DuVall SL, South BR, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J. Am. Med. Inform. Assoc. 2012 doi: 10.1136/amiajnl-2011-000535. http://dx.doi.org/10.1136/amiajnl-2011-000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uzuner Ö, South BR, Shen S, DuVall SL. 2012 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J. Am. Med. Inform. Assoc. 2011;18:554–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark C, Aberdeen J, Coarr M, et al. MITRE system for clinical assertion status classification. J. Am. Med. Inform. Assoc. 2011;18:563–567. doi: 10.1136/amiajnl-2011-000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark C, Good K, Jezierny L, et al. Identifying smokers with a medical extraction system. J. Am. Med. Inform. Assoc. 2008;15:36–39. doi: 10.1197/jamia.M2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Bruijn B, Cherry C, Kiritchenko S, et al. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J. Am. Med. Inform. Assoc. 2011;18:557–562. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J. Am. Med. Inform. Assoc. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. J. Am. Med. Inform. Assoc. 2011;18:580–587. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman WW, Dowling JN. Inductive creation of an annotation schema for manually indexing clinical conditions from emergency department reports. J. Biomed. Inform. 2006;39(2):196–208. doi: 10.1016/j.jbi.2005.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman WW, Dowling JN, Hripcsak G. Evaluation of training with an annotation schema for manual annotation of clinical conditions from emergency department reports. Int. J. Med. Inform. 2008;77:107–113. doi: 10.1016/j.ijmedinf.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. J. Biomed. Inform. 2009;42(5):937–949. doi: 10.1016/j.jbi.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Carrell D, Currier M, Halgrim S. Use and Evaluation of the MIST Open-Source Deidentification Tool. GHRI-IT Poster Session. 2010 Nov [Google Scholar]
- 25.Garla V, Lo Re, 3rd, Dorey-Stein VZ, Kidwai F, Scotch M, Womack J, Justice A, Brandt C. The Yale cTAKES extensions for document classification: architecture and application. J. Am. Med. Inform. Assoc. 2011;18:614–620. doi: 10.1136/amiajnl-2011-000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matheny ME, FitzHenry F, Speroff T, et al. Detection of infectious symptoms from VA emergency department and primary care clinical documentation. Int. J. Med. Inform. 2012;81:143–156. doi: 10.1016/j.ijmedinf.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Roy J, Stewart WF. Prediction modeling using EHR data: challenges, strategies, and a comparison of machine learning approaches. Med. Care. 2010;48(6):S106–S113. doi: 10.1097/MLR.0b013e3181de9e17. [DOI] [PubMed] [Google Scholar]
- 28.IBM. Text Analytics Tools and Runtime for IBM Language Ware, 2011. 2012 Dec; http://www.alphaworks.ibm.com/tech/lrw.
- 29.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat. Lang. Eng. 2004;10(3–4):327–348. [Google Scholar]
- 30.Apache UIMA. http://uima.apache.org/ (December 2012)
- 31.TextSTAT - Simple Text Analysis Tool. http://neon.niederlandistik.fu-berlin.de/static/textstat/TextSTAT-Doku-EN.html.
- 32.Boguraev BK. Towards finite-state analysis of lexical cohesion; Proceedings of the 3rd International INTEX Conference; June, 2000; Liege, Belgium. [Google Scholar]
- 33.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J. Biomed. Inform. 2001;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 34.Harkema H, Dowling JN, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J. Biomed. Inform. 2009;42(5):839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biggs D, de Ville B, Suen E. A method of choosing multiway partitions for classification and decision trees. J. Appl. Stat. 1991;18:49–62. [Google Scholar]
- 36.South BR, Shen S, Barrus R, DuVall SL, Uzuner Ö, Weir C. Qualitative analysis of workflow modifications used to generate the reference standard for the 2010 i2b2/VA challenge. AMIA Annu. Symp. Proc. 2011;2011:1243–1251. [PMC free article] [PubMed] [Google Scholar]
- 37.Vondrick C, Patterson D, Ramanan D. Efficiently scaling up crowdsourced video annotation. Int. J. Comp. Vision. 2012;100 http://dx.doi.org/10.1007/s11263-012-0564-1. [Google Scholar]
- 38.Lin C-Y, Tseng BL, Smith JR. Video collaborative annotation forum: establishing ground-truth labels on large multimedia datasets. Proceedings of the TRECVID 2003 Workshop. 2003 Nov [Google Scholar]
- 39.The Criteria Committee of the New York Heart Association. Nomenclature and Criteria for Diagnosis of Diseases of the Heart and Great Vessels. 9th. Boston, MA: Little, Brown & Co; 1994. pp. 253–256. [Google Scholar]
- 40.Dickstein K, Cohen-Solal A, Filippatos G, et al. ESC guidelines for the diagnosis and treatment of acute and chronic heart failure 2008. Eur. Heart J. 2008;29:2388–2442. doi: 10.1093/eurheartj/ehn309. http://www.escardio.org/guidelines-surveys/esc-guidelines/GuidelinesDocuments/guidelines-HF-FT.pdf. [DOI] [PubMed] [Google Scholar]
- 41. http://www.medicalcriteria.com. 2012 Dec