SUMMARY
Ensuring that gut microbiota respond consistently to prescribed dietary interventions, irrespective of prior dietary practices (DPs), is critical for effective nutritional therapy. To address this, we identified DP-associated gut bacterial taxa in individuals either practicing chronic calorie restriction with adequate nutrition (CRON) or without dietary restrictions (AMER). When transplanted into gnotobiotic mice, AMER and CRON microbiota responded predictably to CRON and AMER diets but with variable response strengths. An individual’s microbiota is connected to other individuals’ communities (‘metacommunity’) by microbial exchange. Sequentially cohousing AMER-colonized mice with two different groups of CRON-colonized mice simulated metacommunity effects, resulting in enhanced responses to a CRON diet intervention and changes in several metabolic features in AMER animals. This response was driven by an influx of CRON DP-associated taxa. Certain DPs may impair responses to dietary interventions, necessitating the introduction of diet-responsive bacterial lineages present in other individuals and identified using the strategies described.
INTRODUCTION
The microbiota of an individual is not an isolated community, but rather exists within a large metacommunity, in which microbes have the potential to disperse between people to colonize new gut habitats and to become locally extinct from others. The diversity and richness of organisms within individuals, as well the similarity in composition between individuals, is governed by several ecological factors, including (i) selective pressures within host habitats, such as diet and antibiotic exposures (Falony et al., 2016; Zhernakova et al., 2016; Bokulich et al., 2016), and (ii) rates of colonization and extinction of specific microbial lineages within and between hosts. Global surveys of human gut microbiota have shown that Westernization is associated with lower taxonomic and functional diversity, which may be partially restored by dietary interventions involving calorie restriction (Le Chatelier et al., 2013; Cotillard et al., 2013). The microbiota of cohabitating individuals develop increased community similarity, indicating that transfer of gut taxa occurs between people with close associations (Song et al., 2013). The relative strengths of selection and dispersal in influencing community structure and diversity can vary across different metacommunities. Comparisons to null models suggest that in the USA, lower dispersal and highly variable selection play major roles in leading to reduced alpha-diversity and greater beta-diversity, whereas, in Sausi and Asaro populations in Papua New Guinea, greater dispersal and more consistent selection lead to greater alpha-diversity and lower beta-diversity (Martínez et al., 2015).
Interpersonal differences in the richness and composition of gut bacterial taxa have been correlated with biomarkers of metabolic and physiological health, as well as variation in metabolic and immunological responses to particular foods (e.g., Le Chatelier et al., 2013; Cotillard et al., 2013; Martínez et al., 2013; Zhu et al., 2016; Zeevi et al., 2015; Stefka et al., 2014). Short-term consumption of animal-or plant-based diets has been reported to reproducibly alter microbial community structure and gene expression in small groups of humans (David et al., 2013). Nonetheless, it is unclear whether individuals practicing different long-term dietary practices (DPs) have the capacity to respond to a given dietary intervention in a similar manner, or whether variation in their community structures leads to marked interpersonal differences in their responses. Thus, understanding how selection and dispersal impact responses to dietary interventions and how diet impacts these processes is important for a number of reasons, including (i) identifying microbes associated with different DPs, (ii) predicting the responses of individuals to prescribed diets based on a consideration of the representation of these microbes in the targeted human population, and (iii) determining the extent to which DP-associated microbes can used therapeutically as probiotics to enhance both the magnitude and durability of responses to prescribed dietary interventions in individuals lacking these taxa.
The current study focuses on two groups of people living in the USA who exemplify distinct DPs: those who practice chronic calorie restriction with optimized intake of nutrients (CRON), and those without prescribed or self-imposed dietary restrictions (AMER). Analysis of the nutrient intake of members of the Calorie Restriction Society has shown that these adherents to a CRON DP exceed the recommended daily intake of all essential nutrients while consuming nearly 50% fewer calories than their AMER counterparts (Fontana et al., 2004). They avoid processed and refined foods rich in (trans) saturated fatty acids and high glycemic carbohydrates. This DP is associated with reductions in risk factors for atherosclerosis and cancer, as well as improvements in several physiological and metabolic parameters (Fontana and Partridge, 2015). Moreover, long-term calorie restriction has been shown to influence gut microbial community structure in C57BL/6J mice (Zhang et al., 2013). We identify distinguishing characteristics of the fecal microbiota of CRON and AMER individuals, use food journals of CRON and AMER individuals to design diets representative of their DPs, and examine the effects of imposing these two disparate DPs on gnotobiotic mice colonized with transplanted fecal microbiota from AMER and CRON individuals. Finally, we examine whether AMER-colonized mice alter their responses to a CRON diet intervention when cohoused sequentially with mice harboring the transplanted microbiota of two different CRON donors. These experiments address several questions. Do transplanted microbiota from different individuals with different DPs take on similar configurations in response to prescribed diets? Do bacteria identified as DP-associated in the human population drive diet-induced reconfigurations in transplanted communities? Does exposure to DP-associated bacteria from other donors (simulating the potential for dispersal within a metacommunity) alter an individual donor’s community and metabolic responses to a diet intervention?
RESULTS
Discriminatory features of the fecal microbiota of CRON and AMER subjects
Single fecal samples were obtained from 34 adult CRON donors with body mass indices (BMIs) from 16.7 to 22.5 kg/m2, and 198 AMER donors. AMER microbiota donors (n=198) were placed into five different BMI ‘bins’ [n=66, lean (BMI ≤ 25 kg/m2); n=29, overweight (BMI 25–30 kg/m2); n=27, obese I (BMI 30–35 kg/m2); n=26, obese II (BMI 35–40 kg/m2); n=50 obese III (BMI >40 kg/m2)]. In the AMER cohort, 170 individuals were related to at least one other AMER donor in either twin sibling or mother-daughter relationships. CRON donors had practiced this DP for 2–21 years and none belonged to the same family. Donors had not used antibiotics for at least four months prior to sampling. The gut microbiota of CRON and AMER subjects were characterized by multiplex sequencing of PCR amplicons generated from variable region 4 (V4) of bacterial 16S rRNA genes present in pulverized fecal samples (Table S1A).
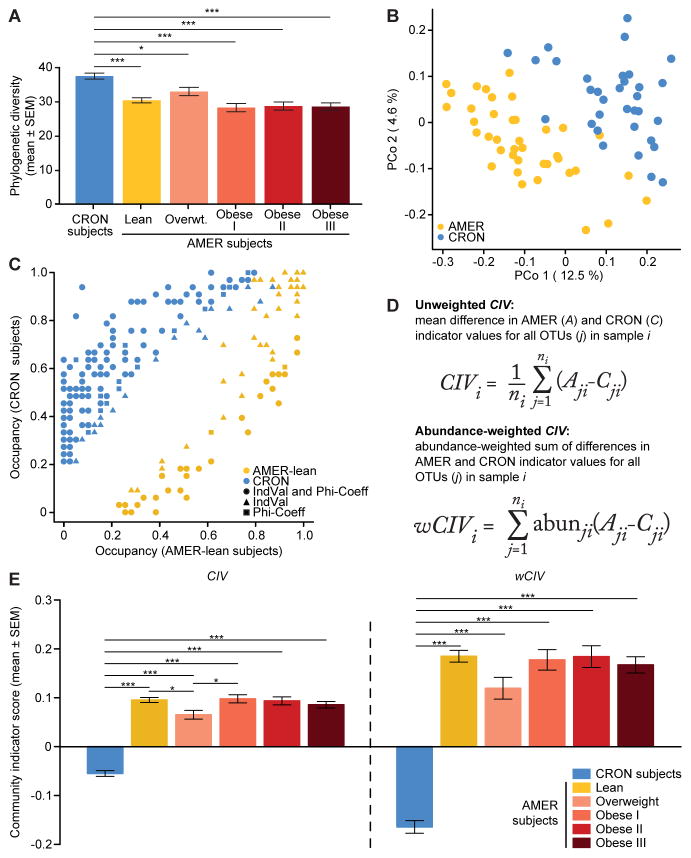
Linear mixed-effects models (LMEs), followed by comparisons of least squares means (LS-means), revealed that CRON fecal microbiota were significantly richer and more diverse than their AMER counterparts (Figure 1A, Table S2A–C). On average, CRON microbiota had greater phylogenetic diversity (PD; P=3.0×10−8), which measures the phylogenetic branch length represented in a community, more OTUs detected (P=0.0001), and greater Shannon’s diversity indices (P=2.7×10−5) and Pielou’s evenness indices (P=4.0×10−5). The increased taxonomic diversity and richness observed in CRON subjects compared to AMER subjects is consistent with the results of a previous study reporting that gut microbiomes with low gene content are associated with lower consumption of fruits and vegetables (Cotillard et al. 2013). While no statistically significant differences in richness or diversity were observed between lean and obese AMER subjects, both phylogenetic diversity (PD, P=0.024) and observed OTUs (P=0.025) were greater in lean versus obese AMER individuals who were members of the twin cohort. Moreover, paired t-tests in twin pairs discordant for obesity also showed significantly greater values in lean co-twins (PD, P=0.011; observed OTUs, P=0.035). These results are consistent with our previous studies (Turnbaugh et al., 2009).
Figure 1. Fecal microbiota of CRON and AMER human subjects.
(A) Faith’s phylogenetic diversity (PD) values in CRON and AMER subjects. (B) PCoA of unweighted UniFrac distances in CRON and lean AMER subjects. (C) Frequency of occurrence (occupancy) of dietary practice (DP)-associated OTUs in CRON and lean AMER subjects. Different colors denote different DPs. Different shapes denote which analyses [indicator value (IndVal) and phi-coefficient] detected significant DP associations for each OTU. (D) Mathematical formulae calculating unweighted and weighted community indicator values (CIV and wCIV). (E) CIV and wCIV values in CRON and AMER subjects. Least-squares means comparisons (following a linear mixed-effects model) identified statistically significant differences in PD, CIV, and wCIV, between subject groups. P-values were adjusted for multiple comparisons according to Holm’s method; *, P<0.05, **, P<0.01, ***, P<0.001. See also Figure S1.
We used UniFrac distances, a phylogenetic measure of the dissimilarity between microbial communities, to examine patterns of beta-diversity. Principal coordinates analysis (PCoA) of unweighted UniFrac distances showed that lean AMER and CRON individuals clustered apart from each other (Figure 1B). Permutational multivariate analysis of variance (PERMANOVA) revealed that DPs explained 7.9% (P=0.0001) of the variance in their unweighted UniFrac distances and 11.6% (P=0.0001) of the variance in their weighted UniFrac distances (Figure S1A, Table S2D–I; to avoid bias due to sampling related AMER individuals, the 39 lean AMER subjects with the most 16S rRNA reads among their respective families were utilized for UniFrac-based analyses, as well as the following Random Forests and indicator species analyses).
Random Forests classification was used to determine how well the compositions and relative abundances of 97%ID OTUs in the human donors’ fecal microbiota discriminated between the AMER and CRON DPs. In 100 independent runs with 1,000 decision trees each, the out-of-box error rate was low (4.4 ± 0.1%; mean ± SEM); the mean percentage of runs in which an individual’s microbiota was successfully classified was 97.2 ± 2.1% for AMER subjects and 93.6 ± 3.7% for CRON individuals (mean ± SEM). Hierarchical clustering of the algorithm’s mean proximity scores placed the sample communities into two large clusters, each dominated by either CRON or AMER subjects (Figure S1B). The 100 Random Forests-generated models also successfully predicted the DPs of the remaining 157 AMER subjects based on their fecal microbiota. Of these subjects, 147 (93.6%) were successfully identified as AMER in every run; this high degree of success was observed across all BMI categories [lean, 99.8 ± 0.2%; overweight, 92.2 ± 5.0%; Obese I, 98.4 ± 1.4%; Obese II, 100.0 ± 0.0%; Obese III, 97.8 ± 2.0% (mean ± SEM)].
To identify bacterial OTUs associated with the CRON and AMER DPs, we combined indicator species analysis and phi-coefficient analysis. Indicator species analysis measures the strength of association between an organism and a habitat type as the product of its fidelity (probability of occurrence in a habitat type), and its specificity mean abundance in that type, normalized to the sum of its mean abundances in all other habitat types observed) (Dufrêne and Legendre, 1997). A taxon is indicative of a particular habitat type if it is much more likely to occur in that type than in another, or if it is much more abundant in that type. Phi-coefficient analysis measures the correlation between a habitat type and the presence or absence of an organism. The significance of these associations was determined by permutation tests, followed by Benjamini-Hochberg corrections for multiple tests. OTUs were considered significantly associated with a DP if they had a significant indicator species value or phi-coefficient, or if both indicated significant associations with the same DP. We compared the results of these analyses to the OTUs’ feature importance scores (mean decreases in accuracy across all Random Forests runs).
Indicator species analysis and phi-coefficient analysis identified 242 DP-associated OTUs, of which 159 (65.7%) were significantly associated with the same DP by both metrics (Figure 1C, Table S3A). Of the significant DP-associated OTUs, 173 (71.5%) were CRON-associated, compared to 69 (28.5%) that were significantly associated with the lean AMER subjects. Of the CRON-associated OTUs, 69 (39.9%) were detected in less than 10% of lean AMER subjects, while only 15 (21.7%) of the lean AMER-associated OTUs were present in less than 10% of the CRON individuals. Random Forests feature selection scores were positively correlated with the association strengths defined by the indicator species analysis (Spearman’s ρ=0.63, P<10−15; Figure S1C). These findings, together with the increased richness and diversity observed in CRON subjects, suggest that the CRON lifestyle is associated with a large number of OTUs that are not commonly detected in people practicing the AMER DP.
We developed a metric, ‘community indicator value’ (CIV), to describe how strongly skewed a microbial community is toward CRON-associated or AMER-associated bacteria. CIV calculates the mean difference between each OTU’s AMER and CRON indicator species values, for all OTUs present in an individual’s gut community. The weighted version (wCIV) is the relative abundance-weighted sum of these differences in indicator species values (Figure 1D). Both metrics range from -1, where all OTUs are completely associated with the CRON DP, to +1, where all are perfectly associated with the AMER DP. Mean values of both CIV and wCIV were significantly lower in CRON individuals than in AMER individuals, indicating that the microbiota of CRON individuals were skewed toward greater representation of bacteria associated with the CRON DP (Figures 1E and S1D, Table S3B–D). These two metrics provide information about different ecological processes. CIV is expected to increase with new colonization by taxa that are more strongly AMER-associated. wCIV is affected by new colonization as well, but is also sensitive to increases in the relative abundance of more AMER-associated taxa that are already present. CIV and wCIV were convenient measures for tracking the responses of CRON and AMER donor microbiota transplanted into gnotobiotic mice in two different experimental contexts: (i) as ‘isolated’ communities exposed to representative CRON and AMER diets monotonously or in diet oscillations, and (ii) as ‘metacommunities’ formed through deliberate sequential cohousings of AMER-and CRON-colonized mice subjected to a CRON diet intervention.
Variability in responses of different human microbiota to CRON and AMER DPs in gnotobiotic mice
To determine whether the gut microbial communities of multiple individuals practicing the AMER or CRON DP mount generalizable responses to a switch in DP, we designed representative AMER and CRON diets and administered them to gnotobiotic mice colonized with fecal microbiota from CRON and AMER individuals. AMER and CRON DPs encompass an enormous set of possible food items and combinations. To identify differences in the diets of AMER and CRON subjects, and then generate diets that embody these differences, we compared food diaries from individuals representing the two DPs (Supplemental Experimental Procedures). On average, on a daily basis, CRON individuals consumed 42.1% fewer kilocalories, 48.6% less total fat, 33.5% less carbohydrates, 37.6% less total protein, and 60.8% less protein from animal sources than their AMER counterparts (ANOVA, p<0.05; Table S4A). We used these food journals to generate randomly selected lists of 41 food entries (the median number consumed daily). We then selected one menu for each DP that was most closely representative of the mean consumption of kilocalories, carbohydrates, proteins, and fats. This approach allowed us to generate representative diets using actual foods consumed by the two human groups without our own biases. The ingredients for each diet were cooked where appropriate, homogenized, and sterilized by gamma-irradiation (Supplemental Experimental Procedures; Table S4B–D).
Ten trios of adult (11–12 week old) male germfree C57BL6/J mice were individually caged and distributed among 10 gnotobiotic isolators and assigned to one of four diet treatments before being colonized, by oral gavage, with the fecal microbiota of one of five AMER donors or one of five CRON donors (Fig. 2A). Each donor’s microbiota was transplanted into mice in a single isolator. Within each isolator, one mouse was fed the AMER diet ad libitum and another the CRON diet restricted to ~60% of ad libitum caloric intake of the AMER diet. Additionally, the third mouse received the diet reflecting microbiota donor’s original DP, but at an altered caloric intake. For AMER donors, the third mouse was fed the AMER diet at a caloric intake equal to that of the CRON restricted treatment. For CRON donors, the third mouse was fed the CRON diet ad libitum (Figure 2A). Mice were maintained on their diet regimens from two weeks prior to colonization until they were sacrificed 11 weeks later. Body weights were recorded weekly throughout the experiment, and fecal samples were taken at the time points shown in Figure 2A.
Figure 2. Effects of diet on the fecal microbiota of gnotobiotic mice colonized with CRON and AMER donor communities.
(A) Experimental design used for monotonously-fed AMER-and CRON-colonized mice. Time points (days) for microbiota sampling are shown. (B,C) The first principal coordinates from PCoA plots of unweighted UniFrac (panel B) and weighted UniFrac (panel C) distances show initial community assembly in fecal microbiota of recipient mice. (D, E) PCoA plots of unweighted UniFrac (panel D) and weighted UniFrac (panel E) distances show the effects of experimental treatments on the phylogenetic structures of the various transplanted donor microbiota. Mean positions in PCoA space are shown for fecal samples collected from each mouse 15 to 56 days following gavage of the human donor’s microbiota. Points connected by yellow lines in both panels are from mice colonized by the same AMER donor, and points connected by blue lines are from the same CRON donor. Circle colors in panel D reflect donor DP, and mouse diet treatment in panel E. (F,G) CIV (panel F) and wCIV (panel G) values (mean ± SEM) in fecal microbiota harvested from each mouse 15 to 56 days after gavage.
Body weight was significantly influenced by diet treatment (P=3.9×10−8; LME) and not by the DPs of microbiota donors (P=0.840; LME); by the end of the experiment, mice on the AMER diet had gained 23.0 ± 3.7% (mean ± SEM) of their initial body weight when fed ad libitum and lost 14.6 ± 1.3% at the restricted caloric level. Likewise, mice fed the CRON diet ad libitum lost 1.0 ± 2.1% while those fed the CRON diet at the restricted level lost 18.4 ± 1.8% (Table S5A). The relative weights of the epididymal fat pads mirrored this pattern and were highly correlated with change in body weight (Pearson’s correlation=0.912, P=3.4×10−11; Table S5A).
Fecal and cecal microbial communities collected over the course of the experiment were characterized by bacterial V4-16S rRNA gene sequencing (Table S1B; Figure 2A). PCoA of unweighted UniFrac distances revealed that the transplanted fecal communities achieved relatively stable compositions by the eighth day after gavage (Figure 2B,C). Therefore, we limited our subsequent analyses to fecal samples taken after the 8-day time point. Beyond this time point, the microbiota of recipient mice captured, on average, 23.9 ± 0.3% (mean ± SEM; range 14.4–35.6%) of their human donors’ OTUs, and 87.5 ± 0.4% (mean ± SEM; range 73.7–96.7%) of the V4-16S rRNA reads in the donors’ microbiota, suggesting that the most prevalent organisms successfully colonized mice (Table S5B).
PERMANOVA revealed that the experimental factors ‘microbiota donor,’ ‘donor DP,’ and ‘mouse diet type,’ influenced unweighted UniFrac and abundance-weighted UniFrac distances differently (Figure 2D,E and Table S5C). For unweighted UniFrac, microbiota donor DP explained 10.5% of the variance, donor explained 51.9%, and the mice’s diets explained 6.5%. For weighted UniFrac, the corresponding values were 5.8%, 39.8% and 27.7%, respectively. Caloric intake did not significantly influence unweighted or weighted UniFrac distances for mice consuming the CRON or AMER diets (Table S5D,E). PCoA of unweighted UniFrac distances also showed that the phylogenetic compositions of the gnotobiotic animals’ fecal microbiota remained highly differentiated from each other according to their human donors and their donors’ DPs (Figure 2D). However, PCoA of weighted UniFrac distances divided the mice’s fecal microbiota according to their diet types (AMER vs. CRON), indicating that the various recipient communities shared similarities in the bacterial lineages that exhibited altered relative abundances in response to the diets (Figure 2E).
CIV and wCIV showed the same pattern, and confirmed that bacteria identified as DP-associated in the human population contributed to the similarity of these community-level responses. Using mean values obtained from all fecal samples collected from each mouse after the 8-day time point, LME models showed that the animals’ diets (F=48.04, P=0.0004) and their microbiota donor’s DPs (F=6.33, P=0.0456) significantly influenced CIVs (Figure 2F, Table S5F,G). In contrast, only the diets significantly influenced wCIV (F=37.61, P=0.0009); for both unweighted CIV and weighted CIV, mice consuming the AMER diet ad libitum consistently had higher values than their counterparts fed the CRON diet at the restricted level (Figure 2G, Table S5F,G). The diet-based responses of individual donors’ microbiota varied in magnitude, but shared predictable features. On a CRON diet, transplanted AMER microbiota manifested an enhanced representation of members that were classified as more strongly CRON-associated (Figure 2F,G, Table S5H–K). Similarly, transplanted CRON microbiota responded to the AMER diet exposure by increasing the representation of more strongly AMER-associated taxa. Paired t-tests revealed no significant differences between groups of mice consuming different amounts of the same diet. Procrustes analyses showed that cecal and fecal microbiota at the end of the experiment showed very similar patterns of beta-diversity, with high correlations for both unweighted UniFrac (M12=0.965, P=0.001) and weighted UniFrac (M12=0.869, P=0.001).
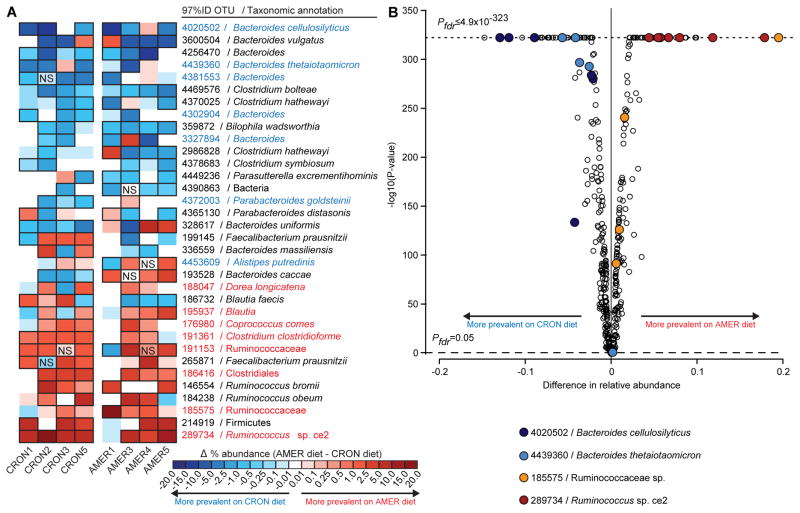
For each donor’s microbiota, Cochran-Mantel-Haenszel tests with strata defined by the sampling days were used to identify OTUs whose relative abundances differed consistently over time between an animal fed the AMER diet ad libitum and an animal consuming the CRON diet at the restricted level. These tests yielded 180 OTUs with significant differences in at least one donor microbiota; 73 were previously identified as significantly DP-associated in the human subjects (46 CRON-associated, 27 AMER-associated; Table S5L). Fifteen of these 73 DP-associated OTUs represented at least 1% of a recipient mouse’s fecal microbiota and exhibited a diet-based difference in relative abundance in at least four donors’ transplanted microbiota. These 15 OTUs accounted for 74 significant diet-based differences observed across the various transplanted donor microbiota, 91.9% of which were in the direction predicted from their DP-association in AMER or CRON population (e.g., the CRON-associated Bacteroides cellulosilyticus OTU 4020502 and the AMER-associated Ruminococcus sp. Ce2 OTU 289734 at the top and bottom of Figure 3A,B). These microbial responses also provided direct evidence that common features of the highly varied diets consumed by a group of unrelated human subjects within a broadly defined DP were captured by our randomized approach to diet design.
Figure 3. Effects of diet on the relative abundances of individual OTUs in the fecal microbiota of AMER-colonized and CRON-colonized mice.
(A) A heatmap shows differences in mean relative abundances of OTUs, for each transplanted human donor community, between recipient mice fed the AMER diet (ad libitum) and those fed the CRON diet (restricted). Cells outlined in black represent significant differences (P<0.05; NS, non-significant differences). AMER-associated and CRON-associated OTUs are highlighted in red and blue text, respectively. (B) A volcano plot shows the significance and magnitude of differences in the relative abundances of OTUs between the diet treatments, as defined Cochran-Mantel-Haenszel tests followed by FDR correction of P-values. PFDR-values of zero are plotted as ≤4.9×10−323 (minimum non-zero value calculated by the R software package). Only OTUs with a mean relative abundance of 0.5% in at least one diet context were included in the analysis. Samples collected from experimental days 15 to 56 following gavage of the indicated donor communities are included in the analysis.
Exposure to multiple CRON microbiota enhances the response of an AMER microbiota to a CRON diet intervention
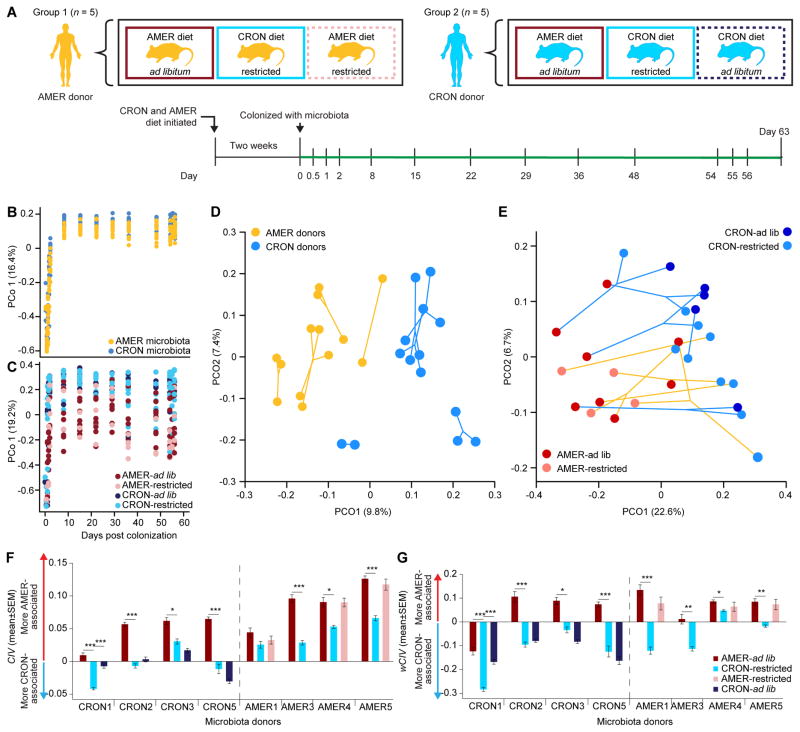
We hypothesized that an AMER donor’s transplanted microbiota would exhibit a greater response to a CRON diet intervention as part of a metacommunity that included CRON microbiota than it would if it remained isolated. Cohousing coprophagic mice has been used in previous studies to determine which bacterial taxa from one community can invade another under various dietary conditions (e.g., Ridaura et al. 2013; Seedorf et al., 2014). We reasoned that cohousing mice harboring different human donor microbiota would allow their ‘isolated’ communities to be placed in a deliberately designed metacommunity. Our experimental design included four groups of mice: two groups colonized with the fecal microbiota of a single AMER donor (AMER5) and two other groups each colonized with microbiota from one of two different CRON donors (Figure 4A,B). The human donor AMER5 was chosen because that subject’s transplanted fecal microbiota had very high CIV values in the gnotobiotic mouse experiment described in Figure 2A (i.e., was the most skewed towards AMER-associated OTUs even in the context of the CRON diet; Figure 2F). The CRON1 and CRON5 donors were chosen due to the very strong responses of their transplanted microbiota to CRON versus AMER diets (Figure 2D–G). Five AMER-colonized mice that would not be exposed to other communities were maintained as controls (AMER-Control). The other five AMER-colonized mice served as the target hosts (AMER-Target) that would be exposed to CRON1 and CRON5 mice. All mice were initially caged individually, with each group of animals maintained in a separate gnotobiotic isolator. All mice were started on the diets of their microbiota donor three days prior to transplantation (Figure 4A). To provide a within-experiment measurement of the response of the transplanted donor communities’ responses to a diet intervention, all mice were maintained on the diet of their microbiota donor for 10 days following transplant (“Diet 1” stage; AMER ad lib, CRON restricted) and were then switched to the opposite diet for 11 days (“Diet 2” stage). Beginning 21 days after initial colonization, all mice in all four groups were maintained on the CRON diet at the restricted feeding level (“CRON diet” stage). At 38 days post colonization, the CRON1 mice were moved to the AMER-Target isolator, and each CRON1 mouse was placed in the cage of an AMER-Target mouse (“Cohousing stage 1” in Figure 4A,B). Upon cohousing, the amount of the CRON diet was doubled in these cages to preserve the level of daily caloric intake the mice had experienced when singly housed. At day 52, the CRON1 mice were returned to their original cages, and the CRON5 mice were moved to the AMER-Target isolator in the same fashion (“Cohousing stage 2” in Figure 4A,B). At day 66, the CRON5 mice were returned to their original cages. All mice remained in their original gnotobiotic isolators until the end of the experiment on day 78 (stage “Post cohousing”). Bedding was changed with every change in diet or housing. This protocol yielded microbiota exposed to one, two, or three communities by the end of the experiment. Fecal microbiota were sampled throughout the diet oscillations and housing changes and profiled by V4-16S rRNA sequencing (Figure 4A,B; Table S1C). Transplantation efficiencies at the ends of the Diet 1 and Diet 2 stages were comparable to those in gnotobiotic mice fed the AMER and CRON diets monotonously (Table S6A; Table S5B).
Figure 4. Responses to a CRON diet intervention in the context of an experimental metacommunity.
(A) AMER-and CRON-colonized mice were subjected to diet oscillations and cohousing in a designed metacommunity, during a CRON diet intervention, as described in the text and Supplemental Experimental Procedures. (B) During cohousing stages, AMER-Control mice were only exposed to their human donor’s microbiota, CRON1 mice were exposed to an AMER and a CRON community, and AMER-Target and CRON5 mice were exposed to microbes from all three. (C,D) CIVs (Panel C) and wCIVs (Panel D) for transplanted fecal bacterial communities are shown as a function of dietary and microbial exposures. More positive CIVs and wCIVs indicate greater representation of more AMER-associated OTUs, while more negative values indicate greater representation of more CRON-associated OTUs. (E) Hierarchical clustering groups the fecal microbiota of mice according to diet and history of microbial exposure. See also Figure S2.
Before cohousing, in isolation, the fecal microbiota of CRON1 and CRON5 mice changed dramatically in response to 10 days on the AMER diet. CIV and wCIV increased in these mice, as did the total relative abundance of AMER-associated OTUs, while the total relative abundance of CRON-associated OTUs decreased. Conversely, AMER-colonized mice showed no significant response in wCIV and only a small increase in the total proportional representation of CRON-associated OTUs after 10 days of exposure to the CRON diet. After an additional 17 days (CRON diet stage), both CIV and wCIV were significantly reduced, but failed to reach the values in either group of CRON-colonized mice while they consumed the CRON diet. The AMER-Control mice did not reach the pre-cohousing CIV and wCIV levels of either group of CRON-colonized mice even after 57 days of consuming the CRON diet (Figure 4C,D; Figure S2; Table S6B–J; also see Figure S2F and Table S7A for OTUs whose relative abundances were significantly altered by the diet interventions as defined by Cochran-Mantel-Haenszel tests).
Having established that the AMER-colonized mice were unable to emulate the configurations of transplanted CRON microbiota in mice fed the CRON diet, we next determined whether sequential cohousing with the CRON1 and CRON5 mice would strengthen the response of the AMER5 community.
During Cohousing stage 1, phylogenetic diversity increased, and both CIV and wCIV decreased in the AMER-Target mice cohoused with CRON1 mice (Figure 4C,D; Figure S2C; Table S6B,C,F,G,J). These changes reflected increases in the numbers and total relative abundances of CRON-associated OTUs in the AMER-Target mice’s fecal microbiota (Figure S2B,D; Table S6E,I,J). In Cohousing stage 2, when AMER-Target mice were caged with CRON5 mice, phylogenetic diversity and the number of CRON-associated OTUs increased further and unweighted CIVs decreased. Nonetheless, the aggregate percent abundance of CRON-associated OTUs and weighted CIVs did not continue to change in AMER-Target animals with this second cohousing (Figure 4C,D; Figure S2B–D; Table S6B,C,E,F,G,I,J). These exchanges of microbes also resulted in convergence in configurations of the fecal microbiota of cohoused mice. Hierarchical clustering of Bray-Curtis dissimilarities between the animals’ fecal microbiota grouped them by their donors and diets. The AMER-Target mice clustered with the AMER-Control mice before cohousing, with the CRON1 mice during the first cohousing, and with CRON5 mice during and after the second cohousing (Figure 4E).
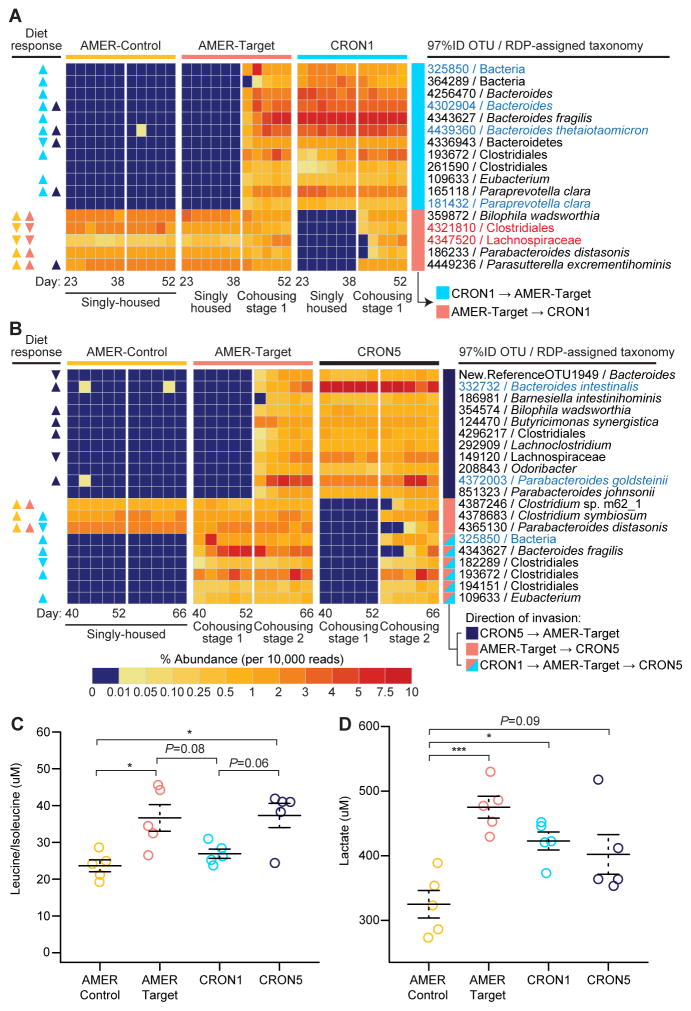
Phi-coefficient-based analysis identified OTUs that were consistent invaders of each group of mice during the two cohousing stages (see Supplemental Experimental Procedures for analytical methods and tests of statistical significance). This analysis identified 27 OTUs that passed from the CRON1 mice into the AMER-Target mice during Cohousing stage 1 (Figure 5A, Table S7B). By the end of Cohousing stage 1, the aggregate relative abundances of these invading OTUs accounted for 27.1% of the total community (24.7–32.1%, range). The 27 OTUs included seven that were CRON-associated in the human population, and 11 that had previously displayed greater relative abundance in the CRON versus AMER diet context in CRON1 mice (i.e., from the comparison of pre-cohousing Diet 1 versus Diet 2 stages) (Figure 5A, Table S7B). Thus, a variety of diet-responsive taxa from the CRON1 community established themselves in an AMER microbiota in the context of a CRON diet. Many (11/27) of these successful invaders belonged to the phylum Bacteroidetes, including OTUs assigned to B. fragilis (4343627), B. thetaiotaomicron (4439360), Paraprevotella clara (165118 and 181432) and the genus Alistipes (213671) (Figure 5A, Figure S3, Table S7B).
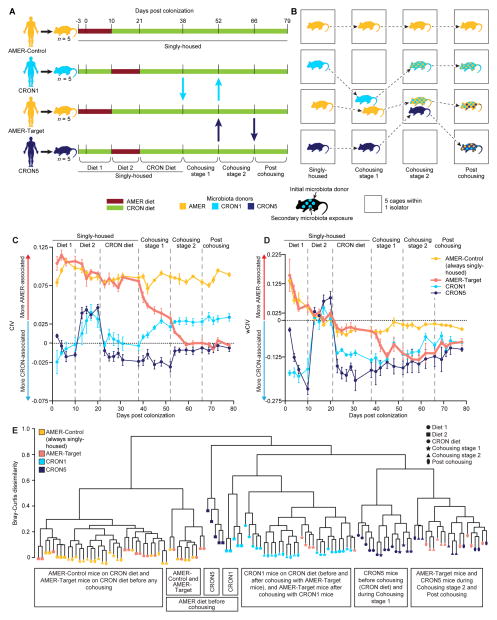
Figure 5. Exchange of bacterial OTUs and changes in liver metabolite concentrations during cohousing of AMER-Target and CRON-colonized mice.
Heatmaps present the mean percent abundances of prominent colonizing OTUs transferred between the (A) AMER-Target mice and CRON1 mice during Cohousing stage 1 and (B) AMER-Target mice and CRON5 mice during Cohousing stage 2 (n=4–5 animals/time point/treatment group). Triangles under the “Diet response” column on the left indicate the direction of significant changes in relative abundance during the pre-cohousing diet oscillation (Diet 1 and Diet 2 stages) in each treatment group (yellow, AMER-Control; salmon, AMER-Target; light blue, CRON1; dark blue, CRON5). Upward-facing triangles indicate that an OTU was more prevalent in the CRON diet (restricted) context while downward facing triangles indicate an OTU that was more prevalent when animals consumed the AMER diet ad libitum. Invading OTUs (see Supplemental Experimental Procedures) with a mean percent abundance of at least 0.5% during the cohousing stages are presented. OTUs that were CRON-associated or AMER-associated in human subjects are highlighted in blue and red, respectively. (C) Individual concentrations and means (± SEM) are presented for leucine/isoleucine and (D) lactate in liver samples taken at the time of euthanasia. Significance of differences in treatment group means was determined by ANOVA (P<0.05 after FDR-correction) followed by Tukey’s post-hoc tests. ***, P<0.001, **, P<0.01, P<0.05. See also Figure S3.
During Cohousing stage 2, an additional 27 OTUs invaded the AMER-Target microbiota, this time from the CRON5-colonized mice (Figure 5B, Figure S3, Table S7C). Six of these OTUs were significantly CRON-associated in humans, and six were significantly more prevalent when the CRON5 mice were fed the CRON diet compared to AMER diet during the pre-cohousing Diet 1 and Diet 2 stages. By the end of Cohousing stage 2, these 27 OTUs comprised a mean of 21.6% (range, 18.9 to 23.6%) of the AMER-Target cagemates’ fecal microbiota. Again, the Bacteroidetes accounted for a large proportion of the invading OTUs (11/27), including Parabacteroides johnsonii (851323), P. goldsteinii (4372003), B. intestinalis (332732), and B. ovatus (4467447) (Figure 5B, Figure S3, Table S7C).
During both cohousing stages, invasion was bidirectional. In Cohousing stage 1, 16 OTUs from the AMER-Target mice migrated to the CRON1 mice (Figure 5A, Table S7D). Seven of these had previously shown an increase in relative abundance in the AMER-Target mice when they were switched to the CRON diet during the pre-cohousing stages. During Cohousing stage 2, 19 OTUs from the AMER5-mice invaded the CRON5 mice, but nearly half of these (9) actually originated in the CRON1 mice, including five OTUs that had increased proportional abundance in the CRON1 mice fed the CRON diet before cohousing (Figure 5B, Table S7E). Thus, much of the influx of OTUs into CRON-colonized mice from their AMER-Target cagemates was made up of OTUs that had shown a preference for the CRON diet in mice and/or humans. Four OTUs originating in the AMER5-colonized mice invaded both the CRON1 and CRON5 mice. The invasion of the CRON mice may be due to several possible factors. CRON donor communities, though more diverse than AMER communities, may possess unoccupied niche space. Taxa that successfully transfer from AMER5 mice to CRON1 mice may displace resident taxa due to better utilization of particular components of the CRON diet than some resident taxa present in the CRON1 community (Table S7D). Moreover, some of these invading taxa may maintain their fitness regardless of the diet by consuming resources available in both diet contexts or derived from the host (e.g., mucosal glycans).
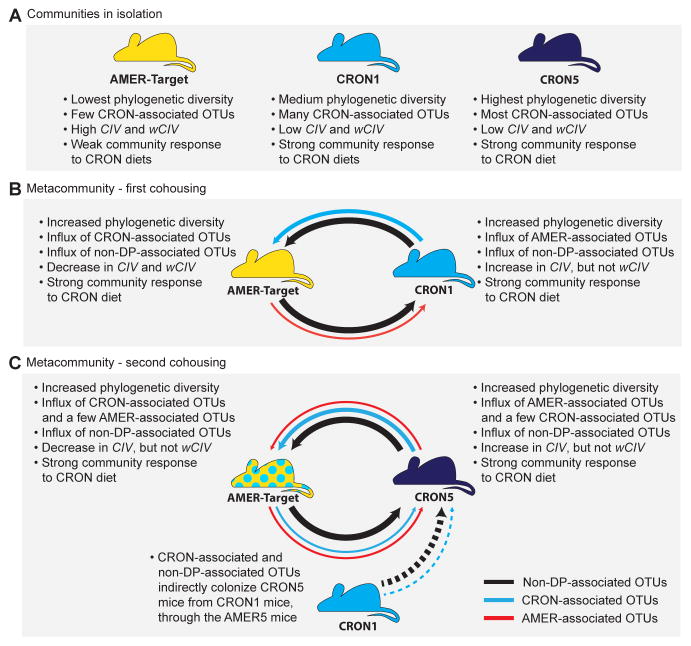
Our experimental metacommunity demonstrated that the exchange of diet-responsive taxa between hosts can enhance the extent of a community’s reconfiguration in response to a diet intervention. This finding raises an obvious question: would similar community configurations arise from cohousing per se, regardless of diet, or must the target community’s host be consuming a CRON diet. One way to determine whether a DP-associated donor microbiota and a diet reflective of that DP are both necessary to enable a response to that DP (i.e., transfer of the relevant OTUs) is to conduct a cohousing experiment using a diet that is not associated with the target or donor microbiota. However, it is not apparent how to design, a priori, a diet that is neutral with respect to the preferences of CRON-associated and AMER-associated taxa; this would require knowledge of the overlap between the CRON and AMER diets at the molecular level as well as the resource requirements of each DP-associated taxon. Nonetheless, we were able to explore this question in part by considering the changes observed in CRON1 mice and CRON5 mice when they came into contact with the AMER-Target mice. During Cohousing stage 1, the mean CIV of CRON1 mice increased (Figure 4C; Tables S6C,G,J), along with an increase in their mean number of AMER-associated OTUs (Figure S2E; Tables S6D,H,J). CRON5 mice exhibited similar increases in CIV and the number of AMER-associated OTUs in their communities during cohousing stage 2 (Figure 4C; Figure S2E; Tables S6C,D,G,H,J). By these metrics, these two CRON communities became more AMER-like despite being fed the CRON diet at the same time. However, metrics incorporating the relative abundances of bacterial lineages (wCIV, total abundances of CRON associated taxa) showed that the CRON mice maintained strong CRON-like structures. We conclude from these observations that the CRON diet did not prevent invasion of AMER-associated taxa into the CRON microbiota. This is consistent with results from the experiment described in Figure 2A, in which transplanted microbiota from five AMER and five CRON subjects maintained community compositions reflective of their donors, but exhibited large differences in the relative abundances of specific taxa in response the AMER and CRON. Figure 6 summarizes the outcomes the diet switching and cohousing experiments.
Figure 6.
Summary of results obtained from microbial communities in isolation and as members of an experimental metacommunity.
Effects of history of microbial exposure on metabolic profiles
There was no statistically significant difference in the percent weight change between AMER-Control and AMER-Target mice from the beginning of cohousing to the end of the Post-cohousing stage (t-test, P=0.082). To characterize the metabolic effects of microbial exchange in the sequential cohousing experiment, we performed mass-spectrometry-based profiling of 81 metabolites in cecal contents and 176 in liver in all animals at the time of euthanasia (Table S8).
Several metabolites present in cecal contents differentiated AMER-Control animals from AMER-Target, CRON1, and CRON5 mice (Table S8A). AMER-Target mice exhibited significantly increased cecal concentrations of propionate compared to AMER-Control mice, as well as moderate, but not significant increases in acetate. AMER-Control mice exhibited elevated concentrations of methylmalonylglycine and succinate. This increase may reflect heightened methylmalonyl-CoA metabolism, in which methylmalonylglycine is produced as a means of purging excess methylmalonyl-CoA that is isomerized to succinyl-CoA. In addition, three inositol metabolites (pinitol, chiro-inositol, and myo-inositol) were significantly higher in AMER-Control animals than in the other three groups. Pinitol was also detectable in the serum of AMER-Control but not in any of the other treatment groups. Inositols are present at high levels in a variety of ingredients that are well represented in the CRON diet, including fruits, nuts, and grains (Clements and Darnell, 1980). Moreover, inositol is a commonly used dietary supplement. Nonetheless, the role of the gut microbiota in metabolizing inositols is not well understood.
AMER-Target mice had significantly increased hepatic concentrations of eight amino acids (leucine/isoleucine, alanine, glutamine/glutamic acid, phenylalanine, tyrosine, ornithine, serine, proline) compared to AMER-Control mice (p<0.05, ANOVA with Tukey’s post-hoc tests). The concentrations of these amino acids in this group were not significantly different from those of CRON5 mice that served as their final cohousing partners (Figure 5C, Table S8B). Likewise, liver lactate concentrations were significantly greater in AMER-Target mice compared to AMER-Controls (P<0.0008, Tukey’s post-hoc test; Figure 5D). Modest decreases in pyruvate, alpha-ketoglutarate and citrate levels were observed in the former compared to latter animals but did not reach statistical significance in liver (Table S8B). Together, these findings suggest that mice harboring an isolated AMER donor community and fed a CRON diet used a greater proportion of glucose oxidatively, with less glycolytic metabolism to lactate and less conversion of pyruvate to alanine and other amino acids, than AMER mice that had been exposed to both CRON microbiota. Conversely, in animals exposed to both the CRON1 and CRON5 microbiota (AMER-Target and CRON5 mice), a greater proportion of glucose was used for lactate and amino acid production. These metabolic features were not observed in the CRON1 group, which had never been exposed to the CRON5 community, and whose intestinal communities were not as enriched in CRON-associated taxa as in either CRON5 or AMER-Target mice (Figure 5C,D).
DISCUSSION
Long-term DPs play a large role in determining the selective environment that the gut microbiota faces, ultimately influencing the composition and diversity of taxa maintained within the gut microbial community. In our study, Americans consuming unrestricted diets maintained less diverse fecal microbiota than those of individuals adhering to plant-rich diet with restricted caloric intake. Moreover, the AMER individuals’ gut communities were marked by a lack of many bacterial lineages that are indicators of the CRON DP. When transplanted into gnotobiotic mice and faced with a CRON diet intervention, some of these less diverse AMER communities mounted community reconfigurations that were weaker than those of their CRON counterparts. By placing these AMER communities into a model meta-community composed of several CRON communities, we were able to utilize the dispersal of organisms between coprophagic mice to enhance the reconfiguration of an AMER individual’s microbiota in response to a CRON diet intervention.
Humans continuously shed microorganisms; a vivid and experimentally validated image is that each individual is literally surrounded by a cloud of his/her microbes (Meadow et al., 2015). More knowledge is needed about the rates of interpersonal exchange of organisms as a function of the ‘health’ of a microbiota and host. To what extent do health status and behavior (e.g., hygiene, dietary practices, antibiotic use) impose selective forces that influence the composition and diversity of an individual’s gut microbiota, as well as the dispersal of organisms between individuals? Experiments in mice have shown that multigenerational exposure to a Western diet poor in “microbiota-accessible carbohydrates” can lead to extirpation of specific bacterial lineages (Sonnenburg et al., 2016). In addition, Cotillard et al. (2013) reported that in humans possessing gut microbiomes with “low gene content”, six weeks of caloric restriction raised gene content to nearly the same levels observed in individuals with high microbiome gene content, although the sources of the microbial gene enrichment were not defined. We may need to evolve our view of ‘social’ diseases to include an understanding of how a given individual’s gut microbial community is connected to those of other hosts in a microbial metacommunity.
The current study illustrates a way of characterizing the extent to which bacterial dispersal under the most permissive conditions (cohousing of coprophagic mice) impacts diet responses. An artificial metacommunity provides an opportunity to mine multiple human microbiota for organisms that are not only reporters of responses to diet interventions but also effectors of host responses to these interventions. Applying this approach may facilitate preclinical identification of (i) diets capable of reprogramming human gut microbial functions in individuals practicing additonal DPs, and (ii) consortia of diet-responsive taxa that may influence the outcomes of given diet interventions. In principle, constructing experimental metacommunities using donors exemplifying contrasting phenotypes of interest may also be useful for capturing taxa that influence other features of host biology in healthy or diseased states.
EXPERIMENTAL PROCEDURES
Human studies
Human fecal samples were obtained from biospecimen collections assembled during previous studies (Turnbaugh et al., 2009; Muegge et al., 2011; Ridaura et al., 2013) as well as from subjects in ongoing studies. Subjects were enrolled and samples were collected using procedures approved by the Washington University Human Studies Committee. Fecal samples were frozen at −20°C within 30 min after collection and placed in a −80°C freezer within 24h.
Animal Studies
Protocols used in experiments involving mice were approved by the Washington University Animal Studies Committee.
Constructing representative diets
Representative CRON and AMER diets were designed based on daily food journals kept by members of the Calorie Restriction Society and a non-restricted control cohort. Briefly, 1,000 randomly selected menus of 41 items from the journals of each cohort were compared to the mean daily intake of kilocalories, carbohydrates, proteins and fats to select the best representative diets for each DP. Ingredients were then homogenized, after cooking eggs and meat, and irradiated before feeding to gnotobiotic mice (Supplemental Experimental Procedures).
Transplanting human fecal microbiota to gnotobiotic mice
Aliquots of pulverized, frozen fecal samples from human donors were resuspended under anaerobic conditions in filter-sterilized PBS supplemented with 0.05% L-cysteine-HCl and 0.1% resazurin. A single 200-μL aliquot of this suspension was administered by oral gavage into the stomach of each mouse within its gnotobiotic isolator. Details of animal husbandry and experimental design are provided in Results and Supplemental Experimental Procedures.
Bacterial 16S rRNA analyses
DNA was isolated from aliquots of pulverized frozen human feces, mouse fecal pellets, and mouse cecal contents by extraction with phenol:chloroform:isoamyl achohol (Supplemental Experimental Procedures). The V4 region of bacterial 16S rRNA genes present in fecal and cecal samples was amplified by PCR and sequenced using an Illumina MiSeq instrument, according to protocols detailed in Bokulich et al. (2013). OTU abundance data were rarefied to 10,000 reads per sample prior to statistical analyses (also see Supplemental Experimental Procedures).
Successfully invading OTUs were identified by a multi-step phi-coefficient analysis performed for each cohousing stage and invasion direction (i.e., CRON→AMER, AMER→CRON). OTUs invading in the AMER→CRON direction were defined as those that were significantly (PFDR<0.05) associated with (i) the AMER-Target mice (versus their CRON cagemates) prior to cohousing, and (ii) samples taken from the CRON-colonized mice during cohousing (versus samples take pre-cohousing) with a phi-coefficient ≥0.6. OTUs invading in the CRON→AMER direction were defined as those that were significantly (PFDR<0.05) associated with (i) the CRON1 or CRON5 mice (versus their AMER-Target cagemates) prior to cohousing, and (ii) samples taken from the AMER-Target mice during cohousing (versus samples take pre-cohousing) with a phi-coefficient ≥0.6, and (iii) the AMER-Target mice (compared to AMER-Control mice) during cohousing (also see Supplemental Experimental Procedures).
Measurement metabolites in mouse cecal contents, serum and liver samples
Cecal contents were analyzed using non-targeted GC-MS of 69 analytes, targeted UPLC-MS of bile acids and targeted GC-MS of short-chain fatty acids. Liver samples were subjected to targeted MS-MS of 176 analytes. Protocols for MS analyses are described in Ridaura et al. (2013; also see Supplemental Experimental Procedures).
Supplementary Material
Acknowledgments
We thank David O’Donnell, Maria Karlsson, and Sabrina Wagoner for help with gnotobiotic mouse husbandry, and Su Deng, Marty Meier and Jessica Hoisington-Lopez for contributions to generating 16S rRNA sequencing data. J.I.G. is a co-founder of Matatu, Inc., a company characterizing the role of diet-by-microbiota interactions in animal health. This work was supported in part by grants from the NIH (DK078669, DK70977, DK30292). N.W.G. received support from an NIH postdoctoral training grant (T32 DK007120). P.P.A is the recipient of a Sir Henry Wellcome Postdoctoral Fellowship from the Wellcome Trust (096100).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, N.W.G., P.P.A., and J.I.G.; Investigation, N.W.G., P.P.A, J.C., O.I.; Resources, A.C.H., L.F.; Data curation, N.W.G., A.C.H., L.F.; Formal analysis, N.W.G.; Writing Original Draft, N.W.G. and J.I.G.; Writing Review and Editing, N.W.G., J.I.G., P.P.A., A.C.H., C.B.N; Supervision, J.I.G.
ACCESSSION NUMBERS
Bacterial V4-16S rRNA datasets have been deposited in the European Nucleotide Archive (ENA) under study accession PRJEB15481.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, Lieber AD, Wu F, Perez-Perez GI, Chen Y, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Ohta M, Richardson PM, Mills DA. Monitoring seasonal changes in winery-resident microbiota. PLoS ONE. 2013;8:e66437. doi: 10.1371/journal.pone.0066437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements RS, Jr, Darnell B. Myo-inositol content of common foods: development of a high-myo-inositol diet. Am J Clin Nutr. 1980;33:1954–1967. doi: 10.1093/ajcn/33.9.1954. [DOI] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne M, Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr. 1997;67:345–366. [Google Scholar]
- Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- Fontana L, Meyer TE, Klein S, Holloszy JO. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci USA. 2004;101:6659–6663. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell. 2015;161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Faloney G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, Louk JA, Rose DJ, Kyureghian G, Pererson DA, et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013;7:269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Stegen JC, Maldonado-Gómez MX, Eren AM, Siba PM, Greenhill AR, Walter J. The gut microbiota of rural Papua New Guineans: composition, diversity patterns and ecological processes. Cell Rep. 2015;11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- Meadow JF, Altrichter AE, Bateman AC, Stenson J, Brown GZ, Green JL, Bohannan BJM. Humans differ in their personal microbial cloud. PeerJ. 2015;3:e1258. doi: 10.7717/peerj.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf H, Griffin NW, Ridaura VK, Reyes A, Cheng J, Rey FE, Smith MI, Simon GM, Scheffrahn RH, Woebken D, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Kights D, Clemente JC, Nakielny S, et al. Cohabiting family members share microbiota with one another and with their dogs. elife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci USA. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li S, Yang L, Huang P, Li W, Wang S, Zhao G, Zhang M, Pang X, Yan Z, et al. Structural modulation of the gut microbiota in life-long calorie-restricted mice. Nat Commun. 2013;4:2163. doi: 10.1038/ncomms3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Faloney G, Vieira-Silva S, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.