Abstract
Purpose
miRNAs are stable and can be extracted from tissues, blood and other body fluid without degradation. miRNAs are abnormally expressed in the presence of a pathological status, including cancer. Therefore, miRNAs are ideal biomarkers for cancer diagnosis and prognosis. Patients with triple negative breast cancer (TNBC) suffer the worst prognosis, although great efforts have been made. Many studies have investigated the role of miRNAs in predicting the outcomes of TNBC patients for better adjustment of treatment. However, results were inconsistent. Thus, we performed a meta-analysis to summarize the published studies for conclusive results.
Methods
Eligible studies from different database were retrieved from the online databases, and we used STSTA 12.0 to analysis the prognostic role of miRNAs in triple negative breast cancer.
Results
Overall high miRNA expression indicated a worse survival with HR value of 1.78 (95% CI: 0.97–3.25). However, subtotal HRs of oncogenic miRNAs and tumor suppressive miRNAs were 2.73 (95% CI: 2.08–3.57; P<0.001) and 0.44 (95% CI: 0.21–0.90; P = 0.024), respectively, and no heterogeneity was observed within the subgroups.
Conclusions
The miRNAs showed a slightly stronger prognostic value for disease-free survival, relapse-free survival and distant metastasis-free survival compared to the overall survival of TNBC patients. Circulating miRNAs could serve as potential biomarkers for the prognosis of TNBC patients and need further investigation.
Introduction
Triple negative breast cancer (TNBC) is an important component of heterogeneous breast cancer, and, according to the new refinement, it can be subdivided into four subgroups [1,2]. Because TNBC lacks the expressions of estrogen receptor (ER), progesterone receptor (PgR) and human epidermal growth factor receptor-2 (HER2), few effective treatments, with the exception of conventional surgery, radiotherapy and chemotherapy, can provide benefit for TNBC patients. Tremendous efforts regarding the treatment, such as tailoring adjuvant chemotherapy regimens, the discovery of emerging drugable pathways and the resensitization to radiotherapy or chemotherapy, have been carried out to improve the overall outcome of TNBC patients. However, TNBC has the worst prognosis among all breast cancer subtypes [3–5]. It’s believed that finding appropriate prognostic biomarkers for TNBC patients would allow an optimized treatment selection of regimens that could eventually benefit the patients.
MicroRNAs (miRNAs) are conserved non-coding RNAs (22–25 nt in length), which negatively regulate messenger RNA (mRNA) expression by partially or completely binding to the 3’ untranslated regions (3’UTRs) of the target mRNA [6]. miRNAs have been shown to mediate the cell fate of TNBC by regulating diverse biological processes, such as cell survival, cell cycle arrest, and differentiation[7–10]. Recent studies have confirmed that miRNAs are differentially expressed in tissues, blood and urine of cancer patients and healthy individuals, and they are stable in the presence of severe conditions [11]. Therefore, miRNAs are endowed with the characteristics of ideal biomarkers, and some studies have shown that miRNAs correlate with poor cancer prognosis, including TNBC [12–20]. However, the opposite results were obtained in several other studies [21–25]. Moreover, with regards to patients with TNBC, many factors, such as the race and age of the patients, the selected miRNA(s), the methodology for miRNA detection and the sample source among different studies, varied, leading to inconsistent outcomes [3,26]. Therefore, the aim of this meta-analysis was to systematically study the related references and yield a convincing outcome on whether miRNAs are ideal prognostic biomarkers for patients with TNBC.
Materials and Methods
Literature retrieval strategy
Meta-analysis of Observational Studies in Epidemiology group (MOOSE) was followed to guide the performance of this meta-analysis [27]. Two reviewers were assigned to independently retrieve literature from the online databases Pubmed, EMBASE, Web of Science and Chinese National Knowledge Infrastructure (CNKI). The time interval was between January 1st, 1993 and January 1st, 2016. The key words for the literature retrieval strategy included “microRNA”, “miRNA”, “Triple negative breast cancer”, “TNBC”, “basal-like breast cancer”, and “BLBC”. References from eligible publications in the literature were also manually screened for further potential literature.
Criteria for inclusion and exclusion
Eligible literature met the following criteria: (1) clear identification of patients with TNBC; (2) analysis of miRNA expression in tissues, blood or other body fluids; and (3) investigation of the association between miRNA expression levels and outcome of the TNBC patients. Publications were excluded if they had one or more of the following criteria: (1) focus on all types of breast cancer instead of TNBC only; (2) human tissues were not used; (3) absence of survival outcomes, such as the hazard ratio (HR), 95% confidence interval (CI), and P values or insufficient data to calculate the HR and 95% CI; and (4) review papers, comments, letters or duplicate publications.
When several publications reported on the same TNBC patient group, the most complete publication was included. Moreover, when one publication reported on two or more different independent TNBC patient groups, and the corresponding HR values and 95% CI were provided or could be calculated by Kaplan-Meier curves, the publication was considered as two or more independent studies.
Definition of oncogenic miRNAs and tumor suppressor miRNAs
According to previous published studies, miRNAs were consideres as tumor suppressive or protective when they were down-regulated compared with normal counterpart, in another word, these miRNAs were associated with an HR value larger than one, otherwise, they were called oncogenic miRNAs or risky miRNAs. In this meta-analysis, each included original study employed different miRNAs, a total HR value was firstly obtained, and then subtotal HR values for oncogenic miRNAs or tumor suppressor miRNAs were calculated.
Quality assessment and data extraction
Two reviewers separately assessed the quality of the included studies using the guideline of the Newcastle-Ottawa Quality Assessment Scale (NOS) [28], and every study was marked with scores ranging from 0 to 9. The studies with scores greater than a 6 were considered high quality and included in this study. Otherwise, they were removed to enhance the quality of the meta-analysis.
Two reviewers separately extracted the following data from all eligible studies: name of the first author, year of publication, sample number, sample source, type of miRNA(s), methodology, definition of cut-off, follow-up, HR values, 95% CI and P value of miRNAs for predicting overall survival (OS), disease-free survival (DFS), relapse-free survival (PFS), and the distant metastasis-free survival (DMFS). If two HR values were provided by univariate and multivariate analyses, the HR values from the latter were selected as this value considers confounding factors, which should yield more precise predictions. However, when the HR values and corresponding 95% CIs were not directly provided, we calculated these values using appropriate summary statistics or Kaplan-Meier curves as described by Tierney et al [29]. Discrepancies or disagreements were resolved by discussion.
Statistical analysis
STATA version 12.0 (Stata Corporation, College Station, TX, USA) was adopted to perform all of the meta-analyses. P values were two-sided and P<0.05 was considered statistically significant. All HR values and their corresponding 95% CIs were used as original data to investigate the collected prognostic value of the overall survival or disease-free survival of TNBC. Generally, a worse prognosis of TNBC was indicated by miRNA over-expression, with pooled HR values over 1.00. Heterogeneity among the HR values was assessed by Cochran’s Q test and Higgins’s I2 statistics. Heterogeneity was taken into consideration when P<0.10 and I2>50%, in which case the random-effect model would be used for the meta-analysis. Otherwise, a fixed-effect model would be used to calculate the pooled HR values. Subgroup analyses or a meta-regression was carried out when necessary. Publication bias was determined by Begg’s funnel plot or Egger’s bias test.
Results
Literature search and study characteristics
We initially acquired a total of 283 relevant items from Web of Science, PubMed, EMBASE, and CNKI, according to the retrieval strategy, and subsequently selected the most relevant 73 articles for a full-text review after the abstract screening. Under the guidance of the inclusive and exclusive criteria, 59 articles were removed because they were systematic reviews, basic preclinical studies, or articles lacking the association of OS or DFS with patients with TNBC. Eventually, 14 articles including 15 studies were included in this meta-analysis (Fig 1).
Fig 1. Flow diagram of the identification and selection of the studies.
The characteristics of the included studies and the enrolled patients are presented in Tables 1 and 2, respectively. The 15 studies were published between 2012 and 2015, and a total of 1473 TNBC patients from China (n = 7), the USA (n = 3), Norway (n = 2), Japan (n = 2) and Latvia (n = 1) were enrolled in the meta-analysis. Most of the studies detected the expression of miRNA(s) in the tissues by RT-PCR, and the definitions of cut-off for miRNA expression were different from one other. All of the studies analyzed the predictive value of miRNAs of OS in TNBC patients, and 8 studies also analyzed the association of miRNA(s) with DFS, RFS and DMFS in TNBC patients. Furthermore, we evaluated the quality of each individual study and obtained a median NOS score of 8, which indicated that the included studies were of high quality.
Table 1. Characteristics of the studies included in this meta-analysis.
| Study ID | Country | Patients | Control | Sample | miRNA | Method | Cut-off | Survival analysis | HR | Follow up (m) | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Yu 2014 | China | 118 | 118 | tissue | miR-301a | RT-PCR | median | OS | Reported | <144 | [12] |
| Cascione 2013 | USA | 39 | 36 | tissue | miR-125b, miR-497, miR-155, miR-16, miR-374a, miR-421, miR-655, miR-374b, | RT-PCR | >2-fold | OS/DFS | Reported | 79 (9–194) | [13] |
| Dong 2014 | China | 72 | 25 | tissue | miR-21 | RT-PCR | >1.5-fold | OS | Reported | >60 | [14] |
| Gasparin 2014 | USA | 160 | - | tissue | miR-155, miR-493, miR-30e, miR-27a | miRNA microarray | median | OS | Reported | 82 | [15] |
| Sahlberg 2014 | Norway | 40 | 63 | serum | miR-18b, miR-103, miR-107, miR-652 | RT-PCR | median | OS/RFS | Reported | >60 | [16] |
| Sahlberg 2014 | Norway | 70 | - | serum | miR-18b, miR-103, miR-107, miR-652 | RT-PCR | median | OS/RFS | Reported | >60 | [16] |
| MacKenzie 2014 | USA | 105 | - | tissue | miR-21 | ISH | >3 | OS | Reported | 124 | [17] |
| Shen 2014 | China | 58 | 31 | tissue | miR-27b-3p | RT-PCR | ROC | OS/DMFS | DE | 68 (60–127) | [18] |
| Toyama 2012 | Japan | 58 | 103 | tissue | miR-210 | RT-PCR | >4 | OS/DFS | Reported | 64.6 (3–149) | [19] |
| Kalniete 2015 | Latvia | 32 | 18 | tissue | miR-214 | RT-PCR | median | OS | DE | 40 | [20] |
| Liu 2015 | China | 456 | - | tissue | miR-126-3p, miR-374b-5p, miR-218-5p, miR-27b-3p, | miRNA microarray | median | OS/DFS | DE | 63.6 (8.4–106.8) | [21] |
| Liu 2015 | China | 41 | 74 | tissue | miR-26a | ISH | >3 | OS | DE | <120 | [22] |
| Shinden 2015 | Japan | 68 | - | tissue | miR-15a | RT-PCR | median | OS/DFS | DE | <60 | [23] |
| Tang 2014 | China | 51 | 51 | tissue | miR-185 | ISH | >2 | OS/DFS | Reported | 74 | [24] |
| Yu 2015 | China | 30 | - | tissue | miR-182 | RT-PCR | median | OS | Reported | <60 | [25] |
OS: overall survival; DFS: disease-free survival; DMFS: distant metastasis-free survival; RFS: relapse-free survival, RT-PCR: real-time polymerase chain reaction; DE data extrapolated; ROC: receiver operating characteristic; ISH: in situ hybridization.
Table 2. Characteristics of the patients enrolled in the included studies.
| Study | age | Grade | Node status | tumor size (cm) | stage | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <50 | >50 | 1+2 | 3+4 | 0 | positive | negative | unknown | <2 | >2 | unknown | 1 | 2 | 3 | 4 | 0 | |
| Dong 2014 | 36.0 | 36.0 | 33 | 39 | 0 | 35 | 37 | 0 | 13 | 59 | 0 | 33 | 39 | 0 | ||
| Gasparin 2014 | 74.0 | 86.0 | 15 | 142 | 3 | 58 | 93 | 9 | - | - | - | - | - | - | ||
| Toyama 2012 | 21.0 | 37.0 | 19 | 39 | 0 | 40 | 17 | 1 | 15 | 43 | 0 | - | - | - | ||
| MacKenzie 2014 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| Yu 2014 | 58.0 | 60.0 | 104 | 4 | 0 | 47 | 51 | 20 | 74 | 43 | 1 | 84 | 34 | 0 | ||
| Tang 2014 | 29.0 | 22.0 | 30 | 21 | 0 | 18 | 33 | 0 | 27 | 24 | ||||||
| Shinden 2015 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| Liu 2015 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| Yu 2015 | 51.1 (31.0–73.0) | 15 | 15 | 0 | 13 | 17 | 0 | 28 | 2 | 0 | 24 | 6 | 0 | |||
| Liu 2015 | 51.6 (26.1–174.3) | 191 | 263 | 2 | - | - | - | - | - | - | 396 | 47 | 13 | |||
| Kalniete 2015 | (27.0–78.0) | 4 | 39 | 7 | 15 | 35 | 0 | 13 | 37 | 0 | 31 | 19 | 0 | |||
| Sahlberg2014 | 60.0 | 14 | 25 | 1 | 18 | 22 | 0 | - | - | - | 27 | 13 | 0 | |||
| Sahlberg 2014 | 58.0 | 27 | 40 | 3 | 35 | 35 | 0 | - | - | - | 36 | 122 | 0 | |||
| Cascione2013 | 43.0 (20.0–50.0) | 7 | 74 | 5 | 30 | 50 | 6 | - | - | - | - | - | - | |||
| Shen 2014 | 46.5 (25.0–79.0) | 19 | 28 | 11 | 36 | 22 | 0 | 26 | 32 | 0 | - | - | - | |||
Meta-analysis of miRNA(s) in predicting the prognosis of TNBC patients
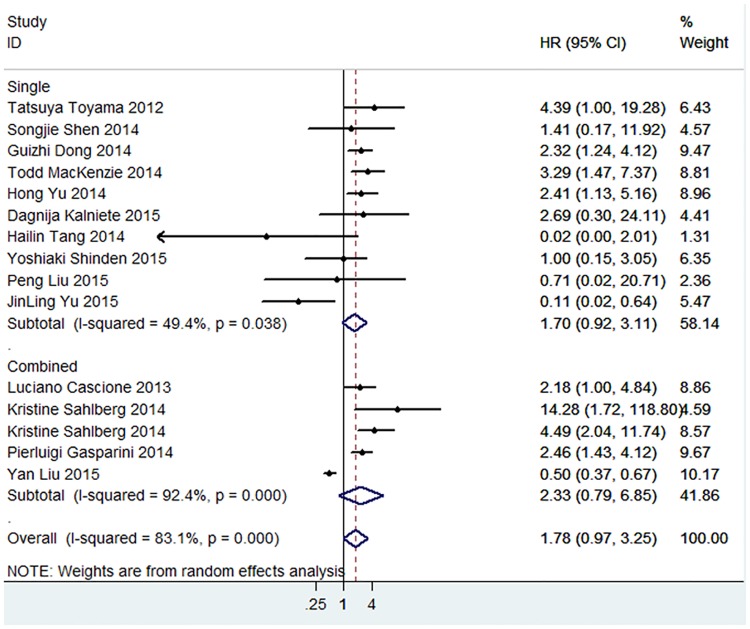
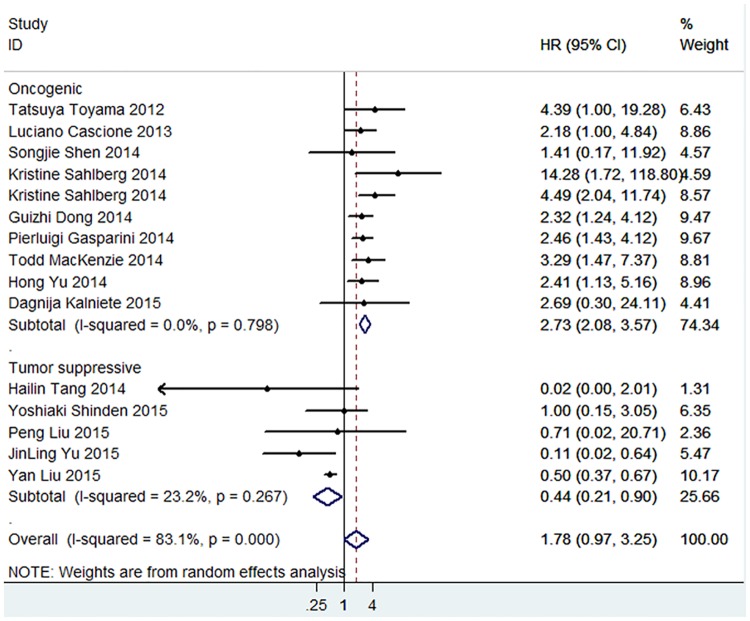
A comprehensive meta-analysis was conducted to investigate the predictive value of total miRNAs in the prognosis of TNBC patients, and the pooled HR (1.78, 95% CI: 0.97–3.25) showed a shorter OS of TNBC patients with significant heterogeneity (P<0.001, I2 = 83.1%). HR values of single miRNA and combined miRNA subgroups were also obtained, and a worse prognosis was shown for the OS of TNBC patients by the detection of combined miRNAs (Fig 2). More importantly, as miRNAs played distinguished roles in the pathogenesis of TNBC, protective or promotive roles were shown in the studies, and a subgroup analysis was performed within the meta-analysis. Of the 15 studies, 10 employed oncogenic miRNAs and 5 tumor suppressor miRNAs. Subtotal HRs were 2.73 (95% CI: 2.08–3.57; P<0.001) and 0.44 (95% CI: 0.21–0.90; P = 0.024) for oncogenic miRNAs and tumor suppressive miRNAs, respectively, and no heterogeneity was observed in the subgroup analysis (Fig 3).
Fig 2. Meta-analysis of subtotal HRs based on single miRNAs and combined miRNAs in predicting the OS of TNBC patients.
Fig 3. Meta-analysis of subtotal HRs based on oncogenic miRNAs and tumor suppressive miRNAs in predicting the OS of TNBC patients.
The same procedure was applied to investigate the pooled HRs for the DFS, RFS and DMFS of TNBC patients, however, we found that However, we then realized that DFS, RFS and DMFS represented different clinical outcomes, it was inappropriate to treat them as equal. And considering that there were too few analogical studies to conclude solid results about DFS, RFS or DMFS, we thought it would be better to not to perform revenant mata-analysis. Subgroups based on patients’ nationality, methodology for miRNA detection, and sample source were also analyzed to investigate the HR values of miRNAs in predicting OS of TNBC patients, as shown in Table 3.
Table 3. The results of the subgroup analysis.
| Subgroup | N | HR | LL | UL | P | I2 | P for heterogeneity |
|---|---|---|---|---|---|---|---|
| Total | 15 | 1.78 | 0.97 | 3.25 | 0.063 | 83.1% | 0.000 |
| Methodology | |||||||
| RT-PCR | 10 | 2.21 | 1.29 | 3.77 | 0.004 | 51.5% | 0.029 |
| miRNA microarray | 2 | 1.09 | 0.23 | 5.21 | 0.913 | 96.2% | 0.000 |
| ISH | 3 | 0.90 | 0.08 | 10.69 | 0.936 | 54.8% | 0.110 |
| Sample source | |||||||
| Tissue | 13 | 1.46 | 0.78 | 2,73 | 0.234 | 82.5% | 0.000 |
| Blood | 2 | 5.32 | 2.37 | 11.93 | 0.000 | 0.00% | 0.322 |
| Country | |||||||
| China | 7 | 0.82 | 0.32 | 2.15 | 0.693 | 47.0% | 0.170 |
| USA | 3 | 2.55 | 1.74 | 3.76 | 0.000 | 0.00% | 0.759 |
| Norway | 2 | 5.32 | 2.37 | 11.93 | 0.000 | 0.00% | 0.322 |
| Japan | 2 | 2.11 | 0.50 | 12.78 | 0.313 | 47.0% | 0.170 |
| Latvia | 1 | 2.69 | 0.30 | 24.12 | 0.377 | - | - |
N: number of studies; HR: hazard ratio; LL: lower limit of 95% CI; UL: upper limit of 95% CI; RT-PCR: real-time polymerase chain reaction; ISH: in situ hybridization.
Sensitivity analysis
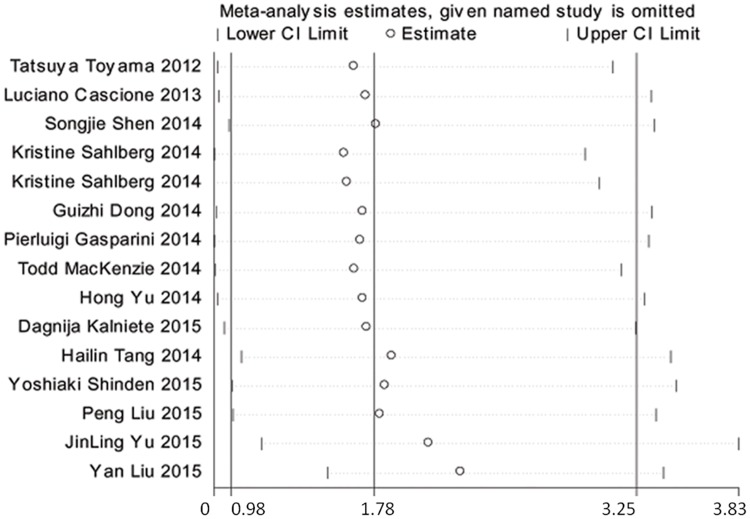
Although no heterogeneity was observed within the subgroups of oncogenic miRNAs and tumor suppressor miRNAs, significant heterogeneity was shown in the comprehensive meta-analysis. Therefore, a sensitivity analysis was performed to explore the source of the heterogeneity, and the results showed that the re-pooled HRs were not influenced when any specific study was omitted. (Fig 4).
Fig 4. Sensitive analysis of meta-analysis for miRNAs in the prediction of OS.
Publication bias
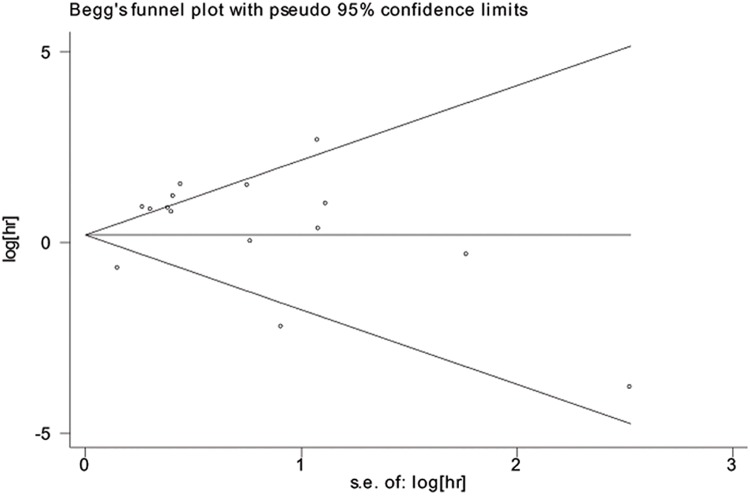
Egger’s and Begg’s tests were used to determine whether publication bias existed in the included studies. The Begg’s funnel plots for the OS meta-analysis and non-OS meta-analysis are shown in Fig 5, and the P values were 0.206 and 0.108, respectively, which suggested that no publication bias existed.
Fig 5. Begg’s funnel plot for publication bias of OS meta-analysis.
Discussion
TNBC accounts for 12–15% of all breast cancers, and it is the most malignant sub-type with the poorest prognosis. Because of the lack of ER, PgR and HER2 expression, TNBC patients are unlikely to benefit from endocrine therapy or anti-HER2 therapy. For these patients, the only therapeutic options are conventional surgery, chemotherapy or radiotherapy. However, even within the same cohort, TNBC patients may respond differently to the same chemotherapy. Thus, it is of great significance to identify suitable prognostic biomarkers for the specific treatment or timely adaption of treatment for TNBC patients [30].
It is well established that miRNAs play important roles in the initiation, development, metastasis and resistance to treatment of TNBC [31,32]. Tumor suppressive miRNAs mediate the degradation or post-transcriptional inhibition of transcripts encoding oncogenes and can target ZEB1/2, HMGB2, Bcl2 and HIF-1α, which contribute to the increased proliferation, invasion, and EMT and reduce the apoptosis of cancer cells. In contrast, oncogenic miRNAs exert the opposite function. Tumor suppressive miRNAs generally tend to be down-regulated in TNBC cells, while oncogenic miRNAs show the opposite change in expression [10]. miRNAs are more stable than mRNA and DNA, are resistant to harsh environments, and can be extracted without degradation from fresh frozen tissues, paraffin embedded tissues and even blood. Because of these specific features, miRNAs have the potential for extensive clinical use [33–35].
Most of the clinical studies focused on miRNAs as potential prognostic or diagnostic biomarkers in cancer patients, including those in TNBC patients with opposite results. To the best of our knowledge, this is the first meta-analysis to summarize previous studies and analyze the association of miRNAs with the prognosis of TNBC patients. Overall, our results revealed that high miRNA expression was associated with shorter OS of TNBC patients, with HR values over 1.00. Meanwhile, significant heterogeneity was observed in the meta-analysis. The source of this heterogeneity was explored by omitting each single study individually and re-pooling the HRs of the remaining studies. However, our results showed that no specific study influenced the overall HR values. Considering that miRNAs play different roles in the pathogenesis of TNBC, we then divided the studies into subgroups based on oncogenic miRNAs and tumor suppressive miRNAs before performing the meta-analysis. As expected, subtotal HRs of oncogenic and tumor suppressive miRNAs for predicting OS of TNBC patients were 2.73 (95% CI: 2.08–3.57) and 0.44 (95% CI: 0.21–0.90), respectively, with no heterogeneity observed within the subgroups. The pooled HR of tumor suppressive miRNAs was significantly lower than that of oncogenic miRNAs, indicating a better OS for TNBC patients with high expression of tumor suppressive miRNAs. Furthermore, the detection of combined miRNAs showed a worse OS compared to the detection of single miRNAs. Hence, we proposed that the combined detection of miRNAs may serve as a stronger prediction methodPrevious meta-analyses confirmed that circulating miRNAs have a great potential in diagnosing human cancers [35,36], as the expression of circulating miRNAs significantly changes in cancer patients compared to healthy control. Circulating miRNAs may serve as ideal prognostic biomarkers, and the results of this meta-analysis support this hypothesis with a high HR value of 5.32 (95% CI: 2.37–11.93, P<0.0001). However, these results should be interpreted with caution because only two studies conducted by one team were included in this analysis [16]. Furthermore, the results from studies on Chinese TNBC patients showed a better OS when miRNAs were highly expressed, and most of the reports focused on the tumor suppressive miRNAs. Whether the different ethnicities enrolled in this meta-analysis influenced the results deserves further investigation.
Through this meta-analysis, we intended to found reliable biomarkers that could guide the clinical doctors with better adjustment of treatment. Effective treatment was considered when the expressions of oncogenic miRNAs were decreased or tumor suppressive miRNAs were increased. On the contrary, proper adjustment should be made when the oncogenic miRNAs or tumor suppressive miRNAs show no significant change treatment.
Furthermore, recognizing the oncogenic miRNAs or tumor suppressive miRNAs showed great application in finding new methods of treating patients with triple negative breast camer. Take miR-21, an oncogenic miRNA as proved by the original study for example, when the drugs based on anti-miR-21were effectively delivered into triple negative breast cancer model with RNA-nano technology, the tumor size and ability to invasion or migration were significantly impaired.
Although this meta-analysis suggested a prognostic role of miRNAs in predicting the outcome of TNBC patients, several limitations should be taken into consideration. First, some of the studies lacked direct HR values, and we had to calculate them from the given data. Although we followed the procedure recommended by Tierney et al., different HR values and corresponding 95% CIs may have been obtained. Second, the miRNAs employed in these studies were quite different, along with the methodologies for miRNA detection and the definition of the cut-off values, which were potentially strong sources of heterogeneity. Third, the number of enrolled TNBC patients was not large enough to obtain solid results, and some detailed information, such as age, tumor size, tumor grade and stage, were missing. Fourth, few studies analyzed the association of miRNAs with DFS, RFS and DMFS of TNBC patients, so it was hard to raise persuasive results about the DFS, RFS and DMFS of included patients. Finally, only two studies focused on circulating miRNAs. Thus, whether circulating miRNAs could serve as powerful biomarkers with a prognostic value for TNBC patients requires more comprehensive and elaborately designed studies.
Conclusion
In summary, this study demonstrates that increased expression of tumor suppressive miRNAs predicted favorable outcomes of TNBC patients, and the increased expression of oncogenic miRNAs was associated with negative outcomes. Further, circulating miRNAs could serve as powerful prognostic biomarkers, and this should be verified by targeted studies.
Supporting Information
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. (2016) Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. J Plos one: 157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA (2014) New strategies for triple-negative breast cancer—deciphering the heterogeneity. Clin Cancer Res 20: 782–790. 10.1158/1078-0432.CCR-13-0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle P (2012) Triple-negative breast cancer: epidemiological considerations and recommendations. Ann Oncol 23 Suppl 6: i7–i12. [DOI] [PubMed] [Google Scholar]
- 4.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13: 4429–4434. 10.1158/1078-0432.CCR-06-3045 [DOI] [PubMed] [Google Scholar]
- 5.Stover DG, Winer EP (2015) Tailoring adjuvant chemotherapy regimens for patients with triple negative breast cancer. Breast 24 Suppl 2: S132–S135. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarzenbacher D, Balic M, Pichler M (2013) The role of microRNAs in breast cancer stem cells. Int J Mol Sci 14: 14712–14723. 10.3390/ijms140714712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phua YW, Nguyen A, Roden DL, Elsworth B, Deng N, Nikolic I, et al. (2015) MicroRNA profiling of the pubertal mouse mammary gland identifies miR-184 as a candidate breast tumour suppressor gene. Breast Cancer Res 17: 83 10.1186/s13058-015-0593-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serpico D, Molino L, Di Cosimo S (2014) microRNAs in breast cancer development and treatment. Cancer Treat Rev 40: 595–604. 10.1016/j.ctrv.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 10.Sui X, Wang X, Han W, Li D, Xu Y, Lou F, et al. (2015) MicroRNAs-mediated cell fate in triple negative breast cancers. Cancer Lett 361: 8–12. 10.1016/j.canlet.2015.02.048 [DOI] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105: 10513–10518. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, Li H, Qian H, Jiao X, Zhu X, Jiang X et al. (2014) Upregulation of miR-301a correlates with poor prognosis in triple-negative breast cancer. Med Oncol 31: 283 10.1007/s12032-014-0283-2 [DOI] [PubMed] [Google Scholar]
- 13.Cascione L, Gasparini P, Lovat F, Carasi S, Pulvirenti A, Ferro A et al. (2013) Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One 8: e55910 10.1371/journal.pone.0055910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong G, Liang X, Wang D, Gao H, Wang L, Wang L et al. (2014) High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol 31: 57 10.1007/s12032-014-0057-x [DOI] [PubMed] [Google Scholar]
- 15.Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci L, et al. (2014) microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 5: 1174–1184. 10.18632/oncotarget.1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahlberg KK, Bottai G, Naume B, Burwinkel B, Calin GA, Børresen-Dale A, et al. (2015) A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res 21: 1207–1214. 10.1158/1078-0432.CCR-14-2011 [DOI] [PubMed] [Google Scholar]
- 17.MacKenzie TA, Schwartz GN, Calderone HM, Graveel CR, Winn ME, Winn M, et al. (2014) Stromal expression of miR-21 identifies high-risk group in triple-negative breast cancer. Am J Pathol 184: 3217–3225. 10.1016/j.ajpath.2014.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen S, Sun Q, Liang Z, Cui X, Ren X, Chen H, et al. (2014) A prognostic model of triple-negative breast cancer based on miR-27b-3p and node status. PLoS One 9: e100664 10.1371/journal.pone.0100664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toyama T, Kondo N, Endo Y, Sugiura H, Yoshimoto N, Iwasa M, et al. (2012) High expression of microRNA-210 is an independent factor indicating a poor prognosis in Japanese triple-negative breast cancer patients. Jpn J Clin Oncol 42: 256–263. 10.1093/jjco/hys001 [DOI] [PubMed] [Google Scholar]
- 20.Kalniete D, Nakazawa-Miklasevica M, Strumfa I, Abolins A, Irmejs A, Gardovskis J, et al. (2015) High expression of miR-214 is associated with a worse disease-specific survival of the triple-negative breast cancer patients. Hered Cancer Clin Pract 13: 7 10.1186/s13053-015-0028-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Cai Q, Bao PP, Su Y, Cai H, Wu J, et al. (2015) Tumor tissue microRNA expression in association with triple-negative breast cancer outcomes. Breast Cancer Res Treat 152: 183–191. 10.1007/s10549-015-3460-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu P, Tang H, Chen B, He Z, Deng M, Wu M, et al. (2015) miR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett 357: 384–392. 10.1016/j.canlet.2014.11.050 [DOI] [PubMed] [Google Scholar]
- 23.Shinden Y, Akiyoshi S, Ueo H, Nambara S, Saito T, Komatsu H, et al. (2015) Diminished expression of MiR-15a is an independent prognostic marker for breast cancer cases. Anticancer Res 35: 123–127. [PubMed] [Google Scholar]
- 24.Tang H, Liu P, Yang L, Xie X, Ye F, Wu M, et al. (2014) miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol Cancer Ther 13: 3185–3197. 10.1158/1535-7163.MCT-14-0243 [DOI] [PubMed] [Google Scholar]
- 25.Yu J, Shen W, Gong B, Xu J, Gao B, Zhao H, et al. (2015) Expression of microRNA-182 in triple-negative breast cancer and its clinicopathological significance. Journal of Tongji University(Medical Science).: 29–32. [Google Scholar]
- 26.Daly B, Olopade OI (2015) A perfect storm: How tumor biology, genomics, and health care delivery patterns collide to create a racial survival disparity in breast cancer and proposed interventions for change. CA Cancer J Clin 65: 221–238. 10.3322/caac.21271 [DOI] [PubMed] [Google Scholar]
- 27.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. (2000) Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 283: 2008–2012. [DOI] [PubMed] [Google Scholar]
- 28.Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25: 603–605. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 29.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR (2007) Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8: 16 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esposito A, Criscitiello C, Curigliano G (2015) Highlights from the 14(th) St Gallen International Breast Cancer Conference 2015 in Vienna: Dealing with classification, prognostication, and prediction refinement to personalize the treatment of patients with early breast cancer. Ecancermedicalscience 9: 518 10.3332/ecancer.2015.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin L, Lim M, Zhao S, Sano Y, Simone BA, Savage JE, et al. (2014) The metastatic potential of triple-negative breast cancer is decreased via caloric restriction-mediated reduction of the miR-17~92 cluster. Breast Cancer Res Treat 146: 41–50. 10.1007/s10549-014-2978-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaca-Paniagua F, Alvarez-Gomez RM, Maldonado-Martinez HA, Perez-Plasencia C, Fragoso-Ontiveros V, et al. (2015) Revealing the Molecular Portrait of Triple Negative Breast Tumors in an Understudied Population through Omics Analysis of Formalin-Fixed and Paraffin-Embedded Tissues. PLoS One 10: e126762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gamez-Pozo A, Berges-Soria J, Arevalillo JM, Nanni P, Lopez-Vacas R, Navarro H, et al. (2015) Combined Label-Free Quantitative Proteomics and microRNA Expression Analysis of Breast Cancer Unravel Molecular Differences with Clinical Implications. Cancer Res 75: 2243–2253. 10.1158/0008-5472.CAN-14-1937 [DOI] [PubMed] [Google Scholar]
- 34.Shin VY, Siu JM, Cheuk I, Ng EK, Kwong A (2015) Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br J Cancer 112: 1751–1759. 10.1038/bjc.2015.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Wang S, Cao X, Liu J (2014) Analysis of circulating microRNA biomarkers for breast cancer detection: a meta-analysis. Tumour Biol 35: 12245–12253. 10.1007/s13277-014-2533-5 [DOI] [PubMed] [Google Scholar]
- 36.Cui Z, Lin D, Song W, Chen M, Li D (2015) Diagnostic value of circulating microRNAs as biomarkers for breast cancer: a meta-analysis study. Tumour Biol 36: 829–839. 10.1007/s13277-014-2700-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.