Abstract
Cellular senescence, an irreversible growth arrest triggered by a variety of stressors, plays important roles in normal physiology and tumor suppression, but accumulation of senescent cells with age contributes to the functional decline of tissues. Senescent cells undergo dramatic alterations to their chromatin landscape that affect genome accessibility and their transcriptional program. These include the loss of DNA-nuclear lamina interactions, the distension of centromeres, and changes in chromatin composition that can lead to the activation of retrotransposons. Here we discuss these findings, as well as recent advances in microscopy and genomics that have revealed the importance of the higher-order spatial organization of the genome in defining and maintaining the senescent state.
Keywords: cellular senescence, chromatin, chromosome structure, Hi-C, aging, DNA damage
Cellular senescence performs important roles in both normal physiology and disease
Cellular senescence is an irreversible growth arrest that can be induced by genotoxic stress (See Glossary), telomere shortening, and overexpression of oncogenes [1]. Several senescence effector pathways are shared across multiple types of senescence, but some are stimulus dependent [2]. Senescence plays a key role in many physiological processes such as tissue patterning during development [3, 4], wound healing [5, 6] and tissue repair [7]. Because senescence can restrict cell proliferation, it is thought to function as a tumor suppressor mechanism in response to the activation of oncogenes, a phenotype named oncogene-induced senescence (OIS) [8, 9]. Senescent cells also acquire a senescence-associated secretory phenotype (SASP) by secreting cytokines that can result in pro-inflammatory signaling and ultimately in their clearance by the immune system [10]. A notable exception is apparently senescent human naevi, caused by oncogene activation in melanocytes, which are not cleared and can persist in the skin for many years [11]. With age, senescent cells accumulate in tissues [12] due to a hypothesized decreased capacity for immune system clearance [2], and can cause chronic inflammation [1] and tissue dysfunction [13]. Further supporting their key role in aging is evidence that humans born with abnormally short telomeres due to inherited defects in telomere maintenance experience accelerated age-associated pathologies due to impaired tissue renewal capacity [14, 15]. Telomeres are key structural components protecting the ends of chromosomes. Successive cell divisions lead to progressively shorter telomeres and, ultimately, to exposed chromosome ends. Cells detect this as irreparable DNA damage and permanently exit the cell cycle, a phenomenon called replicative senescence (RS) that was first described in serial passaged human diploid fibroblasts in culture [16].
Senescent cells are now recognized as one of the key hallmarks of human aging [17]. It was recently shown that senescent cell clearance in mouse aging models improves healthspan and extends lifespan [18, 19]. Thus, there is much interest in targeting senescent cells for clearance via pharmacological interventions, referred to as senolytic drugs, to alleviate pathologies of aging and improve healthspan [20, 21]. A better understanding of the senescent phenotypes will improve our chances of developing such treatments.
In recent years, extensive evidence has emerged that senescent cells accumulate profound chromatin structural alterations. The idea that changes in chromatin are a key component of cellular senescence has a long history [22]. Although several of these initial ideas now have widespread support, whether these changes are causal or incidental is still an active area of research. Here we present a review of some recent advances in our understanding of the role of chromatin and chromosome organization in cellular senescence.
Senescent cells display extensive changes in chromatin structure and chromosome organization
Senescence-associated changes in chromatin structure and function
Senescent cells display profound chromatin changes, which can differ depending on how senescence is induced. While the role of some of these changes is not yet fully understood, some are driven by the stressors and others may be secondary changes. For example, in response to DNA damage cells activate the DNA damage response (DDR). Irreparable DNA damage triggers a persistent DDR activation and permanent cell cycle arrest [23]. This trigger of cellular senescence is associated with dynamic changes to chromatin including a transient chromatin decondensation at the site of DNA damage and subsequent compaction, which helps to promote ATM/ATR signaling [24]. Besides initiating the canonical signaling cascade of the DDR, ATM/ATR phosphorylates the histone H2A variant H2AX [25], which acts to hold double-strand break (DSB) ends together to facilitate repair and to recruit DNA repair proteins to the site of damage [26, 27].
Beyond these focal chromatin changes, microscopic methods have identified visible morphological alterations to the chromatin of senescent cells. One important alteration, which is most commonly found in OIS, is the emergence of senescence-associated heterochromatin foci (SAHF) [28, 29] (Figure 1). SAHF are dense, repressive chromatin foci, visible via DAPI staining, that contain the High-Mobility Group A (HMGA1 and HMGA2) proteins, the histone variant mH2A, and HP1 Protein [28, 30]. They are enriched for heterochromatic marks such as H3K9me3 [30, 31] and H4K20me3 [32], with the facultative heterochromatin mark H3K27me3 lining its periphery [33]. This phenotype has been extensively reviewed elsewhere, but does not appear to be a universal feature of all types of senescent cells [34].
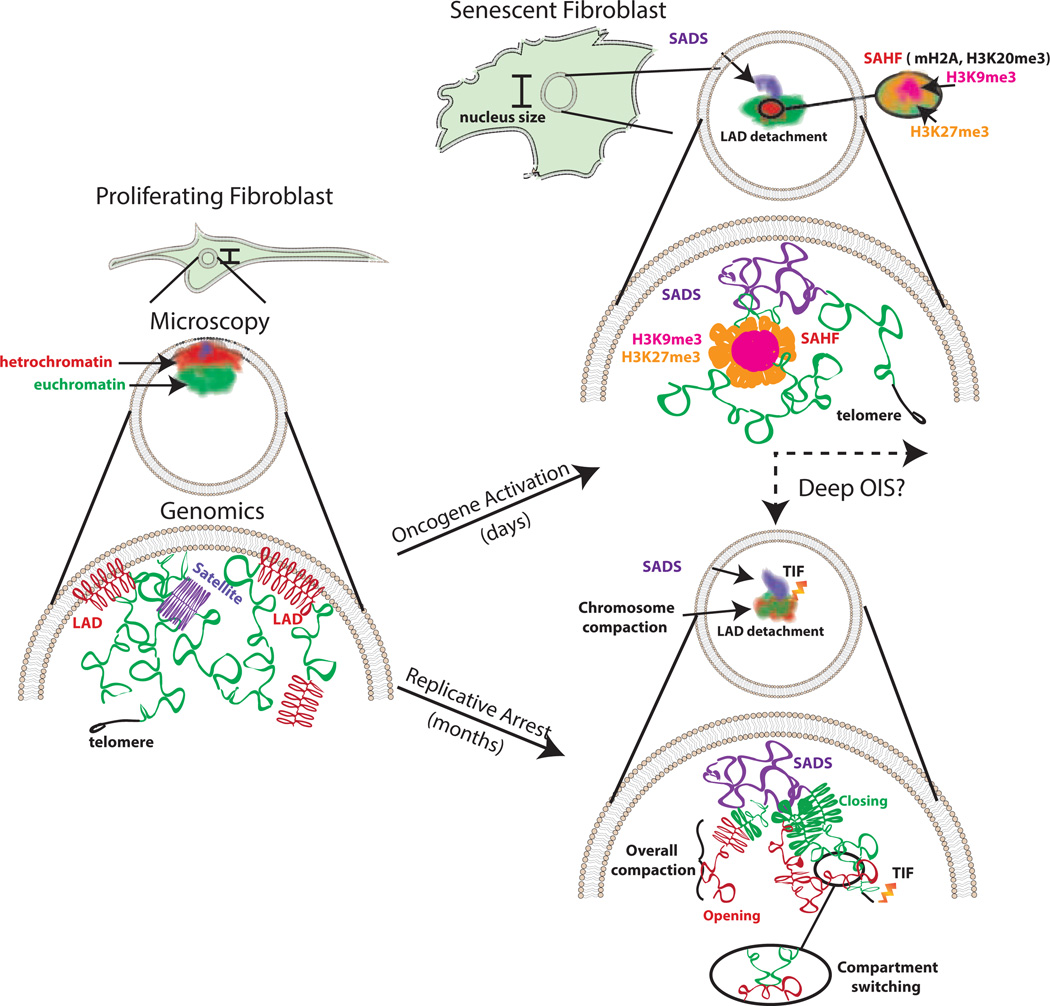
Figure 1. Key Figure: Changes in chromosome organization in OIS and RS.
Summary of the alterations to chromatin that occur in senescent cells observed by microscopy and genomics techniques. In proliferating cells, heterochromatin and the centromere tend to associate with the nuclear periphery and the euchromatic regions are found in the nuclear interior. The centromere is densely packed. Euchromatin is accessible for transcription. Peripheral heterochromatin is tethered to the nuclear lamina in regions called LADs. In cellular senescence, conserved and distinct features are observed depending on the stimuli. Shared features include SADS formation and the loss of interaction with the nuclear periphery or LADs detachment. SAHF appear to be predominantly induced in OIS and show foci with an H3K9me3 interior and an H3K27me3 halo. Other features, such as chromosome shrinkage, focal hypermethylation/decreased accessibility of euchromatic regions, hypomethylation/increased accessibility of constitutive heterochromatin and compartment switching, have only been investigated in RS but might be present in other types of senescence as well. Telomere Dysfunction-Induced Foci (TIF) are prominent in RS [98–100]; however, some evidence suggests this feature may be present in OIS [101].
Genomic approaches have also identified alterations to chromatin regulation including global changes to DNA methylation and chromatin accessibility. In RS, euchromatic gene rich regions display focal hypermethylation and lose accessibility, while regions of constitutive heterochromatin become more accessible and hypomethylated [35–37].
Chromatin alteration is also mediated by changes in the abundance of some of its constitutive components. Late-passage cells approaching senescence display reduced expression of the core H3 and H4 histone components of the nucleosome [38]. Another chromatin component, the linker histone H1, is also depleted in senescent cells [39]. In addition to histone depletion, a recent line of evidence points to a role for autophagy, a protein recycling mechanism, in the degradation of chromatin fragments in the cytoplasm in both RS and OIS [40], and of proteins associated with the nuclear lamina in OIS, including LMNB1 and LMNA [41, 42]. However, it is not completely clear whether H1 depletion is a consequence of the ejection of chromatin into the cytoplasm also known as cytoplasmic chromatin fragments), a decrease in histone synthesis, or other mechanisms [40, 43, 44].
Loss of DNA association with the nuclear envelope (NE)
The nuclear lamina is a protein structure that surrounds the interior of the nuclear membrane and is composed of A- and B-type lamins and associated proteins. The nuclear lamina interacts with gene-poor, heterochromatic regions of chromosomes named lamina-associated domains (LADs), ranging from 0.1 to 10 megabases in size [45]. Mutations in genes encoding lamins or associated proteins lead to pathologies with a large variety of clinical symptoms [46]. For example, most patients with the rare premature aging disorder Hutchinson-Gilford Progeria Syndrome (HGPS) carry a point mutation that leads to the activation of a cryptic splice site in the lamin A gene (LMNA) [47, 48]. This results in the production of a shortened form of lamin called progerin. HGPS cells have defective nuclear morphology and functions (reviewed in [49]). Similar to HGPS patients, cells from normal aging individuals also display nuclear lamina alterations and an increase in the amount of progerin in advanced age due to the sporadic use of the cryptic splice sites [50]. HGPS patient fibroblasts possess several attributes shared with cellular senescence, such as decreased proliferative capacity [51], disrupted nuclear morphology, loss of peripheral heterochromatin [52], DNA damage [53] and telomere shortening [54].
A more direct indication that the lamina plays a role in senescence and aging was provided by studies showing that lamin B1 (LMNB1) is downregulated in both senescent cells and aged cells in human skin; its silencing in normal proliferating cells can also induce a premature senescence phenotype [44, 55]. Loss of nuclear lamina components also causes the detachment of LADs and their relocalization to the nuclear interior in normal cells [56]. Furthermore, knockdown of LMNB1 in proliferating cells recapitulates many alterations to the chromatin landscape observed in RS cells, such as the formation of regions enriched for the activating H3K4me3 and repressive H3K27me3 marks (‘mesas’), and H3K27me3-depleted regions (‘canyons’) [57]. In OIS cells, LMNB1 depletion at LADs leads to H3K9me3-marked heterochromatin spatial (or 3D) redistribution, a step thought to be necessary for senescence-associated heterochromatin foci (SAHF) formation [58]. While LMNB1-DNA interactions decrease in many LADs in OIS cells, there are some regions that display increased LMNB1 binding, including gene-rich regions where formation of a new LAD might play a role in gene-silencing [58]. These same regions show a change in spatial organization as discussed below [59].
Changes in chromosome organization
Within the context of aging and senescence, Hi-C (Box 1) was first applied to study chromosome organization in fibroblasts from HGPS patients [60]. This study revealed that at late passages, compartmentalization separating active euchromatic A compartments and repressive B compartments in HGPS fibroblasts is significantly lost. A subset (12%) of the regions that could still be assigned to a specific compartment in later passaged HGPS cells changed their compartment association with respect to normal cells, a phenomenon termed compartment switching [60]. These regions also displayed significant correlation with alterations in the repressive histone mark H3K27me3 and in LADs. Compartment switching has been linked to gene expression changes, and it is likely to play a role in transcriptional regulation as evidenced by the fact that it features prominently during stem cell differentiation [61] and early B cell development [62].
Box 1. Genome-wide chromosome conformation capture.
High-resolution maps of 3D interactions along the chromosome are possible due to new developments in high-throughput conformation capture (or Hi-C) [95]. The Hi-C method uses formaldehyde to crosslink chromatin, which physically links two pieces of DNA that, while potentially linearly far apart, are actually in close proximity in 3D space. By capturing many of these long-range interactions along a chromosome, a 3D map of interactions can be constructed called a contact map [96]. Hi-C has revealed that, at super-megabase resolution, there are compartments within the interphase chromosome that correlate with markers of euchromatin (A compartment) and heterochromatin (B compartment) and vary across cell types [96]. At sub-megabase resolution there are features called topological associated domains (TADs), regions containing foci with strong cis interactions, that are invariant across cell types [97].
In OIS, Hi-C revealed a loss in shorter range interactions (on the order of 0.2–5 megabases) within chromosomes specifically at heterochromatic regions and a gain in long-range contacts with other repressive regions [59]. One study associated these changes with SAHF formation in OIS, which could be explained as a spatial redistribution of heterochromatic compartments to form closely interacting regions, rather than as an increase of heterochromatin at new regions [33]. Based on this and prior work in the field, a model was proposed for SAHF formation in which LAD regions with strong local interactions become dissociated from the NE and reassociate in the nuclear interior to form the core of the SAHF. By comparing the above mentioned data to embryonic stem cells (ESCs) and somatic cells, the authors concluded that local chromatin connectivity is progressively lost, with senescent cells representing a potential endpoint of chromosome remodeling during differentiation.
Changes in chromosome organization identified in RS cells by Hi-C were intermediate between the OIS and HGPS cells [63]. Genome-wide, RS cells displayed a global increase in short-range contacts. This feature appears similar to the increase in short range contacts in OIS cells for euchromatin domains, but it also extends into the heterochromatic regions in RS. In contrast to HGPS, which showed a dramatic loss of compartmentalization, the overall 3D organization of chromosomes was maintained in RS cells. By conducting a variety of complementary experiments including chromosome painting and 3D DNA FISH, it was concluded that the increased short-range contacts could be attributed to a decrease in the volume occupied by chromosome arms (Figure 1) [63]. This was in stark contrast to the behavior of centromeres, which showed significant decondensation as described below. In silico modeling of the chromosome 3D structure was also consistent with increased short-range contacts caused by a decreased chromosome volume. Finally, similar to HGPS, a small subset of regions underwent compartment switching, which was generally consistent with the direction of the change in expression of genes in these regions. For example, several cell-cycle genes were found to switch to a repressive compartment and were downregulated in RS.
Changes in chromatin structure directly affect the transcriptional program of senescent cells
Cellular senescence is a state of permanent growth arrest and, unsurprisingly, many of the genes that are downregulated during senescence are shared with the quiescent state and involve the cell-cycle pathway [64]. Beyond those, senescent cells display additional alterations to the transcriptional program. Apart from a small subset of well-characterized effector genes, which of these transcriptional changes are necessary to establish and maintain the senescent state, and which are simply a byproduct thereof, is still an open question. Some examples, such as alterations to the expression of genes in compartment switching domains, were mentioned in the previous section. We discuss here some additional examples in which transcriptional regulation is achieved via chromatin changes.
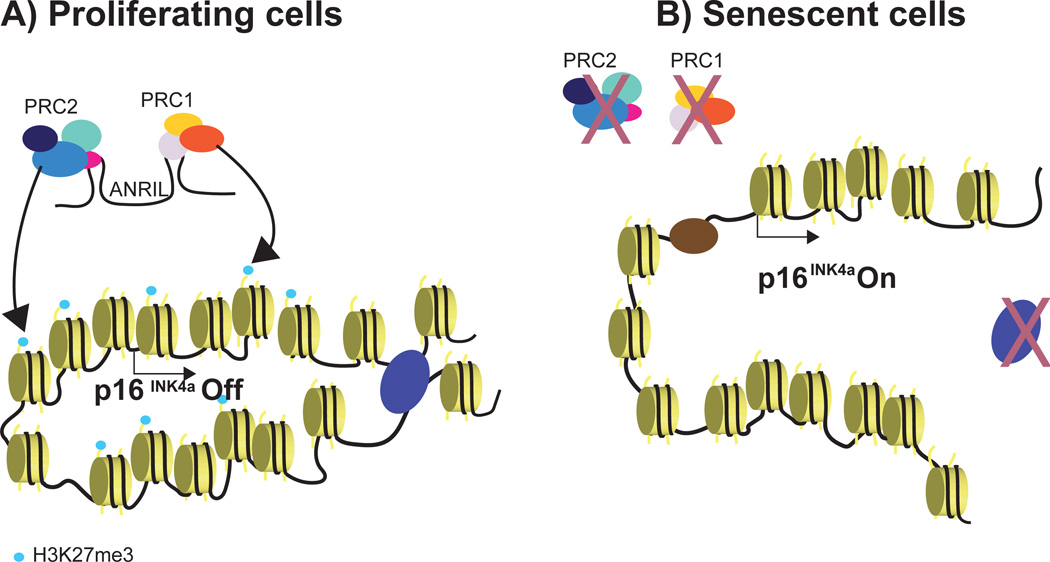
Regulation of the senescence effector p16 (CDKN2A gene at the INK4A locus) occurs at the level of chromatin and has proven to be quite complex. In proliferative cells, the INK4A locus is kept in a polycomb repressed state by polycomb repressive complexes (PRC1 and PRC2) first identified by PRC-group protein BMI1 binding to the INK4A locus [65, 66] (Figure 2a). The EZH2 histone methyltransferase component of the PRC2 complex is responsible for deposition of the repressive H3K27me3 histone mark in normal cells [67]. During cellular senescence, a transcriptional downregulation of EZH2 causes a loss of H3K27me3 and the activation of p16 [67]. One mechanism that could cause the PRC-group proteins to redistribute in senescence is that they have recently been characterized to localize to sites of DNA damage [68]. In addition to PRC-group proteins, a long-noncoding RNA called ANRIL has an important role in mediating PRC recruitment and repression of p16 [69]. In many types of cancer, the INK4A locus is aberrantly silenced by DNA methylation, and this aberrant repression is associated with the loss of the CCCTC-binding factor (CTCF) binding to a nearby boundary element [70]. CTCF, originally described as a boundary element separating euchromatin and heterochromatin, is now known to mediate chromatin looping together with cohesin proteins [70] through a process called chromatin extrusion [71]. The role of CTCF at the INK4A locus would require precise regulation to avoid aberrantly silencing p16 and causing cancer. There are three sites of CTCF enrichment found near the INK4A locus [72]. In normal proliferative cells, CTCF expression is high and forms compact loops at INK4A [72]. In OIS cells, CTCF expression decreases and the CTCF-mediated looping appears less compact, potentially allowing for transcription of p16 [72] (Figure 2b). The transcriptional derepression of p16 could also be aided by the pioneer transcription factor FOXA1, traditionally functioning to open repressive chromatin. FOXA1 binds the INK4A locus, is also a target of PRC repression itself, and is thought to function as an activator of p16 by decreasing nucleosome density [73]. CTCF binding sites are widespread in the genome, and it remains to be seen if a loss of CTCF-mediated looping might be responsible for other transcriptional changes in senescence.
Figure 2. Chromatin-based control of the INK4A locus.
A) In proliferating cells, polycomb repressive complexes (PRC1 and PRC2) are recruited by a long non-coding RNA, ANRIL, to the INK4A locus. This leads to the trimethylation of H3K27 by PRC2 and the repression of p16. CTCF forms chromatin looping that further maintains the gene repression state. B) In cellular senescence, PRC delocalized from the INK4A locus, leading to a loss of the H3K27me3 mark. CTCF expression also decreases, allowing the decompaction of chromatin. FOXA1, a transcription factor, binds to the INK4A locus and is proposed to decrease nucleosome density, leading to p16 expression.
Another way chromatin regulation can alter gene expression in senescence is through histone modification. For instance, H3K4me3 at cell-cycle genes is lost in senescence and this leads to the repression of cell-cycle genes [74]. Specifically, the loss of H3K4me3 is mediated in two ways: through the cleavage of the histone H3 tail by a protease [40], Cathepsin L, and through the demethylation of H3K4 by JARID1A and JARID1B H3K4 demethylases [74]. In OIS, the loss of H3K9me3 and the gain of H3K4me3 are found to activate Bone morphogenic protein 2 (BMP2) [75, 76] and subsequently the BMP-SMAD pathway. This pathway is involved in several biological processes, such as differentiation and development [77, 78]. Importantly, the BMP-SMAD pathway has been implicated in several types of cancers, including lung cancer, in which BMP2 is reported to promote tumor growth by inducing angiogenesis [79, 80].
Higher order chromatin organization at the telomeres has also been reported to alter gene expression in cells undergoing telomere shortening, a phenomenon known as telomere position effect over long distances (TPE-OLD) [81]. Specifically, long telomeres form loops and interact with genes at a distance of up to 10 Mb, such as Desmoplakin (DSP) and phenylalanyl-tRNA synthetase 2 (FARS2), as revealed by Hi-C on human chromosome 6p [81]. As telomeres shorten during subsequent rounds of cell division, loss of gene-telomere interactions leads to alterations in the expression of genes near telomeres. For instance, the DSP gene has been shown to have increased gene expression in cells that have shorter telomeres. Some other genes that have been identified to have increased expression when telomeres become shorter are Cyclin D2 (CCND2) and complement component 1s subcomplement (C1S) [81].
Chromosome centromeres are an additional class of heterochromatic region displaying dramatic structural alterations in senescence. These regions harbor a class of repeats named satellites, which are normally constitutively repressed, but in RS cells pericentric satellite HSATII distends and becomes accessible [36] (Figure 1). The distension of centromeres, or senescence-associated distension of satellites (SADS), was then extensively observed and confirmed via 3D DNA fluorescent in situ hybridization (FISH) experiments on pericentric satellite II and centromeric alpha satellite in both RS and OIS [63, 82]. The satellite sequences are also hypomethylated, consistent with distension and derepression [35]. It has been proposed that SADS formation is one of the early modifications to the chromatin landscape in senescence [63, 82], but whether it is required to induce or maintain the senescent state is still unclear. Recently, it was shown that SIRT6, a histone deacetylase, removes H3K18 acetylation in normal proliferative cells to maintain silencing of pericentric satellites, and that SIRT6 depletion leads to senescence and accumulation of pericentric satellite transcripts [83, 84]. Because SIRT6 is one of the first factors recruited at the sites of DSB (within 5s) [85], it is possible that in the presence of extensive irreparable DNA damage SIRT6 sequestration away from pericentric satellite DNA rapidly initiates the centromere unraveling.
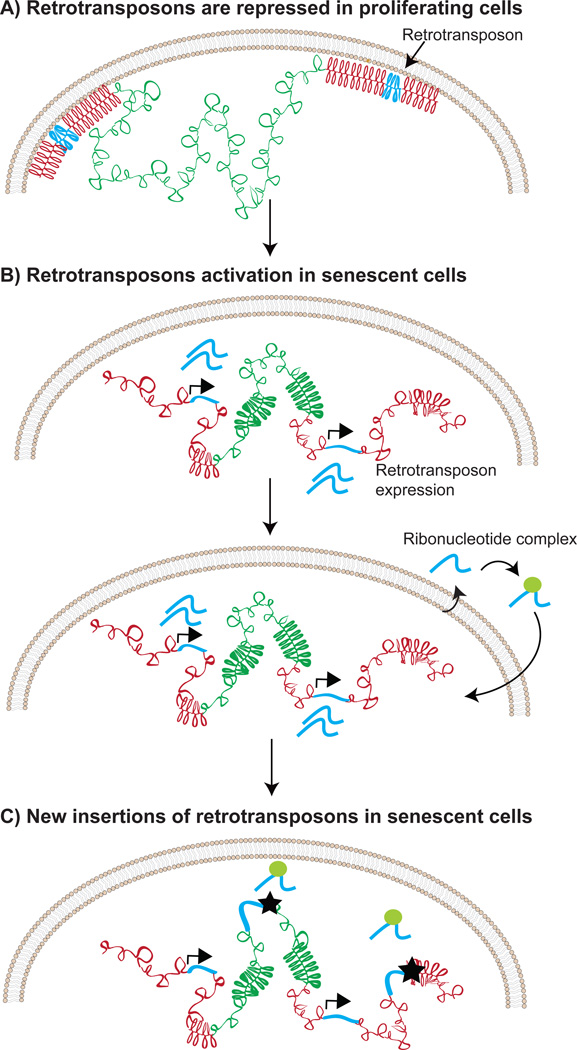
Similar to the distension of satellites, retrotransposons, mobile genetic elements normally repressed in heterochromatin, have also been documented to increase chromatin accessibility in senescent cells [36]. The derepression of retrotransposons causes increased RNA expression and mobilization of retrotransposons in senescent cells [36]. A similar activation of retrotransposons is observed in cancer and in aged cells using mouse models [86, 87]. There is evidence linking SIRT6 depletion to retrotransposon activation in aged cells, which closely mirrors the implicated role for SIRT6 in repressing satellites [88]. Thus, loss of peripheral heterochromatin has profound implications for the cell, clearly represented by the activation of transposons, which are capable of propagating genome instability by insertional mutagenesis (Figure 3). The depletion of SIRT6 at retrotransposons is triggered by genotoxic stress, however, the molecular mechanism for depletion is still unclear [88]. Furthermore, it is not clear whether SIRT6 depletion at satellites precedes transposons or whether these two processes are linked.
Figure 3. Retrotransposon mobilization in senescent cells.
A) The transcription of retrotransposons is repressed in proliferating cells. B) Changes in chromatin structure, such as the decondensation of heterochromatin and the loss of interaction between DNA and lamin allow the transcriptional activation of retrotransposons. The transcribed retrotransposon RNAs are reverse-transcribed into DNA in a nucleoprotein complex. Note that only a small subset of retrotransposons is competent for autonomous retrotransposition in humans (~100 copies of L1Hs elements) [102]. C) Retrotransposon DNA attempts to integrate into the genome (shown by the stars).
Concluding remarks
Ample evidence exists for profound changes in the chromatin landscape characterizing senescent cells. Some of these changes appear to be shared across different types of senescence. These include early SADS formation and the loss of interaction with the nuclear periphery. Others depend on how senescence is induced, such as SAHF formation in OIS. Other features, such as activation of retrotransposons, chromosome shrinkage, focal hypermethylation of euchromatic regions, hypomethylation of constitutive heterochromatin and compartment switching, have only been investigated in RS but might be present in other types of senescence as well. Indeed, SAHF, which have been shown to consist of a single chromosome [29, 33, 39], might represent an extreme case of chromosome shrinkage primarily driven by the condensation of heterochromatic LADs in the nuclear interior after they dissociate from the lamina. It is tempting to speculate that the observed differences between them could be primarily attributed to the large difference in the time scale over which RS and OIS occur (months vs. weeks). Time course experiments that combine complementary experimental techniques to measure 3D chromosome organization, DNA accessibility, and transcriptional changes would allow us to perform a better comparison between them (See Outstanding Questions). In particular, increased resolution provided by the latest advances in Hi-C technology and microscopy, such as capture Hi-C [89] and superresolution microscopy [90], will allow us to better understand the relation between changes in chromosome folding and transcriptional activity of individual genes. For example, it was recently reported that in OIS, the enhancer landscape undergoes global remodeling, with recruitment of BRD4 to newly activated super-enhancers proximal to SASP genes [91]. In this case, promoter capture Hi-C [89] could be applied to elucidate the changes in chromosome looping leading to the activation of these enhancers and of their target genes.
Box 3. Outstanding Questions.
To what extent are the changes to chromatin observed in cell culture conserved in senescent cells in vivo?
The time-scale for studying OIS in cell culture is on the order of days to weeks. Chromatin is dynamic and can change over time. Are the observed changes to chromatin in OIS their endpoint or do they continue to change over time?
Of the diverse chromatin alterations in cellular senescence, which ones are necessary and sufficient for driving or maintaining the senescence program?
Do proteins involved in defining the 3D organization of the genome such as cohesins, condensins, and CTCF drive the chromatin alterations in cellular senescence?
What is the temporal ordering of chromatin changes in senescent cells? Are some changes necessary for others to occur?
Can recently developed techniques such as high resolution capture Hi-C, enhancer promoter capture Hi-C, and super resolution microscopy reveal additional details about chromatin alterations in senescent cells?
Is there significant variability in chromatin organization at the single-cell level in senescent cells?
Does the chromatin structure of a senescent cell promote tumor formation upon senescence bypass?
Although we are still far from an integrated functional model accounting for all the changes discussed above, some of its components are starting to emerge. In particular, multiple lines of evidence point to the presence of two concurrent and interdependent processes: 1) a stochastic process of random DNA damage, and 2) a regulated transcriptional program leading to cell cycle exit and proinflammatory signaling. DNA damage-induced redistribution of chromatin-modifying factors, such as SIRT6, is likely to mediate the loss of heterochromatin responsible for SADS and activation of retrotransposons. A similar chromatin redistribution model has also been proposed in organismal aging [88, 92, 93]. Increased retrotransposition activity can lead to abortive retrotransposition events that promote new DNA damage and further redistribution of chromatin-modifying factors. At the same time, some of the observed structural changes appear to be part of the regulated transcriptional program of senescent cells, such as the INK4A locus, cell cycle genes in compartment switching, and SASP gene activation via remodeling of the super-enhancers landscape.
Structural changes in chromatin organization, such as SADS and detachment of LADs might be responsible, at least in part, for the irreversibility of the senescent state. However, it was recently reported that some OIS cells acquire chromatin changes that promote the expression of the human telomerase reverse transcriptase gene (hTERT) and, as a consequence, escape senescence [94]. This can have important consequences for tumor formation, as senescent cells share features of the cancer epigenome [35]. These results also point to the idea that not all senescent cells are made equal, and different degrees of senescence might exist in the presence of the same stimulus. Recent advances in single cell technology (e.g. single-cell Hi-C, ATAC-seq, DNA-seq and RNA-seq) have created an opportunity to tackle this problem. In addition, single cell technology could be applied to the study of senescent cells in vivo, which has been hampered by the difficulty of isolating a sufficient number of senescent cells from tissues to perform assays that routinely require millions of cells.
Box 1. Trends.
Novel methods to investigate genome organization (Hi-C) have revealed that senescent chromosomes undergo a global spatial reorganization that can alter transcription and can explain some of the observed chromatin changes (e.g. SAHFs).
Loss of interaction between the lamina and constitutive heterochromatin induces large-scale changes in chromosome organization in cellular senescence.
Centromere decondensation is an early event shared by many types of senescence and potentially a new marker for the identification of senescent cells.
SIRT6 is an early responder to DNA damage and is involved in satellite distension and derepression of retrotransposons.
Change in accessibility of heterochromatic regions leads to the mobilization of retrotransposons, which might contribute to the propagation of genomic instability.
Acknowledgments
We would like to thank John Sedivy and Andrew Leith for their help in the preparation of this manuscript. This work was supported in part by the following NIH grants: R56 AG050582-01 and P20 GM109035-01A1 to N.N., and F31AG050365 to S.W.C..
Glossary
- Euchromatin
The accessible portion of chromatin that is less repetitive, gene-dense, and active for transcription.
- Heterochromatin
The inaccessible chromatin that is repetitive, gene-poor, and repressive to transcription.
- Histone Marks
Histones possess long peptide tails that protrude from the nucleosomes and are modified post-translationally to change the properties of chromatin.
- Hi-C
High-throughput whole genome conformation capture sequencing is a technique that when combined with paired-end sequencing can identify long-range spatial interactions within and between chromosomes.
- Retrotransposon
A mobile genetic element that can mobilize and insert into new locations in the genome (a process known as retrotransposition) using a copy and paste mechanism via an RNA intermediate.
- Satellite DNA
Prevalent repetitive DNA in eukaryotic genomes that comprises tandemly-repeated sequences.
- Centromere
The region of the chromosome that connects the sister chromatids during mitosis, composed of satellite DNA and typically tightly compacted.
- Telomere
The protective cap at the end of linear chromosomes composed of satellite repeat DNA that can be extended by the enzyme telomerase.
- Fluorescent In Situ Hybridization (FISH)
A technique that uses sequence homology to precisely label regions within the chromosome with complementary DNA labelled with a fluorescent molecule.
- Boundary element
Regions that serve as spatial separators between heterochromatin and euchromatin compartments along the chromosome.
- TADs
Topological associated domains are sub-megabase regions identified by Hi-C that show strong cis interaction preference and are conserved between cell-types.
- SASP
The senescence-associated secretory phenotype is a secretory program initiated by senescent cells, characterized by the secretion of proinflammatory cytokines, chemokines, proteases and other soluble factors.
- SAHF
Senescence-associated heterochromatic foci are regions of dense compact DNA (by DAPI staining) that form during oncogene induced senescence.
- SADS
Senescence-associated distension of satellites is the distension of satellite DNA sequences including the centromeres observed in oncogene induced senescence and replicative senescence.
- LADs
Lamina-associated domains are regions of the chromatin that interact physically with the nuclear lamina that are typically heterochromatic.
- Quiescence
A reversible arrest to the cell-cycle that is sometimes referred to as G zero phase (triggered by serum starvation in cell culture).
- Replicative senescence
An irreversible arrest to the cell-cycle that is caused by telomere attrition due to the accumulation of cell-divisions.
- Oncogene-induced senescence
An arrest to the cell-cycle caused by the sudden activation of an oncogene.
- Genotoxic stress
A toxic insult that damages the DNA of the cell and, depending on the severity, may also trigger cell arrest (e.g. ionizing radiation).
- Oncogene
A gene that, when mutated or expressed at aberrant levels, can lead to the development of cancer.
- Autophagy
The breakdown of cellular components, which serves multiple purposes including the removal of damaged cellular machinery and the recycling of cellular building blocks.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509(7501):439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muñoz-Espín D, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155(5):1104–1118. doi: 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Storer M, et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell. 2013;155(5):1119–1130. doi: 10.1016/j.cell.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 5.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell. 2014;31(6):722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jun J-I, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat. Cell Biol. 2010;12(7):676–685. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134(4):657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salama R, et al. Cellular senescence and its effector programs. Genes Dev. 2014;28(2):99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courtois-Cox S, et al. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27(20):2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 10.Coppe JP, et al. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 12.Waaijer MEC, et al. The number of p16INK4a positive cells in human skin reflects biological age. Aging Cell. 2012;11(4):722–725. doi: 10.1111/j.1474-9726.2012.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coppé J-P, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alder JK, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc. Natl. Acad. Sci. U. S. A. 2008;105(35):13051–13056. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armanios MY, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007;356(13):1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 16.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 17.López-Otín C, et al. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22(1):78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, et al. The Achilles' heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015;14(4):644–658. doi: 10.1111/acel.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macieira-Coelho A, Puvion-Dutilleul F. Genome reorganization during aging of dividing cells. Adv. Exp. Med. Biol. 1985;190:391–419. doi: 10.1007/978-1-4684-7853-2_20. [DOI] [PubMed] [Google Scholar]
- 23.Fumagalli M, et al. Stable cellular senescence is associated with persistent DDR activation. PLoS One. 2014;9(10):e110969. doi: 10.1371/journal.pone.0110969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess RC, et al. Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 2014;9(5):1703–1717. doi: 10.1016/j.celrep.2014.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogakou EP, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 26.Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3(2):149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- 27.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123(7):1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 28.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113(6):703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang R, et al. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 2007;27(6):2343–2358. doi: 10.1128/MCB.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang R, et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell. 2005;8(1):19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Narita M, et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126(3):503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 32.Nelson DM, et al. Mapping H4K20me3 onto the chromatin landscape of senescent cells indicates a function in control of cell senescence and tumor suppression through preservation of genetic and epigenetic stability. Genome Biol. 2016;17(1):1–20. doi: 10.1186/s13059-016-1017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandra T, et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol. Cell. 2012;47(2):203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parry AJ, Narita M. Old cells, new tricks: chromatin structure in senescence. Mamm. Genome. 2016;27(7–8):320–331. doi: 10.1007/s00335-016-9628-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cruickshanks HA, et al. Senescent cells harbour features of the cancer epigenome. Nat. Cell Biol. 2013;15(12):1495–1506. doi: 10.1038/ncb2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Cecco M, et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013;12(2):247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hänzelmann S, et al. Replicative senescence is associated with nuclear reorganization and with DNA methylation at specific transcription factor binding sites. Clin. Epigenetics. 2015;7(1):19. doi: 10.1186/s13148-015-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Sullivan RJ, et al. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010;17(10):1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Funayama R, et al. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 2006;175(6):869–880. doi: 10.1083/jcb.200604005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov A, et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 2013;202(1):129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dou Z, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527(7576):105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenain C, et al. Autophagy-mediated degradation of nuclear envelope proteins during oncogene-induced senescence. Carcinogenesis. 2015;36(11):1263–1274. doi: 10.1093/carcin/bgv124. [DOI] [PubMed] [Google Scholar]
- 43.Freund A, et al. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012;23(11):2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimi T, et al. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011;25(24):2579–2593. doi: 10.1101/gad.179515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 46.Worman HJ. Nuclear lamins and laminopathies. J. Pathol. 2012;226(2):316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Sandre-Giovannoli A, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300(5628):2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 48.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423(6937):293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arancio W, et al. Epigenetic involvement in Hutchinson-Gilford progeria syndrome: a mini-review. Gerontology. 2014;60(3):197–203. doi: 10.1159/000357206. [DOI] [PubMed] [Google Scholar]
- 50.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312(5776):1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benson EK, et al. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J. Cell Sci. 2010;123(Pt 15):2605–2612. doi: 10.1242/jcs.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. U. S. A. 2006;103(23):8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu B, et al. Genomic instability in laminopathy-based premature aging. Nat. Med. 2005;11(7):780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 54.Decker ML, et al. Telomere length in Hutchinson-Gilford progeria syndrome. Mech. Ageing Dev. 2009;130(6):377–383. doi: 10.1016/j.mad.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 55.Dreesen O, et al. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J. Cell Biol. 2013;200(5):605–617. doi: 10.1083/jcb.201206121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solovei I, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152(3):584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 57.Shah PP, et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013;27(16):1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadaie M, et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev. 2013;27(16):1800–1808. doi: 10.1101/gad.217281.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chandra T, et al. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015;10(4):471–483. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCord RP, et al. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23(2):260–269. doi: 10.1101/gr.138032.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol. 2012;13(12):1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Criscione SW, et al. Reorganization of chromosome architecture in replicative cellular senescence. Sci Adv. 2016;2(2):e1500882. doi: 10.1126/sciadv.1500882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lackner DH, et al. A genomics approach identifies senescence-specific gene expression regulation. Aging Cell. 2014;13(5):946–950. doi: 10.1111/acel.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacobs JJ, et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 66.Maertens GN, et al. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One. 2009;4(7):e6380. doi: 10.1371/journal.pone.0006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21(5):525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ginjala V, et al. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol. Cell. Biol. 2011;31(10):1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol. Cell. 2009;34(3):271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U. S. A. 2015;112(47):E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirosue A, et al. Quantitative assessment of higher-order chromatin structure of the INK4/ARF locus in human senescent cells. Aging Cell. 2012;11(3):553–556. doi: 10.1111/j.1474-9726.2012.00809.x. [DOI] [PubMed] [Google Scholar]
- 73.Li Q, et al. FOXA1 mediates p16(INK4a) activation during cellular senescence. EMBO. J. 2013;32(6):858–873. doi: 10.1038/emboj.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duarte LF, et al. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat. Commun. 2014;5:5210. doi: 10.1038/ncomms6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujimoto M, et al. Epigenetic alteration to activate Bmp2-Smad signaling in Raf-induced senescence. World J. Biol. Chem. 2016;7(1):188–205. doi: 10.4331/wjbc.v7.i1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaneda A, et al. Activation of Bmp2-Smad1 signal and its regulation by coordinated alteration of H3K27 trimethylation in Ras-induced senescence. PLoS Genet. 2011;7(11):e1002359. doi: 10.1371/journal.pgen.1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abe E, et al. Essential requirement of BMPs-2/4 for both osteoblast and osteoclast formation in murine bone marrow cultures from adult mice: antagonism by noggin. J. Bone Miner. Res. 2000;15(4):663–673. doi: 10.1359/jbmr.2000.15.4.663. [DOI] [PubMed] [Google Scholar]
- 78.Wang RN, et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014;1(1):87–105. doi: 10.1016/j.gendis.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langenfeld EM, et al. Bone morphogenetic protein 2 stimulation of tumor growth involves the activation of Smad-1/5. Oncogene. 2006;25(5):685–692. doi: 10.1038/sj.onc.1209110. [DOI] [PubMed] [Google Scholar]
- 80.Langenfeld EM, Langenfeld J. Bone morphogenetic protein-2 stimulates angiogenesis in developing tumors. Mol. Cancer Res. 2004;2(3):141–149. [PubMed] [Google Scholar]
- 81.Robin JD, et al. Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 2014;28(22):2464–2476. doi: 10.1101/gad.251041.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Swanson EC, et al. Higher-order unfolding of satellite heterochromatin is a consistent and early event in cell senescence. J. Cell Biol. 2013;203(6):929–942. doi: 10.1083/jcb.201306073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nagai K, et al. Depletion of SIRT6 causes cellular senescence, DNA damage, and telomere dysfunction in human chondrocytes. Osteoarthritis Cartilage. 2015;23(8):1412–1420. doi: 10.1016/j.joca.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 84.Tasselli L, et al. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016;23(5):434–440. doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toiber D, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell. 2013;51(4):454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Criscione SW, et al. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 2014;15:583. doi: 10.1186/1471-2164-15-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Cecco M, et al. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging. 2013;5(12):867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van Meter M, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015;47(6):598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 90.Wang S, et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016 doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tasdemir N, et al. BRD4 Connects Enhancer Remodeling to Senescence Immune Surveillance. Cancer Discov. 2016;6(6):612–629. doi: 10.1158/2159-8290.CD-16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Imai S, Kitano H. Heterochromatin islands and their dynamic reorganization: a hypothesis for three distinctive features of cellular aging. Exp Gerontol. 1998;33(6):555–570. doi: 10.1016/s0531-5565(98)00037-0. [DOI] [PubMed] [Google Scholar]
- 93.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patel PL, et al. Derepression of hTERT gene expression promotes escape from oncogene-induced cellular senescence. Proc Natl Acad Sci U S A. 2016;113(34):E5024–E5033. doi: 10.1073/pnas.1602379113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 2012;9(10):999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.d'Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426(6963):194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 99.Sedelnikova OA, et al. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol. 2004;6(2):168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 100.Takai H, et al. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13(17):1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 101.Suram A, et al. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 2012;31(13):2839–2851. doi: 10.1038/emboj.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100(9):5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]