SUMMARY
The Mre11 complex (Mre11, Rad50 and Nbs1) is integral to both DNA repair and ATM-dependent DNA damage signaling. All three Mre11 complex components are essential for viability at the cellular and organismal level. To delineate essential and non-essential Mre11 complex functions that are mediated by Nbs1, we used TALEN-based genome editing to derive Nbs1 mutant mice (Nbs1mid mice) which harbor mutations in the Mre11 interaction domain of Nbs1. Nbs1mid alleles that abolished interaction were incompatible with viability. Conversely, a 108 amino acid Nbs1 fragment comprising the Mre11 interface was sufficient to rescue viability and ATM activation in cultured cells and to support differentiation of hematopoietic cells in vivo. These data indicate that the essential role of Nbs1 is via its interaction with Mre11, that most of the Nbs1 protein is dispensable for Mre11 complex functions, and suggest that Mre11 and Rad50 directly activate ATM.
INTRODUCTION
The DNA damage response (DDR) is important for maintaining genomic integrity. It comprises pathways that mediate DNA repair, DNA damage signaling, cell cycle regulation and apoptosis. Impairment of the DDR is associated with diverse human pathologies such as cancer, neurodegenerative disorders, immune deficiency, and premature aging (Ciccia and Elledge, 2010).
The Mre11 complex—Mre11, Rad50, and Nbs1 (Xrs2 in S. cerevisiae)—influences all aspects of the DDR via its role as DNA double strand break (DSB) sensor as well as its enzymatic and structural roles in DSB repair (Stracker and Petrini, 2011). Each member of the complex has been identified as the underlying basis of chromosome instability syndromes associated with immunodeficiency, radiosensitivity, cell cycle checkpoint defects and cancer predisposition (Stracker and Petrini, 2011). These disorders each exhibit decrements in ATM activation or activity, consistent with the idea that the Mre11 complex is required for the activation of ATM. This conclusion is supported by biochemical and genetic analyses in mice, yeast, and human cells (Cerosaletti and Concannon, 2004; Difilippantonio et al., 2005; Schiller et al., 2012; Shull et al., 2009; Stracker et al., 2008; Theunissen et al., 2003; Waltes et al., 2009; Williams et al., 2002).
Whereas Mre11 and Rad50 orthologs are present in Bacteria, Archea, and Eukaryota, Nbs1 appears to be restricted to Eukaryota. Accordingly, the protein appears to influence functions that are unique to eukaryotic cells. Unlike Mre11 and Rad50, Nbs1 does not appear to bind DNA, nor does it specify enzymatic activities relevant to DNA repair. Nbs1 primarily influences Mre11 complex function via mediating protein interactions via its N and C terminal domains that influence DNA repair, subcellular localization, and ATM-dependent checkpoint and apoptotic functions (Cerosaletti and Concannon, 2003; Desai-Mehta et al., 2001; Larsen et al., 2014; Lloyd et al., 2009; Saito Y, 2013; Stracker and Petrini, 2011; Williams et al., 2008). The mechanistic basis for Mre11 complex dependent ATM activation remains unclear. It is notable that the appearance of Nbs1 in eukaryotes coincides with the Mre11 complex's role in promoting DNA damage signaling, as does the Mre11 domain with which Nbs1 interacts. The Mre11 interaction interface of Nbs1 is a bipartite structure comprising Mid1 and Mid 2 (Mre11 interaction domain) that is conserved among Nbs1 orthologs (Schiller et al., 2012).
Having previously established that the N and C termini alone and in combination are dispensable for ATM activation (Stracker and Petrini, 2011), we undertook mutagenesis of the Mre11-Nbs1 interface with the goal of impairing Nbs1-Mre11 interaction while leaving the Nbs1 protein otherwise intact in an effort to define the role of Nbs1 in ATM activation. Using a transcription activator like effector nuclease (TALEN)-based genome editing, we created an allelic series in mice consisting of six mutations (Nbs1mid) within Mid2 that impair the interaction between Nbs1 and Mre11 to varying degrees. The most severe mutants abolished Mre11-Nbs1 interaction which resulted in the loss of cellular and organismal viability. These data indicate that Mre11-Nbs1 interaction is essential and therefore required for ATM activation.
Complementation of Nbs1 deficient cells with Nbs1 fragments spanning Mid1 and Mid2 rescued the viability of cultured cells and hematopoietic cells in vivo. Cells rescued in this manner also exhibited some indices of ATM function. In vitro, the Nbs1 fragments that rescued viability promoted Mre11 dimerization and DNA binding. In addition, they restored the ability of CtIP to activate Mre11 endonuclease activity, a function shown to be dependent on Nbs1. Collectively, these data suggest that Nbs1-Mre11 interaction is required for proper assembly of the Mre11 complex. Accordingly, that interaction is required for the concerted activities of Mre11 and Rad50 that govern DNA repair and DNA damage signaling and promote viability.
RESULTS
Evolutionarily conserved NFKxFxK motif in Nbs1 is essential for mouse embryogenesis
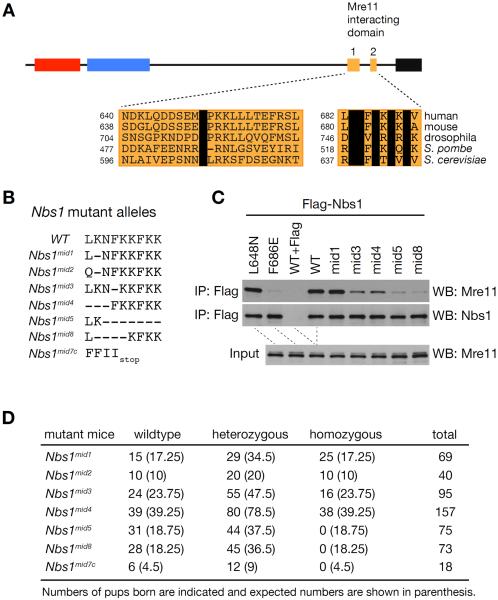
The mammalian Nbs1 protein interacts with Mre11 via a bipartite domain near the C-terminus, comprising Mid1 and Mid2 (Figure 1A). Mid2 includes a highly conserved NFKxFxK motif, whereas Mid1 is characterized by a single conserved leucine at position 648 of the mouse protein (Figure 1A). We carried out mutagenesis of the NBS1 cDNA to identify alleles that weakened the Nbs1-Mre11 interaction. Mutation of L648 had a minimal effect on Mre11-Nbs1 interaction, whereas the interaction was severely impaired by mutation of F686 in Mid2 (Figure 1C). On that basis, we carried out TALEN-based gene editing in mice to induce small deletions within Mid2 and thereby compromise the interaction between Mre11 and Nbs1 (Figure S1A).
Figure 1. NFKxFxK motif in Nbs1 is essential for mouse embryogenesis.
(A) Schematic structure of mammalian Nbs1 protein and sequence alignment of Mre11-interacting domain 1 and 2 among different species. Invariable residues are shown in black box. FHA, BRCT and AIM denote forkhead-associated domain, BRCA1 C terminal domain and ATM interacting motif, respectively. (B) Sequence of Nbs1mid mutant alleles created by TALEN-mediated gene editing. Nbs1mid7c is a truncated mutant allele. (C) Mre11 interaction of different Nbs1mid mutants was assessed by immunoprecipitation (IP) by Flag antibodies followed by Western blot for Mre11. Flag-tagged WT or mutant full-length Nbs1 were transiently expressed in MEFs. Flag peptide (100 μg/ml) was added to WT sample during IP for a Flag IP control. (D) Intercrosses of heterozygous Nbs1mid mutants. Numbers of pups born are indicated and expected numbers are shown in parenthesis.
We generated seven new Nbs1mid mutant mice (Nbs1mid1, Nbs1mid2, Nbs1mid3, Nbs1mid4, Nbs1mid5, and Nbs1mid8) in which the NFKxFxK motif of Mid2 is altered (Figure 1B). The genomic sequences of Nbs1 Exon 13 from founder mutant lines are listed (Figure S1B). These mutations were modeled in cDNA expression constructs. The ability of the corresponding protein products to interact with Mre11 was assessed by co-immunoprecipitation. Nbs1mid1 was indistinguishable from WT, whereas Mre11 interaction was moderately (Nbs1mid3 and Nbs1mid4) to severely impaired (Nbs1mid5 and Nbs1mid8) (Figure 1C). The binding of Nbs1mid2 is similar to that of wild type protein (data not shown). Whereas Nbs1mid1/mid1, Nbs1mid2/mid2, Nbs1mid3/mid3 and Nbs1mid4/mid4 mice were born at expected Mendelian frequencies, homozygosity for either Nbs1mid5 or Nbs1mid8 mutations was lethal, indicating that the Mre11-Nbs1 interaction is essential for embryonic viability (Figure 1D).
Disruption of Mre11-Nbs1 interaction compromises DDR and promotes tumorigenesis
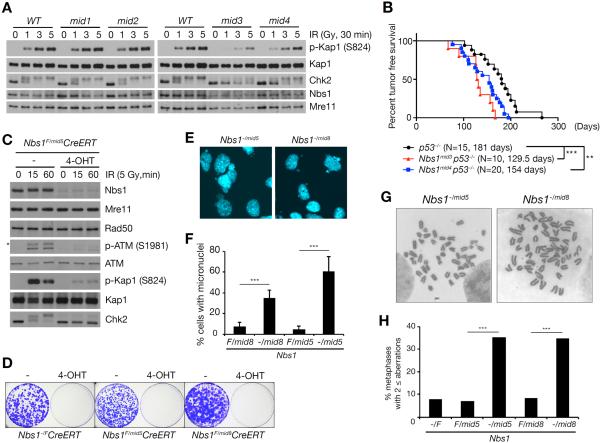
The extent to which Nbs1-Mre11 interaction was impaired scaled with the phenotypic severities observed. We assessed the effects of Nbs1mid mutations on ATM activation by examining phosphorylation of the ATM substrates Kap1 (S824) and Chk2 following ionizing radiation (IR). In homozygous Nbs1mid1 and Nbs1mid2 cells, those end points were indistinguishable from WT, whereas Nbs1mid3 and Nbs1mid4 homozygotes exhibited defects in phosphorylation (Figure 2A). Those mutants also exhibited defects in the G2/M checkpoint, indicative of reduced ATM activation. One hour following treatment with three Gy IR, the mitotic index of WT cells decreased by 85–87% (Figure S2A), whereas the decrease was only 64% for Nbs1mid3/mid3 and 72% for Nbs1mid4/mid4, consistent with the more severe impairment of Kap1 and Chk2 phosphorylation in Nbs1mid3/mid3 than Nbs1mid4/mid4. Similarly, colony formation assays indicated that IR resistance was reduced in Nbs1mid3/mid3 and Nbs1mid4/mid4 cells (Figure S2B).
Figure 2. Mre11-Nbs1 interaction is important for DDR and tumor suppression.
(A) IR-induced ATM signaling in WT, Nbs1mid1, Nbs1mid2, Nbs1mid3, and Nbs1mid4 thymocytes. ATM signaling was assessed by Western blot for the phosphorylation of KAP1 (S824) and hyperphosphorylation of Chk2, which are ATM substrates. Note that Chk2 migrates slowly when hyperphosphorylated. (B) Mouse tumor free survival. Each data point represents the percent survival of mice with each genotype at a given age. N denotes total number of mice for each genotype and the average age in death in days is shown. P-value of double mutants compared to p53−/− was determined by Wilcoxon rank sum test (**p<0.01 and ***p<0.001). (C) ATM signaling in Nbs1−/mid5 MEFs was assessed by Western blot for phospho-ATM (S1981) and the phosphorylation of ATM substrates, KAP1 (S824) and Chk2. The asterisk indicates nonspecific band. (D) Colony formation assay to assess cell viability of Nbs1−/mid5 and Nbs1−/mid8 alleles in MEFs. (E) Nuclei were stained with DAPI (4',6-diamidino-2-phenylindole). (F) % of nuclei with micronuclei was counted from randomly taken images (mean ± s.d., 100<N from seven images). P-value was determined by unpaired t-test (***p<0.001). (G) Representative chromosome metaphase images of Nbs1−/mid5 and Nbs1−/mid8 MEFs. (H) % of metaphases with aberrations. (***p<0.001, Fisher's exact test, 40< metaphases).
The defects imparted by Nbs1mid3 and Nbs1mid4 increased cancer risk. In our colony, p53 deficient mice present with thymic lymphoma at approximately 180 days of age. Over a ten month time course, Nbs1mid3/mid3 and Nbs1mid4/mid4 mice did not present with malignancy (data not shown); however when combined with p53 deficiency, the mean tumor-free survival decreased by 27–50 days relative to p53−/− mice (***p<0.001; Nbs1mid3 p53−/− vs. p53−/− and **p<0.01; Nbs1mid4 p53−/− vs. p53−/−) (Figure 2B). In addition to reduced latency, the spectrum of tumors arising in double mutant mice expanded to include leiomyosarcoma, squamous cell carcinoma, hemangiosarcoma, rhabdomyosarcoma, and histiocytic sarcoma (Table S1). We propose that destabilization of the Mre11-Nbs1 interface impairs ATM activation and modifies the p53−/− phenotype.
We were unable to establish embryonic fibroblasts homozygous for the Nbs1mid5 or Nbs1mid8 alleles. To define the cellular phenotypes of Nbs1mid5 and Nbs1mid8, these mice were crossed with Nbs1F mice in which cre expression inactivates NBS1 (Demuth et al., 2004). Following transduction of a tamoxifen (4-OHT) regulated cre recombinase into immortalized MEFs from Nbs1F/mid5 and Nbs1F/mid8 mice, 4-OHT was added to the media for 24 hrs. cre-mediated deletion of Nbs1F was evident within 24–48 hrs (data not shown), and the remaining Nbs1mid5 and Nbs1mid8 proteins were present at markedly reduced levels, whereas Mre11 and Rad50 levels were unchanged (Figure 2C and Figure S2C). Colonies of Nbs1−/mid5 or Nbs1−/mid8 MEFs were not recovered (Figure 2D), indicating that as in the case of mouse embryos, the Mre11-Nbs1 interaction is essential. At four days following the induction of cre activity (prior to cell death), indices of genome destabilization were evident in both Nbs1−/mid5 or Nbs1−/mid8 cells, including micronuclei and chromosome aberrations (Figure 2E–H). These outcomes resemble those observed upon genetic ablation of RAD50, MRE11, or NBS1 (Adelman et al., 2009; Buis et al., 2008; Reina-San-Martin et al., 2005).
ATM activation was assessed in Nbs1−/mid5 and Nbs1−/mid8 cells at four days following cre introduction. IR-induced Kap1 S824 and Chk2 phosphorylation was nearly undetectable in Nbs1−/mid5 (Figure 2C) and severely attenuated in Nbs1−/mid8 (Figure S2C) cells, suggesting that impairing Mre11-Nbs1 interaction compromised ATM activation. Accordingly, IR-induced ATM S1987 autophosphorylation, a direct index of ATM activation (Bakkenist and Kastan, 2003; Paull, 2015) was also sharply decreased in Nbs1−/mid5 cells relative to Nbs1F/mid5 controls (Figure 2C). As expected, both Nbs1mid5 and Nbs1mid8 alleles exhibit defects in the G2/M checkpoint that were considerably more severe than Nbs1mid3/mid3 or Nbs1mid4/mid4 (Figure S2D). These assessments may underestimate the severity of the Nbs1−/mid5 and Nbs1−/mid8 phenotypes due to the possible presence of residual Nbs1 protein.
Nbs1 minimal fragment rescues Nbs1 deficiency
Previously, a C terminal truncation of 100 amino acids of human Nbs1 that included Mid1 and Mid2 was unable to rescue viability of Nbs1-deficient mouse cells (Difilippantonio et al., 2005). Data presented here indicate that the presence of an essentially complete Nbs1 protein that is unable to interact with Mre11 was not sufficient for viability or for ATM activation. Given that the N and C termini are dispensable singularly or in combination for cell viability and ATM activation (Stracker and Petrini, 2011) (data not shown), we used deletional mutagenesis to define the “minimal Nbs1” required to support viability.
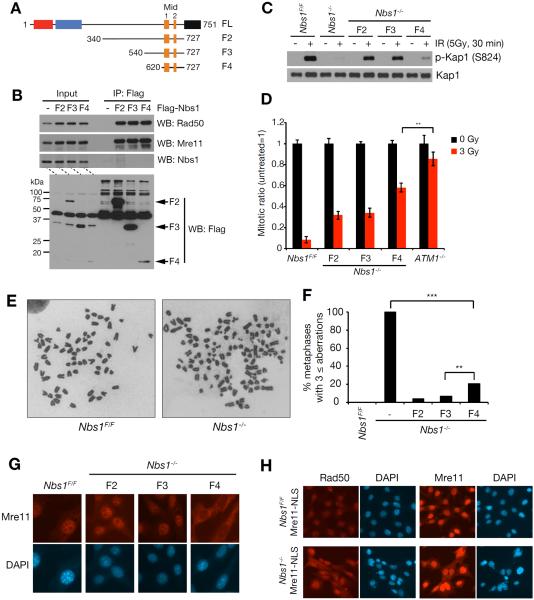
Three NBS1 gene segments encoding N terminal truncation fragments, all of which also lacked the C terminal 24 amino acids of Nbs1 were constructed in a retroviral expression vector. The constructs encoded fragments of 388, 188, and 108 amino acids (F2, F3 and F4, respectively) fused to a Flag epitope and SV40 nuclear localization signal (NLS) at their N termini for nuclear localization (Figure 3A). The Nbs1 gene segments were transduced into Nbs1F/F MEFs, and the ability of the encoded fragments to interact with Mre11 was assessed via Flag immunoprecipitation. All fragments co-immunoprecipitated with Mre11 and Rad50 (Figure 3B). Moreover, F4 which spans just 108 amino acids inclusive of Mid1 and Mid2 displaced full length Nbs1 from Mre11 and Rad50, arguing that Nbs1 is unlikely to interact with other domains of the Mre11 complex (Figure 3B).
Figure 3. Nbs1 minimal fragment rescues Nbs1 deficiency.
(A) Structure of Nbs1 fragments used in rescue experiments. Domains are indicated in Figure 1A. (B) Mre11 interaction of Nbs1 minimal fragments. Flag-tagged Nbs1 fragments (F2, F3 and F4) were expressed in Nbs1F/F MEFs and immunoprecipitation with Flag antibodies was performed followed by Western blot for Mre11, Rad50 and Nbs1. (C) IR-induced ATM signaling in Nbs1−/− MEFs expressing Nbs1 minimal fragments. As an ATM substrate, phosphorylation of KAP1 (S824) was assessed. (D) IR-induced G2/M cell cycle checkpoint of Nbs1−/− MEFs expressing Nbs1 minimal fragments. Mitotic cells were detected by measuring mitosis-specific phosphorylation of histone H3 (Ser10). ATM−/− MEFs was used for controls. P-value was determined by unpaired t-test (**p<0.01, mean ± s.d., three independent experiments). (E and F) Metaphase spread of Nbs1−/−-MEFs expressing Nbs1 minimal fragments. (E) Representative chromosome metaphase images of Nbs1F/F and Nbs1−/− MEFs. (F) The graph indicates the percent ratio of metaphases with aberrations (**p<0.01 and ***p<0.001, Fisher's exact test, 65< metaphases from two independent experiments). (G) Immunofluorescence cell staining of Mre11 in Nbs1−/− MEFs expressing Nbs1 minimal fragments. Nuclei are shown by DAPI (4',6-diamidino-2-phenylindole) staining. (H) Immunofluorescence cell staining of Rad50 and Mre11 in Nbs1−/− MEFs expressing Mre11-NLS. For Nbs1−/−-Mre11-NLS cells, Nbs1F/FcreERT2-Mre11-NLS cells was treated with 4-OHT for 24 hrs and stained at day 3. Nuclei are shown by DAPI (4',6-diamidino-2-phenylindole) staining.
Subsequently, cre activity was induced with 4-OHT and the ability of fragment-containing cells to form colonies was assessed. Whereas no colonies formed from control (vector-transduced) cells, each of the cells expressing F2, F3, and F4 were able to form colonies after ten days in culture. PCR genotyping and Western blot confirmed that the introduced fragments were the sole source of Nbs1 protein remaining in the Nbs1−/− cells—the fragments are hereafter designated “rescue fragments” (Figure S3A–C). By cloning the cells in this manner and by propagation in culture over the course of several weeks, any contribution from residual Nbs1 proficient (Nbs1F) cells to colony formation or to subsequent phenotypic assessments was excluded. These data indicated that as few as 108 amino acids of Nbs1 spanning the Mre11-Nbs1 interaction interface is sufficient to sustain the viability of cultured cells.
In addition to sustaining viability, the rescue fragments were able to promote ATM activation in Nbs1−/− cells. The phenotypes of Nbs1 fragment-expressing cells were compared to a culture of Nbs1F/F cells at four days post cre induction. Whereas Kap1 S824 phosphorylation was sharply reduced at 0.5 hour post 5 Gy IR in Nbs1−/−, it was readily evident in F2- and F3-containing cells, and to a lesser extent in F4 cells (Figure 3C). IR-induced Kap1 S824 phosphorylation of F4 cells was confirmed as ATM dependent activity by pretreatment with ATM inhibitor (Figure S3D). Those complemented cells exhibited restoration the G2/M checkpoint. One hour following treatment with three Gy IR, the mitotic index of F2-, F3-, and F4-containing Nbs1−/− cells decreased by 68%, 66%, and 44%, respectively, while the decrease was only 14% for ATM−/− cells (Figure 3D). These data indicate that a substantial degree of ATM-dependent checkpoint function was retained in rescue fragment-expressing cells. A fragment of human Nbs1 spanning residues 401 to 754 was previously shown to suppress ATM activation and nuclear localization defects in NBS patient cells (Cerosaletti and Concannon, 2004).
To obtain a quantitative assessment of DDR function in rescue fragment-containing cells, the frequency of spontaneous chromosome aberrations was assessed. 100 % of Nbs1−/− cells exhibit widespread chromosome fragility, with greater than three aberrations per metaphase spread (Figure 3E and F). In contrast, fewer than 21% of F2-, F3-, and F4-containing cells exhibited three or more aberrations (Figure 3F), indicating a substantial degree of residual function.
Nevertheless, rescue fragment-expressing cells did not phenocopy cells expressing wild type Nbs1. We observed sharply reduced nuclear localization of Mre11 in F4-containing cells relative to F2 or F3-containing cells (Figure 3G), likely accounting for reduced Kap1 phosphorylation in F4-containing cells. Data from budding yeast and human cells indicate that Xrs2 and Nbs1 are required for nuclear localization of Mre11 and Rad50 (Cerosaletti and Concannon, 2004; Cerosaletti et al., 2006), and enforced nuclear localization of Mre11 in S. cerevisiae partially restored function to xrs2Δ mutants (Oh et al., 2016; Tsukamoto et al., 2005). F4-containing cells were more sensitive to IR than F2 cells (Figure S3E), and we reasoned that a contributing factor to the reduced efficiency of F4-dependent restoration of ATM activation may be aberrant localization of Mre11 and Rad50. To determine whether enforced nuclear localization of Mre11 would mitigate the effects of Nbs1 deficiency, we expressed a SV40 nuclear localization signal-Mre11 cDNA (MRE11-NLS) in Nbs1F/F cells. No viable Nbs1−/− MRE11-NLS-expressing cells were recovered at three weeks, and at 4 days post cre induction, Mre11-NLS did not restore IR-induced G2/M checkpoint functions (Figure S4). Consistent with data from human NBS cells (Lakdawala et al., 2008), enforced nuclear localization of Mre11 in Nbs1−/− cells did not restore nuclear localization of Rad50 (Figure 3H).
Biochemical effects of Nbs1 minimal fragment
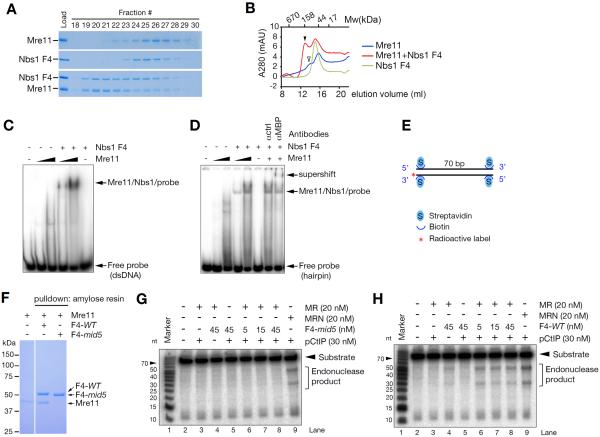
These data suggested that Nbs1 influences the stability and assembly of the Mre11 complex in addition to its subcellular localization. To test that interpretation, we examined the effect of human Nbs1 F4 on the biochemical properties of Mre11 in vitro. The human Mre11 core (1-411 aa) (Park et al., 2011) was purified and subjected to size exclusion chromatography. Individually, Mre11 and Nbs1 F4 fused to a maltose binding protein (F4-MBP) primarily appeared as single, monomeric peaks (apparent MW: 42 kDa for Mre11 and 59 kDa for F4-MBP). When mixed at 1:1 ratio, we observed a new peak with an apparent molecular weight of 184 kDa, consistent with the co-elution of Mre11 and Nbs1 dimers (i.e., two F4-MBP and two Mre11 cores) (Figure 4A and B). The new peak was not observed in F4-mid5-MBP in which the NFKKFKK motif is deleted (Figure S5).
Figure 4. Nbs1 minimal fragment promotes Mre11 dimer formation, Mre11 DNA binding and nuclease activity.
(A). Gel filtration of Mre11 and Nbs1 complex by Superdex 200 in the presence of 1 mM MnCl2. Equimolar mixture of human Mre11 (1-411aa) and Nbs1 (F4) protein were used at 0.5 μM. (B) Elution traced of gel filtration experiment. Molecular weight was estimated by gel filtration standard. Arrowheads indicate higher molecular weight complex; Mre11/Nbs1 dimers (closed) and Mre11 dimer (open). (C and D) DNA binding of Mre11. Electrophoretic mobility shift assay (EMSA) was performed using purified human Mre11 (1-411aa, 1 μM and 5 μM) and Nbs1 (F4, 5 μM) proteins, and end labeled double stranded or hairpin DNA probe (10 nM). Supershift was performed using 1 μg of MBP antibody. All EMSA reactions were performed in the absence of Mn2+. (E) Mre11 endonuclease assay probe. The 3' end labeled 70 bp dsDNA are blocked at both ends with streptavidin. (F) Amylose pull-down assay for Mre11 binding of F4 and F4-mid5 protein. Equimolar mixture of human Mre11 (1-411aa) and Nbs1 (MBP-F4-WT or MBP-F4-mid5) protein were incubated with amylose resin at 1 μM in the presence of 1 mM MnCl2 and the interaction was visualized by Coomassie blue stained SDS-PAGE. (G and H) Endonuclease assay with Mre11/Rad50/phosphorylated CtIP with F4-mid5 (G) or F4-WT (H). M, R, and pCtIP denote Mre11, Rad50 and phosphorylated CtIP, respectively.
F4-MBP also stimulated DNA binding by the Mre11 core. Mre11 binds DNA as a dimer (Williams et al., 2008). We found that the binding of Mre11 to dsDNA (Figure 4C) or a hairpin (Figure 4D) substrate was stimulated by Nbs1 F4 in EMSA assays. Supershifting induced by MBP antisera confirmed that Mre11/DNA complex contains Nbs1 (Figure 4D).
We next examined the effect of F4-MBP on Mre11 nuclease activity. Phosphorylated CtIP has recently been shown promote Mre11 endonuclease in a manner that depends on Nbs1 (Anand et al., 2016). Mre11-Rad50 complex was incubated with a 70 bp radiolabeled dsDNA substrate the ends of which were blocked by streptavidin to prevent exonucleolytic degradation (Figure 4E). In the absence of F4-MBP, or in the presence of the non-interacting F4-mid5-MBP fragment (Figure 4F), CtIP did not promote endonucleolytic cleavage by Mre11 (Figure 4G and H). In contrast, wild type F4-MBP with phosphorylated CtIP promoted the endonuclease of MRE11-RAD50 (Figure 4H).
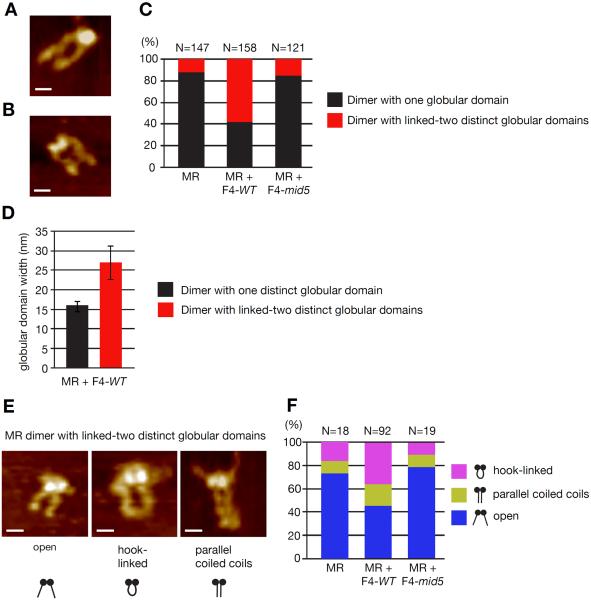
To further examine the hypothesis that Nbs1 mediates proper assembly of Mre11 and Rad50, we carried out scanning force microscopy (SFM). Previous SFM analysis, revealed a stoichiometry of two or four Nbs1 proteins per M2R2 complex (van der Linden et al., 2009). However the addition of two or four Nbs1 proteins to the globular domain of M2R2 obscured possible structural rearrangement. The minimal Nbs1 fragment identified here allowed analysis of Mre11 changes in complex architecture by SFM imaging. The M2R2 complex is characterized by a single globular domain (Mre11 + Rad50 ATPase domains) with two protruding coiled coils (Figure 5A). The coiled coils appear are usually apart, and 32% of the time appear linked by the Zinc-hook domains at their apexes (de Jager et al., 2004; Moreno-Herrero et al., 2005).
Figure 5. Nbs1 minimal fragment causes rearrangement of MR architecture.
(A and B) SFM image examples of MR complexes appearing with one globular domain (A) or appearing with globular domain with two distinguishable parts (B). (C) Distribution of MR molecules based on globular domain arrangement, alone and in the presence of Nbs1 F4-WT or -mid5. (D) The two forms of MR illustrated in (A) and (B) can be distinguished based on the width of their globular domain. The width for molecules with one globular domain and two distinct linked globular domains is plotted (average of n=66 and n=92 respectively, ± s.d.). (E) SFM image examples of coiled coil arrangements classified for complexes with two distinct linked globular domains (Scale bar = 20 nM). (F) Distribution of the coiled coli arrangement among MR complexes with two distinct linked globular domains, alone and in the presence of Nbs1 F4-WT or -mid5.
To assess the effect of F4 on Mre11 complex assembly, the full length human M2R2 complex was incubated at a 1:1 molar ratio with F4-MBP or F4-mid5-MBP (non-binding mutant) before imaging. A striking rearrangement of the Mre11 globular domain was induced in the presence of the F4 peptide, with the globular domain appearing as two distinct but linked globular objects (Figure 5B). The proportion of Mre11 complexes with this conformation increases from 12% to 58% in the presence of the F4 peptide but does not significantly change in the presence of the control F4-mid5 peptide (Figure C). This seperation into two distinct globular objects is accompanied by an increase in width of the gloublar domain (Figure 5D), consistent different conformations of related Mre11 complexes captured in X-ray crystallography studies (Wyman et al., 2011). Incubation with the F4 peptide also notably changed the conformatin of the coiled coils. In the presence of the F4 peptide, Mre11 complexes exhibited a notable increase in complexes with coiled coils linked via the zinc-hook apexes (Figure 5E and F). These data suggest that that Nbs1 influences nanoscale arrangement of Rad50 and Mre11 globular domains with consequent influence on conformational flexibility of the Rad50 coiled-coils favoring dimerization of the Zinc-hooks. Collectively these data strongly support the view that Nbs1's influence on the physical disposition of the Mre11 complex constitutes its essential function, and that this influence underlies its requirement for nuclear localization, cell viability and ATM activation.
Nbs1−/− fetal liver cells reconstitute hematopoietic system upon Nbs1 minimal fragment expression
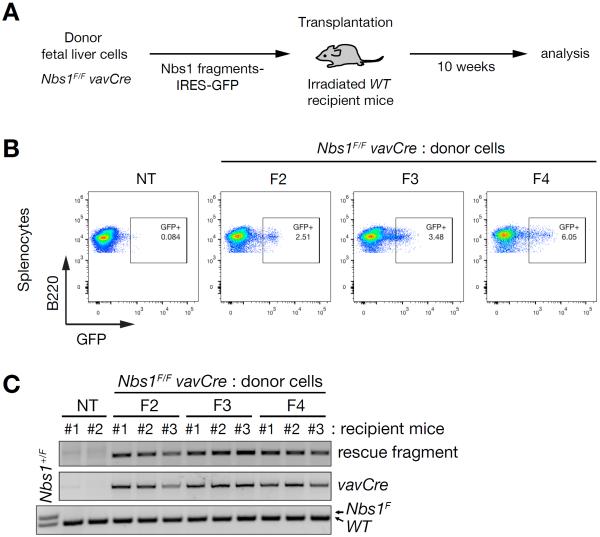
Immortalized cells are likely more tolerant of genotoxic stress than constituents of tissues in vivo. To determine whether the rescue fragments would sustain viability in vivo, we assessed their ability to support the differentiation of lymphocytes. Previous analyses indicated that the Mre11 complex is required for lymphocyte development (Balestrini et al., 2015; Callen et al., 2007; Deriano et al., 2009; Reina-San-Martin et al., 2004). Nbs1F/F mice were crossed to vavCre mice, which express cre recombinase in hematopoietic stem cells (HSCs) (Stadtfeld and Graf, 2005). Hematopoietic Nbs1-deficiency resulted in perinatal lethality due to lack of bone marrow development (Figure S6).
Fetal liver cells (FLCs) from E13.5 embryos were isolated and transduced with Nbs1 rescue fragment encoding in an IRES-GFP MSCV retrovirus prior to transplantation into lethally irradiated mice as depicted (Figure 6A). Ten weeks after transplantation, spleen was isolated and assessed for GFP positive cells. Although we did not observe complete reconstitution, GFP-positive, B220-positive cells comprised six percent of splenocytes (Figure 6B). The percentages of GFP positive, B220-positive cells may underestimate the degree of reconstitution by rescue fragment-containing HSCs due to silencing of the MSCV retrovirus during hematopoietic differentiation as observed previously (Cherry et al., 2000). The Mre11 complex is dispensable for viability of quiescent cells (Adelman et al., 2009); hence silencing of F4 expression may be tolerated because splenocytes are largely quiescent.
Figure 6. Hematopoietic reconstitution by Nbs1 minimal fragment.
(A) Experimental scheme of fetal liver cell transplantation. (B) Flow cytometry analysis of splenocytes from recipient mice 10 weeks post transplantation. For B cells, B220+ cells were gated from total splenocytes and plotted by GFP signal. Plot shown here are representative of recipient mice from each group of Nbs1 fragments. (C) PCR genotyping of splenocytes for exogenous Nbs1 fragment, vavCre and Nbs1F allele, which are donor FLCs-specific. Nbs1+/F MEFs were used for PCR control for Nbs1F allele. NT denotes control mouse.
PCR genotyping confirmed that recipient mice contain differentiated cells derived from Nbs1 fragment containing Nbs1−/− FLCs and that the fragment-encoding construct is present; the Nbs1F allele was not detected (Figure 6C). This confirms that this rescue is not by remaining undeleted Nbs1F allele in donor FLCs. No reconstitution was observed in control mice transduced with FLCs lacking the rescue fragments (data not shown). These data indicate that as with transformed cells, the 108 amino acids spanning the Mre11 interaction interface of Nbs1 (F4; Figure 3A) is sufficient to promote viability in vivo. Moreover, that minimal Nbs1 fragment was sufficiently functional to support differentiation of HSCs into splenic B cells.
DISCUSSION
To examine the role of Nbs1 in Mre11 complex functions, we undertook mutagenesis of Nbs1 in an attempt to weaken the interaction with Mre11, and thereby examine the functionality of the core Mre11-Rad50 complex disassociated from Nbs1. Mutations that disrupted Mre11 interaction caused inviability. Hence, the presence of a non-interacting but otherwise intact Nbs1 protomer was not sufficient for viability, establishing that Nbs1 interaction per se is essential. Conversely, we found that 108 amino acid of Nbs1 spanning the Mre11 interaction domain was sufficient to promote cell viability. And, though ATM activation was reduced in that setting, it was not abolished. Collectively, the data strongly argue that ATM activation is not directly dependent on Nbs1. Instead, we propose that essential functions of Nbs1 are to ensure proper assembly and subcellular localization of the Mre11 complex which in turn promotes viability and influences ATM activation by Mre11 and Rad50.
The Role of Nbs1 in the Mre11 complex: Essential Functions
Nbs1 clearly mediates essential as well as non-essential functions. With respect to the former, the data presented here indicate that the Mre11-Nbs1 interaction is specifically required for cellular and organismal viability. The levels of Mre11 and Rad50 protein were not changed in the Nbs1mid mutants cells. Hence, it is the loss of interaction, rather than global destabilization of Mre11 complex components that accounts for the loss of viability in non-interacting Nbs1mid mutants. Further, this emphasizes the fact that Mid 2 (the conserved NFKxFxK motif, which was the target of the TALEN-based mutagenesis) is the major determinant of Nbs1's association with the Mre11 complex. These data are consistent with the finding that the human Nbs1tr645 allele in which the C terminal 100 amino acids of Nbs1 were deleted was unable to support viability of mouse embryos (Difilippantonio et al., 2005).
What are the essential functions of Nbs1 in the Mre11 complex? First, it is clear that the nuclear localization of Mre11 is influenced by Nbs1, but this does not solely depend on the Mre11 interaction domain. Mre11 complex mislocalization is observed in human NBS and A-TLD cells, as well as in Nbs1ΔB/ΔB and Mre11ATLD1/ATLD1 mouse models of those human mutations, neither of which harbor alterations in their respective interaction domains (Carney et al., 1998; Difilippantonio et al., 2005; Reina-San-Martin et al., 2005; Stewart et al., 1999; Williams et al., 2002). Nbs1 and Mre11 levels are reduced in those settings, raising the possibility that the stoichiometry of complex components may also influence nuclear localization. Alternatively, those mutations may disrupt as yet undescribed interactions required for nuclear localization.
Nevertheless, promoting nuclear localization of Mre11 and Rad50 is likely not the only function of Nbs1 required for viability. Enforced nuclear localization by Mre11-NLS could not restore nuclear localization of Rad50 in human NBS cells (Lakdawala et al., 2008), nor did it compensate for Nbs1 deficiency in MEFs (Figure 3H). The data presented here are most consistent with the interpretation that Nbs1 influences the assembly and disposition of the complex. Supporting that view, we showed that dimerization and DNA binding by the N terminal 411 amino acid core of Mre11 was enhanced by the F4 fragment (Figure 4A–D), although it is likely that dimeric assemblies of full length Mre11 may exhibit greater stability. Further support comes from the fact that only in the presence of Nbs1 or the minimal fragment is CtIP able to stimulate Mre11 endonucleolytic cleavage (Figure 4H). Finally, scanning force microscopy analysis reveals that in the presence of Nbs1—whether minimal fragment F4 or the full length Nbs1 protein— the Mre11-Rad50 complex exhibits substantial structural differences in the disposition of Mre11 and Rad50 in the globular domain, in the path of the coiled coils, and in the association of the apical (presumably hook) domains of Rad50. These physical data resonate with genetic data indicating that mutations in Rad50 exert effects on regions distal to the altered residues (Al-Ahmadie et al., 2014; Hohl et al., 2015; Hohl et al., 2011; Hopfner et al., 2002). On the basis of these findings, we favor the view that Nbs1 functions as a chaperone to promote proper assembly of the complex, which is a prerequisite for its enzymatic and DNA binding functions as well as for determining its subcellular localization.
The Role of Nbs1 in the Mre11 complex: Non-essential Functions
The non-essential functions of Nbs1 have been illuminated by genetic analyses in human cells and mice. Nbs1 contains N terminal FHA and BRCT domains which are disrupted in the canonical Nbs1657Δ5 allele inherited by NBS patients and the corresponding Nbs1Δ B allele in mice. Those mutants exhibit defects in DSB end resection, DNA repair, and cell cycle checkpoint activation, presumably due to the loss of protein interactions mediated by those domains (Alt et al., 2005; Chapman and Jackson, 2008; Kobayashi et al., 2002; Larsen et al., 2014; Lloyd et al., 2009; Maser et al., 2001; Melander et al., 2008; Morishima et al., 2007; Spycher et al., 2008; Williams et al., 2009; Wu et al., 2012; Wu et al., 2008). However, human and mouse cells lacking those domains are viable and retain the ability to activate ATM (Difilippantonio et al., 2005; Williams et al., 2002). Deletion of the Nbs1 C-terminus (the Nbs1Δ C allele) which has been reported to bind ATM (Falck et al., 2005; You et al., 2005) had no effect on ATM activation or cell viability, but is required for ATM-dependent apoptosis (Stracker et al., 2007).
The Nbs1Δ BC allele is a composite of the Nbs1Δ B and Nbs1Δ C. The outcomes of Nbs1Δ B and Nbs1Δ C are simply additive in Nbs1Δ BC/Δ BC mice rather than synergistic; the phenotypic outcomes attributable to Nbs1Δ B and Nbs1Δ C unchanged in the composite Nbs1Δ BC (Shull et al., 2009; Stracker et al., 2007). Therefore, we propose that Nbs1 serves as a platform for Mre11 complex assembly and for the recruitment of ATM substrates to enhance access of the activated kinase to substrates that govern ATM- and Nbs1-dependent functions. In this context, we draw a distinction between ATM activation and ATM activities: in the former circumstance, a properly assembled and localized complex is required for ATM activation whereas in the latter, Nbs1 potentiates ATM activity by promoting access of the active kinase to its downstream effectors.
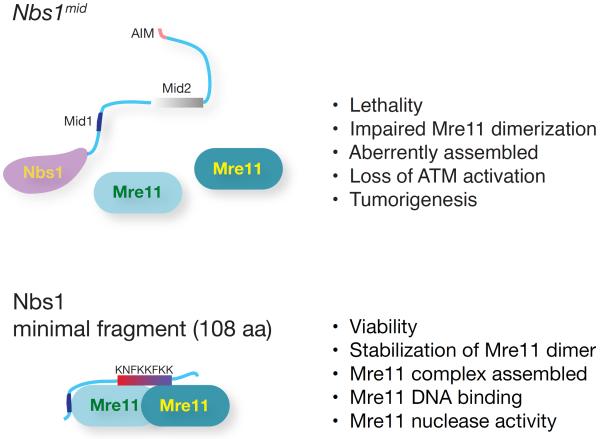
Although viability and ATM activation are lost upon genetic ablation of Nbs1, the protein does not appear to influence those outcomes directly. Rather, its association with Mre11 (and so Rad50) via its conserved interaction interface appears to influence both the subcellular localization and the proper assembly of the Mre11 complex, which in turn accounts for its influence on viability as well as ATM activation (Figure 7). Indeed, most of the Nbs1 protein, save for the Mre11 interaction interface is dispensable for viability as well as ATM activation. Therefore, the mechanism(s) underlying ATM activation would appear to be mediated by Mre11 and Rad50. Notably, ATM appears to interact with Rad50 in vitro (Lee and Paull, 2005), and recent genetic and biochemical analyses have shown that Rad50 influences the activation of ATM or its budding yeast ortholog Tel1 (Al-Ahmadie et al., 2014; Deshpande et al., 2014; Hohl et al., 2015). These data underlie the speculation that Rad50 is likely to be the proximal effector of ATM activation.
Figure 7. The Mre11 interaction domain of Nbs1 is necessary and sufficient for Mre11 complex functions.
Disruption Mre11-Nbs1 interaction results in cellular and organismal lethality and increased tumorigenesis due to a defects in Mre11 complex function. Nbs1 minimal fragment spanning just Mre11-Nbs1 interaction interface is sufficient to sustain the viability of cells and stabilize Mre11 dimer and Mre11 DNA binding and nuclease activity. These data indicate that the essential role of Nbs1 is via its interaction with Mre11, and that most of the Nbs1 protein is dispensable for Mre11 complex functions. Mid1, Mid2, and AIM denote Mre11-interacting domain 1, Mre11-interacting domain 2, and ATM interacting motif, respectively.
Manipulation of the DDR for therapeutic benefit offers significant potential (O'Connor, 2015). Accordingly, understanding of ATM activation is an important issue. This study thus provides important mechanistic insight toward that goal by defining the role of Nbs1 in promoting Mre11 complex functions in the DDR.
EXPERIMENTAL PROCEDURES
For detailed protocols of cell lines, cellular assay, Fetal liver cell transplantation, Immunofluorescence staining, Histopathology, see Supplemental Experimental Procedures
Mice
Nbs1mid mice were generated by help of Memorial Sloan-Kettering Mouse Genetics core. Detailed protocol will be provided upon request. Nbs1ΔB mice were previously described (Williams et al., 2002) and Nbs1F and vavCre mice were kindly provided by Zhao-Qi Wang (Fritz Lipmann Institute, Germany) and Hans-Guido Wendel (Memorial Sloan-Kettering Cancer Center, USA), respectively. The Institutional Animal Care and Use Committee of Memorial Sloan Kettering Cancer Center approved all protocols for animal use.
Protein purification and analysis
Bacterial expression vector for N-terminal his-tagged human Mre11 (2-411aa) was gifted from Dr. John Tainer (Lawrence Berkeley National Laboratory, USA). With C-terminal his-tag, human Nbs1 (F4, 622-729aa) was constructed in pMAL vector (New England BioLabs) for N-terminal MBP-tag for its solubility. See Supplemental Experimental Procedures for the purification, electrophoretic mobility shift assay (EMSA), and nuclease assay.
SFM analysis
For SFM analysis of MR/NBS1 F4-WT or -mid5 complexes, MR protein and Nbs1 F4 fragment were mixed in reaction buffer (20 mM Tris-HCl pH8, 0.1 M EDTA, 0.2 M NaCl, 2 mM DTT) at a molar ratio 1:1 (8 nM of MR to 8 nM F4-WT or -mid5) in total volume of 20 μl, and incubated for 5 min incubation on ice. The samples were then diluted 5 times in final volume of 20 μl in reaction buffer and deposited on freshly cleaved mica. After 1 min incubation at room temperature the mica was washed with milliQ water and dried with filtered air. Samples were imaged at room temperature and humidity with a Nanoscope VIII (Digital Instruments) operating in tapping mode. Type NHC-W, silicon tips with resonance frequency 310–372 kHz, were obtained from Nanosensors (Veeco Instruments, Europe). Images were collected at 2.5 × 2.5 μm, standard resolution 512 lines × 512 rows, and processed only by flattening to remove background slope. Images were quantified first by identifying MR complexes by visual inspection where molecules consisting a large globular domain with two protruding coiled coils were identified as M2R2. These were further classified based on the arrangement of globular domains as dimers with one globular domain or dimers with two distinct linked globular domains. The latter were further categorized according to the arrangement of coiled coils as: open, parallel or hook-linked. The frequency of the different forms was expressed as percentage of total molecules counted. The width of individual globular domains was determined using SFMetrics V4e software (Sanchez and Wyman, 2015) by manually measuring the longest axis across the globular domain.
Statistical Analysis
Statistical significance was analyzed by unpaired t-test, Fisher's exact test, or Wilcoxon rank sum test, and expressed as a p-value as indicated in the figure legends.
Supplementary Material
Acknowledgements
We thank John Tainer, Julien Lafrance-Vanasse, Titia de Lange, Hans-Guido Wendel and Zhao-Qi Wang for reagents, cells and mice. Members of the Petrini laboratory including Thomas J. Kelly provided helpful comments and suggestions throughout the course of this study. We are very grateful to Willie Mark and Peter Romanienko for designing and carrying out TALEN-based mutagenesis. This work was supported by the Geoffrey Beene Center at MSKCC and NIH grant GM59413 to J.H.J.P, 1F32 GM105296 to J.H.K, and the MSK Cancer Center Core Grant P30 CA008748.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions J.H.K., M.G., and R.A. performed the experiments and analyzed the data. J.H.K., C.W., P.C., and J.H.P. designed the experiments. J.H.K., and J.H.P. wrote the paper.
REFERENCES
- Adelman CA, De S, Petrini JH. Rad50 is dispensable for the maintenance and viability of postmitotic tissues. Mol Cell Biol. 2009;29:483–492. doi: 10.1128/MCB.01525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ahmadie H, Iyer G, Hohl M, Asthana S, Inagaki A, Schultz N, Hanrahan AJ, Scott SN, Brannon AR, McDermott GC, et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer discovery. 2014;4:1014–1021. doi: 10.1158/2159-8290.CD-14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem. 2005;280:18771–18781. doi: 10.1074/jbc.M413387200. [DOI] [PubMed] [Google Scholar]
- Anand R, Ranjha L, Cannavo E, Cejka P. Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol Cell. 2016 doi: 10.1016/j.molcel.2016.10.017. doi: 10.1016/j.molcel.2016.10.017. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Balestrini A, Nicolas L, Yang-Lott K, Guryanova OA, Levine RL, Bassing CH, Chaudhuri J, Petrini JH. Defining ATM-independent Functions of the Mre11 Complex with a Novel Mouse Model. Mol Cancer Res. 2015 doi: 10.1158/1541-7786.MCR-15-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Cerosaletti K, Concannon P. Independent roles for nibrin and Mre11-Rad50 in the activation and function of Atm. J Biol Chem. 2004;279:38813–38819. doi: 10.1074/jbc.M404294200. [DOI] [PubMed] [Google Scholar]
- Cerosaletti K, Wright J, Concannon P. Active role for nibrin in the kinetics of atm activation. Molecular and cellular biology. 2006;26:1691–1699. doi: 10.1128/MCB.26.5.1691-1699.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerosaletti KM, Concannon P. Nibrin forkhead-associated domain and breast cancer C-terminal domain are both required for nuclear focus formation and phosphorylation. J Biol Chem. 2003;278:21944–21951. doi: 10.1074/jbc.M211689200. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, Trujillo KM, Sung P, Hopfner KP, Carney JP, Tainer JA, Connelly JC, Leach DR, Kanaar R, Wyman C. Differential arrangements of conserved building blocks among homologs of the Rad50/Mre11 DNA repair protein complex. Journal of molecular biology. 2004;339:937–949. doi: 10.1016/j.jmb.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Demuth I, Frappart PO, Hildebrand G, Melchers A, Lobitz S, Stockl L, Varon R, Herceg Z, Sperling K, Wang ZQ, et al. An inducible null mutant murine model of Nijmegen breakage syndrome proves the essential function of NBS1 in chromosomal stability and cell viability. Hum Mol Genet. 2004;13:2385–2397. doi: 10.1093/hmg/ddh278. [DOI] [PubMed] [Google Scholar]
- Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Molecular and cellular biology. 2001;21:2184–2191. doi: 10.1128/MCB.21.6.2184-2191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Williams GJ, Limbo O, Williams RS, Kuhnlein J, Lee JH, Classen S, Guenther G, Russell P, Tainer JA, et al. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014;33:482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, Yang YP, Petukhova GV, Eckhaus M, Feigenbaum L, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Hohl M, Kochanczyk T, Tous C, Aguilera A, Krezel A, Petrini JH. Interdependence of the rad50 hook and globular domain functions. Mol Cell. 2015;57:479–491. doi: 10.1016/j.molcel.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Kwon Y, Galvan SM, Xue X, Tous C, Aguilera A, Sung P, Petrini JH. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nature structural & molecular biology. 2011;18:1124–1131. doi: 10.1038/nsmb.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol. 2002;12:1846–1851. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- Lakdawala SS, Schwartz RA, Ferenchak K, Carson CT, McSharry BP, Wilkinson GW, Weitzman MD. Differential requirements of the C terminus of Nbs1 in suppressing adenovirus DNA replication and promoting concatemer formation. J Virol. 2008;82:8362–8372. doi: 10.1128/JVI.00900-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DH, Hari F, Clapperton JA, Gwerder M, Gutsche K, Altmeyer M, Jungmichel S, Toledo LI, Fink D, Rask MB, et al. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat Cell Biol. 2014;16:792–803. doi: 10.1038/ncb3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel RA, Farnham PJ, Petrini JH. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Molecular and cellular biology. 2001;21:6006–6016. doi: 10.1128/MCB.21.17.6006-6016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- Morishima K, Sakamoto S, Kobayashi J, Izumi H, Suda T, Matsumoto Y, Tauchi H, Ide H, Komatsu K, Matsuura S. TopBP1 associates with NBS1 and is involved in homologous recombination repair. Biochem Biophys Res Commun. 2007;362:872–879. doi: 10.1016/j.bbrc.2007.08.086. [DOI] [PubMed] [Google Scholar]
- O'Connor MJ. Targeting the DNA Damage Response in Cancer. Mol Cell. 2015;60:547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- Oh J, Al-Zain A, Cannavo E, Cejka P, Symington LS. Xrs2 Dependent and Independent Functions of the Mre11-Rad50 Complex. Mol Cell. 2016;64:1–11. doi: 10.1016/j.molcel.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Chae J, Kim YC, Cho Y. Crystal structure of human Mre11: understanding tumorigenic mutations. Structure. 2011;19:1591–1602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Paull TT. Mechanisms of ATM Activation. Annu Rev Biochem. 2015 doi: 10.1146/annurev-biochem-060614-034335. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Nussenzweig MC, Nussenzweig A, Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc Natl Acad Sci U S A. 2005;102:1590–1595. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y FH, Kobayashi J. Role of NBS1 in DNA damage response and its relationship with cancer development. Transl Cancer Res. 2013;2:178–189. [Google Scholar]
- Sanchez H, Wyman C. SFMetrics: an analysis tool for scanning force microscopy images of biomolecules. BMC bioinformatics. 2015;16:27. doi: 10.1186/s12859-015-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Mockel C, Schele A, Strasser K, Jackson SP, et al. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nature structural & molecular biology. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull ER, Lee Y, Nakane H, Stracker TH, Zhao J, Russell HR, Petrini JH, McKinnon PJ. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009;23:171–180. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 2005;132:203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Couto SS, Cordon-Cardo C, Matos T, Petrini JH. Chk2 suppresses the oncogenic potential of DNA replication-associated DNA damage. Mol Cell. 2008;31:21–32. doi: 10.1016/j.molcel.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Morales M, Couto SS, Hussein H, Petrini JH. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature. 2007;447:218–221. doi: 10.1038/nature05740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen JW, Kaplan MI, Hunt PA, Williams BR, Ferguson DO, Alt FW, Petrini JH. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol Cell. 2003;12:1511–1523. doi: 10.1016/s1097-2765(03)00455-6. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Mitsuoka C, Terasawa M, Ogawa H, Ogawa T. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol Biol Cell. 2005;16:597–608. doi: 10.1091/mbc.E04-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden E, Sanchez H, Kinoshita E, Kanaar R, Wyman C. RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucleic acids research. 2009;37:1580–1588. doi: 10.1093/nar/gkn1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Mirzoeva OK, Morgan WF, Lin J, Dunnick W, Petrini JH. A murine model of Nijmegen breakage syndrome. Curr Biol. 2002;12:648–653. doi: 10.1016/s0960-9822(02)00763-7. [DOI] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhang X, Zhang L, Wu CY, Rezaeian AH, Chan CH, Li JM, Wang J, Gao Y, Han F, et al. Skp2 E3 ligase integrates ATM activation and homologous recombination repair by ubiquitinating NBS1. Mol Cell. 2012;46:351–361. doi: 10.1016/j.molcel.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci U S A. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C, Lebbink J, Kanaar R. Mre11-Rad50 complex crystals suggest molecular calisthenics. DNA repair. 2011;10:1066–1070. doi: 10.1016/j.dnarep.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Molecular and cellular biology. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.