Abstract
Purpose of Review
Evidence has linked neuropsychiatric disorders with epigenetic marks as either a biomarker of disease, biomarker of exposure, or mechanism of disease processes. Neuropsychiatric epidemiologic studies using either target brain tissue or surrogate blood tissue each have methodological challenges and distinct advantages.
Recent findings
Brain tissue studies are challenged by small sample sizes of cases and controls, incomplete phenotyping, post-mortem timing, and cellular heterogeneity, but the use of a primary disease relevant tissue is critical. Blood-based studies have access to much larger sample sizes and more replication opportunities, as well as the potential for longitudinal measurements, both prior to onset and during the course of treatments. Yet, blood studies also are challenged by cell-type heterogeneity, and many question the validity of using peripheral tissues as a brain biomarker. Emerging evidence suggests that these limitations to blood-based epigenetic studies are surmountable, but confirmation in target tissue remains important.
Summary
Epigenetic mechanisms have the potential to help elucidate biology connecting experiential risk factors with neuropsychiatric disease manifestation. Cross-tissue studies as well as advanced epidemiologic methods should be employed to more effectively conduct neuropsychiatric epigenetic research.
Keywords: Neuropsychiatric disorders, DNA methylation, Epigenetics, Tissue, Blood
Introduction
The study of epigenetic variation, which is well established in cancer research and plant biology, has become increasingly integrated into the epidemiology and potential etiology of other common human diseases. Epigenetics, which refers to regulatory information not contained in the DNA sequence itself, includes DNA methylation and hydroxymethylation, histone modifications, chromatin structure, and some forms of RNA [1]. These marks are part of a gene expression regulation system that developmentally controls the spatial and temporal regulation of gene expression [2] and provides complementary information to the DNA sequence. Importantly, epigenetic marks can be dynamic, reversible, and susceptible to environmental insult or nutritional supply [3, 4]. This area of research has stimulated the fields of epidemiology and medicine as potential mechanisms mediating gene-environment interaction and for phenotype heterogeneity among genetic disorders [5]. Even in cases where epigenetics is not mechanistically related to disease, epigenetic marks may be an important biomarker of historic exposure [6], as has been shown with famine exposure during the Dutch Hunger Winter [7]. Alternatively, epigenetic marks may represent an early biomarker of pathogenesis that can be used to target early interventions with clinical applications [8]. In particular, neuropsychiatric disease research has been stimulated by the potential for epigenetic mechanisms to help elucidate the biology connecting experiential risk factors with psychiatric disease manifestation [9], in addition to the potential for understanding phenotype heterogeneity for a given genetic risk. The emerging field of neuroepigenetics has identified epigenetic underpinnings of learned behavior and central nervous system development [10]. Further, many psychiatric disorders are thought to be neurodevelopmental in origin, and given the role of epigenetic processes in cellular differentiation and development, the study of epigenetic variation may inform understanding of disease mechanisms and risk.
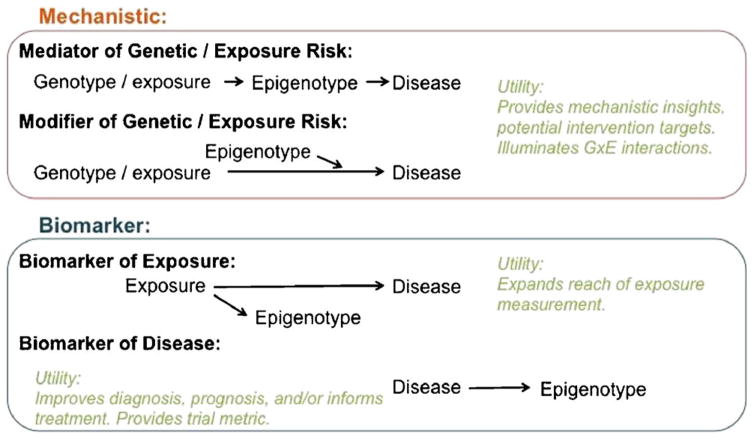
Given this potential, studies have pursued epigenetic measurement in epidemiologic and clinical studies, as summarized in recent reviews specific to autism spectrum disorder [11], bipolar disorder [12], schizophrenia [13, 14], post-traumatic stress disorder [15], substance abuse disorder [16], and other neuropsychiatric disorders. Epigenetic marks are being investigated for their potential mechanistic disease role as either a mediator or a modifier of environmental or genetic risk, or as a biomarker of exposure or disease (Fig. 1). These cutting-edge projects are also forging the path for what is and what is not feasible and fruitful in the epidemiology of neuropsychiatric disease. A major debate in this regard has been the utility of peripheral tissue samples, such as blood, for the study of disorders that primarily manifest in the brain, a challenge we refer to as the “tissue issue.” Peripheral tissues provide many opportunities for large sample sizes and multiple replication opportunities, while target-tissue brain studies are limited to post-mortem sampling and relatively small sample sizes. While epigenetics involves many potential mechanisms of gene regulation, including histone modification, miRNA expression, and chromatin remodeling, this article focuses on DNA methylation. DNA methylation is most frequently investigated in epidemiology because DNA is easy to collect and is already archived in large numbers, allowing researchers to leverage existing genetic epidemiology samples. In this article, we address the potential benefits of epigenetics research in neuropsychiatric disorders, the pragmatics as well as advantages and limitations of using brain vs peripheral tissues, the importance of sample timing, which is feasible in human studies, and suggest practical approaches moving forward.
Fig. 1.
Utility of epigenetic marks for neuropsychiatric disease research. Figure adapted from [83–85]
Utility of Brain Tissue
Epigenetic marks are critical features of cellular differentiation and cellular phenotype; therefore, epigenomic signatures are specific to types of tissues and cells. Neuropsychiatric disorders are primarily diseases of the brain; thus, brain samples are logically the most appropriate tissue source for epigenetic study. For this reason, many studies examining epigenetic marks for neuropsychiatric disease have focused on brain material (see Table 1 for examples). In fact, neuropsychiatric diseases may affect a specific subset of cells in a very specific region of the brain. It is not yet possible to perform in vivo epigenetic studies of the brain to capture information during the critical time points of development, especially in humans. Only post-mortem brain samples are available for epigenetic measurement, and even post-mortem brain samples for research are scarce for most developmental psychiatric disorders including autism, schizophrenia, and bipolar disorder and do not even exist for more common conditions like anxiety. Perhaps the biggest challenge in using brain tissues is in obtaining control tissue samples from an adequate number of unaffected individuals matched for factors such as age, sex, and exposures. The recent PsychENCODE project recognizes these issues and aims to compile a public resource of multi-layered “omic” and regulatory data on healthy and diseased human brains throughout different stages of development. Despite these efforts, the current availability and applicability of primary tissue for epigenetic analysis is limited for most neuropsychiatric disorders.
Table 1.
Examples of human genome-wide neuropsychiatric epigenetics publications using target (brain) or surrogate (blood) tissues with evidence for association.
| Approach | Source | Tissue | Neurospychiatric disorder | Sample | Methylation assay |
|---|---|---|---|---|---|
| Target | (Nagarajan et al. 2006) [86] | Prefrontal cortex | Autism spectrum disorders | 19 cases, 14 controls (males) | Candidate gene |
| Target | (Mill et al. 2008) [87] | Frontal cortex | Schizophrenia | 35 cases, 35 controls | Genome-wide (12,000 CpG islands) |
| Target | (Abdolmaleky et al. 2011) [88] | Frontal lobe | Bipolar and schizophrenia | 35 BPD, 35 SCZ, 35 control | Candidate gene (Bisulfite sequencing HTR2A) |
| Target | (James et al. 2013) [89] | Cerebellar cortex | Autism spectrum disorders | 13 cases, 13 controls | Candidate gene |
| Target | (Ladd-Acosta et al. 2014) [90] | Temporal cortex, prefrontal cortex, cerebellum | Autism spectrum disorders | 7 cases, 10 controls | Genome-wide (Illumina 450 k) |
| Target | (Zhu et al. 2014) [91] | Cerebellum, cerebral cortex | Autism spectrum disorders | 54 cases, 43 controls | Candidate gene |
| Target | (Nardone et al. 2014) [92] | Prefrontal cortex, cerebral cortex | Autism spectrum disorders | 12 cases, 12 controls | Genome-wide (Illumina 450 k) |
| Target | (Wockner et al. 2014) [93] | Frontal cortex | Schizophrenia | 24 cases, 24 controls | Genome-wide (Illumina 450 k) |
| Target | (Pidsley et al. 2014) [94] | Prefrontal cortex, matched cerebellum | Schizophrenia | 21 cases, 23 controls, 179 fetal sanokes | Genome-wide (Illumina 450 k) |
| Target | (Jaffe et al. 2016) [95•] | Dorsolateral prefrontal cortex | Schizophrenia | 191 cases, 35 fetal, 300 controls | Genome-wide (Illumina 450 k) |
| Target | (Hannon et al. 2016) [61] | Fetal whole brain | Schizophrenia | 173 fetal brain samples (testing for enrichment of meQTLs at SCZ genetic loci) | Genome-wide (Illumina 450 k) |
| Target | (Pandey et al. 2016) [96] | Prefontal cortex | Suicide | 52 suicide victims, 27 non-suicidal psychiatric, 24 normal controls | Gene and protein expression of SKA2 |
| Surrogate | (Nguyen et al. 2010) [38] | Lymphoblastoid cell line | Autism spectrum disorders | 3 discordant MZ twin pairs, 2 discordant sib pairs | Genome-wide (CpG island array 8.1 K) |
| Surrogate | (Carrard et al. 2011) [97] | Peripheral blood | Bipolar and schizophrenia | 58 BPD, 40 SCZ, 67 controls | Candidate gene (High resolution melt of 5HTR1A) |
| Surrogate | (Dempster et al. 2011) [98] | Peripheral blood | Schizophrenia | 22 discordant twin pairs | Genome-wide (Illumina 27 K) |
| Surrogate | (Rusiecki et al. 2013) [99] | Peripheral blood | PTSD | 75 cases, 75 controls | Candidate gene (Pyrosequencing n = 5) |
| Surrogate | (Nishioka et al. 2013) [100] | Peripheral blood | Schizophrenia | 18 first-episode cases, 15 controls | Genome-wide (Illumina 27 K array) |
| Surrogate | (Zhang et al. 2013) [101] | Peripheral blood | Alcohol dependence | 518 cases, 369 controls | Genome-wide (Illumina Golden Gate CpG island) |
| Surrogate | (Aldinger et al. 2013) [102] | Lymphoblastoid cell line | Autism spectrum disorders | Women, 4 cases, 6 controls (discovery), 13 cases, 13 controls (replication) | Genome-wide (Illumina 27 K) |
| Surrogate | (Wong et al. 2014) [103] | Peripheral blood | Autism spectrum disorders | 25 discordant MZ twin pairs | Genome-wide (Illumina 27 K) |
| Surrogate | (Dempster et al. 2014) [104] | Buccal cell | Adolescent depression | 18 Discordant MZ twin pairs | Genome-wide (Illumina 450 K) |
| Surrogate | (Fisher et al. 2015) [105] | Buccal cell | Childhood psychotic symptoms | 24 Discordant MZ twin pairs | Genome-wide (Illumina 450 K) |
| Surrogate | (Kang et al. 2015) [106] | Peripheral blood | Major or minor depressive disorder | 309 breast cancer patients | Candidate gene (Pyrosequencing of BDNF) |
| Surrogate | (Kim et al. 2016) [107] | Peripheral blood | Anxiety, depression, hostility | 538 community dwelling older men | Candidate gene (Pyrosequencing n = 7 immune genes) |
| Surrogate | (Kahl et al. 2016) [108] | Peripheral blood | Major depressive disorder | 52 cases, 18 controls, longitudinal 6 weeks | Candidate gene (Bisulfite sequencing n = 2) |
| Surrogate | (Montano et al. 2016) [109••] | Peripheral blood | Schizophrenia | Discovery: 689 cases, 645 controls; Replication: 247 cases, 250 controls | Genome-wide (Illumina 450 k) |
| Combined | (Gregory et al. 2009) [79] | Superior temporal gyrus, Peripheral blood | Autism spectrum disorders | 8 cases, 8 controls brain, 20 cases, 20 controls blood | Candidate gene |
| Combined | (Sabunciyan et al. 2012) [78] | Prefrontal cortex discovery, lymphoblastoid cell line follow-up | Major depressive disorder | 39 cases, 27 controls | Genome-wide (CHARM); Candidate gene (Pyrosequencing validation n = 17 DMRs, cell line n = 1 DMR) |
| Combined | (Walton et al. 2016) [65] | Filter by correlated sites in paired temporal lobe and blood, discovery in blood | Schizophrenia | 12 brain and blood, 111 cases, 122 controls | Genome-wide (Illumina 27 K) |
Challenges of Brain-Based Research
While brain tissue may at first glance appear to be the “go ld standard” for epigenetic analysis in neuropsychiatric disorders, the timing of acquisition, cell type, and sample sizes actually available present many limitations. First, there is limited information about the specific brain regions (and cell types) across developmental time points that are likely to be of primary importance in the pathogenesis of particular disorders. There are marked functional differences across brain regions, and these are reflected in epigenetic differences that impact gene function [17•]. Also, for some disorders such as autism and schizophrenia, connectivity between brain regions may be of primary importance, rather than specific regional states [18]. Therefore, an argument for tissue specificity should really be cast as an argument for brain region—and cell type—specificity, and current approaches to brain epigenetic measurement are only beginning to reach this granularity [19, 20]. With respect to cell type, a portion of epigenomic marks are known to be cell-type specific, and the brain is a heterogeneous mix of neurons and glia, including astrocytes, oligodendrocytes, and microglia. Thus, if individuals vary in the proportion of various cell types, this will result in DNA methylation differences when bulk sections of tissues are homogenized. Methods have been developed to deconvolute brain tissue DNA methylation data into coarse cell-type proportions (neuron vs glia) [21, 22], helping in part to address this source of confounding. Further, emerging research shows that single neurons or subtypes may have highly specific transcriptional profiles [23, 24] as well as individual DNA sequences from accumulated damage during active transcription [25], potentially reflecting developmental lineage and function [26] or memory [27, 28]. This implies that epigenetic measurement from aggregated cells even of the same type may not be fully informative. Currently, microdissection or sorting of brain tissue into pure cell populations, or further into single cells, is challenging and not practical for large-scale studies, though new technical developments may increase the feasibility across populations. In addition, epigenetic marks are analyzed in one brain region at a time (see Table 1), and efforts to map as well as match epigenetic patterns across the brain with particular behaviors have not been possible.
Second, the nature of post-mortem sampling has implications for interpretations of epigenetic measurement, similar to those encountered in the gene expression literature [29]. Fundamentally, these brains are sampled after the disease has occurred, which distorts the prospective timing clarity needed for etiological studies and raises concerns that the epigenetic patterns detected are a consequence of disease rather than part of the cause. Further, there may be bias in biological signals due to cause of death [30], tissue pH [31], or pre-mortem agonal state [32]. The effect of dying itself may have unknown effects on the gene regulation process. The limitations imposed by post-mortem sampling can be somewhat mitigated by comparison with non-human models where pre-symptomatic or even embryonic brain samples can be obtained, but causal connections may simply not be possible from current human brain tissue designs. Careful sampling and storage and integration of measures of pre- and postmortem factors in epigenetic analyses [33, 34] may also help distinguish cause of death from disease-related patterns.
Third, even if the limitations above can be overcome, a major problem is the limited availability of suitable material and a lack of replication potential. Most studies therefore have low power to detect or replicate effects, especially if disease-associated epigenetic changes are subtle. A related methodological issue is the suitability of case and control brains included in epigenetic studies. Often in brain biorepositories, case definitions are loose and deeper diagnostic phenotype data are not available. Case classification may often be based on symptoms obtained from family members post-mortem, rather than direct clinician confirmation as is available in studies with living participants. In addition, the absence of disease symptoms or related co-morbidities is not always confirmed in controls. Parsing neuropsychiatric disorders into subphenotypes with potentially shared features and etiology is an emerging area of research that is often not a possibility for post-mortem annotations when the sample sizes are often low. Further, important potential confounder variables or effect modifiers are typically limited. Controls are often skewed to older individuals, a potentially important problem for epigenetics where age appears to be highly correlated with at least DNA methylation [35]. Environmental exposure data are typically not collected in brain bank archives, and our tools to estimate retrospective exposures from archived samples, pinpointing a pre-disease state, are limited. To be viable moving forward, brain-based epigenetic work in neuropsychiatric disorders will need large time-consuming efforts to collect and compile large numbers of appropriate brain samples from affected individuals and controls with available clinical, demographic, and risk factor data, which require coordination across multiple groups and brain repositories for different neuropsychiatric disorders.
Many creative alternatives to brain-based models of epigenetics have been explored. Transformed lymphoblastoid cell lines that have been archived for genetic studies are an important existing source of genetic material linked with phenotype. The transformation process of B cells with Epstein-Barr virus (EBV) involves epigenetic reprogramming; however, so, the results of these studies must be interpreted cautiously [36]. Nonetheless, many neuropsychiatric disease DNA methylation associations, including autism, have been identified in transformed lymphoblastoid cell lines [37, 38], and these observations warrant further inquiry. Another convenient source for epigenetic discovery in neuropsychiatric disease research are buccal or saliva samples. Buccal cells are derived from the ectoderm during development, similar to the central nervous system, are non-invasive to collect, and a recent study suggests that buccal cells are more informative of tissue differential methylation signatures than blood [39]. Saliva samples are mixtures of white blood cells and buccal epithelial cells and may be arduous to collect from children with neurodevelopmental disorders, as several milliliters are required. The most mechanistically promising option is the use of induced Pluripotent Stem Cells (iPSCs) derived from cases and sex- and age-matched controls which can be differentiated into neuronal progenitor cells (NPCs) and neuroblasts. This model allows for direct experimentation of pharmaceutical agents, environmental exposures, and nutrients on human neural cells and model brain organoids, customized to the genetic backbone relevant for a disorder [40]. The generation of neuroblasts and other target tissue models from iPSCs, however, is mediated by epigenetic reprogramming, which is often incomplete, meaning the cells retain epigenetic information from their cell type of origin [41, 42]. The iPSC reprogramming process would also erase exposure or disease epigenetic marks from the original patient, which would restrict neuroblast studies of early exposure or disease. Extensive heterogeneity has been observed between iPSC lines and even within clones of the same line [43]; thus, replication across studies may be challenging. Currently, the production of iPSCs is very labor intensive, although cell culture models could potentially be developed for investigation across an epidemiologic sample. In terms of epidemiologic scale, whole blood remains the most obvious surrogate tissue for epigenetics studies.
Practicality of Leveraging Blood in Epidemiology
Peripheral blood is currently the most abundant type of sample used for epidemiologic and clinical research. A clear advantage of blood samples is the relatively non-invasive collection that is easy to obtain, often at the time patients are undergoing blood draws for clinical evaluations. The amount of DNA extracted from standard pediatric peripheral blood samples (3 mL) is generally sufficient for multiple genetic and DNA methylation experiments. One advantage of blood samples is that many neuropsychiatric disorder research laboratories and large specimen biorepositories, such as the Psychiatric Genomics Consortia, several NIH repositories, the Autism Genetic Resource Exchange (AGRE), and the Simons Simplex Collection for example, already have DNA samples isolated from blood with paired detailed phenotyping data, often on multiple family members. The convenience and availability of blood as a tissue for epigenetic inquiry in existing epidemiologic studies with strong neuropsychiatric phenotyping and existing environmental exposure data are an important practical research consideration.
The use of peripheral blood in epigenetic epidemiologic studies affords several methodological advantages. First, blood can be obtained prior to disease onset to establish appropriate timing for testing epigenetic marks as an etiologic factor in disease. Observation of early marks is potentially actionable through future intervention studies. Second, blood can be sampled serially prior to disease and during the disease process. The use of longitudinal samples allows for an estimation of change in epigenetic marks over time, potentially related to disease risk. Third, causal modeling options are available for studies with appropriate temporal sequence. Fourth, large sample sizes are often achievable, yielding the statistical power to observe modest effect sizes. Fifth, as the most common type of tissue measured in epidemiologic settings, results can be compared or even combined across studies. Sixth, family-based design options are possible, which is particularly useful for relatively rare psychiatric disorders that are clustered in families [44]. Similarly, blood samples can be used in studies targeting and following individuals at high or low risk of disease. Finally, the identification of blood-based biomarkers of disease may have translational utility in clinical settings.
Challenges of Blood-Based Neurospychiatric Epigenetic Research
Epigenetic marks are critical features of cellular phenotype such that epigenetic differences across tissue types are expected [45, 46]. Thus, the relevance of blood epigenetic findings must be carefully considered when trying to make inferences about specific mechanistic pathways for brain-related disorders. Levels of DNA methylation at specific CpG sites are often not comparable across tissues, and inter-individual variation in DNA methylation at the majority of CpG sites on the Illumina 450 K array, the most widely used tool in epigenetic epidemiology, is only weakly correlated across blood and brain tissues (cortex and cerebellum) [47••]. Further, principal component analysis of paired brain and blood DNA methylation data from unaffected individuals separates tissue type in the first component, explaining more of the variance than between-person variability, although both intra- and inter-individual variability are detectible via such an analysis [48]. However, epidemiologic data are typically looking to identify associations between epigenetic marks and disease (or environmental exposures), such that the actual level of methylation is less relevant than the conservation of inter-individual variation across tissues [47••]. Another concern is that hydroxymethylcytosine, another DNA modification that is of increasing interest for neuropsychiatric disease because it is particularly enriched in the brain, occurs at extremely low levels in the blood [49]. Because common DNA methylation quantitation methods do not distinguish between DNA methylation and DNA hydroxymethylation, observed methylation levels in blood may not reflect the methylation state relevant in the brain.
As in studies of post-mortem brain tissue, cellular heterogeneity is an important potential confounder when quantifying DNA methylation in blood samples. A whole blood sample is composed of a heterogeneous population of neutrophils, lymphocytes, monocytes, and other cell types. The proportion of white cells in an individual at any time can be influenced by factors including bacterial or viral infection, inflammation, dietary intake, stress, medication, or other environmental exposures. Ideally, epigenetic epidemiology analyses would be performed on sorted cell populations, but this is not possible from frozen archived blood samples or previously isolated DNA from blood derivatives. Cell-type estimation algorithms have been developed for adult and cord blood [50•, 51, 52] and can be used to partially control for such heterogeneity in association analyses [53]. Expanding blood cell reference panels to populations with variable age, sex, and race would further improve estimation strategies.
Argument for Blood-Based Neuropsychiatric Epigenetic Research
Epigenetic studies are performed to investigate both disease mechanisms as well as to identify associated biomarkers of either exposure or disease (Fig. 1). Blood-based epigenetics studies in neuropsychiatric disorders have shown promising associations with respect to both mechanisms and identification of disorder biomarkers (Table 1) and continue to be useful in each regard for multiple reasons.
First, neuropsychiatric disorders may have mechanistic underpinnings in tissues beyond the brain, including blood. Many neuropsychiatric disorders, such as autism spectrum disorder [54], schizophrenia [55], and bipolar disorder [56], have been associated with inflammation and altered immune response. Furthermore, if one looks at a neuropsychiatric disorder as a “systems” disorder (especially as individuals with autism spectrum disorder often present with gastrointestinal and immune system problems), peripheral tissues may be used as a window to detect disrupted pathways (metabolic or signaling) that we may carry to the brain. Altered immune processes and similar phenotypes that are associated with neuropsychiatric diseases may be well measured in blood samples.
With additional respect to mechanistic insights, blood-based epigenetics may be a proxy for brain epigenetics in certain circumstances. There are now examples where inter-individual variation in DNA methylation is correlated between blood and regions of the brain, although this varies across the genome and may be driven in part by underlying DNA sequence variation [17•, 47••]. Genetic influences on DNA methylation—e.g., via methylation quantitative trait loci (mQTLs) [47••, 57, 58] and genotype-driven allele-specific DNA methylation [59]—are often consistent across tissues [60], although even these effects can be tissue-specific [61]. Disease-specific studies have probed target-surrogate tissue correlations at specific loci. For example, frontal cortex DNA methylation differences in Parkinson’s disease are associated with those found in blood [62]. There are also useful cross-tissue proxy examples from the non-neurological literature, such as cancer [63] and loss of imprinting [64]. Future studies will continue to refine our understanding of cross-tissue epigenetic similarities over developmental time and in specific cell types.
Information on correlation between target and surrogate tissue DNA methylation can be exploited for epidemiologic study design and analyses. For example, among variably methylated CpG sites in the brain, a subset (7.9 %) of sites was correlated (rho > 0.59) between temporal lobe and blood [65]. These blood-brain correlated sites were used to a priori filter blood CpG discovery analysis in a schizophrenia case-control study. Although these specific CpG sites may be markers of DNA sequence variation, future studies may use similar methods to reduce the number of attempted comparisons and focus analyses on potentially brain informative regions. A module of CpG sites is commonly susceptible to aging in both brain and blood [66], which in addition to indicating that aging processes are consistent across tissues, may represent another subset of informative surrogate tissue markers. Many DNA methylation patterns are highly conserved across tissues [67], and recent studies propose statistical methods to predict methylation of target tissue using methylation profiles measured in surrogate tissues [68], particularly among CpG sites with substantial variation. With further understanding of locations of non-methyl modifications at CpG sites (e.g., hydroxymethyl), we may better understand and predict cross-tissue differences and similarities and leverage that knowledge in our epidemiologic design and analyses.
Surrogate tissue epigenetics research has important applications for disease biomarkers. Early biomarkers of neuropsychiatric disease may be particularly useful where early interventions are possible [69]. Cord blood or placenta DNA methylation may predict later neurodevelopmental disorders in childhood, and monitoring these early biomarkers of disease could help target at risk children for treatment. Blood-based markers may also be informative for monitoring disease progression and treatment effectiveness.
Epigenetic marks as biomarkers of exposure are relevant for determining exposure-disease associations in situations where direct chemical exposures have already been flushed from the body [6]. Many environmental toxicants have been associated with global or gene-specific epigenetic change [70]. Work is underway to identify unique epigenetic signatures of chemical exposure, including exposures to air pollution particulate matter [71] and mercury [72], or nutrient intake, such as folate supplements [73]. A recent meta-analysis across 13 cohorts demonstrated that the in utero exposure to smoking is associated with a consistent DNA methylation signature at infancy and in childhood [74]. Future studies could measure DNA methylation in children and estimate smoking exposure during gestation. In mice, glucocorticoid exposure was associated with corresponding DNA methylation differences in blood and hippocampus at the Fkbp5 candidate gene [75], while bisphenol A exposure is associated with parallel hippocampus and blood changes in Bdnf [76]. Stress-responsive methylation at the COMT gene in rat pre-frontal cortex has been further correlated with methylation in lymphocytes [77]. Chemical and CpG site specificity of environmental epigenetic biomarkers will continue to be investigated and refined.
Conclusions
Going forward, it is important that epigenetic epidemiology studies recognize the pragmatic reality of their tissue sample and be straightforward about the intention, utility, and scope of possible inferences. Blood and brain-based epidemiologic studies can provide potential insights to neuropsychiatric disease research, though each has limitations (Table 2). Where possible, we advocate for a combined approach of paired blood and brain research. This can be effectively demonstrated in relatively few examples to date [65, 78, 79]. Cross-tissue analyses should be encouraged in human and animal models. More tissue banks may consider archiving blood alongside brain tissues to increase these opportunities. Indeed the recent TARGET initiative from the National Institute for Environmental Health Sciences has been developed to better understand surrogate versus target tissue epigenetic measures. We further advocate for researchers to exploit epidemiologic design options available for blood-based research settings. These include taking advantage of longitudinal samples, adequate phenotype information, environmental exposure and covariate data, large sample sizes, and replication opportunities. In addition, methodological approaches to enhance causal inference can be used [80], such as mediation and moderation analyses that examine specific roles of epigenetic marks in exposure-based or genetic causes of disease [81, 82]. Ideally, epidemiologic studies would obtain either directly measured cell-type counts from whole blood, which can be done using routine hospital assays, or prioritize sorting blood samples into specific cell types at time of collection, which may require specialized training and equipment. In cases where neither is possible, we recommend cell-type estimation and adjustment using cell-type epigenetic reference panels. Future studies may also consider the impact of aggregate (global) epigenetic differences versus genome-wide site-specific features that have rarely been estimated in the same samples.
Table 2.
Rationale/challenges for tissue type
| Tissue type | Disease relevance | Timing/inherent design bias | Availability/sample size | Technical challenges |
|---|---|---|---|---|
| Blood |
|
|
|
|
| Brain |
|
|
|
|
Multiple lines of evidence across neuropsychiatric disorders point to a role for epigenetic alterations in these diseases. There are pragmatic reasons to examine blood epigenomes and evidence that this approach can be informative for multiple purposes. Future studies in blood and brain will hopefully replicate and expand upon the current literature, taking advantage of emerging methodological developments, though these studies will likely maintain some of the current limitations. In our first “at bat” in conducting neuropsychiatric epigenetic epidemiology research, we may have only reached first base, but the potential and opportunities are such that we should keep improving and swinging for a home run.
Supplementary Material
Acknowledgments
Funding Sources
Drs. Bakulski and Fallin were supported by the National Institute of Environmental Health Sciences (ES017646). Dr. Hu was supported by the National Institute of Environmental Health Sciences (ES023061). Dr. Mill was supported by the UK Medical Research Council (MRC; MR/K013807/1) and the US National Institutes of Health (AG036039).
Footnotes
Compliance with Ethical Standards
Conflict of Interest
Dr. Kelly M. Bakulski, Dr. Alycia Halladay, Dr. Valerie W. Hu, Dr. Jonathan Mill, and Dr. M. Daniele Fallin declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–8. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–40. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 4.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23(8):853–9. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Gen. 2004;20(8):350–8. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Ladd-Acosta C. Epigenetic signatures as biomarkers of exposure. Curr Environ Health Rep. 2015;2(2):117–25. doi: 10.1007/s40572-015-0051-2. [DOI] [PubMed] [Google Scholar]
- 7.Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun. 2014;5:5592. doi: 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodenhiser D, Mann M. Epigenetics and human disease: translating basic biology into clinical applications. Can Med Assoc J. 2006;174(3):341–8. doi: 10.1503/cmaj.050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh J, Eisch AJ. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: Unraveling the genome to understand the mind. Neurobiol Dis. 2010;39(1):73–84. doi: 10.1016/j.nbd.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80(3):624–32. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loke YJ, Hannan AJ, Craig JM. The Role of Epigenetic Change in Autism Spectrum Disorders. Front Neurol. 2015;6:107. doi: 10.3389/fneur.2015.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdolmaleky HM, Zhou JR, Thiagalingam S. An update on the epigenetics of psychotic diseases and autism. Epigenomics. 2015;7(3):427–49. doi: 10.2217/epi.14.85. [DOI] [PubMed] [Google Scholar]
- 13.Ibi D, Gonzalez-Maeso J. Epigenetic signaling in schizophrenia. Cell Signal. 2015;27(10):2131–6. doi: 10.1016/j.cellsig.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shorter KR, Miller BH. Epigenetic mechanisms in schizophrenia. Prog Biophys Mol Biol. 2015;118(1–2):1–7. doi: 10.1016/j.pbiomolbio.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinkers CH, Kalafateli AL, Rutten BP, Kas MJ, Kaminsky Z, Turner JD, et al. Traumatic stress and human DNA methylation: a critical review. Epigenomics. 2015;7(4):593–608. doi: 10.2217/epi.15.11. [DOI] [PubMed] [Google Scholar]
- 16.Cadet JL, McCoy MT, Jayanthi S. Epigenetics and Addiction. Clin Pharmacol Ther. 2016;99(5):502–511. doi: 10.1002/cpt.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13(6):R43. doi: 10.1186/gb-2012-13-6-r43. Blood and brain DNA methylation comparison, including mQTLs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohammad-Rezazadeh I, Frohlich J, Loo SK, Jeste SS. Brain connectivity in autism spectrum disorder. Curr Opin Neurol. 2016;29(2):137–47. doi: 10.1097/WCO.0000000000000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods. 2016;13(3):229–32. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11(3):499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8(3):290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montano CM, Irizarry RA, Kaufmann WE, Talbot K, Gur RE, Feinberg AP, et al. Measuring cell-type specific differential methylation in human brain tissue. Genome Biol. 2013;14(8):R94. doi: 10.1186/gb-2013-14-8-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19(2):335–46. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112(23):7285–90. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350(6256):94–8. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuzik J, Zeisel A, Mate Z, Calvigioni D, Yanagawa Y, Szabo G, et al. Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat Biotechnol. 2016;34(2):175–83. doi: 10.1038/nbt.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci. 2016;19(1):102–10. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- 28.Heyward FD, Gilliam D, Coleman MA, Gavin CF, Wang J, Kaas G, et al. Obesity Weighs down Memory through a Mechanism Involving the Neuroepigenetic Dysregulation of Sirt1. J Neurosci. 2016;36(4):1324–35. doi: 10.1523/JNEUROSCI.1934-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barton AJ, Pearson RC, Najlerahim A, Harrison PJ. Pre- and postmortem influences on brain RNA. J Neurochem. 1993;61(1):1–11. doi: 10.1111/j.1471-4159.1993.tb03532.x. [DOI] [PubMed] [Google Scholar]
- 30.Harrison PJ, Heath PR, Eastwood SL, Burnet PW, McDonald B, Pearson RC. The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vulnerability and comparison with their encoded proteins. Neurosci Lett. 1995;200(3):151–4. doi: 10.1016/0304-3940(95)12102-a. [DOI] [PubMed] [Google Scholar]
- 31.Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, et al. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet. 2004;13(6):609–16. doi: 10.1093/hmg/ddh065. [DOI] [PubMed] [Google Scholar]
- 32.Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, et al. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55(4):346–52. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernst C, McGowan PO, Deleva V, Meaney MJ, Szyf M, Turecki G. The effects of pH on DNA methylation state: In vitro and postmortem brain studies. J Neurosci Methods. 2008;174(1):123–5. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 34.Pidsley R, Mill J. Epigenetic studies of psychosis: current findings, methodological approaches, and implications for postmortem research. Biol Psychiatry. 2011;69(2):146–56. doi: 10.1016/j.biopsych.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 35.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen KD, Sabunciyan S, Langmead B, Nagy N, Curley R, Klein G, et al. Large-scale hypomethylated blocks associated with Epstein-Barr virus-induced B-cell immortalization. Genome Res. 2014;24(2):177–84. doi: 10.1101/gr.157743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarachana T, Xu M, Wu RC, Hu VW. Sex hormones in autism: androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PLoS One. 2011;6(2):e17116. doi: 10.1371/journal.pone.0017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J. 2010;24(8):3036–51. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowe R, Gemma C, Beyan H, Hawa MI, Bazeos A, Leslie RD, et al. Buccals are likely to be a more informative surrogate tissue than blood for epigenome-wide association studies. Epigenetics. 2013;8(4):445–54. doi: 10.4161/epi.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prilutsky D, Palmer NP, Smedemark-Margulies N, Schlaeger TM, Margulies DM, Kohane IS. iPSC-derived neurons as a higher-throughput readout for autism: promises and pitfalls. Trends Mol Med. 2014;20(2):91–104. doi: 10.1016/j.molmed.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(12):1117–9. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat Rev Mol Cell Biol. 2013;14(6):357–68. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newschaffer CJ, Croen LA, Fallin MD, Hertz-Picciotto I, Nguyen DV, Lee NL, et al. Infant siblings and the investigation of autism risk factors. J Neurodev Disord. 2012;4(1):7. doi: 10.1186/1866-1955-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roadmap Epigenomics C. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47••.Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10(11):1024–32. doi: 10.1080/15592294.2015.1100786. Cross-tissue blood and brain DNA methylation comparisons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farre P, Jones MJ, Meaney MJ, Emberly E, Turecki G, Kobor MS. Concordant and discordant DNA methylation signatures of aging in human blood and brain. Epigenetics Chromatin. 2015;8:19. doi: 10.1186/s13072-015-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15(3):R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. Estimate cell proportions from a mixed cell DNA methylation measure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15(2):R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, McKenney S, et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–362. doi: 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2):142–7. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Estes ML, McAllister AK. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci. 2015;16(8):469–86. doi: 10.1038/nrn3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muller N, Weidinger E, Leitner B, Schwarz MJ. The role of inflammation in schizophrenia. Front Neurosci. 2015;9:372. doi: 10.3389/fnins.2015.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenblat JD, McIntyre RS. Bipolar Disorder and Inflammation. Psychiatr Clin North Am. 2016;39(1):125–37. doi: 10.1016/j.psc.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 57.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gamazon ER, Badner JA, Cheng L, Zhang C, Zhang D, Cox NJ, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry. 2013;18(3):340–6. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marzi SJ, Meaburn EL, Dempster EL, Lunnon K, Paya-Cano JL, Smith RG, et al. Tissue-specific patterns of allelically-skewed DNA methylation. Epigenetics. 2016;11(1):24–35. doi: 10.1080/15592294.2015.1127479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith AK, Kilaru V, Kocak M, Almli LM, Mercer KB, Ressler KJ, et al. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics. 2014;15:145. doi: 10.1186/1471-2164-15-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hannon E, Spiers H, Viana J, Pidsley R, Burrage J, Murphy TM, et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci. 2016;19(1):48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics. 2013;8(10):1030–8. doi: 10.4161/epi.25865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barault L, Ellsworth RE, Harris HR, Valente AL, Shriver CD, Michels KB. Leukocyte DNA as surrogate for the evaluation of imprinted Loci methylation in mammary tissue DNA. PLoS One. 2013;8(2):e55896. doi: 10.1371/journal.pone.0055896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walton E, Hass J, Liu J, Roffman JL, Bernardoni F, Roessner V, et al. Correspondence of DNA Methylation Between Blood and Brain Tissue and Its Application to Schizophrenia Research. Schizophr Bull. 2016;42(2):406–14. doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MP, van Eijk K, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13(10):R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18(24):4808–17. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma B, Wilker EH, Willis-Owen SA, Byun HM, Wong KC, Motta V, et al. Predicting DNA methylation level across human tissues. Nucleic Acids Res. 2014;42(6):3515–28. doi: 10.1093/nar/gkt1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zwaigenbaum L, Bauman ML, Choueiri R, Fein D, Kasari C, Pierce K, et al. Early Identification and Interventions for Autism Spectrum Disorder: Executive Summary. Pediatrics. 2015;136(Suppl 1):S1–9. doi: 10.1542/peds.2014-3667B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bollati V, Baccarelli A. Environmental epigenetics. Heredity. 2010;105(1):105–12. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf K, et al. A Genome-Wide Analysis of DNA Methylation and Fine Particulate Matter Air Pollution in Three Study Populations: KORA F3, KORA F4, and the Normative Aging Study. Environ Health Perspect. 2016 doi: 10.1289/ehp.1509966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bakulski KM, Lee H, Feinberg JI, Wells EM, Brown S, Herbstman JB, et al. Prenatal mercury concentration is associated with changes in DNA methylation at TCEANC2 in newborns. Int J Epidemiol. 2015;44(4):1249–62. doi: 10.1093/ije/dyv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joubert BR, Herman T, Felix JF, Bohlin J, Ligthart S, Beckett E, et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun. 2016 doi: 10.1038/ncomms10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joubert Bonnie R, Felix Janine F, Yousefi P, Bakulski Kelly M, Just Allan C, Breton C, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98(4):680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ewald ER, Wand GS, Seifuddin F, Yang X, Tamashiro KL, Potash JB, et al. Alterations in DNA methylation of Fkbp5 as a determinant of blood-brain correlation of glucocorticoid exposure. Psychoneuroendocrinology. 2014;44:112–22. doi: 10.1016/j.psyneuen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kundakovic M, Gudsnuk K, Herbstman JB, Tang D, Perera FP, Champagne FA. DNA methylation of BDNF as a biomarker of early-life adversity. Proc Natl Acad Sci U S A. 2015;112(22):6807–13. doi: 10.1073/pnas.1408355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ursini G, Bollati V, Fazio L, Porcelli A, Iacovelli L, Catalani A, et al. Stress-related methylation of the catechol-O-methyltransferase Val 158 allele predicts human prefrontal cognition and activity. J Neurosci. 2011;31(18):6692–8. doi: 10.1523/JNEUROSCI.6631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE, et al. Genome-wide DNA methylation scan in major depressive disorder. PLoS One. 2012;7(4):e34451. doi: 10.1371/journal.pone.0034451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, Markunas CA, et al. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Med. 2009;7:62. doi: 10.1186/1741-7015-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rothman KJ, Greenland S. Causation and causal inference in epidemiology. Am J Public Health. 2005;95(Suppl 1):S144–50. doi: 10.2105/AJPH.2004.059204. [DOI] [PubMed] [Google Scholar]
- 81.Relton CL, Davey SG. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41(1):161–76. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mackinnon DP. Integrating Mediators and Moderators in Research Design. Res Soc Work Pract. 2011;21(6):675–81. doi: 10.1177/1049731511414148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bakulski KM, Fallin MD. Epigenetic epidemiology: promises for public health research. Environ Mol Mutagen. 2014;55(3):171–83. doi: 10.1002/em.21850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ladd-Acosta C, Fallin MD. The role of epigenetics in genetic and environmental epidemiology. Epigenomics. 2016;8(2):271–83. doi: 10.2217/epi.15.102. [DOI] [PubMed] [Google Scholar]
- 85.Houseman EA, Kim S, Kelsey KT, Wiencke JK. DNA Methylation in Whole Blood: Uses and Challenges. Curr Environ Health Rep. 2015;2(2):145–54. doi: 10.1007/s40572-015-0050-3. [DOI] [PubMed] [Google Scholar]
- 86.Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1(4):e1–11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abdolmaleky HM, Yaqubi S, Papageorgis P, Lambert AW, Ozturk S, Sivaraman V, et al. Epigenetic dysregulation of HTR2A in the brain of patients with schizophrenia and bipolar disorder. Schizophr Res. 2011;129(2–3):183–90. doi: 10.1016/j.schres.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 89.James SJ, Shpyleva S, Melnyk S, Pavliv O, Pogribny IP. Complex epigenetic regulation of engrailed-2 (EN-2) homeobox gene in the autism cerebellum. Transl Psychiatry. 2013;3:e232. doi: 10.1038/tp.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP. Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry. 2014;19(8):862–71. doi: 10.1038/mp.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu L, Wang X, Li XL, Towers A, Cao X, Wang P, et al. Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Hum Mol Genet. 2014;23(6):1563–78. doi: 10.1093/hmg/ddt547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nardone S, Sams DS, Reuveni E, Getselter D, Oron O, Karpuj M, et al. DNA methylation analysis of the autistic brain reveals multiple dysregulated biological pathways. Transl Psychiatry. 2014;4:e433. doi: 10.1038/tp.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wockner LF, Noble EP, Lawford BR, Young RM, Morris CP, Whitehall VL, et al. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry. 2014;4(1):e339. doi: 10.1038/tp.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pidsley R, Viana J, Hannon E, Spiers H, Troakes C, Al-Saraj S, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15(10):483. doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95•.Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19(1):40–7. doi: 10.1038/nn.4181. Large brain tissue study considering development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pandey GN, Rizavi HS, Zhang H, Bhaumik R, Ren X. The Expression of the Suicide-Associated Gene SKA2 is Decreased in the Prefrontal Cortex of Suicide Victims, but Not of Non-Suicidal Patients. Int J Neuropsychopharmacol. 2016 doi: 10.1093/ijnp/pyw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carrard A, Salzmann A, Malafosse A, Karege F. Increased DNA methylation status of the serotonin receptor 5HTR1A gene promoter in schizophrenia and bipolar disorder. J Affect Disord. 2011;132(3):450–3. doi: 10.1016/j.jad.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 98.Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20(24):4786–96. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rusiecki JA, Byrne C, Galdzicki Z, Srikantan V, Chen L, Poulin M, et al. PTSD and DNA Methylation in Select Immune Function Gene Promoter Regions: A Repeated Measures Case-Control Study of U.S. Military Service Members. Front Psychiatry. 2013;4:56. doi: 10.3389/fpsyt.2013.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nishioka M, Bundo M, Koike S, Takizawa R, Kakiuchi C, Araki T, et al. Comprehensive DNA methylation analysis of peripheral blood cells derived from patients with first-episode schizophrenia. J Hum Genet. 2013;58(2):91–7. doi: 10.1038/jhg.2012.140. [DOI] [PubMed] [Google Scholar]
- 101.Zhang H, Wang F, Kranzler HR, Zhao H, Gelernter J. Profiling of childhood adversity-associated DNA methylation changes in alcoholic patients and healthy controls. PLoS One. 2013;8(6):e65648. doi: 10.1371/journal.pone.0065648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aldinger KA, Plummer JT, Levitt P. Comparative DNA methylation among females with neurodevelopmental disorders and seizures identifies TAC1 as a MeCP2 target gene. J Neurodev Disord. 2013;5(1):15. doi: 10.1186/1866-1955-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wong CC, Meaburn EL, Ronald A, Price TS, Jeffries AR, Schalkwyk LC, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2014;19(4):495–503. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dempster EL, Wong CC, Lester KJ, Burrage J, Gregory AM, Mill J, et al. Genome-wide methylomic analysis of monozygotic twins discordant for adolescent depression. Biol Psychiatry. 2014;76(12):977–83. doi: 10.1016/j.biopsych.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fisher HL, Murphy TM, Arseneault L, Caspi A, Moffitt TE, Viana J, et al. Methylomic analysis of monozygotic twins discordant for childhood psychotic symptoms. Epigenetics. 2015;10(11):1014–23. doi: 10.1080/15592294.2015.1099797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kang HJ, Kim JM, Kim SY, Kim SW, Shin IS, Kim HR, et al. A Longitudinal Study of BDNF Promoter Methylation and Depression in Breast Cancer. Psychiatry Investig. 2015;12(4):523–31. doi: 10.4306/pi.2015.12.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim D, Kubzansky LD, Baccarelli A, Sparrow D, Spiro A, 3rd, Tarantini L, et al. Psychological factors and DNA methylation of genes related to immune/inflammatory system markers: the VA Normative Aging Study. BMJ Open. 2016;6(1):e009790. doi: 10.1136/bmjopen-2015-009790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kahl KG, Georgi K, Bleich S, Muschler M, Hillemacher T, Hilfiker-Kleinert D, et al. Altered DNA methylation of glucose transporter 1 and glucose transporter 4 in patients with major depressive disorder. J Psychiatr Res. 2016;76:66–73. doi: 10.1016/j.jpsychires.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 109••.Montano C, Taub MA, Jaffe A, Briem E, Feinberg JI, Trygvadottir R, et al. Association of DNA methylation differences with schizophrenia in an epigenome-wide association study. JAMA Psychiatry. 2016;73(5):506–14. doi: 10.1001/jamapsychiatry.2016.0144. Large, well replicated surrogate tissue study. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.