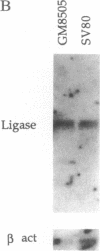
Abstract
Alteration of DNA ligase I activity is a consistent biochemical feature of Bloom's syndrome (BS) cells. DNA ligase I activity in BS cells either is reduced and abnormally thermolabile or is present in an anomalously dimeric form. To assess the role of DNA ligase function in the etiology of BS, we have cloned the DNA ligase I cDNA from normal human cells by a PCR strategy using degenerate oligonucleotide primers based on conserved regions of the Saccharomyces cerevisiae and Schizosaccharomyces pombe DNA ligase genes. Human DNA ligase I cDNAs from normal and BS cells complemented a S. cerevisiae DNA ligase mutation, and protein extracts prepared from S. cerevisiae transformants expressing normal and BS cDNA contained comparable levels of DNA ligase I activity. DNA sequencing and Northern blot analysis of DNA ligase I expression in two BS human fibroblast lines representing each of the two aberrant DNA ligase I molecular phenotypes demonstrated that this gene was unchanged in BS cells. Thus, another factor may be responsible for the observed reduction in DNA ligase I activity associated with this chromosomal breakage syndrome.
Full text
PDF


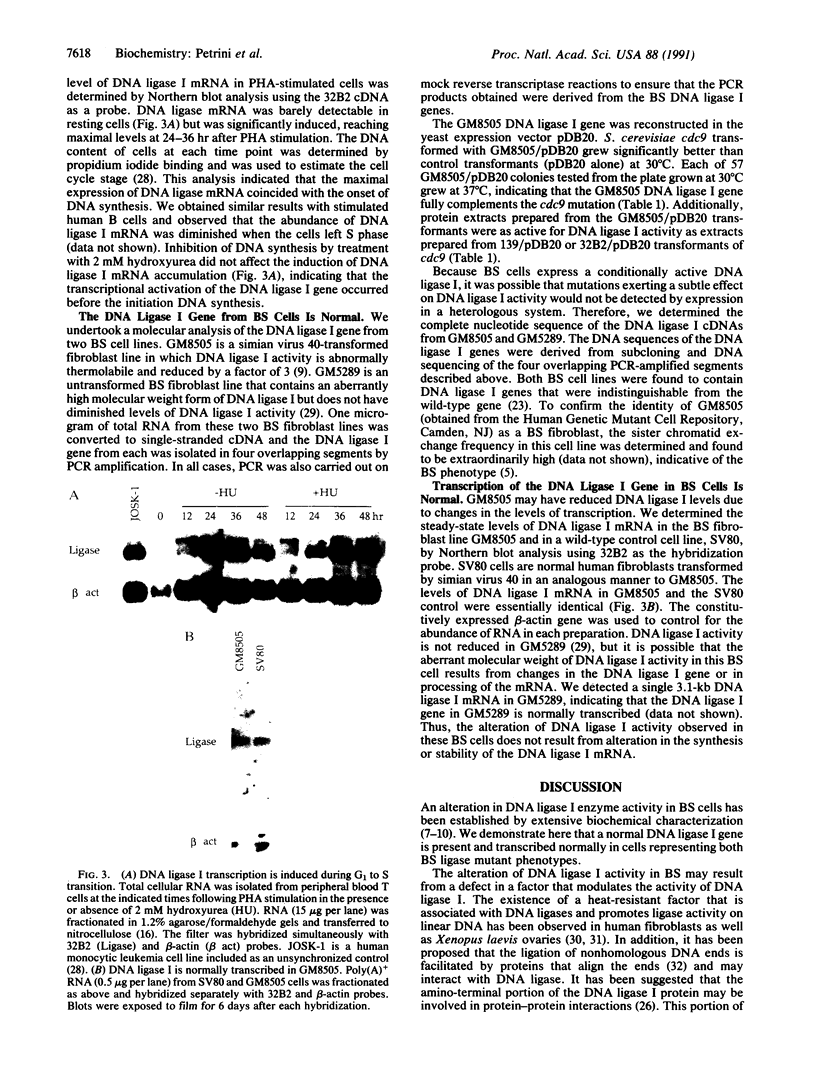

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arrand J. E., Willis A. E., Goldsmith I., Lindahl T. Different substrate specificities of the two DNA ligases of mammalian cells. J Biol Chem. 1986 Jul 15;261(20):9079–9082. [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. Molecular characterisation of the DNA ligase gene, CDC17, from the fission yeast Schizosaccharomyces pombe. Eur J Biochem. 1987 Feb 2;162(3):659–667. doi: 10.1111/j.1432-1033.1987.tb10688.x. [DOI] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. The nucleotide sequence of the DNA ligase gene (CDC9) from Saccharomyces cerevisiae: a gene which is cell-cycle regulated and induced in response to DNA damage. Nucleic Acids Res. 1985 Dec 9;13(23):8323–8337. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D. E., Johnston L. H., Kodama K., Tomkinson A. E., Lasko D. D., Lindahl T. Human DNA ligase I cDNA: cloning and functional expression in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6679–6683. doi: 10.1073/pnas.87.17.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne M. L., Alexander R. F., Benbow R. M. DNA binding protein from ovaries of the frog, Xenopus laevis which promotes concatenation of linear DNA. J Mol Biol. 1984 Jan 5;172(1):87–108. doi: 10.1016/0022-2836(84)90416-9. [DOI] [PubMed] [Google Scholar]
- Becker D. M., Fikes J. D., Guarente L. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1968–1972. doi: 10.1073/pnas.88.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti R. S., Schonberg S., German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. Y., Becker F. F., German J., Ray J. H. Altered DNA ligase I activity in Bloom's syndrome cells. Nature. 1987 Jan 22;325(6102):357–359. doi: 10.1038/325357a0. [DOI] [PubMed] [Google Scholar]
- Dehazya P., Sirover M. A. Regulation of hypoxanthine DNA glycosylase in normal human and Bloom's syndrome fibroblasts. Cancer Res. 1986 Aug;46(8):3756–3761. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Furukawa Y., Piwnica-Worms H., Ernst T. J., Kanakura Y., Griffin J. D. cdc2 gene expression at the G1 to S transition in human T lymphocytes. Science. 1990 Nov 9;250(4982):805–808. doi: 10.1126/science.2237430. [DOI] [PubMed] [Google Scholar]
- GERMAN J., ARCHIBALD R., BLOOM D. CHROMOSOMAL BREAKAGE IN A RARE AND PROBABLY GENETICALLY DETERMINED SYNDROME OF MAN. Science. 1965 Apr 23;148(3669):506–507. doi: 10.1126/science.148.3669.506. [DOI] [PubMed] [Google Scholar]
- German J. Bloom's syndrome. I. Genetical and clinical observations in the first twenty-seven patients. Am J Hum Genet. 1969 Mar;21(2):196–227. [PMC free article] [PubMed] [Google Scholar]
- German J., Schonberg S., Louie E., Chaganti R. S. Bloom's syndrome. IV. Sister-chromatid exchanges in lymphocytes. Am J Hum Genet. 1977 May;29(3):248–255. [PMC free article] [PubMed] [Google Scholar]
- Goulian M., Richards S. H., Heard C. J., Bigsby B. M. Discontinuous DNA synthesis by purified mammalian proteins. J Biol Chem. 1990 Oct 25;265(30):18461–18471. [PubMed] [Google Scholar]
- Gupta P. K., Sirover M. A. Altered temporal expression of DNA repair in hypermutable Bloom's syndrome cells. Proc Natl Acad Sci U S A. 1984 Feb;81(3):757–761. doi: 10.1073/pnas.81.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hütteroth T. H., Litwin S. D., German J. Abnormal immune responses of Bloom's syndrome lymphocytes in vitro. J Clin Invest. 1975 Jul;56(1):1–7. doi: 10.1172/JCI108058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Nasmyth K. A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978 Aug 31;274(5674):891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- Kenne K., Ljungquist S. Expression of a DNA-ligase-stimulatory factor in Bloom's syndrome cell line GM1492. Eur J Biochem. 1988 Jun 15;174(3):465–470. doi: 10.1111/j.1432-1033.1988.tb14121.x. [DOI] [PubMed] [Google Scholar]
- Langlois R. G., Bigbee W. L., Jensen R. H., German J. Evidence for increased in vivo mutation and somatic recombination in Bloom's syndrome. Proc Natl Acad Sci U S A. 1989 Jan;86(2):670–674. doi: 10.1073/pnas.86.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko D. D., Tomkinson A. E., Lindahl T. Eukaryotic DNA ligases. Mutat Res. 1990 Sep-Nov;236(2-3):277–287. doi: 10.1016/0921-8777(90)90011-s. [DOI] [PubMed] [Google Scholar]
- Lehmann A. R., Willis A. E., Broughton B. C., James M. R., Steingrimsdottir H., Harcourt S. A., Arlett C. F., Lindahl T. Relation between the human fibroblast strain 46BR and cell lines representative of Bloom's syndrome. Cancer Res. 1988 Nov 15;48(22):6343–6347. [PubMed] [Google Scholar]
- Nasmyth K. A. Temperature-sensitive lethal mutants in the structural gene for DNA ligase in the yeast Schizosaccharomyces pombe. Cell. 1977 Dec;12(4):1109–1120. doi: 10.1016/0092-8674(77)90173-8. [DOI] [PubMed] [Google Scholar]
- Rünger T. M., Kraemer K. H. Joining of linear plasmid DNA is reduced and error-prone in Bloom's syndrome cells. EMBO J. 1989 May;8(5):1419–1425. doi: 10.1002/j.1460-2075.1989.tb03523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Seal G., Brech K., Karp S. J., Cool B. L., Sirover M. A. Immunological lesions in human uracil DNA glycosylase: association with Bloom syndrome. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2339–2343. doi: 10.1073/pnas.85.7.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thode S., Schäfer A., Pfeiffer P., Vielmetter W. A novel pathway of DNA end-to-end joining. Cell. 1990 Mar 23;60(6):921–928. doi: 10.1016/0092-8674(90)90340-k. [DOI] [PubMed] [Google Scholar]
- Tomkinson A. E., Totty N. F., Ginsburg M., Lindahl T. Location of the active site for enzyme-adenylate formation in DNA ligases. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):400–404. doi: 10.1073/pnas.88.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollberg T. M., Seal G., Sirover M. A. Monoclonal antibodies detect conformational abnormality of uracil DNA glycosylase in Bloom's syndrome cells. Carcinogenesis. 1987 Nov;8(11):1725–1729. doi: 10.1093/carcin/8.11.1725. [DOI] [PubMed] [Google Scholar]
- Weksberg R., Smith C., Anson-Cartwright L., Maloney K. Bloom syndrome: a single complementation group defines patients of diverse ethnic origin. Am J Hum Genet. 1988 Jun;42(6):816–824. [PMC free article] [PubMed] [Google Scholar]
- Willis A. E., Lindahl T. DNA ligase I deficiency in Bloom's syndrome. Nature. 1987 Jan 22;325(6102):355–357. doi: 10.1038/325355a0. [DOI] [PubMed] [Google Scholar]
- Willis A. E., Weksberg R., Tomlinson S., Lindahl T. Structural alterations of DNA ligase I in Bloom syndrome. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8016–8020. doi: 10.1073/pnas.84.22.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. W., Becker F. F., Chan J. Y. Fingerprinting of near-homogeneous DNA ligase I and II from human cells. Similarity of their AMP-binding domains. J Biol Chem. 1990 Oct 25;265(30):18130–18134. [PubMed] [Google Scholar]