Abstract
Background
Polyomavirus nephropathy (PVAN) is a common cause of kidney allograft dysfunction and loss. To identify PVAN-specific gene expression and underlying molecular mechanisms we analyzed kidney biopsies with and without PVAN.
Methods
The study included 168 posttransplant renal allograft biopsies (T cell mediated rejection=26, PVAN=10, normal functioning graft (STA) =73, and interstitial fibrosis/tubular atrophy (IF/TA) =59) from 168 unique kidney allograft recipients. We performed gene expression assays and bioinformatics analysis to identify a set of PVAN-specific genes. Validity and relevance of a subset of these genes are validated by QPCR and IHC.
Results
Unsupervised hierarchical clustering analysis of all the biopsies revealed high similarity between PVAN and TCMR gene expression. Increased statistical stringency identified 158 and 252 unique PVAN and TCMR injury-specific gene transcripts respectively. While TCMR-specific genes were overwhelmingly involved in immune response costimulation and TCR signaling, PVAN-specific genes were mainly related to DNA replication process, RNA polymerase assembly and pathogen recognition receptors. A principal component analysis using these genes further confirmed the most optimal separation between the 3 different clinical phenotypes. Validation of 4 PVAN-specific genes (RPS15, CFD, LTF, and NOSIP) by QPCR and confirmation by immunohistochemistry of 2 PVAN-specific proteins with anti-viral function (LTF and IFITM1) was done.
Conclusions
In conclusion, even though PVAN and TCMR kidney allografts share great similarities on gene perturbation, PVAN-specific genes were identified with well-known anti-viral properties that provide tools for discerning PVAN and AR as well as attractive targets for rational drug design.
Introduction
Polyomavirus-associated nephropathy (PVAN) remains an important opportunistic infection after renal transplantation. Asymptomatic viremia may be observed in 10-30% of transplant recipients, and 4-10% may develop PVAN with allograft loss occurring in approximately 50% of cases 1-4. Even though immunosuppressive condition is the main cause for viral reactivation, not all latent infections in Kidney transplant patients lead to the development PVAN. This suggests that while the increased burden of immunosuppression appears to be crucial for viral reactivation, individual immune-susceptibility to viral infection and type of intragraft inflammation are likely to be involved in the pathogenesis of PVAN 5.
BKV replication can rapidly be monitored by nucleic acid testing (q-PCR) analysis of urine or plasma samples. The diagnosis of PVAN requires histological assessment showing typical viral cytopathic changes in tubular epithelial cells and a positive immunostaining against the LT antigen of simian polyomavirus (SV40). Importantly, accompanying these lesions, there is an important tubulo-interstitial inflammatory cell infiltrate, which is indistinguishable from typical histological patterns of that observed in acute T cell mediated rejection (TCMR) 6,7. This is of clinical relevance since the therapeutic approaches are opposed; PVAN treatment fundamentally focuses on a progressive reduction of immunosuppression with eventual adjuvant medication, whereas TCMR requires of additional rescue immunosuppressive therapy (8). Therefore, an accurate recognition of the PVAN-associated molecular fingerprint could help to better discriminate these 2 processes and provide new insight of the pathogenic mechanisms of the disease.
Gene expression studies in biopsy samples of PVAN and TCMR focusing at specific genes by PCR analysis have reported a similar over-expressed transcripts associated with similar T cell activation and costimulation pathways such as IFN-ɣ, perforin, CXCR3 or CD40/CD40L, respectively 8,9. Likewise, in the urine, increased inflammatory cytokines similar to TCMR have also been shown in PVAN patients 10,11. However, the evaluation using high-throughput microarray analysis of PVAN/TCMR tissue allograft samples to significantly enrich the genomic picture of these pathological features is scarce. Recently, Lubetzky et al 12 investigated the genomics of PVAN, mainly in whole blood and in a reduced number of tissue allograft samples by microarray analysis. Authors reported a significantly increased pathogenesis-based transcript activity of cytotoxic T cells and natural killer cells in PVAN resembling TCMR, suggesting the involvement of adaptive and innate immunity in both settings.
With the aim of obtaining a deeper understanding of the main molecular mechanisms taking place during the inflammatory process both in PVAN and TCMR, we employed a high throughput microarray analysis of kidney allograft biopsies. Here, we report that while both pathological features overlap with a relevant number of inflammatory and cytotoxic pathogenesis-related transcripts, PVAN does also show differentially up-regulated gene-transcripts related to immune response to organisms and particularly to viral infection. Importantly, significantly over-expressed genes and gene products (proteins) in PVAN patients were further confirmed by qPCR and IHC.
Materials and Methods
Patients and biopsies
The study comprised of 168 posttransplant renal allograft biopsies (TCMR=26, PVAN=10, normal functioning graft without subclinical rejection or other injury (STA) =73, and interstitial fibrosis/tubular atrophy (IF/TA) =59) from 168 unique pediatric and adolescent kidney allograft recipients (1 to 21 years of age) (Table 1). All patients received an immunosuppressive regimen consisting of a combination of tacrolimus (Prograf, Astellas Pharma), mycophenolate mofetil (Cellcept, Hoffman-La Roche) and daclizumab (Zenapax, Hoffman-La Roche) or thymoglobulin (Sanofi) induction. Some patients received a steroid-avoidance regimen, while others received a steroid-based immunosuppressive regimen, as previously described 13. Clinical and histological demographic characteristics were collected for all the biopsies. A subset of biopsies with PVAN, TCMR, and STA phenotypes, matched for major clinical variables such as recipient and donor age, % living donor kidneys, time posttransplant, immunosuppression usage, which was used as selected case-controls for more stringent analysis for PVAN biology (Table 1). Diagnosis of TCMR and IFTA was made by biopsy histology Banff classification 14. All IF/TA samples showed Banff scores grade II or higher, without showing any other specific accompanying lesions or AR. PVAN was defined as positivity of polyomavirus PCR in peripheral blood, together with a positive SV40 stain in the concomitant renal allograft biopsy according to Banff criteria15. A small number of patients had BK DNA replication but no evidence of PVAN on biopsy; these patients are categorized as BKVB (BK viremia in blood only) and have been included in the global gene expression first phase analysis in Table 1 and Figure 1. The Ethics Committee of Stanford University Medical School and UCSF Medical Center approved the study. All patients/guardians provided informed consent to participate in the research, in full adherence to the Declaration of Helsinki. The clinical and research activities being reported are consistent with the Principles of the Declaration of Istanbul as outlined in the ‘Declaration of Istanbul on Organ Trafficking and Transplant Tourism.
Table 1.
Relevant Clinical Demographics.
| Main clinical variables (n=168 patients and biopsies) | PVAN (n=10) | TCMR (n=26) | IFTA (n=59) | STA* (n=73) | P value (>0.05=NS) |
|---|---|---|---|---|---|
| CNI-based (%) | 100 | 100 | 100 | 100 | NS |
| Mean donor age (yr±SD) | 24±10 | 30±11 | 32±11 | 31±11 | NS |
| Induction % IL2RmAb (induction useage is IL2RmAb or Thymoglobulin) | 80 | 75 | 85 | 80 | NS |
| %Steroid-free maintenance immunosuppression | 60 | 58 | 65 | 70 | NS |
| % prior history of acute rejection | 15% | 10% | 8% | 4% | NS |
| Recipient gender (%F) | 20 | 21 | 57 | 36 | NS |
| Mean recipient age (yr±SD) | 14±4 | 11±6 | 11±6 | 11±6 | NS |
| Type of transplant (% living donor) | 66 | 71 | 63 | 70 | NS |
| Time post-transplant (mo±SD) | 10±9 | 11±7 | 13±6 | 13±19 | NS |
| Type of Biopsy (%Cause) | 60 | 21 | 5 | 0 | <0.0001 |
| Serum creatinine at the timeof biopsy (mg/dL) | 1.48±0.56 | 1.84±0.50 | 0.88±0.42 | 0.72±0.36 | <0.0001 |
| Kidney histology lesions (Banff score for each renal compartment;0-3) (mean±sd; range) | |||||
| Acute Banff scores (range) | |||||
| Acute tubuli (t) | 1.7±1 (1-3) | 1.5±0.9 (1-3) | 0.03±0.18(0-1) | 0.03±0.18 (0-1) | #NS |
| Acute Interstitium (i) | 1.5±1.5 (1-2) | 2.2±0.9 (1-3) | 0.02±0.13(0-1) | 0 | |
| Acute glomeruli (ag) | 0.1±0.3 (0-1) | 0.2±0.3 (0-1) | 0 | 0 | |
| Acute vascular (av) | 0 | 0 | 0 | 0 | |
| Peritubular capilaritis (ptc) | 0 | 0.3±0.5 (0-1) | 0 | 0 | |
| SV40 | Positive | Negative | Negative | Negative | |
| C4d | 0 | 0 | 0 | 0 | |
| Chronic Banff scores (range) | $NS | ||||
| Chronic tubuli (ct) | 1.3±0.9 (0-3) | 0.7±0.5 (0-1) | 1.3±1.1 (0-3) | 0.32±0.54 (0-1) | μp<0.0001 |
| Chronic interstitium (ci) | 1.2±0.7 (0-2) | 0.7±0.5 (0-1) | 1.1±1.2 (0-2) | 0 | |
| Chronic vascular (cv) | 0.3±0.5 (0-1) | 0.2±0.4 (0-1) | 0.5±0.8 | 0.28±0.46 (0-1) | |
| Chronic glomeruli (cg) | 0.3±0.7 (0-2) | 0 | 0.0±0.0 | 0 | |
| BK viremia (copies/ml) (mean) | 21262±38342 | 0 | 0 | 0 | |
NS refers to a p>0.05
5 BKVB patients had stable graft function
PVAN and TCMR samples were not statistically significant regarding t and i scores (#NS).
PVAN and IFTA samples were not statistically significant regarding the ct and ci scores ($NS)
PVAN and IFTA were statistically different for ct and ci when compared to TCMR and STA (μp<0.0001) samples. All patients on steroid-free immunosuppression were not on steroids at the time of biopsy.
Figure 1.
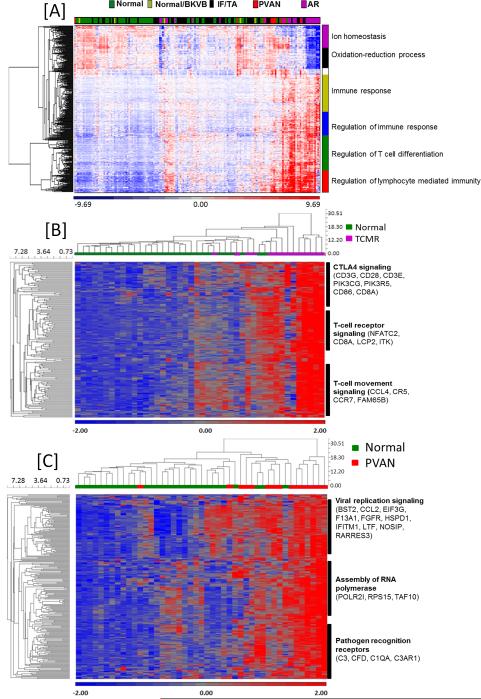
(A)- Global gene expression pattern of samples classified as PVAN were largely similar to TCMR samples when compared to normal kidney biopsies, BK viremia (BKVB), and IF/TA. (B)- Gene expression clustering demonstrates TCMR-specific gene signature. (C)- Gene expression clustering demonstrates PVAN-specific gene signature compared to normal (STA). A few genes (n=16) were also differentially expressed in the IF/TA patients compared to PVAN but this analysis is not as clean as many patients with PVAN also have associated IF/TA changes in the graft.
RNA Extraction, Quality Control, Amplification and Microarray Hybridization
Needle biopsies were collected at the time of biopsy procedure and immediately submerged in RNAlater (Qiagen, Valencia, CA) and stored at −80 °C until use. For the gene expression analysis purpose, total RNA was extracted and hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays as previously described in Naesens et al 16. For processing and normalization of the scanned images, dChip 2006 software was used, with perfect match [PM]/mismatch [MM] difference modeling and invariant set normalization 17.
QPCR verification
A total of 250 ng total RNA was first reversed transcribed into complementary DNA (cDNA) using Superscript II (Invitrogen, Life Technologies, Foster City, CA, USA) according to the manufacture's protocol in the Eppendorf vapo.protect thermal cycler (Eppendorf, Hauppauge, NY, USA). From each sample 1.56 ng cDNA were amplified in a target specific amplification step for 4 genes (RPS15, CFD, LTF, NOSIP) TaqMan PreAmp Master Mix (Cat. No. 4488593, Life Technologies, Foster City, CA, USA and TaqMan Primers and Probes (RPS15- Cat. No. Hs01358643_g1; CFD- Cat. No. Hs00157263_m1; LTF- Cat. No. Hs00914334_m1; NOSIP- Cat. No. Hs00211028_m1; Life Technologies, Foster City, CA, USA) for a total of 18 amplification cycles. QPCR reactions were performed in the BioMark RT PCR system using 18S gene (Primer and probe Cat. No. Hs03003631_g1, Life Technologies, Foster City, CA, USA) as a house keeping gene and Human XpressRef Universal Total RNA (Qiagen Inc, Valencia, CA, Cat. No. 338112) as a reference RNA for a total of 40 cycles (Fluidigm, San Francisco, CA, USA). Resulting chip data was initially analyzed for QC using the BioMark Analysis Software Version 2.0 (Fluidigm, San Francisco, CA, USA) and Ct values were exported into Excel.
Immunohistochemistry of kidney allograft biopsies
Nine representative cases of kidney allograft biopsies of PVAN (n=3), TCMR (n=3) and STA (n=3) were stained for 2 over-expressed transcripts in PVAN kidney biopsies. To determine the extent of renal damage and classify them into the 3 different phenotypes (PVAN, TCMR and STA), all renal biopsies were analyzed by 2 blinded pathologists. The biopsies were probed with rabbit polyclonal to Lactoferrin (Abcam, Cambridge, MA), rabbit polyclonal to IFN-inducible transmembrane 1 (IFITM-1) (Abcam, Cambridge, MA) and SV40 (Becton Dickinson, Madrid, Spain). Immune staining in formalin-fixed, paraffin-embedded tissues was performed as described previously 18-20. As positive controls for LTF, IFITM1 and SV40 human tonsil, liver carcinoma and kidney allograft paraffin-embedded tissues were used as positive controls, respectively. To quantify LTF and IFITM1 expression a semi quantitative score from 0 to 3 in the different compartments of the kidney (glomeruli, vessels, tubuli and interstitium) was used.
Data Processing and Analysis
Differentially expressed genes between biopsy groups were identified using an empirical Bayes moderated t-test using a Benjamini-Hochberg adjusted adjustment in the program AltAnalyze version 2.0.8 21. In all comparisons an FDR adjusted p<0.05 was used for filtering. After identifying the differentially expressed genes, these probe sets were analyzed using GO-Elite version 1.2.6 with all available default annotation resources 21-24 to identify enriched biological pathways, Ontologies and gene-sets. To evaluate immune cell infiltration, immune cell-type specific markers were computationally inferred using a new marker identification algorithm (LineageProfiler) applied to 2 large published microarray studies (GSE22886, GSE15907) to be used by GO-Elite. Principal component analysis (PCA), expression clustering (hierarchical or HOPACH), pathway filtering and visualization were also performed in AltAnalyze using the default parameters. The raw data sets for the 168 biopsies included are deposited at the Gene Expression Omnibus under GSE72925.
Results
Demographics
During the time-frame period of the study, 168 unique pediatric kidney allograft biopsies indicated either for cause or protocol, were initially included. Subsequently, 55 out of the 168 sample-set with matched demographics were included for the gene expression analysis. 10 (~18%) patients were found to have Polyomavirus-associated nephropathy (PVAN) (70% Stage C and 30% Stage B)15, 15 of 55 (~27%) pure T cell mediated rejection (TCMR) and 30 of 55 (~55%) were considered as patients with stable functioning graft (STA) as no abnormalities were observed in their biopsies. As shown in Table 1, there were no differences regarding main clinical demographic characteristics such as donor and recipient age and gender, type of maintenance and induction immunosuppression, number of previous transplants and type of transplant. All biopsies were performed during the first 24 months after transplantation, either for protocol (at 6 or 24 months) or for cause because of allograft dysfunction or presence of BK DNA replication in peripheral blood. At the time of assessment, allograft function was not different between the 3 groups. Only the acute inflammatory Banff scores in the tubuli and interstitium of renal allograft compartments were significantly higher among PVAN and TCMR as compared to STA patients. The mean serum creatinine value of selected 15 TCMR patients was significantly higher (1.6 mg/dL) than the mean serum creatinine value of selected 30 STA patients (0.85 mg/dL) (p<0.001). The relatively lower serum creatinine seen in AR in this cohort is a function of this being a pediatric cohort. As per definition, only PVAN patients showed positivity for SV40 immunostaining and BK DNA replication in peripheral blood. Five patients with Bk virus presence only in the blood (BKVB) were included in the cohort of normal biopsies.
Unsupervised hierarchical clustering of all kidney allograft biopsies
First, unsupervised hierarchical clustering analysis was performed among all 168 posttransplant renal allograft biopsies with the aim of having a broad gene expression picture using the 500 top ranked probesets based on an FDR adjusted f-test p-value (p<0.05 and 2-fold increase expression). This analysis indicated that the global gene expression pattern of samples classified as PVAN were largely similar to TCMR samples as both compared to normal kidney biopsies (Figure 1). The 5 biopsies with BKVB aligned with normal biopsies and were not studies as a separate phenotype. The top 500 genes were enriched in immune system process (p=3.38E-21) and immune response (p=3.38E-21).
Distinctive gene set expression for PVAN and TCMR as compared to STA in well-matched kidney transplant patients
Next, in order to have the most comparable study population, we selected for each PVAN (n=10) and TCMR (n=15) patients, the best demographically matched STA patient for this analysis (n=30). With a FDR adjusted p-value criteria of P<0.05, a total of 4047 probesets showed TCMR specific regulation of that 2483 probesets were significantly upregulated and 1564 were significantly downregulated). With the same FDR adjusted p-value criteria of P<0.05, a total of 11594 probsets showed PVAN specific regulation of that 5450 probesets were significantly upregulated and 6144 were significantly downregulated). Among upregulated genes 241 probesets were common in both TCMR and PVAN and among downregulated genes 332 probesets were common in both TCMR and PVAN tissues.
Using Principle Component Analysis (PCA) and taking into account the 150 top ranked probesets based on a FDR adjusted p-value (p<0.05 and 2-fold increase expression), a significant clustering of genes were observed among TCMR and STA as well as PVAN and STA. PVAN and TCMR samples clustered together when all 3 sample types were used in the PCA plot (Supplemental Figure 1).
With the aim of identifying critical PVAN injury-specific genes and in TCMR, we increased stringency of the specificity to the p value <0.01 with >2 fold increased expression that resulted in 209 unique genes increased in PVAN (Supplemental Table 1) and 252 unique genes increased in TCMR as compared to STA kidney biopsies. The TCMR-specific probesets that were not significant in PVAN and STA individuals were basically involved in CTLA4 (CD3G, CD28, CD3E, PIK3CG, PIK3R5, CD86, CD8A) and T cell receptor (NFATC2, CD8A, LCP2, ITK) signaling in cytotoxic T lymphocytes as well as related to cellular movement (CCL4, CCR5, CCR7, FAM65B) of T lymphocytes. Regarding the 115 unique probesets most significantly enriched in PVAN and not in TCMR and STA patients, were mainly involved in DNA replication and RNA binding (BST2, EIF3G, F13A1, FGFR1, HSPD1, IFITM1, LTF, RPS15, NOSIP, and RARRES3), assembly of RNA polymerase (POLR2I, TAF10) and pathogen recognition receptors (C3, C1QA, C3AR1 and CFD). As indicated by both principal component analysis and HOPACH gene clustering of these PVAN specific genes, the most optimal separation between the 30 STA, 15 TCMR and 10 PVAN samples was achieved, relative to any of the prior gene sets (Figures 1B and 1C).
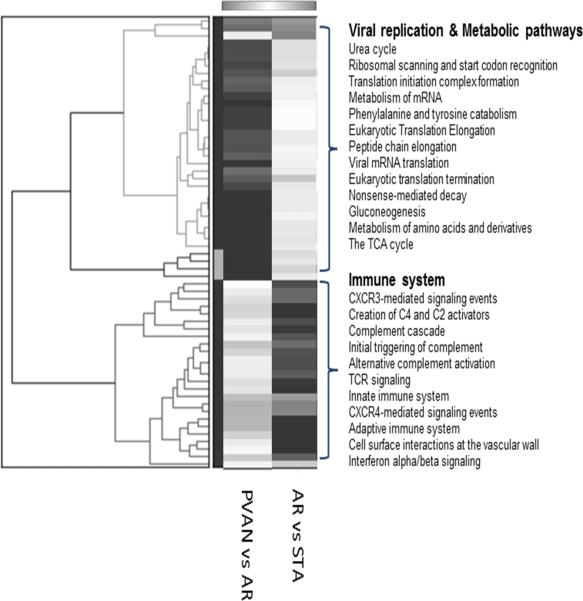
To determine the functional relevance of genes enriched in PVAN but not TCMR, we performed a comprehensive pathway/gene-set analysis using the software GO-Elite. This analysis showed enrichment of 2 distinct set of pathways. Immune system related pathways such as complement cascade, TCR signaling, innate immune system, adaptive immune system were specific to TCMR associated genes whereas PVAN associated genes were enriched with DNA replication pathways such as mRNA processing, ribosomal scanning, viral mRNA translation etc. and metabolic pathways such as urea cycle, gluconeogenesis, TCA cycle etc. demonstrating 2 distinct molecular events occurring at the time of TCMR and PVAN (Figure 2).
Figure 2.
A comprehensive pathway/gene-set analysis using the software GO-Elite was performed to identified molecular pathways enriched in PVAN compared to TCMR. This resulted in enrichment of 2 distinct set of pathways. Immune system related pathways such as complement cascade, TCR signaling, innate immune system, adaptive immune system were specific to TCMR whereas PVAN associated genes were enriched with DNA replication pathways such as mRNA processing, ribosomal scanning, viral mRNA translation etc. and metabolic pathways such as urea cycle, gluconeogenesis, TCA cycle etc. were specific to PVAN demonstrating 2 distinct molecular events occurring at the time of TCMR and PVAN.
RT-PCR validation of selected genes
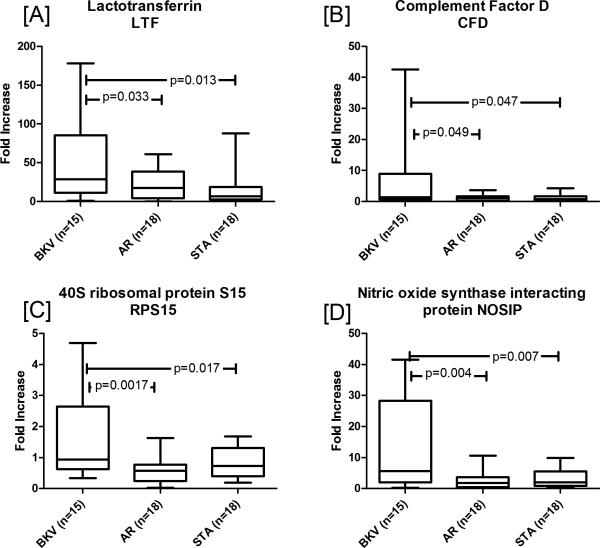
In order to validate PVAN specific gene expression data, 4 genes (LTF, CFD, RPS15, and NOSIP) were selected for QPCR validation. As is illustrated in Figure 3, over-expression of all 4 genes in PVAN vs. STA and also in PVAN vs. TCMR was confirmed. In an independent set of PVAN (n=15), AR (n=18) and STA (n=18) fold increase in gene expression for; (i) lactotransferrin (LTF) in PVAN was significant when compared to AR (p=0.04) and STA (p=0.02), (ii) complement factor D (CFD) in PVAN was significant when compared to AR (p=0.05) and STA (p=0.05), (iii) 40 ribosomal protein S15 (RPS15) in PVAN was significant when compared to AR (p=0.002) and STA (p=0.02), (iv) nitric oxide synthase interacting protein (NOSIP) in PVAN was significant when compared to AR (p=0.004) and STA (p=0.007).
Figure 3.
QPCR validations of PVAN specific genes. In order to validate PVAN specific gene expression data 4 genes (LTF, CFD, RPS15, and NOSIP) were selected for QPCR validation. Over-expression of all 4 genes in PVAN vs. STA and also in PVAN vs. TCMR was confirmed. Gene expression for; (A) lactotransferrin (LTF) in PVAN was significant when compared to AR (p=0.04) and STA (p=0.02), (B) complement factor D (CFD) in PVAN was significant when compared to AR (p=0.05) and STA (p=0.05), (C) 40 ribosomal protein S15 (RPS15) in PVAN was significant when compared to AR (p=0.002) and STA (p=0.02), (D) nitric oxide synthase interacting protein (NOSIP) in PVAN was significant when compared to AR (p=0.004) and STA (p=0.007). The first and third quartiles are at the ends of the box, the median is indicated with a horizontal line in the interior of the box, and the maximum and minimum are at the ends of the whiskers.
Immunohistochemistry analysis of PVAN-associated gene products
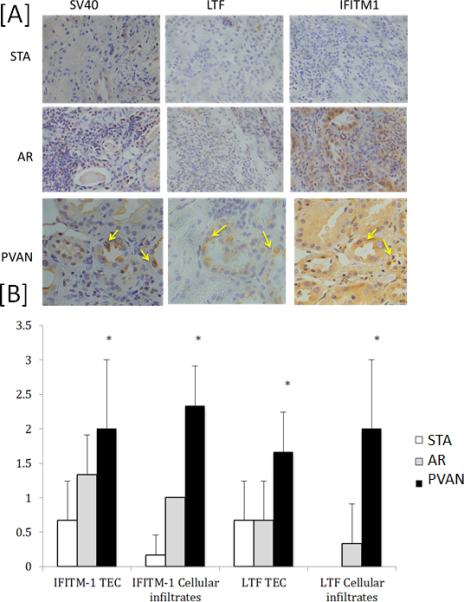
Using immunohistochemistry (IHC) analysis, we assessed the expression of 2 gene transcripts (LTF and IFITM-1) at the protein level. These transcripts were highly upregulated in kidney biopsies with PVAN as compared to TCMR and STA and have been previously reported to have anti-viral properties, LTF (24,25) and IFITM1 (92,30) (Figure 4a). As shown in Figure 4b, LTF and IFITM1 expression was significantly higher in both in tubulo-epithelial cells and within the mononuclear cellular infiltrates in PVAN patients as compared to STA and TCMR kidney transplant recipients.
Figure 4.
Using immunohistochemistry (IHC) validation of PVAN specific expression of LTF and IFITM1 in kidney biopsies with PVAN. We assessed the expression of 2 gene transcripts (LTF and IFITM-1) at the protein level. These transcripts were highly upregulated in kidney biopsies with PVAN as compared to TCMR and STA. A) Three representatives phenotypes from 3 representative transplant patient biopsies evaluated for the different protein stains. TCMR and STA are shown at 10× and the PVAN samples are shown at 40× magnification. Co-localization of IFITM-1 with BK viral inclusions are marked with yellow arrows in the PVAN patient. LTF also localizes in proximity to the SV40 and IFITM-1 stains in the renal tubule. (B) Semi quantitative analysis of protein expression at tubulo-epithelial cells (TEC) and mononuclear cells of patients with PVAN, TCMR and STA.
Discussion
The advent of PVAN is still a major concern in kidney transplantation, as it accounts for the main cause of allograft loss. This is explained, in great part, to the rather poor understanding of the dominant mechanisms of the disease. While it is a widely accepted the fact that recognizing PVAN as early as possible is a key factor to increase the likelihood of success 5, it still remains unclear, which is the best treatment approach to follow. In this study, using high-throughput microarrays analysis we demonstrate that the inflammatory process occurring both in PVAN and TCMR, either due to a protective anti-viral or an allogeneic immune response, respectively, merges in a remarkably similar transcriptional gene pattern, essentially associated with effector immune pathways of both the adaptive and the innate immunity.
As reported earlier by other researchers 8,12, PVAN and TCMR share important similarities at the gene expression level, further helping to explain the difficulty of differentiating both types of diseases. In this regard, Mannon et al 8 evaluated target genes by qPCR in kidney allograft biopsies from recipients with PVAN, TCMR or patients with stable allograft function and showed that despite the significantly high resemblance of gene expression between PVAN and TCMR, transcription of certain molecules associated with graft fibrosis and markers of epithelial-mesenchymal transformation (EMT) were significantly higher in PVAN specimens, suggesting a higher profibrogenic transcriptional profile in PVAN than in TCMR patients. The higher tissue chronicity in the PVAN patient samples may also explain some of the observed differences between TCMR and PVAN relating to glucose and protein metabolism genes. Furthermore, in a recent study, Lubetzky and coworkers 12 evaluated gene expression in the whole blood as well as few kidney tissue allograft samples in PVAN and TCMR kidney transplant recipients. These studies have shown high levels of proinflammatory molecules in both settings, such as the interferon gamma-induced chemokines CXCL9 and CXCL10 11, as well as other Th-2-induced cytokines such as sIL-1RA, IL-3, IL-6, and sIL-6R, being particularly high in patients with high BK DNA replication 25.
Our study validated 4 PVAN-specific genes by qPCR and further confirmed using IHC. This illustrates the potential functional relevance of some of the particular transcripts uniquely expressed in PVAN biopsies as compared to rejecting patients. Firstly, Lactotransferrin (LTF), a member of the transferrin family found in mucous epithelial cells and secondary granules of polymorphonuclear neutrophils was highly differentially expressed among PVAN patients. Its expression in PVAN biopsies was significantly higher both within cellular infiltrates and in tubulo-epithelial cells as compared to biopsies with TCMR and STA. in fact, LTF has been shown to play a key role in the defense against various pathogenic microorganisms, by inhibiting different enveloped 26,27 and naked 28,29 virus in different virus-cell systems. In addition, in experimental animal models it has been shown that oral administration of LTF or peptides thereof is effective in reducing bacterial infections and inflammation in the urinary tract, possibly through transfer of LTF or its peptides to the site of infection via renal secretion30. Of note, LTF treatment has been evaluated and shown to prevent early steps of BK virus infection in vitro, most likely through the interaction with BK viral capsidic structures 31. This finding is of relevance, since the high expression of LTF transcripts in kidney allografts infected by BK, might be the expression of a physiological protective response of the host against the virus in order to overcome the viral infection. Observation of increased expression of genes associated with TCA cycle, urea cycle, and gluconeogenesis is biologically plausible. Even though there is no report of increased metabolism in BKV infection, alteration of metabolism especially glucose metabolism and TCA cycle has been observed in case of cytomegalovirus infection32. It is intuitive to assume that successful replication of virus in the infected cells requires an environment that is suitable for increased supply of nutrient, energy, and macromolecular synthesis, which is reflected in the upregulated gene expression of genes related with metabolic pathways. Complment Factor D (CFD) was also upregulated in PVAN, which is a serine protease that cleaves C3b-bound factor B, resulting in the generation of Bb and formation of the alternative pathway C3 convertase (C3bBb) a key system for immune surveillance and homeostasis33. Increased expression of CFD and other complement genes shows activation of complement system at the time of viral infection. There is increased generation of nitric oxide (NO) due to viral infection, which is harmful34. NOSIP is 1 of the genes upregulated in PVAN, negatively regulates nitric oxide production by inducing translocation of NOS1 and NOS3 to actin cytoskeleton and inhibiting their enzymatic activities35. Furthermore, another highly up-regulated gene observed within PAVN patients as compared to TCMR and stable individuals was the IFN-inducible transmembrane 1 (IFITM-1) transcript. Interestingly, IFITM proteins are a family of ubiquitously expressed restriction factors that mediate potent IFN-induced antiviral activity by inhibiting viral entry, particularly the step of membrane fusion 36-38. Nevertheless, while IFITM-1 antiviral activity has been well characterized against RNA viruses 37,39,40, it has been shown to induce the opposite effect, that is to enhance infection of several DNA viruses. Indeed, it has been demonstrated that the antiviral activity of IFITM proteins is likely mediated by preventing endosome fusion and viral entry into the cytosol36-38, as well as inhibit viral entry by preventing escape from the endocytic pathway particularly among DNA virus41. Therefore, over-production of IFITM-1 molecules in PVAN patients could be hypothesized to represent a signal of persistent DNA replication, illustrating the aggressive nature of BKV infection in kidney transplantation. Indeed the co-localization of IFITM-1 in close proximity to LTF in tubular cells showing BK viral inclusions further corroborate these previously reported data (Figure 4).
In summary, even though PVAN and TCMR kidney allografts share great similarities on gene perturbation, particular PVAN-specific transcripts are differentially expressed, some of them encoding for molecules with well-known anti-viral properties. Further tracking such effector molecules in the context of BK virus infection may lead to the discovery of novel potential therapeutic targets that may eventually overcome the development and persistence of BKV infection after kidney transplantation.
Supplementary Material
Acknowledgments
We would like to acknowledge Anyou Wang and Mark Nguyen for manuscript preparation and Cristian Varela, Nuria Bolaños and Dr. Benjamin Torreón from the SCT at UB for the methodical help with all the IHC experiments.
Funding:
1. R01 DK083447 (NIDDK) to Dr. Minnie Sarwal
2. Spanish Society of Nephrology grant, Spanish Grant (FIS PI15/01263), and European grant from the BIODRIM consortium (12CEE014) to Dr. Oriol Bestard.
Abbreviations
- PVAN
Polyomavirus nephropathy
- STA
stable and normal functioning graft without subclinical rejection or other injury
- IF/TA
interstitial fibrosis/tubular atrophy
- TCMR
T cell mediated rejection
- QPCR
quantitative polymerase chain reaction
- IHC
Immunohistochemistry
- PCA
principal component analysis
- LTF
Lactoferrin
- NOSIP
Nitric oxide synthase interacting protein
- IFITM1
Interferon induced transmembrane protein 1
Footnotes
Author contributions: MMS and TKS conceived the study, TKS, SS,TQT, and JT performed the experiments, TKS, OB, NS, MN and SR analyzed the data, TKS, MMS, OB, and MN wrote the manuscript.
Disclosure: The authors declare no conflicts of interest.
References
- 1.Dharnidharka VR, Cherikh WS, Abbott KC. An OPTN analysis of national registry data on treatment of BK virus allograft nephropathy in the United States. Transplantation. 2009;87(7):1019–1026. doi: 10.1097/TP.0b013e31819cc383. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch HH, Randhawa P. Practice ASTIDCo. BK polyomavirus in solid organ transplantation. Am J Tranplant. 2013;13(Suppl 4):179–188. doi: 10.1111/ajt.12110. [DOI] [PubMed] [Google Scholar]
- 3.Ramos E, Drachenberg CB, Wali R, Hirsch HH. The decade of polyomavirus BK-associated nephropathy: state of affairs. Transplantation. 2009;87(5):621–630. doi: 10.1097/TP.0b013e318197c17d. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch HH, Knowles W, Dickenmann M, et al. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N J Engl Med. 2002;347(7):488–496. doi: 10.1056/NEJMoa020439. [DOI] [PubMed] [Google Scholar]
- 5.Babel N, Volk HD, Reinke P. BK polyomavirus infection and nephropathy: the virus-immune system interplay. Nat Rev Nephrol. 2011;7(7):399–406. doi: 10.1038/nrneph.2011.59. [DOI] [PubMed] [Google Scholar]
- 6.Drachenberg CB, Papadimitriou JC, Hirsch HH, et al. Histological patterns of polyomavirus nephropathy: correlation with graft outcome and viral load. Am J Transplant. 2004;4(12):2082–2092. doi: 10.1046/j.1600-6143.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch HH, Randhawa P. Practice ASTIDCo. BK virus in solid organ transplant recipients. Am J Transplant. 2009;9(Suppl 4):S136–146. doi: 10.1111/j.1600-6143.2009.02904.x. [DOI] [PubMed] [Google Scholar]
- 8.Mannon RB, Hoffmann SC, Kampen RL, et al. Molecular evaluation of BK polyomavirus nephropathy. Am J Transplant. 2005;5(12):2883–2893. doi: 10.1111/j.1600-6143.2005.01096.x. [DOI] [PubMed] [Google Scholar]
- 9.Girmanova E, Brabcova I, Klema J, et al. Molecular networks involved in the immune control of BK polyomavirus. Clin Dev Immunol. 2012;2012:972102. doi: 10.1155/2012/972102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dadhania D, Snopkowski C, Ding R, et al. Validation of noninvasive diagnosis of BK virus nephropathy and identification of prognostic biomarkers. Transplantation. 2010;90(2):189–197. doi: 10.1097/TP.0b013e3181e2a932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson JA, Kim EJ, Begley B, et al. Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am J Transplant. 2011;11(10):2228–2234. doi: 10.1111/j.1600-6143.2011.03680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubetzky M, Bao Y, P OB, et al. Genomics of BK viremia in kidney transplant recipients. Transplantation. 2014;97(4):451–456. doi: 10.1097/01.TP.0000437432.35227.3e. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Chang A, Naesens M, et al. Steroid-Free Immunosuppression since 1999: 120 pediatric renal transplants with sustained graft and patient benefits. Am J Transplant. 2009;9:1362–1372. doi: 10.1111/j.1600-6143.2009.02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sis B, Mengel M, Haas M, et al. Banff '09 meeting report: antibody mediated graft deterioration and implementation of Banff working groups. Am J Transplant. 2010;10(3):464–471. doi: 10.1111/j.1600-6143.2009.02987.x. [DOI] [PubMed] [Google Scholar]
- 15.Sar A, Worawichawong S, Benediktsson H, Zhang J, Yilmaz S, Trpkov K. Interobserver agreement for Polyomavirus nephropathy grading in renal allografts using the working proposal from the 10th Banff Conference on Allograft Pathology. Hum Pathol. 2011;42(12):2018–2024. doi: 10.1016/j.humpath.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Naesens M, Khatri P, Li L, et al. Progressive histological damage in renal allografts is associated with expression of innate and adaptive immunity genes. Kidney Int. 2011;80(12):1364–1376. doi: 10.1038/ki.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2(8):RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ieni A, Barresi V, Branca G, Giuffre G, Rosa MA, Tuccari G. Immunoexpression of lactoferrin in bone metastases and corresponding primary carcinomas. Oncology letters. 2013;5(5):1536–1540. doi: 10.3892/ol.2013.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parra-Herran CE, Yuan L, Nucci MR, Quade BJ. Targeted development of specific biomarkers of endometrial stromal cell differentiation using bioinformatics: the IFITM1 model. Mod Pathol. 2014;27(4):569–579. doi: 10.1038/modpathol.2013.123. [DOI] [PubMed] [Google Scholar]
- 20.Schmid H, Burg M, Kretzler M, Banas B, Grone HJ, Kliem V. BK virus associated nephropathy in native kidneys of a heart allograft recipient. Am J Transplant. 2005;5(6):1562–1568. doi: 10.1111/j.1600-6143.2005.00883.x. [DOI] [PubMed] [Google Scholar]
- 21.Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38(Web Server issue):W755–762. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31(1):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 23.Salomonis N, Hanspers K, Zambon AC, et al. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zambon AC, Gaj S, Ho I, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28(16):2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadeghi M, Daniel V, Schnitzler P, et al. Urinary proinflammatory cytokine response in renal transplant recipients with polyomavirus BK viruria. Transplantation. 2009;88(9):1109–1116. doi: 10.1097/TP.0b013e3181ba0e17. [DOI] [PubMed] [Google Scholar]
- 26.Andersen JH, Jenssen H, Gutteberg TJ. Lactoferrin and lactoferricin inhibit Herpes simplex 1 and 2 infection and exhibit synergy when combined with acyclovir. Antiviral Res. 2003;58(3):209–215. doi: 10.1016/s0166-3542(02)00214-0. [DOI] [PubMed] [Google Scholar]
- 27.Andersen JH, Jenssen H, Sandvik K, Gutteberg TJ. Anti-HSV activity of lactoferrin and lactoferricin is dependent on the presence of heparan sulphate at the cell surface. J Med Virol. 2004;74(2):262–271. doi: 10.1002/jmv.20171. [DOI] [PubMed] [Google Scholar]
- 28.Drobni P, Naslund J, Evander M. Lactoferrin inhibits human papillomavirus binding and uptake in vitro. Antiviral Res. 2004;64(1):63–68. doi: 10.1016/j.antiviral.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Pietrantoni A, Di Biase AM, Tinari A, et al. Bovine lactoferrin inhibits adenovirus infection by interacting with viral structural polypeptides. Antimicrob Agents Chemother. 2003;47(8):2688–2691. doi: 10.1128/AAC.47.8.2688-2691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haversen LA, Engberg I, Baltzer L, Dolphin G, Hanson LA, Mattsby-Baltzer I. Human lactoferrin and peptides derived from a surface-exposed helical region reduce experimental Escherichia coli urinary tract infection in mice. Infect Immun. 2000;68(10):5816–5823. doi: 10.1128/iai.68.10.5816-5823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longhi G, Pietropaolo V, Mischitelli M, et al. Lactoferrin inhibits early steps of human BK polyomavirus infection. Antiviral Res. 2006;72(2):145–152. doi: 10.1016/j.antiviral.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y, Clippinger AJ, Alwine JC. Viral effects on metabolism: changes in glucose and glutamine utilization during human cytomegalovirus infection. Trends Microbiol. 2011;19(7):360–367. doi: 10.1016/j.tim.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akaike T, Maeda H. Nitric oxide and virus infection. Immunology. 2000;101(3):300–308. doi: 10.1046/j.1365-2567.2000.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dedio J, Konig P, Wohlfart P, Schroeder C, Kummer W, Muller-Esterl W. NOSIP, a novel modulator of endothelial nitric oxide synthase activity. FASEB J. 2001;15(1):79–89. doi: 10.1096/fj.00-0078com. [DOI] [PubMed] [Google Scholar]
- 36.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13(1):46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brass AL, Huang IC, Benita Y, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. 2013;425(24):4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang IC, Bailey CC, Weyer JL, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raychoudhuri A, Shrivastava S, Steele R, Kim H, Ray R, Ray RB. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol. 2011;85(24):12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren CJ, Griffin LM, Little AS, Huang IC, Farzan M, Pyeon D. The antiviral restriction factors IFITM1, 2 and 3 do not inhibit infection of human papillomavirus, cytomegalovirus and adenovirus. PLoS One. 2014;9(5):e96579. doi: 10.1371/journal.pone.0096579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.