Abstract
CENP-A is a histone H3 variant key to epigenetic specification of mammalian centromeres. Using transient overexpression of CENP-A mutants, two recent reports in Developmental Cell proposed essential centromere functions for post-translational modifications of human CENP-A. Phosphorylation at Ser68 was proposed to have an essential role in CENP-A deposition at centromeres. Blockage of ubiquitination at Lys124 was proposed to abrogate localization of CENP-A to the centromere. Following gene inactivation and replacement in human cells, we demonstrate that CENP-A mutants that cannot be phosphorylated at Ser68 or ubiquitinated at Lys124 are assembled efficiently at centromeres during G1, mediate early events in centromere establishment at an ectopic chromosomal locus, and maintain centromere function indefinitely. Thus, neither post-translational modification of Ser68 or Lys124 is essential for long-term centromere identity, propagation, cell cycle-dependent deposition, maintenance, function, or mediating early steps in centromere establishment.
eTOC blurb
CENP-A is a histone H3 variant important for centromere specification. In this Matters Arising article, using CENP-A gene replacement strategies, Fachinetti, Logsdon, et al. provide evidence that the Ser68phos and Lys124ub modifications of CENP-A, previously proposed to regulate CENP-A function, are not required for long-term centromere identity, function, or maintenance.
Introduction
The centromere is the chromosomal locus that orchestrates connections to the microtubule-based mitotic spindle, recruits signaling components to avoid catastrophic missegregation of entire chromosomes at cell division, and serves as the point of final cohesion prior to the moment of sister chromatid separation (Westhorpe and Straight, 2015). Although centromeres are typically found in genomic locations of highly repetitive sequences, it has been clear for nearly twenty years that centromere location in mammals and many other eukaryotes does not depend on a particular DNA sequence (Eichler, 1999; Karpen and Allshire, 1997). The sequences that are typically found at centromeres are paradoxically neither necessary nor sufficient for specifying centromere location. It is now clear that the histone H3 variant, CENP-A, is key to specifying centromere location (Black and Cleveland, 2011; Fachinetti et al., 2013). Most attractive models for centromere specification involve an epigenetic mechanism for centromere propagation, and at the heart of these models is a self-propagation step where CENP-A chromatin marks centromere location as the site for assembly of new CENP-A chromatin following centromeric DNA replication.
CENP-A chromatin assembly is cell cycle-regulated, occurring only after mitotic exit and restricted to an early portion of G1 (Jansen et al., 2007; Schuh et al., 2007). Nascent CENP-A is expressed across the cell cycle but with elevated synthesis in late S-phase and G2 (Shelby et al., 1997), incorporated into a complex containing its partner histone H4 and a dedicated histone chaperone named HJURP (Dunleavy et al., 2009; Foltz et al., 2009), but only assembled into centromeric chromatin at mitotic exit (Jansen et al., 2007; Schuh et al., 2007) in a process that requires the centromere targeting of Mis18 and its partner molecules (comprising the ‘Mis18 complex’) (Barnhart et al., 2011; Fujita et al., 2007; Moree et al., 2011; Silva et al., 2012). The sub-cellular/sub-chromosomal targeting and cell cycle regulation of CENP-A and its chromatin assembly components may be controlled by post-translational modifications, and recent reports have indeed described candidate post-translational modifications of CENP-A (Bade et al., 2014; Bailey et al., 2013; Bui et al., 2012; Niikura et al., 2015; Yu et al., 2015; Zeitlin et al., 2001), HJURP (Bailey et al., 2016; Kato et al., 2007; Müller et al., 2014; Wang et al., 2014), and the Mis18 complex (McKinley and Cheeseman, 2014; Silva et al., 2012).
Regarding the modification of CENP-A itself, two recent reports in Developmental Cell (Niikura et al., 2015; Yu et al., 2015) each proposed essential roles for modifiable residues. In one case, Ser68 of CENP-A was reported to be phosphorylated, with phosphorylation proposed to have an “essential physiological role in CENP-A deposition at centromeres” (Yu et al., 2015). Experiments using overexpressed versions of CENP-A in which Ser68 was replaced with either a non-phosphorylatable glutamine (CENP-AS68Q) (the side-chain in this position in histone H3) or a phosphomimetic glutamate (CENP-AS68E) were reported to lead to defects in centromere assembly and/or function. From this evidence, the authors claimed that CENP-A Ser68 is “absolutely required for the recognition of CENP-A by HJURP” and “critical for the functions of CENP-A” (Yu et al., 2015). In the other case, overexpression of a version of CENP-A in which Lys124 was mutated to an arginine (CENP-AK124R), an amino acid that cannot be ubiquitinated, was reported to “abrogate centromere localization of CENP-A” (Niikura et al., 2015). A corresponding substitution from histone H3 (akin to S68Q) could not be chosen because H3 also has a lysine at this position, but arginine maintains the positive charge. The authors concluded that ubiquitination of Lys124 is “required for efficient interaction with HJURP” and “essential for CENP-A deposition at the centromere” (Niikura et al., 2015).
Our own work has focused on defining the essential residues of CENP-A necessary for centromere establishment, inheritance, and function (Bassett et al., 2012; Black et al., 2004, 2007; Fachinetti et al., 2013, 2015; Logsdon et al., 2015). We developed three strategies, two described in our prior work to assess the functional requirements for centromere identity and maintenance mediated by CENP-A (Fachinetti, et al., 2013; Logsdon et al., 2015) and one new one using gene replacement at the endogenous CENP-A locus. The first of these is complete genetic replacement of endogenous CENP-A with CENP-A mutants, followed by assessment of long-term cell survival and fidelity of centromere-dependent chromosome segregation (Fachinetti et al., 2013). Long-term functional assessment of centromere function is essential because we showed that once both endogenous CENP-A alleles are inactivated, 9 cell cycles are required to reduce the endogenous CENP-A [which is stably centromere-bound (Bodor et al., 2013; Falk et al., 2015; Jansen et al., 2007)] to less than 1 molecule per centromere (Fachinetti et al., 2013). Our second strategy involves the replacement of the CENP-A gene with an auxin inducible degron- (AID-) or SNAP-tagged CENP-A harboring relevant mutations at the endogenous CENP-A loci, followed by assessment of cell viability and nascent chromatin assembly at centromeres. Finally, our third strategy assesses CENP-A function in centromere establishment by targeting it (or mutants of it or H3/CENP-A chimeric histones) to an ectopic chromosomal locus (Logsdon et al., 2015). The function of each CENP-A mutant or H3/CENP-A chimera is then assessed at this naïve locus by measuring early events prior to subsequent recruitment of detectable endogenous CENP-A protein.
Here, we use all three of the approaches described above to directly test the proposed requirement for phosphorylation of Ser68 (to be referred to as Ser68phos; Yu et al., 2015) or ubiquitination of Lys124 (i.e., Lys124ub; Niikura et al., 2015) in long-term centromere maintenance or centromere establishment.
Results & Discussion
We first tested if Ser68phos or Lys124ub were required for maintenance of centromere identity and function in the short- or long-term. To do this (see schematic in Fig. 1A), we exploited diploid human (RPE-1) cells which we had engineered to carry one disrupted and one “floxed” CENP-A allele (CENP-A−/F), the latter of which could be inactivated by action of Cre recombinase (Fachinetti et al., 2013). CENP-A rescue gene constructs encoding EYFP fused to the amino-terminus of wild type CENP-A, CENP-AS68Q, or CENP-AK124R were stably expressed by retroviral integration (Fig. 1A, B). [We note that while the choice of substitution of S68Q to maintain a residue found at this position in bulk histone H3 is not likely to affect the structure and stability of the nucleosomes into which the mutant assembles, substitution to a residue not found at that position in canonical or variant histones, such as the S68E mutation, may disrupt normal nucleosome structure and/or stability. Therefore, replacement experiments with an acidic mutant would test only whether chronic negative charge at this position is tolerated, a very different question than testing if transient CENP-A phosphorylation is essential to CENP-A function.]
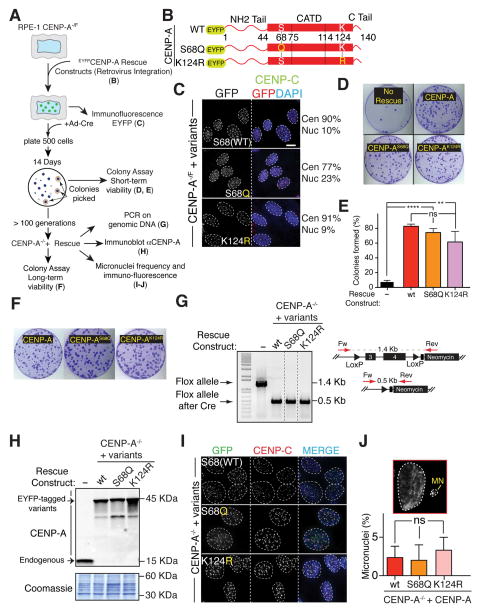
Figure 1. CENP-A post-translational modifications on Ser68 and Lys124 are not required for long-term centromere maintenance and function.
(A) Schematic of the experiments described in B–J.
(B) Schematic representing the different CENP-A rescue constructs amino terminally-tagged with EYFP (enhanced yellow fluorescent protein). Yellow letters highlight the amino acid substitutions used in each of the indicated constructs. Amino acids positions are also indicated.
(C) Representative immunofluorescence shows centromeric and/or nuclear localization of the of EYFP-rescue constructs for the indicated CENP-A−/F cell lines. The percentage of their intensities (>25 centromeres/nuclear signal) is also indicated. Unpaired t test for all variants: **** p < 0.0001. Yellow letters highlight the amino acid substitution used in each of the indicated constructs. Scale bar = 5 μm.
(D) Representative images of clonogenic survival experiments for the indicated cell lines from the colony formation assay in Panel A (short-term survival).
(E) Bar graphs represent quantitation of colony survival of the experiments in Panel D. Error bars represent the SEM of four independent experiments. Unpaired t test: **** p < 0.0001, ** p = 0.0098.
(F) Images of representative crystal violet–stained colonies from the colony formation assay in Panel A (long-term survival). Cells were grown for about 3 months and successively tested for the colony assay formation in the complete absence of the endogenous CENP-A.
(G) PCR analysis on DNA extracted from the surviving colonies in the CENP-A−/− cells expressing the different rescue constructs. Schematic of the PCR strategy is also shown.
(H) Immunoblots of cell extracts with antibodies against CENP-A to determine the level of expression of the indicated rescue constructs (45 kDa) and the absence of the endogenous CENP-A protein (15 kDa) in the CENP-A−/− cell lines. The corresponding coomassie gel is also shown as a loading control.
(I) Representative immunofluorescence shows localization of the of EYFP-tagged rescue constructs for the indicated CENP-A−/− cell lines. CENP-C staining is also shown. Scale bar = 5 μm.
(J) Quantification of micronuclei frequency in the indicated cell lines. Bars represent the mean of >100 cells per condition. Error bars represent the SEM of three independent experiments. A representative image is also shown.
CENP-A mutants that cannot be phosphorylated on Serine 68 (CENP-AS68Q) or ubiquitinated on Lysine 124 (CENP-AK124R) both localized to centromeres in a similar manner as wild type CENP-A (Fig. 1C). The remaining “floxed” CENP-A allele was then inactivated by expression of Cre recombinase. Short-term viability was determined by colony formation 14 d after inactivation of the remaining endogenous CENP-A allele. The CENP-AS68Q and CENP-AK124R mutants yielded a comparable number of “rescued” colonies visualized 14 d after inactivation of the remaining endogenous CENP-A gene (Fig. 1D, E). Hence, survival could not be the result of selection for a rare mutation that permitted cell viability. Thus, despite the claims derived exclusively from evidence using transient overexpression of either CENP-A mutant by Yu et al., 2015 and Niikura et al., 2015 for the essentialness of Ser68 phosphorylation or Lys124 ubiquitination for recognition and loading by HJURP, neither centromeric localization nor short-term viability (up to 14 generations) was affected by mutations that completely blocked Ser68 phosphorylation or Lys124 ubiquitination (Figs. 1C–E).
Furthermore, since CENP-A is a long-lived protein (Shelby et al., 2000) and persists at centromeres (Bodor et al., 2013; Falk et al., 2015; Jansen et al., 2007; Fachinetti et al., 2013), the ability of the phosphorylation and ubiquitination-deficient CENP-A mutants to maintain long-term centromere function in the complete absence of endogenous CENP-A was assessed by scoring continued viability of the individual colonies from the short-term assay for growth for >100 generations. CENP-A mutants that prevented constitutive phosphorylation of Ser68 (CENP-AS68Q) or ubiquitination of Lys124 (CENP-AK124R) produced a comparable number and size of colonies as did wild type CENP-A (Fig. 1F). Excision of the floxed allele and the lack of any detectable endogenous CENP-A was confirmed after >100 cell generations by PCR and immunoblot (Fig. 1G, H). Each non-phosphorylatable or non-ubiquitinatable mutant localized to centromeres (Fig. 1I). In addition, there was no detectable increase in micronuclei formation with any of the mutant versions that supported long-term viability (Fig. 1J), indicating that mitotic centromere function is similarly maintained by each of the CENP-A mutants compared with unmodified CENP-A. Altogether, our evidence demonstrates that these CENP-A mutants retain full capability for incorporation into centromeric chromatin and conferring and maintaining long-term centromere function.
We next investigated if the non-phosphorylatable or non-ubiquitinatable CENP-A mutants (CENP-AS68Q and CENP-AK124R) are able to maintain centromere function and identity when expressed from the endogenous CENP-A locus. The advantage of this system over the previous gene replacement strategy is that CENP-A expression is kept under its endogenous promoter, which is known to be subjected to cell-cycle regulation (Shelby et al., 1997). To this end (Fig. 2A), we utilized CRISPR/Cas9 technology to replace both endogenous CENP-A alleles such that one allele encodes for EGFP-tagged wild type CENP-A with an auxin inducible degron tag (which allows for its rapid degradation), and the other allele encodes for SNAP-tagged CENP-A (wild type or mutant). The SNAP-tagged CENP-A proteins expressed from the endogenous CENP-A promoter include wild type CENP-A, CENP-AS68Q, CENP-AK124R, and CENP-Aα2.2 (Fig. 2B), the last of which harbors three amino acid substitutions that convert CENP-A to the residues found at the same positions in canonical H3 and which prevent targeting to centromeres (Bassett et al., 2012). All cell lines were generated in the diploid human DLD-1 cell line that stably expresses the F-box protein TIR1-9xMyc (Holland et al., 2012), allowing the inducible degradation of AID-tagged proteins upon addition of the auxin hormone indole-3-acetic acid (IAA).
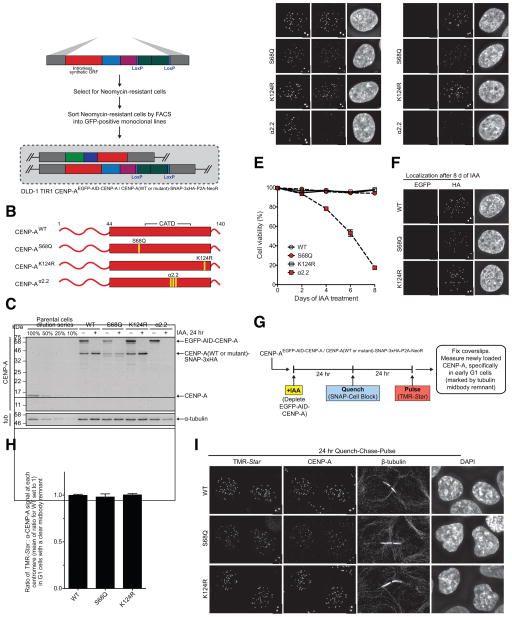
Figure 2. Expression of CENP-AS68Q or CENP-AK124R from the endogenous CENP-A locus is sufficient for centromere function, maintenance, and propagation in the early part of the G1 phase of the cell cycle. See also Figure S1.
(A) Schematic of the biallelic gene replacement approach used to replace the endogenous CENP-A gene with EGFP-AID-tagged CENP-A on one allele and CENP-A (wild type or mutant) tagged with SNAP-3xHA-P2A-NeoR on the other allele in DLD-1 TIR-1 cells via CRISPR/Cas9-mediated gene editing.
(B) Schematic showing the indicated mutants of CENP-A tagged with SNAP-3xHA-P2A-NeoR, which replace the endogenous CENP-A gene on one allele, as indicated in Panel A.
(C) Immunoblot of whole cell lysates from each of the indicated cell lines. Relevant cell lines were treated with 500 μM IAA for 24 hr to degrade EGFP-AID-tagged CENP-A. The blot was probed with anti-CENP-A and anti-tubulin antibodies.
(D) Representative images showing localization of EGFP-AID-tagged CENP-A and CENP-A(wild type or mutant)-SNAP-3xHA at centromeres. Upon treatment with 500 μM IAA for 24 hr, the EGFP-AID-tagged CENP-A is no longer detected.
(E) Quantification of the percentage of viable cells in the indicated cell lines upon treatment with 500 μM IAA for 8 d. Every 2 d, cells were collected and stained with Trypan Blue and counted on a hemocytometer to calculate the percentage of viable cells based on Trypan Blue uptake. Mean +/− SEM is shown for each time point.
(F) Representative images of the indicated cell lines after 8 d of treatment with 500 μM IAA. The SNAP-3xHA-tagged CENP-A mutants are still present at endogenous centromeres.
(G) Schematic for the quench-chase-pulse experiment in which the existing pool of CENP-A is quenched with SNAP-Cell Block, new CENP-A is synthesized, and newly loaded CENP-A is labeled with TMR-Star 24 hr later.
(H) Quantification of the quench-chase-pulse experiment in which TMR-Star and total CENP-A signals are measured at centromeres in G1 cells (marked by a tubulin midbody remnant). Mean +/− SEM is shown.
(I) Representative images showing that TMR-Star-labeled CENP-A is loaded at centromeres for each of the cell lines. The tubulin midbody remnant is shown between daughter G1 cells. Cells in which TMR-Star-labeled CENP-A is not detected at centromeres are shown in each representative image. Scale bar: 5 μm. Insets show magnification of the boxed region.
We first assessed the expression and localization of the tagged CENP-A histones by immunoblot and immunofluorescence microscopy (Fig. 2C, D). The CENP-AWT, CENP-AS68Q, and CENP-AK124R cell lines all expressed EGFP-AID-tagged CENP-A and the relevant SNAP-tagged CENP-A mutant (Fig. 2C) and localized to endogenous centromeres (Fig. 2D). Upon treatment with IAA for 24 hr, the AID-tagged CENP-A was degraded in all cell lines and no longer detected by immunoblot or immunofluorescence microscopy (Fig. 2C, D). Before and after IAA treatment, SNAP-tagged CENP-AS68Q levels were slightly lower, in terms of total protein (Fig. 2C) and centromere accumulation (Fig. 2D), than for CENP-AWT and CENP-AK124R. The CENP-Aα2.2 cell line also expressed IAA-sensitive EGFP-AID-CENP-A (Fig. 2C, D), but SNAP-tagged CENP-Aα2.2 was not detected by either immunoblot or immunofluorescence microscopy (Fig. 2C, D). Since the CENP-Aα2.2 mutant is not compatible with centromere targeting, (Bassett et al., 2012), we conclude that it fails to assemble into chromatin and is rapidly degraded. Thus, CENP-Aα2.2 serves as an effective null in our experiments.
Next, we assessed the ability of these cell lines to propagate in short-term culture upon depletion of EGFP-AID-tagged CENP-A. We treated cells with IAA for 8 d and quantified the percentage of viable cells during this time course. Cell lines expressing SNAP-tagged CENP-AWT, CENP-AS68Q, and CENP-AK124R were all viable, showing no significant change in the percentage of living cells (Fig. 2E). However, the cell line containing SNAP-tagged CENP-Aα2.2 showed significant cell death starting on day 2 and increasing until the end of the time course (Fig 2E). Even after 8 d of IAA treatment, both CENP-AS68Q and CENP-AK124R were targeted to centromeres (Fig. 2F), further confirming our findings with our approach using transgenes (Fig. 1).
We next performed a quench-chase-pulse chromatin assembly experiment (Jansen et al., 2007) to determine whether or not the mutant versions are efficiently assembled at centromeres during the G1 phase of the cell cycle (Fig. 2G). We treated cells with IAA to first deplete EGFP-AID-tagged CENP-A (Fig. S1A), quenched the existing pool of the SNAP-tagged CENP-A protein (Fig. S1B), and then chased the cells to allow for new CENP-A synthesis and deposition. TMR-Star labeling then exclusively occurred on newly synthesized CENP-A, and we measured its abundance relative to the total CENP-A pool at centromeres in early G1 cells identified by the presence of a clearly defined tubulin-positive midbody remnant (Fig. 2G). We found that both SNAP-tagged CENP-AS68Q and CENP-AK124R mutants assembled at centromeres similarly to wild type CENP-A (Fig. 2H, I). Thus, neither phosphorylation of Ser68 nor ubiquitylation of Lys124 are required for efficient centromere propagation. Additionally, we note that since the TMR-Star labeling is performed on living cells, there is no indication that fixation artifacts before staining could explain discrepancies between our findings and those published by either Yu et al., 2015 or Niikura et al., 2015.
We next employed our recently described approach (Logsdon et al., 2015; Fig. 3A) to assess the ability of CENP-A (wild type or mutant versions) or H3/CENP-A chimeric histones fused to the Lac repressor (LacI) to interact and recruit HJURP for nascent centromere chromatin assembly at an array of Lac operator (LacO) sequences present on chromosome 1 of U20S cells (Janicki et al., 2004). Wild type CENP-A fused to LacI robustly recruited HJURP in this assay. In contrast, HJURP was undetectable at LacO arrays bound by LacI-H3 (Fig. 3B, C). CENP-AS68Q yielded HJURP recruitment to LacO at an average level of about 1/4th that of unmodified CENP-A (Fig. 3B, C). HJURP was also recruited to about 1/4th the level of wild type LacI-CENP-A by LacI-H3CATD, a chimeric variant that localizes at centromeres (Black et al., 2004) and recognized by HJURP (Foltz et al., 2009) but which cannot nucleate successful kinetochore assembly (Fachinetti et al., 2013). These findings are in line with a model where CENP-AS68Q generates a less rigid ternary complex with H4 and HJURP, as observed with hydrogen/deuterium exchange experiments with purified components (Logsdon et al., 2015), and that this, in turn, does not allow CENP-AS68Q to stabilize HJURP at the LacO array to the same extent as does LacI-CENP-A (Fig. 3B, C).
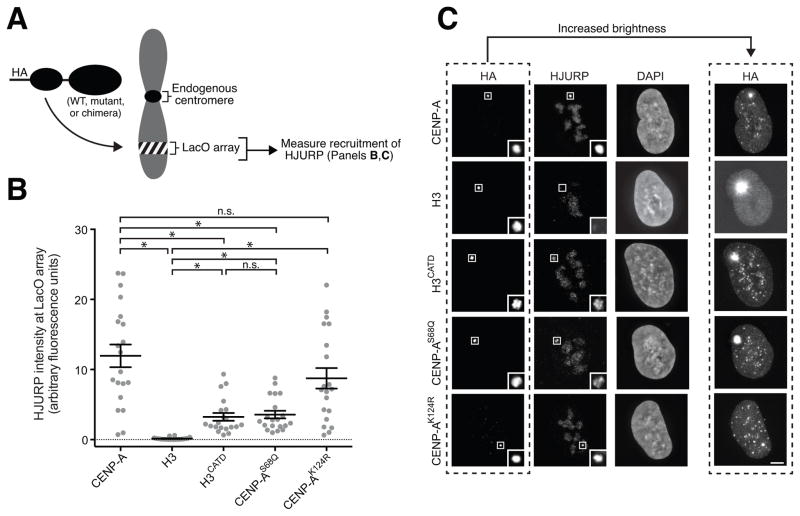
Figure 3. LacI-fused versions of CENP-A harboring mutations at Ser68 or Lys124 recruit HJURP to the LacO array to varying degrees, but each version efficiently assembles into nucleosomes at the centromere. See also Figure S2.
(A) Schematic of the experiments described in Panels B, C where either wild type CENP-A (as depicted), a mutant version of CENP-A, or an H3/CENP-A chimeric histone is used to assess HJURP recruitment to a stably integrated LacO array.
(B) Quantitation of HJURP fluorescence intensity at the LacO array for the indicated HA-LacI-fused histone. Each data point represents the intensity measured at the LacO array within a single cell with the mean +/− SEM shown for each chimera. An asterisk denotes significant differences between the means of two data sets (*, p < 0.05), with others marked as not significant (n.s.).
(C) Representative immunofluorescence images of HJURP recruitment to the LacO array by the indicated HA-LacI-fused histone. Scale bar: 5 μm. Insets show magnification of the boxed region.
Regarding an essential role for Lys124ub for the interaction of CENP-A with HJURP (Niikura et al., 2015), our findings are to the contrary. CENP-AK124R produced no significant change in its ability to recruit endogenous HJURP (Fig. 3B, C). Furthermore, LacI-fused CENP-AS68Q and CENP-AK124R, as well as LacI-H3CATD, were primarily and strongly localized to the LacO array but were also found at centromeres (Fig. 3C; see anti-HA immunofluorescence images with increased brightness). Thus, even in the absence of selection, CENP-A mutants without Ser68 phosphorylation or Lys124 ubiquitination continue to interact with endogenous HJURP.
We next used the LacI tagging and LacO array assay to test the ability of LacI-fused CENP-A mutants to mediate early steps in establishing a centromere, namely the recruitment of endogenous proteins of the constitutive centromere associated network (CCAN), including CENP-C and CENP-T (Fig. S2A–D). We have previously used this assay to establish that the N-terminal 14 amino acid residues, the CATD, and the C-terminal 6 amino acids of CENP-A are required in an H3 chimera to enable early steps in centromere establishment (Logsdon et al., 2015). This histone H3 chimera recruits CENP-C and CENP-T to the same extent as does wild type CENP-A at a time point before any detectable endogenous CENP-A is recruited to the LacO array (Logsdon et al., 2015). Likewise, a CATD-containing H3 chimera with a Q68S mutation recruits the same level of HJURP to the LacO array as does wild type CENP-A without co-recruiting any endogenous CENP-A (Logsdon et al., 2015). Thus, early centromere establishment steps in our assay are most simply attributed to the LacI-fused CENP-A protein we introduce instead of endogenous CENP-A that would be recruited in later cell cycles. Using this assay, LacI-fused mutant versions of CENP-A (S68Q and K124R) were determined to recruit both CENP-C (Fig. S2A, B) and CENP-T (Fig. S2C, D) to extents similar to a LacI fusion to wild type CENP-A. Even though LacI-fused CENP-AS68Q has a lower steady state accumulation of HJURP at the LacO array than wild type CENP-A (Fig. 3B, C), this mutant efficiently assembles centromere chromatin that supports normal recruitment of both CENP-C and CENP-T. Our results are consistent with a higher dissociation rate of HJURP from the LacO-tethered CENP-AS68Q relative to wild type CENP-A. While it is possible that a decrease in the kinetics of association with HJURP could be detrimental in some contexts by destabilizing a limited pool of CENP-A (e.g., in cell types with exceptionally long G2 phases), it is inconsequential in typically used cultured cell examples (Figs. 1–3). All versions of CENP-A that we tested are expressed at similar levels and only generate a slight (~35%) excess of total CENP-A levels in the cells in which they are expressed (Fig. S2E–H). Thus, neither Ser68phos nor Lys124ub are required for efficiently mediating the first steps in centromere establishment. We also note that our results with CENP-AK124R are inconsistent with an important role for the acetylation that was detected by others (Bui et al., 2012) at this site.
Our evidence has established that, in presence or complete absence of endogenous CENP-A, CENP-A mutants expressed from an endogenous CENP-A locus and which cannot be phosphorylated on Ser68 (CENP-AS68Q) or ubiquitinated at Lys 124 (CENP-AK124R) are: 1) recognized by HJURP, 2) incorporated into centromeric chromatin, 3) retain normal cell cycle-dependent deposition, presumably mediated by HJURP, 4) capable of driving early events in establishing new centromere identity, and 5) sufficient for maintaining human centromeres in the short and long-term that function with high fidelity. We also note that our results with CENP-AK124R are inconsistent with an important role for the acetylation that was detected by others (Bui et al., 2012) at this site. While it is possible that these modifications do occur in some contexts, our evidence refutes the central claims of Yu et al., 2015 and Niikura et al., 2015 that either of these modifications plays an essential role in generating or maintaining functional centromeric chromatin. Specifically to the claims of Yu et al., 2015, we note that Ser68 is not among the CENP-A-specific residues that we found are required for centromere establishment (Logsdon et al., 2015), identity (Fachinetti et al., 2013), long-term function (Fachinetti et al., 2013), or access into the HJURP-mediated pathway for centromeric chromatin assembly (Bassett et al., 2012; Foltz et al., 2009; Logsdon et al., 2015).
Experimental Procedures
Cell culture
Cells were maintained at 37°C in a 5% CO2 atmosphere. hTERT RPE-1 cells were maintained in DMEM:F12 medium containing 10% tetracycline-free fetal bovine serum (Clontech), 0.348% sodium bicarbonate, 100 U/ml penicillin, 100 U/ml streptomycin and 2 mM L-glutamine. DLD-1 TIR1 cells and U2OS-LacO cells (a gift from S. Janicki, Wistar Institute, Philadelphia, PA) were maintained in DMEM medium containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. DLD-1 TIR1 stable cell lines were maintained in media listed above supplemented with 750 μg/mL G418-S and 2 μg/mL Puromycin.
Constructs
For plasmids used in the generation of RPE-1 stable cell lines, full-length of the human CENP-A open reading frame tagged with EYFP at the amino-terminal or CENP-A mutants were cloned into a pBabe-based vector for retrovirus generation. CENP-A mutations were generated via the Gibson assembly technique. For plasmids used in the generation of the DLD-1 TIR1 cell lines, EGFP-AID-CENP-A and CENP-A(WT)-SNAP-3xHA-P2A-NeoR repair templates were constructed using NEBuilder HIFI DNA Assembly Master Mix (NEB E2621). Briefly, pUC19 was digested with EcoRI and HindIII. The 5′UTR and 3′UTR CENP-A gene regions (~800 bp each) were PCR-amplified from DLD-1 TIR1 genomic DNA. EGFP was PCR-amplified from a derivative of pBabePuro-LAP-CENP-N (Foltz et al., 2006), and AID was PCR-amplified from pcDNA5-FRT-TO-H2B-AID-YFP (Holland et al, PNAS, 2012). The intronless CENP-A gene was designed using IDT’s codon optimization tool, which chooses codons with a bias similar to the natural bias in the human genome, and then ordered as a gBlock gene fragment (IDT). SNAP-3xHA-P2A-NeoR was also ordered as a gBlock gene fragment. The 5′UTR, EGFP, AID, intronless CENP-A, 3′UTR, and pUC19 fragments were assembled with the NEBuilder HIFI DNA Assembly Master Mix. Similarly, the 5′UTR, codon-optimized CENP-A, SNAP-3xHA-P2A-NeoR, 3′UTR, and pUC19 fragments were also assembled using NEBuilder HIFI DNA Assembly Master Mix. To generate the indicated versions of CENP-A-SNAP-3xHA-P2A-NeoR repair templates, QuikChange site-directed mutagenesis (Agilent) was performed on CENP-A(WT)-SNAP-3xHA-P2A-NeoR to convert S68 to a Q, K124 to an R, and AEAFLVH to CEAYLVG (spanning amino acids 98-104 in CENP-A) to generate the α2.2 mutant. The sgRNA/Cas9 plasmids targeting the 5′UTR and 3′UTR of the CENP-A gene were constructed by annealing oligos and then ligating them into pX330 (Cong et al., 2013) at the BbsI cut sites. For the 5′ UTR CENP-A sgRNA, the following oligos were annealed: 5′-CACCGgtgtcatgggcccgcgccgc -3′ and 5′-AAACgcggcgcgggcccatgacacC -3′. For the 3′ UTR CENP-A sgRNA, the following oligos were annealed: 5′-CACCGctgacagaaacactgggtgc-3′ and 5′-AAACgcacccagtgtttctgtcagC-3′. All LacI-fusion plasmids were constructed in a pcDNA3.1 vector containing a hemagglutinin (HA) tag and LacI tag fused to the N-terminus of the CENP-A, H3, H3/CENP-A chimera, or CENP-A mutant histone. Construction of pcDNA3.1-HA-LacI-CENP-A, -H3CATD, and -H3 were performed as described previously (Logsdon et al., 2015). To construct pcDNA3.1-HA-LacI-CENP-AS68Q, -CENP-AS68A, -CENP-AS68D, and -CENP-AK124R, PCR site-directed mutagenesis using Quikchange (Agilent Technologies, Inc.) was performed on pcDNA3.1-HA-LacI-CENP-A. All plasmids were verified by sequencing.
Generation of stable cell lines
The CENP-A transgenes used in this study were introduced into RPE-1 cells by retroviral delivery as described previously (Shah et al., 2004). Stable integration was selected with 10 μg/ml blasticidin S and single clones were isolated using fluorescence activated cell sorting (FACS Vantage; Becton Dickinson, Franklin Lakes, NJ). To generate DLD-1 TIR1 stable cell lines in which both alleles of CENP-A were replaced, 400 ng of each repair template (EGFP-AID-CENP-A plasmid and CENP-A(WT or mutant)-SNAP-3xHA-P2A-NeoR plasmid) and 100 ng of each sgRNA/Cas9 plasmid (5′UTR sgRNA/Cas9 plasmid and 3′UTR sgRNA/Cas9 plasmid) were co-transfected into DLD-1 TIR1 cells (Holland et al., 2012) using Lipofectamine 2000 (Invitrogen). Five days after transfection, 750 μg/mL G418-S was added, and cells were cultured with G418-S for 2–3 weeks. GFP-positive cells were FACS-sorted into monoclonal lines in 96-well plates, and surviving clones were assessed by immunofluorescence microscopy. Monoclonal cell lines of the indicated genotypes were verified by PCR and sequencing of genomic DNA.
Immunoblotting
Whole cell lysates were collected from the described cell lines after the indicated cell generations and/or with the indicated treatments. For immunoblot analysis, protein samples were separated by SDS-PAGE, transferred onto nitrocellulose membranes (BioRad), blocked with 5% milk, and then probed with the following antibodies: GFP (Cell Signaling, 1:1000), α-tubulin (Clone DM1A, 1:5000), CENP-A (Cell Signaling #2186, 1:1000), and ACA (Antibodies Incorporated, 2 μg/mL).
Clonogenic colony assay and Adeno(Ad)-Cre treatment
4 × 104 cells were plated in a 12-well plate. The next day, cells were washed 3 times in DMEM:F12 medium containing 2% fetal bovine serum. Ad-Cre virus was added at MOI 250 in 400 μL of DMEM:F12 medium containing 2% fetal bovine serum. After 3.5 hr, cells were washed 3 times with DMEM:F12 medium containing 10% fetal bovine serum. After 2 d, 500 cells were plated in triplicate on a 10 cm2 dish. After 14 d, colonies were fixed 10 min in methanol and stained for 10 min using a crystal violet staining solution (1% crystal violet, 20% EtOH). For long-term survival, after 14 d of Ad-Cre infection individual colonies were collected by colony picking, screened for the presence of endogenous CENP-A via PCR and western blot, and the selected clones were grown for 3 months (> 100 generations).
Cell viability assay
The DLD-1 TIR1 stable cell lines were seeded at 1.5 × 105 cells/well in triplicate in 6-well plates, and 500 μM IAA was added every other day. Cells were collected at the indicated time points, stained with Trypan Blue (Corning), and counted on a hemocytometer to calculate the percentage of viable cells out of total cells based on Trypan Blue uptake.
SNAP labeling experiments
The DLD-1 TIR1 stable cell lines were seeded at 6 × 104 cells/well onto poly-lysine-coated coverslips in a 24-well plate in growth medium supplemented with 500 μM IAA. 24 hr after seeding, cells were quenched with 10 μM SNAP-Cell Block (NEB, S9106S) for 30 min at 37°C, washed several times with warm growth medium, and incubated in growth medium supplemented with IAA for 2 hr to allow excess block to diffuse out of the cells. One coverslip was labeled with 2 μM SNAP-Cell TMR-Star (NEB, S9105S) for 30 min at 37°C, washed several times with warm growth medium, incubated in the growth medium supplemented with IAA for 2 hr, washed with warm growth medium again, and fixed in 4% formaldehyde to ensure that the block had gone to completion (t=0 time point). 24 hr after block, the remaining coverslips were labeled with 2 μM SNAP-Cell TMR-Star for 30 min at 37°C, washed several times with warm growth medium, incubated in the growth medium supplemented with IAA for 2 hr, washed with warm growth medium again, and fixed in 4% formaldehyde (t=24 hr time point). Coverslips were stained with the indicated antibodies and imaged with a 100x objective. All images were deconvolved, and CRaQ analysis (Bodor et al., 2012) was performed to quantify the intensity of TMR-Star relative to the intensity of total CENP-A at centromeres.
Indirect immunofluorescence
For experiments involving hTERT RPE-1 stable cell lines, cells were fixed in 4% formaldehyde at room temperature. Incubations with primary antibodies were conducted in blocking buffer for 1 hr at room temperature using the following antibodies: CENP-A (Abcam, 1:1500), CENP-C (MBL, 1:1000). For experiments involving the DLD-1 TIR1 stable cell lines, cells were fixed in 4% formaldehyde in PBS for 10 mins, permeabilized with 0.5% Triton X-100 in PBS for 5 min, and then washed in 0.1% Tween in PBS for 5 min 3 times prior to blocking and antibody incubation. Cells were blocked in 2% FBS, 2% BSA, and 0.1% Tween in PBS, and then incubated with primary and secondary antibodies. An anti-HA.11 mouse monoclonal antibody (16B12; Covance) and CENP-A (3–19) mouse monoclonal antibody (ADI-KAM-CC006-E; Enzo) were used at 1 μg/mL. β-tubulin rabbit monoclonal antibody conjugated to AlexaFluor488 (9F3; Cell Signaling; #3623) was used at 1:100 dilution. FITC- and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used at a 1:200 dilution. For experiments involving LacI-fusion proteins, U2OS-LacO cells were transfected with vectors expressing HA-LacI-CENP-A, -H3, -H3/CENP-A, or -CENP-A mutant using FuGENE6 (Roche) and processed for indirect immunofluorescence 48 hr later. Immunofluorescence experiments were performed as described previously (Logsdon et al., 2015). For experiments involving HJURP immunofluorescence, cells were pre-extracted with 0.5% Triton X-100 in PBS for 3 min, fixed with 4% formaldehyde in PBS for 10 min, and then washed with 0.1% Tween in PBS for 5 min. For experiments involving CENP-C and CENP-T immunofluorescence, cells were fixed in 4% formaldehyde in PBS for 10 min, extracted with 0.5% Triton X-100 in PBS for 5 min, and then washed in 0.1% Tween in PBS for 5 min 3 times prior to blocking and antibody incubation. All cells were blocked in 2% FBS, 2% BSA, and 0.1% Tween in PBS, and then incubated with primary and secondary antibodies. An anti-HA.11 mouse monoclonal antibody (HA.11; Covance; 16B12), anti-HJURP rabbit polyclonal antibody (Bassett et al., 2012), and anti-CENP-T rabbit polyclonal antibody (a gift from I. Cheeseman, Whitehead Institute and Massachusetts Institute of Technology, Cambridge, MA) was used at 1 μg/mL. Affinity-purified anti-CENP-C rabbit polyclonal antibody (Bassett et al., 2012) was used at 0.75 μg/mL. FITC-, Cy3-, and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used at a 1:200 dilution. Cells were DAPI-stained and mounted with Vectashield mounting medium (Vector Laboratories, Inc.) onto glass slides prior to imaging.
Image acquisition, quantification, and micronuclei assay
For experiments in hTERT RPE-1 cell lines, immunofluorescence images were collected using a Deltavision Core system (Applied Precision). Quantification of centromere signal intensity on interphase cells was done manually as described (Fachinetti et al., 2015). In summary, un-deconvolved 2D maximum intensity projections were saved as un-scaled 16-bit TIFF images and signal intensities determined using MetaMorph (Molecular Devices). A 10 × 10 pixel circle was drawn around a centromere and an identical circle drawn adjacent to the structure (nuclear). The integrated signal intensity of each individual centromere and nuclear signal was calculated by subtracting the fluorescence intensity of the background (measured outside the nucleus). To measure the number of micronuclei, cells were plated on a 12-well glass slide, fixed with formaldehyde and stained with DAPI. The frequency of the number of micronuclei per cell versus the total number of cells was then measured. For DLD-1 TIR1 and U2OS-LacO experiments, cells were imaged on an inverted fluorescence microscope (Leica DMI6000B) equipped with a charge-coupled device camera (Hamamatsu Photonics ORCA AG) and a 100x 1.4 NA objective lens. For the U2OS-LacO experiments, HA-LacI fusion protein expression varies from cell to cell for each transfection, as expected; cells with moderate expression were chosen, while cells with very high expression were specifically excluded. Furthermore, to ensure that none of the differences in centromere establishment behavior of the histones reported in this study are caused by wide variations in copy number of HA-LacI fusion proteins at the LacO array, all HA-LacI proteins in this study expressed to high enough levels to have many cells in the moderate expression range (1,000–3,000 maximal pixel intensity in a scale of 0–4,095 for a 50-ms exposure in the channel used for anti-HA detection). At least 20 individual cells were imaged for each individual experiment, with experiments repeated multiple times in all cases. Data are shown from one or more experiments that are representative of two or more independent experimental repeats. Images were acquired in 0.2 μm z-sections and deconvolved using LAS software (Leica). Images were max-projected into single two-dimensional images using ImageJ (Version 1.46r). Deconvolved and max-projected images were used for all fluorescence intensity measurements. The fluorescence intensity at the LacO array was measured by placing a 20 × 20 pixel box around the LacO array (as indicated by the HA signal) and measuring the total pixel intensity within the box in the channel of the relevant centromere or kinetochore protein. The mean fluorescence intensity at centromeres was measured using an ImageJ macro, CRaQ (Bodor et al., 2012), in which a 7 × 7 pixel box was placed around the centroid position of each centromere and the total pixel intensity within the box was measured and averaged over the total number of centromeres in each cell. In both cases, a mean background fluorescence intensity was measured and subtracted from the LacO array fluorescence intensity and mean centromeric fluorescence intensity. Mean background intensity was measured at nuclear sites that were not coincident with the LacO array or centromeres. In the case of HJURP, which does not localize to centromeres during most of the cell cycle, the intensity at the LacO array was measured by summing the total pixel intensity in a 20 × 20 pixel box and subtracting the mean background intensity in the nucleoplasm.
Supplementary Material
Highlights.
CENP-A gene replacement demonstrates Ser68phos and Lys124ub are dispensable
Ser68phos and Lys124ub are not required to maintain centromeres indefinitely
CENP-A without Ser68phos or Lys124ub mediates centromeric chromatin assembly in G1
Ser68phos and Lys124ub are dispensable for early steps in centromere establishment
Acknowledgments
We thank S. Janicki (Wistar) and I. Cheeseman (MIT) for reagents, V. Barra (I. Curie) for technical help, and L. Guo and K. Haislop (UPenn) for helping develop new gene editing strategies. This work was supported by National Institutes of Health Grants R01-GM074150 (D.W.C.) and R01-GM082989 (B.E.B), T32-GM007229 (UPenn Cell and Molecular Biology Training Grant; G.A.L) and by Labex ≪CelTisPhyBio≫, the Institut Curie and the ATIP-Avenir 2015 program (D.F.). D.W.C. receives salary support from the Ludwig Institute for Cancer Research, D.F. from the ATIP-Avenir 2015 program. The authors declare no competing financial interests.
Footnotes
Author Contributions
D.F. and A.A. performed experiments in Fig. 1. G.A.L. performed experiments in Figs. 2, 3, S1, and S2. E.B.S. developed essential tools for CRISPR/Cas9-mediated gene editing. D.F., G.A.L., B.E.B., and D.W.C. conceived the experimental design, analyzed the data, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bade D, Pauleau AL, Wendler A, Erhardt S. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev Cell. 2014;28:508–519. doi: 10.1016/j.devcel.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci. 2013;110:11827–11832. doi: 10.1073/pnas.1300325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AO, Panchenko T, Shabanowitz J, Lehman SM, Bai DL, Hunt DF, Black BE, Foltz DR. Identification of the post-translational modifications present in centromeric chromatin. Mol Cell Proteomics MCP. 2016;15:918–931. doi: 10.1074/mcp.M115.053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell. 2012;22:749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LET, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Bodor DL, Rodríguez MG, Moreno N, Jansen LET. Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. In: Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM, editors. Current Protocols in Cell Biology. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2012. [DOI] [PubMed] [Google Scholar]
- Bodor DL, Valente LP, Mata JF, Black BE, Jansen LET. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell. 2013;24:923–932. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M, Dimitriadis EK, Hoischen C, An E, Quénet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP Is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Eichler EE. Repetitive conundrums of centromere structure and function. Hum Mol Genet. 1999;8:151–155. doi: 10.1093/hmg/8.2.151. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LET, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev Cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Duyne GDV, Vinogradov SA, et al. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348:699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, Cleveland DW. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci. 2012;109:E3350–E3357. doi: 10.1073/pnas.1216880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of Holliday Junction–Recognizing Protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- Logsdon GA, Barrey EJ, Bassett EA, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, Black BE. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J Cell Biol. 2015;208:521–531. doi: 10.1083/jcb.201412011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. Polo-like Kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3/CENP-A loading. Cell Rep. 2014;8:190–203. doi: 10.1016/j.celrep.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Niikura Y, Kitagawa R, Ogi H, Abdulle R, Pagala V, Kitagawa K. CENP-A K124 ubiquitylation is required for CENP-A deposition at the centromere. Dev Cell. 2015;32:589–603. doi: 10.1016/j.devcel.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of Centromere and Kinetochore Proteins. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores Is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MCC, Bodor DL, Stellfox ME, Martins NMC, Hochegger H, Foltz DR, Jansen LET. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, et al. Mitotic regulator Mis18β interacts with and specifies the centromeric assembly of molecular chaperone Holliday Junction Recognition Protein (HJURP) J Biol Chem. 2014;289:8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Straight AF. The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, et al. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell. 2015;32:68–81. doi: 10.1016/j.devcel.2014.11.030. [DOI] [PubMed] [Google Scholar]
- Zeitlin SG, Barber CM, Allis CD, Sullivan KF. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J Cell Sci. 2001;114:653–661. doi: 10.1242/jcs.114.4.653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.