Abstract
The clinical demand on rapid microbiological diagnostic is constantly increasing. PCR coupled to electrospray ionization-mass spectrometry, PCR/ESI-MS, offers detection and identification of over 750 bacteria and Candida species directly from clinical specimens within 6 hours. In this study, we investigated the clinical performance of the IRIDICA BAC LRT Assay for detection of bacterial pathogens in 121 bronchoalveolar lavage (BAL) samples that were received consecutively at our bacterial laboratory for BAL culture. Commensal or pathogenic microorganisms were detected in 118/121 (98%) BAL samples by PCR/ESI-MS, while in 104/121 (86%) samples by routine culture (P<0.01). Detection of potentially pathogenic microorganisms by PCR/ESI-MS was evaluated in comparison with conventional culture-based or molecular methods. The agreement between positive findings was overall good. Most Staphylococcus aureus-positive PCR/ESI-MS results were confirmed by culture or species-specific PCR (27/33, 82%). The identity of Streptococcus pneumoniae could however be confirmed for only 6/17 (35%) PCR/ESI-MS-positive samples. Non-cultivable and fastidious pathogens, which were not covered by standard culture procedures were readily detected by PCR/ESI-MS, including Legionella pneumophila, Bordetella pertussis, Norcadia species and Mycoplasma pneumoniae. In conclusion, PCR/ESI-MS detected a broad range of potential pathogens with equal or superior sensitivity compared to conventional methods within few hours directly from BAL samples. This novel method might thus provide a relevant tool for diagnostics in critically ill patients.
Introduction
Pneumonia and other infections of the lower respiratory tract (LRT) are serious health problems associated with high morbidity and mortality worldwide [1, 2]. Early optimal antimicrobial treatment is linked to improved patient outcome, reduced risk for resistance development and adverse side effects by the antimicrobials [3]. Although Streptococcus pneumoniae is the most common bacterial pathogen isolated from community-acquired pneumonia (CAP), a large proportion of CAP is caused by other pathogens, with considerable geographical variations [4, 5]. In contrast, Gram-negative species and Staphylococcus aureus dominate among bacteria isolated from hospital-acquired pneumonia (HAP) [6, 7]. The wide range of possible pathogens challenge the development of methods for rapid detection and identification, needed to ensure timely appropriate antimicrobial therapy.
Conventional culture-based investigation of LRT secretions is currently the method of choice for detection of bacterial pathogens in critically ill patients with suspected pneumonia [3, 8]. Culture-based approaches however often fail in detecting a causative agent. Prior antimicrobial therapy decreases the sensitivity of methods based on bacterial viability [9, 10], and certain pathogens are generally not cultivable with standard methods. In addition, these methods are time-consuming and labor-intensive. During the recent years, novel culture-independent techniques have been proposed as promising alternatives for detection and identification of pathogens from severe pneumonia. Different to samples from sterile body sites, commensal and colonizing microorganisms complicate the analysis in LRT secretions. While multiplex PCR assays have short turn-around times and are suited for detection of multiple pathogens simultaneously, the target panel is limited and information regarding commensal microorganisms is generally lacking. Diagnostic PCR-kits designed for sepsis diagnosis, e.g. LightCycler SeptiFast from Roche Diagnostics [11] have broader coverage but lack important respiratory pathogens. Similarly, poor performance and insufficient coverage has been reported for the Unyvero multiplex PCR device (Curetis AG) for direct point-of-care detection of respiratory pathogens [12, 13]. Assays such as the RB5 Anyplex II kit (Seegene) are designed for detection of few atypical bacterial pathogens only. There is thus an obvious need for a rapid method covering both common and atypical microorganisms directly from LRT specimens.
PCR/ESI-MS overcomes these limitations by combining multiple broad range PCR reactions with electrospray ionization-mass spectrometry (PCR/ESI-MS), which rapidly provides sequence information from the generated amplicons [14]. Subsequent bioinformatical analysis with an integrated database enables identification of species and resistance determinants. An internal calibrant molecule in each PCR reaction permits semi-quantitation of the detected genomes. The first PCR/ESI-MS version for clinical research, the Ibis T5000 universal biosensor platform (Ibis Biosciences, an Abbott company), used an assay kit for identification of over 600 bacteria and Candida species. Evaluation of this assay on brochoalveolar lavage (BAL) samples showed however suboptimal performance, with a concordance to standard culture-based methods of 66% for detection of bacterial pathogens [15]. Further development led to the PLEX-ID instrument (Abbott). The concordance rate of this analysis with conventional methods was 77% for detection of bacterial pathogens and 45% when microorganisms from the normal flora were included [16]. It should be noted that the applied assay was intended for use in sterile fluids and not BAL. A re-designed, research-use-only version of the instrument was recently tested in LRT secretions. This study demonstrated the improved detection of bacterial pathogens in patients under antibiotic treatment with 49/104 (47%) hits in culture-negative samples [17]. However, the study considered only primary detections for evaluation and performance on polymicrobial samples was neglected.
The lack of a gold standard method hampers evaluation of discrepant findings originated from conventional culture-dependent methods and PCR/ESI-MS. In the hitherto published studies, detections from PCR/ESI-MS were not systematically confirmed by complementing methods when culture results were missing. This situation makes it extremely difficult to interpret the quality of the PCR/ESI-MS results reported previously.
In November 2014, a PCR/ESI-MS platform was CE-marked and became commercially available with the product name IRIDICA. The aim of the present study was to evaluate its performance in detection of respiratory pathogens in clinical BAL samples, using the IRIDICA BAC LRT Assay (Ibis Biosciences, Abbott Molecular, Des Plaines, IL). Discrepant results for selected bacterial species were further analyzed with species-specific or broad range-PCR. As the IRIDCA BAC LRT Assay occasionally reported “Fungus no ID”, we also aimed to study if such result was linked to any positive fungal culture results.
Materials and Methods
Study material
All BAL samples included in this study (n = 121) were part of standard hospital care and taken as per routine practice. Median age of the patients was 60 years (range 0–90 years) and the majority was male (83/121, 69%). Most samples were collected at the clinic for pulmonary diseases/allergy (72/121, 60%), intensive care units, hematology or transplantation (29/121, 24%), or at the clinic for infectious diseases (12/121, 10%). Consecutive BAL samples sent to the Division of Clinical Microbiology, Karolinska University Laboratory Huddinge, Stockholm, Sweden, for standard microbiological diagnostic between May 2014 and March 2015 were subjected to routine culture-based diagnostic and the remaining samples was stored at -20°C for subsequent analysis by PCR/ESI-MS.
IRIDICA PCR/ESI-MS
Bacterial and Candida DNA was detected and semi-quantified using the IRIDICA BAC LRT Assay (Abbott), designed to detect 780 bacterial and Candida species and four antibiotic resistance markers, mecA, vanA/vanB and blaKPC. For a defined panel of non-Candida fungal DNA, the assay reports “Fungus detected—No ID can be provided”. The analysis requires approximately six hours, and up to six samples including a negative control can be run simultaneously. From each sample, an aliquot of 100 μl BAL was analyzed. Samples were processed according to the manufacturer’s recommendation, including mechanical and chemical lysis; extraction of DNA followed by PCR, using 18 primer pairs in 16-well pre-coated PCR strips; desalting and purification of the amplicons; and analysis by ESI-MS. Species identification was achieved by bioinformatical analysis with an integrated software and database. Quantitation of detected genomes was based on internal calibrant molecules in each reaction and was reported as a semi-quantitative “level” for each detection.
Lower detection limit for Staphylococcus aureus by PCR/ESI-MS
The methicillin-resistant S. aureus (MRSA) strain CCUG 31966 was used to establish the lower detection level PCR/ESI-MS using the IRIDICA BAC LRT Assay for this species as well as for the mecA resistance determinant. Bacterial colonies were suspended in PBS to a McFarland 0.5. A 10-fold dilution series was prepared in PBS and dilutions 10−3, 10−4, 10−5 and 10−6 were analyzed in 4–6 replicates. The dilution, for which both S. aureus and mecA was detected in all runs was regarded as limit for reliable detection (lower detection limit).
Routine culture-based microbiological diagnostics
A quantitative culture technique was employed for routine culture-based analysis of the clinical samples. This included aerobic and anaerobic culture on non-selective agar (blood agar); a crystal violet-containing blood agar plate with an optochin disc at 5% CO2 for selection and identification of S. pneumoniae; a chocolate agar plate with an oleandomycin disc at 5% CO2 for selection and identification of Haemophilus influenzae; and a cysteine-lactose-electrolyte-deficient (CLED) agar plate for identification of Enterobacteriaceae. All agar plates were inoculated with 10 μl BAL each and were incubated for a total of 2 days at 37°C. The lower limit of detection was 100 CFU/ml, and growth of >104 CFU/ml was regarded significant. Primary respiratory pathogens were reported regardless of quantity, while potential pathogens, colonizers or contaminants were reported when dominating and/or present in significant quantities. Relevant microorganisms were identified by MALDI-TOF MS (Bruker Daltonik) or the Vitek 2 system (bioMérieux). A confidence score ≥2.0 or a probability score >93% were considered reliable species identification by MALDI-TOF MS or Vitek 2, respectively, as recommended by the manufacturers. Complementing analyses were carried out when indicated. Antimicrobial susceptibility testing was performed by disc diffusion according to EUCAST standards (EUCAST, http://www.eucast.org/). For verification of results from PCR/ESI-MS, results from related patient samples were recorded if available, including results from routine fungal culture.
Definition of normal respiratory flora
Following bacterial species were in general regarded as part of the normal respiratory flora: α-hemolytic streptococci, coagulase-negative staphylococci, Enterococcus species, Neisseria species, Rothia species, Corynebacterium species and Candida species. Growth of these species was usually summarized and reported as “normal respiratory flora” from routine microbiological diagnostics.
Verification of detections and species identification
Selected samples were analyzed for the presence of S. aureus and S. pneumonia by PCR targeting the nuc or lytA gene, respectively. DNA was extracted from a 500-μl aliquot of BAL using the MagNA Pure LC DNA Isolation Kit III (Roche Diagnostics, Mannheim, Germany) following the manufacturer’s protocol. PCR for lytA was performed on a LightCycler 2.0 (Roche Diagnostics) as described [18]; PCR for nuc was performed according to a previously described protocol [19] but adopted to the Rotorgene Q system (Qiagen) using 0.3 μM of each primer. The presence of other pathogens was verified by standard diagnostic PCR assay targeting respiratory tract pathogens (RB5 Anyplex II, Seegene; detecting Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, Bordetella pertussis and Bordetella parapertussis), or by 16S rRNA sequencing.
Statistical analysis
Differences were evaluated by the Mann Whitney or Kruskal-Wallis test as appropriate, categorical results were compared using Fisher’s exact test. P-values below 0.05 were considered statistically significant.
Ethics statement
This study was performed in accordance with the Declaration of Helsinki. An ethical permission was not required since the samples were anonymized an deidentified before being obtained by the authors.
Results
PCR/ESI-MS detects overall more microorganisms than routine culture
A total of 245 microorganisms were detected by PCR/ESI-MS in 118/121 (98%) BAL samples included in the study (Fig 1). The microorganisms belonged to 60 different species, with 18 species regarded as primary or potential pathogens (referred to as pathogens hereafter, Table 1) for LRT infections. The remaining 42 species were considered part of the normal respiratory flora, colonizers or contaminants (Table 2). Pathogens were detected in 86/121 (71%) BAL samples, with two different pathogenic species in 15/86 (17%) samples. No detection was made 3/121 (2%) BAL samples. In contrast, “no growth” was reported for 17/121 (14%) samples after routine culture (P<0.01).
Fig 1. Detection of multiple microorganisms in bronchoalveolar lavage samples by PCR/ESI-MS.
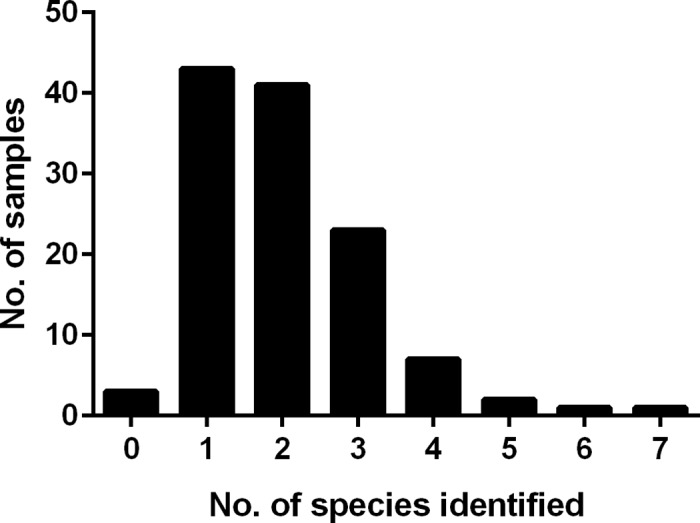
A total of 121 bronchoalveolar lavage samples were analyzed. The number of samples is depicted against the number of bacterial and Candida species identified per sample; various α-hemolytic streptococcal species are counted as one species.
Table 1. Detection of bacterial pathogens in bronchoalveolar lavage by PCR/ESI-MS (IRIDICA).
| Primary and potential pathogens | IRIDICA-positive | |
|---|---|---|
| Total | Confirmed by culture and/or PCR | |
| Gram-positive bacteria | ||
| Staphylococcus aureus | 33 | 27a |
| Streptococcus pneumoniae | 17 | 6 |
| Corynebacterium pseudodiphteriticum | 1 | - |
| Gram-negative bacteria | ||
| Haemophilus influenzae | 20 | 16 |
| Klebsiella pneumoniae | 4 | 4 |
| Enterobacter cloacae-complex | 5b | 4 |
| Escherichia coli | 2c | 2 |
| Escherichia vulneris | 1 | - |
| Proteus mirabilis | 2 | 1 |
| Proteus vulgaris | 1 | - |
| Klebsiella oxytoca | 1 | - |
| Pseudomonas aeruginosa | 8 | 7 |
| Stenotrophomonas maltophilia | 2 | 1 |
| Chryseobacterium indologenes | 1 | - |
| Atypical/ fastidious speciesd | ||
| Nocardia species | 1 | 1 |
| Bordetella pertussis | 1 | 1 |
| Legionella pneumophila | 1 | 1 |
| Mycoplasma pneumoniae | 1 | 1 |
aPositive in culture or for nuc; one negative sample was not available for further investigation.
bFor one sample, both E. cloacae-complex and E. cancerogenus was reported by IRIDICA; this result was counted as one isolate belonging to the E. cloacae-complex.
cFor one sample, both E. coli and E. coli/Shigella was reported by IRIDICA; this result was counted as one E. coli isolate.
dPresence of these bacteria was confirmed by conventional PCR assays (L. pneumophila, B. pertussis, M. pneumoniae), sequencing (Nocardia species), special culture (L. pneumophila) or detection of urinary antigen (L. pneumophila).
Table 2. Detection of commensal flora in bronchoalveolar lavage by PCR/ESI-MS (IRIDICA).
| Commensal microorganisms | Total |
|---|---|
| Candida species | |
| Candida albicans | 21 |
| Other Candida species | 7 |
| Gram-positive cocci | |
| α-hemolytic streptococci | 42 |
| Coagulase-negative staphylococci | 16 |
| Enterococcus species | 12 |
| Rothia species | 10 |
| Granulicatella adiacens | 8 |
| Gemella species | 3 |
| Gram-positive rods | |
| Corynebacterium species | 3 |
| Lysinibacillus sphaericus | 1 |
| Gram-negative cocci | |
| Neisseria species | 2 |
| Moraxella catarrhalis/nonliquefaciensa | 3 |
| Gram-negative rods | |
| Leclercia adecarboxylata | 1 |
| Actinobacillus capsulatus | 2 |
| Aggregatibacter segnis | 1 |
| Pseudomonas fluorescens | 1 |
| Anaerobe bacteria | |
| Lactobacillus species | 3 |
| Propionibacterium acnes | 2 |
| Fusobacterium nucleatum | 2 |
| Porphyromonas endodontalis | 1 |
| Veillonella species | 2 |
aIn one of these samples, a Moraxella species was isolated by routine culture and identified as Moraxella (Branhamella) catarrhalis by MALDI-TOF MS.
Performance of PCR/ESI-MS for detection of Staphylococcus aureus
S. aureus was among the most frequently detected potential pathogen, with 33 positive samples by PCR/ESI-MS. S. aureus was reported for 15/33 (45%) samples from routine culture, indicating dominating growth and significant numbers of colonies. Interestingly, semi-quantitative PCR/ESI-MS levels for S. aureus-specific DNA were significantly higher for these 15 specimens (Fig 2). Dose-response experiments with spiked samples defined a concentration of approximately 0.4×104 CFU/ml as lower level for detection of this species, corresponding to PCR/ESI-MS levels of 35±6.9. All 15 samples with significant growth of S. aureus yielded levels above this threshold. In 12/18 (67%) culture-negative samples, detection of S. aureus by PCR/ESI-MS was confirmed by species-specific PCR. Five of the six remaining samples were tested negative by PCR; one was not available for further investigation. Together, the presence of S. aureus was verified by culture and/or PCR in 27/33 (82%) PCR/ESI-MS-positive samples. Among the studied material, there were no PCR/ESI-MS-negative samples with recorded growth of S. aureus.
Fig 2. Semi-quantitative detection of Staphylococcus aureus by PCR/ESI-MS.
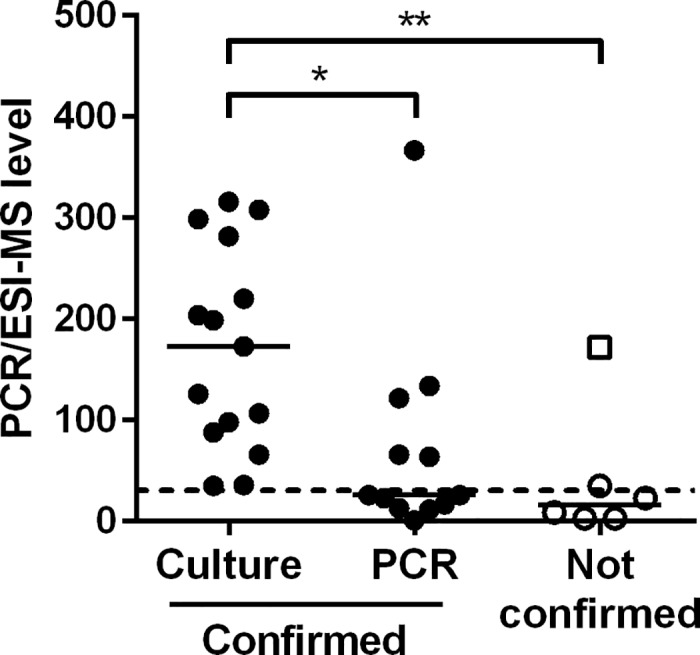
S. aureus was detected in a total of 33 bronchoalveolar lavage samples. In 27 samples, the results were confirmed by culture (n = 15) or PCR for nuc (n = 12). Individual values with median are shown, comparison was made by Kruskal-Wallis with Dunn’s multiple comparison post-test; *P<0.05, **P<0.01. The dashed line indicates the mean level for the lower detection limit of S. aureus by PCR/ESI-MS (0.4×104 CFU/ml), the open square indicates a sample not available for confirming tests.
Performance of PCR/ESI-MS for detection of streptococcus pneumoniae
S. pneumoniae was detected in 17 BAL samples by PCR/ESI-MS. The presence of S. pneumoniae was however confirmed in only 6/17 (35%) samples, in two by positive culture and in another four samples by species-specific PCR (Fig 3). Semi-quantitative PCR/ESI-MS levels did not significantly differ between lytA-positive and lytA-negative samples (P = 0.35). High PCR/ESI-MS levels were however obtained for the two samples which were both lytA- and culture-positive. In addition, the CT-values for lytA indicated higher levels of the pathogen for culture-positive compared to culture-negative samples and overall, values correlated with semi-quantitative levels from the PCR/ESI-MS analysis (S1 Fig). Three culture-positive samples did not yield positive results for S. pneumoniae or any other streptococcal species by PCR/ESI-MS.
Fig 3. Detection of Streptococcus pneumoniae with PCR/ESI-MS.
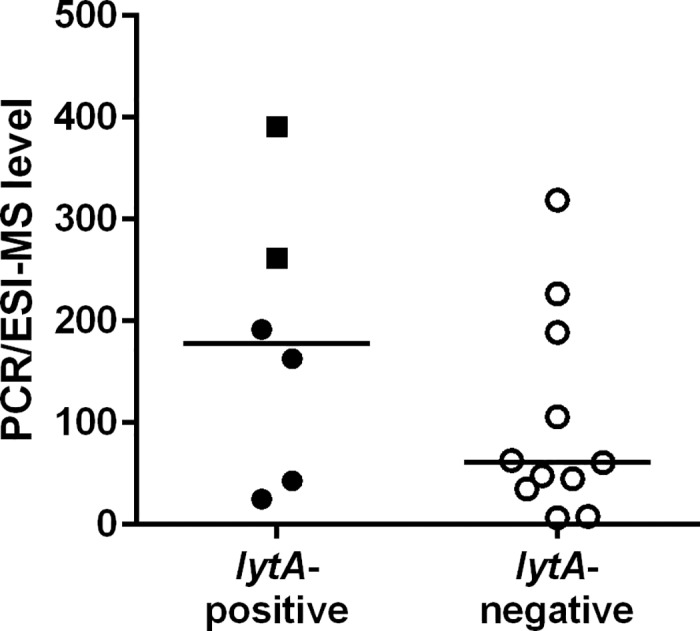
S. pneumoniae was detected in a total of 17 bronchoalveolar lavage samples. In 6 samples, the results were confirmed by PCR for lytA. Individual values with median are shown, the squares indicate culture-positive samples.
Detection of haemophilus influenza
Detection of H. influenzae by PCR/ESI-MS was confirmed by culture in 16/20 (80%) BAL samples (Table 1). Semi-quantitative PCR/ESI-MS levels for 3/4 culture-negative samples were low or moderate (S2 Fig). Interestingly, two of these patients had been culture-positive for H. influenzae two weeks before the study sample was taken. Heavy growth of Klebsiella pneumoniae might have obscured the presence of H. influenzae in the fourth sample which yielded high PCR/ESI-MS levels (Table 3). Similarly, CFU levels were significantly higher in samples with positive result by PCR/ESI-MS, while sparse growth of H. influenzae was detected in three PCR/ESI-MS-negative samples (Table 3).
Table 3. Data for patients with discrepant results with respect to respiratory pathogens obtained by PCR/ESI-MS and routine culture-based analysis.
| Patient | Culture reporta (CFU/ml) | PCR/ESI-MS (semi-quantitative levels) |
|---|---|---|
| Male, 36 yrs | Haemophilus influenzae (102−103) | Proteus mirabilis (127) |
| Female, 70 yrs | Haemophilus influenzae (103−104), Streptococcus pneumoniae (103−104) | Streptococcus pneumoniae (262) |
| Male, 35 yrs | Haemophilus influenzae (102−103), Group G streptococcus (102−103) | Viridans/Salivarius Group Streptococcus (31), Viridans Group Streptococcus (18), Gemella sanguinis (11), Bacteria no ID (11) |
| Male, 84 yrs | Normal respiratory flora | Haemophilus influenzae (47)b |
| Male, 74 yrs | No growth | Haemophilus influenzae (37), Candida albicans (33) |
| Male, 41 yrs | Normal respiratory flora; mold | Haemophilus influenzae (156)b, Streptococcus species (76), Granulicatella adiacens (52) |
| Male, 46 yrs | Streptococcus pneumoniae (>104), Klebsiella pneumoniae (>104) | Haemophilus influenzae (503)c, Klebsiella pneumoniae (14) |
| Male, 33 yrs | Normal respiratory flora | Corynebacterium pseudodiphteriticum (120) |
| Male, 6 yrs | Moraxella (Branhamella) catarrhalis (>104), Streptococcus pneumoniae (103−104) | Moraxella catarrhalis/nonliquefaciens (547)d, Streptococcus species (49) |
| Male, 72 yrs | Yeast (102−103) | Enterobacter cloacae-complex (169), Granulicatella adiacens (29), Candida dubliniensis (70), Candida albicans (109) |
| Female, 72 yrs | Normal respiratory florae | Escherichia vulneris (165), Streptococcus pseudopneumoniae (694), Candida albicans (302) |
| Female, 68 yrs | No growth | Proteus vulgaris (461), Staphylococcus lugdunensis (5) |
| Male, 48 yrs | Normal respiratory flora | Klebsiella oxytoca (11)f, Streptococcus pneumoniae (45), Moraxella catarrhalis/nonliquefaciens (32), Candida albicans (17) |
| Male, 56 yrs | Streptococcus pneumoniae (102−103) | Pseudomonas aeruginosa (237), Bacteria no ID (37) |
| Female, 70 yrs | No growth | Stenotrophomonas maltophilia (304), Candida albicans (9) |
| Male, 29 yrs | Normal respiratory flora | Streptococcus pneumoniae (43), Chryseobacterium indologenes (17), Granulicatella adiacens (77), Bacteria no ID (47) |
| Male, 49 yrs | Klebsiella pneumoniae | Staphylococcus aureus (17)g, Streptococcus species (10), Granulicatella adiacens (90), Bacteria no ID (40) |
a Primary or potentially pathogenic species are indicated in bold; for results regarding S. aureus and S. pneumoniae see Fig 1 and Fig 2.
b Positive culture for H. influenzae 2 weeks before study samples.
c Positive culture for H. influenzae 2 weeks after study sample.
d Moraxella species with comment “normal respiratory flora” from PCR/ESI-MS.
e Positive fungal culture.
f Positive culture for Klebsiella oxytoca in a sample taken 1 day before study sample.
g Staphylococcus aureus in another respiratory sample taken the same day.
PCR/ESI-MS allows detection of non-cultivable and fastidious pathogens
In four BAL samples, atypical pathogens were detected which were not covered by the routine culture-based method, namely Legionella pneumophila, Bordetella pertussis, Mycoplasma pneumoniae and Nocardia species. The detection of these microorganisms was confirmed by other methods in the identical or in related patient samples (Table 1).
Resistance determinants mecA, vanA and vanB and blaKPC
The methicillin resistance-determinant mecA was detected in 21/121 (17%) BAL samples. Only 8/21 (38%) of these samples were also positive for S. aureus, indicating the potential presence of methicillin-resistant S. aureus (MRSA). Dose-response experiments showed that PCR/ESI-MS levels for S. aureus and the mecA gene were of similar range. An overall correlation between mecA and S. aureus-specific signals in these eight samples could however not been established, suggesting other origin of the mecA in at least part of the samples. Moreover, MRSA was not detected in any of the samples by culture, and significant growth of S. aureus was only reported for one sample. In one patient with high mecA-specific PCR/ESI-MS levels, however, a MRSA was isolated from another respiratory sample. Of the remaining mecA-positive samples, 12/13 displayed high PCR/ESI-MS levels (>100), and coagulase-negative staphylococci were detected in all of them, suggesting an associated of mecA to a none-S. aureus staphylococcal species.
There was no BAL sample that was vanA/vanB-positive. Similarly, no carbapenem-resistant or blaKPC-positive BAL sample was detected in the studied material.
Report of fungal DNA without identification
While the BAC LRT Assay identifies Candida to genus level and eight Candida species furthermore to species level, a large panel of fungal DNA can be detected but not species-identified. The assay is moreover not designed for detection of fungi. However, for seven samples, the presence of fungal DNA was reported. Six of these samples were also sent for routine fungal diagnostics. In five of them, growth of fungi other than Candida was detected, including Aspergillus fumigatus, Aspergillus versicolor, Penicillium species, Saccharomyces cerevisiae and Trichosporon ashii.
Discussion
PCR/ESI-MS is one of the most recent methods developed for detection and identification of microorganisms and selected resistance markers directly from clinical specimens. The aim of the present study was to evaluate the analytical performance of the PCR/ESI-MS platform IRIDICA in identification of microorganisms from BAL samples.
PCR/ESI-MS could identify 60 different microorganisms in 121 BAL samples and demonstrated an overall higher sensitivity compared to routine culture-based microbiological diagnostics, with identification of microorganisms in 15/17 (88%) culture-negative samples. In only one PCR/ESI-MS-negative sample, growth of normal respiratory flora was reported. Detailed microbiological diagnostics of BAL samples may be valuable information to the clinician depending on the clinical status of the patient.
When the analytical performance of PCR/ESI-MS in detection of primary or potential pathogens was analyzed, we found that the method in general was not inferior to culture-based methods in detection of these microorganisms. The vast majority of pathogens detected by culture, 52/60 (87%), could be detected by PCR/ESI-MS.
Identification of S. pneumoniae and differentiation from other α-hemolytic streptococci is notoriously difficult [20, 21]. Among the sample material investigated in this study, only 6/17 (35%) detections of S. pneumoniae were confirmed by culture or additional molecular analysis. In contrast, S. pneumoniae could not be identified in three samples with positive culture results. Interestingly, multiple matches were reported for the two samples with positive culture results, specified as Streptococcus mitis/pneumoniae and S. pneumoniae. This result was not displayed for any of the culture-negative samples. The present data indicates that PCR/ESI-MS has limited specificity and sensitivity in detection of S. pneumoniae. The low number of clinical isolates detected in BAL cultures does however not allow solid conclusions and further studies are needed. Based on the findings presented here, other tests complementary to PCR/ESI-MS are recommended to determine the presence or absence of S. pneumoniae if clinically indicated.
We experienced a similar problem in identification of Moraxella catarrhalis, which was reported as Moraxella catarrhalis/nonliquefaciens belonging to the normal flora. Problems to differentiate the two Moraxella species by molecular methods have previously been reported and been related to limited genomic sequence data for design of specific primers [13].
While all detected microorganisms are reported as positive finding by PCR/ESI-MS, culture reports include some degree of evaluation by a clinical microbiologist. Based on a low number of colonies or the presence of other fast-growing microorganisms, opportunistic or slow-growing pathogens might not be reported. This may explain the majority of the discrepancies observed for detection of S. aureus, a colonizer of the upper respiratory tract in up to 30% of adults [22, 23]. Samples with S. aureus-negative culture reports yielded overall significantly lower PCR/ESI-MS levels. Moreover, bacteria belonging to the normal flora were detected in 12/18 (67%) of these low-levels samples, but only in 4/15 (27%) samples, for which S. aureus was considered relevant by culture (P<0.05). These observations allow the suggestion that most discrepancies between culture reports and PCR/ESI-MS results are based on clinical interpretation of culture results rather than on false-positive reactions by PCR/ESI-MS. The present data also suggests that semi-quantitative levels and co-detection of commensal flora are relevant information for clinical interpretation of results from PCR/ESI-MS, in particular with respect to detection of S. aureus. PCR/ESI-MS results from BAL samples should thus be interpreted similarly to culture results. Microorganisms that are regarded as normal respiratory flora in BAL culture should reasonably be indicated as being part of the normal respiratory flora even when reporting results from PCR/ESI-MS on BAL.
In the present study, we were able to detect non-cultivable and fastidious pathogens with PCR/ESI-MS, i.e. L. pneumophila, M. pneumonia, Nocardia species and B. pertussis, which were overlooked by routine standard culture. Rapid detection of atypical pathogens from BAL samples by PCR/ESI-MS might be important in the clinical routine.
In addition, detection of fungal DNA by PCR/ESI-MS proved to be a reliable indication for the presence of non-Candida fungal species. Although no identification can be provided by the IRIDICA BAC LRT Assay, detection of fungal DNA in a sample may suggest that fungal-specific investigation is indicated and that antifungal treatment could be considered. It should however be noted that the IRIDICA BAC LRT Assay is not designed for detection of fungal species other than Candida and sensitivity for detection of molds by this assay has not been studied.
The IRIDCA BAC LRT Assay panel also includes selected major resistance determinants, i.e. mecA, vanA, vanB, and blaKPC. The only gene detected in the samples investigated here was mecA. In Sweden, only 1–3% of S. aureus from bloodstream infections are currently mecA-positive (http://ecdc.europa.eu) [24, 25]. Coagulase-negative staphylococci are however commonly detected as part of the normal flora or contaminant, and are frequently positive for mecA. In regions with relatively low prevalence of MRSA, the detection of mecA without physical linkage to a S. aureus isolate is thus of no diagnostic value in this kind of sample.
One of the limitations of the present study is the lack of clinical data. The clinical value of a comprehensive microbiological response to the clinicians could therefore not be assessed. In addition, the analysis was performed on consecutive BAL samples. In clinical practice however, samples from only a selected group of patients will be chosen for analysis by PCR/ESI-MS. The array of pathogens might thus probably differ and influence the overall performance of PCR/ESI-MS. A recent study on BAL samples from mechanically ventilated patients [26] indicated promising clinical performance of PCR/ESI-MS in comparison with other microbiological diagnostic approaches.
In conclusion, our study demonstrates overall reliable detection of respiratory pathogens by PCR/ESI-MS directly from BAL samples. PCR/ESI-MS thus provides an interestingly tool for the rapid detection of a wide range of pathogens from polymicrobial samples. This might be especially interesting for critically ill patients undergoing antimicrobial therapy at the time of sampling. Studies including clinical data are warranted to improve interpretation of the data.
Supporting Information
(A) Culture results for S. pneumoniae in relation to CT-values for the lytA gene. (B) Relation between semi-quantitative PCR/ESI-MS levels and CT-values for the lytA gene.
(TIF)
In the sample indicated by a triangle, heavy growth of K. pneumoniae was detected.
(TIF)
Acknowledgments
Abbott provided an IRIDICA PCR/ESI-MS instrument and IRIDICA test kits for the study. However, Abbott had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors received no specific financial funding for this work.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Abbott provided an IRIDICA PCR/ESI-MS instrument and IRIDICA test kits for the study. However, Abbott had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors received no specific financial funding for this work.
References
- 1.Welte T, Torres A, Nathwani D. Clinical and economic burden of community-acquired pneumonia among adults in Europe. Thorax. 2012;67: 71–9. 10.1136/thx.2009.129502 [DOI] [PubMed] [Google Scholar]
- 2.Arnold FW, Wiemken TL, Peyrani P, Ramirez JA, Brock GN, authors C. Mortality differences among hospitalized patients with community-acquired pneumonia in three world regions: results from the Community-Acquired Pneumonia Organization (CAPO) International Cohort Study. Respir Med. 2013;107: 1101–11. 10.1016/j.rmed.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 3.Prina E, Ranzani OT, Torres A. Community-acquired pneumonia. Lancet. 2015;386: 1097–108. 10.1016/S0140-6736(15)60733-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown JS. Geography and the aetiology of community-acquired pneumonia. Respirology. 2009;14: 1068–71. 10.1111/j.1440-1843.2009.01641.x [DOI] [PubMed] [Google Scholar]
- 5.Cilloniz C, Ewig S, Polverino E, Marcos MA, Prina E, Sellares J, et al. Community-acquired pneumonia in outpatients: aetiology and outcomes. Eur Respir J. 2012;40: 931–8. 10.1183/09031936.00168811 [DOI] [PubMed] [Google Scholar]
- 6.Torres A, Ewig S, Lode H, Carlet J. Defining, treating and preventing hospital acquired pneumonia: European perspective. Intensive Care Med. 2009;35: 9–29. 10.1007/s00134-008-1336-9 [DOI] [PubMed] [Google Scholar]
- 7.Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis. 2010;51 Suppl 1: S81–7. [DOI] [PubMed] [Google Scholar]
- 8.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44 Suppl 2: S27–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Souweine B, Veber B, Bedos JP, Gachot B, Dombret MC, Regnier B, et al. Diagnostic accuracy of protected specimen brush and bronchoalveolar lavage in nosocomial pneumonia: impact of previous antimicrobial treatments. Crit Care Med. 1998;26: 236–44. [DOI] [PubMed] [Google Scholar]
- 10.Kottmann RM, Kelly J, Lyda E, Gurell M, Stalica J, Ormsby W, et al. Bronchoscopy with bronchoalveolar lavage: determinants of yield and impact on management in immunosuppressed patients. Thorax. 2011;66: 823. [DOI] [PubMed] [Google Scholar]
- 11.Baudel JL, Tankovic J, Dahoumane R, Carrat F, Galbois A, Ait-Oufella H, et al. Multiplex PCR performed of bronchoalveolar lavage fluid increases pathogen identification rate in critically ill patients with pneumonia: a pilot study. Ann Intensive Care. 2014;4: 35 10.1186/s13613-014-0035-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunze N, Moerer O, Steinmetz N, Schulze MH, Quintel M, Perl T. Point-of-care multiplex PCR promises short turnaround times for microbial testing in hospital-acquired pneumonia—an observational pilot study in critical ill patients. Ann Clin Microbiol Antimicrob. 2015;14: 33 10.1186/s12941-015-0091-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadsby NJ, McHugh MP, Russell CD, Mark H, Conway Morris A, Laurenson IF, et al. Development of two real-time multiplex PCR assays for the detection and quantification of eight key bacterial pathogens in lower respiratory tract infections. Clin Microbiol Infect. 2015;21: 788 e1–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordana-Lluch E, Gimenez M, Quesada MD, Ausina V, Martro E. Improving the diagnosis of bloodstream infections: PCR coupled with mass spectrometry. Biomed Res Int. 2014;2014: 501214 10.1155/2014/501214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeng K, Hardick J, Rothman R, Yang S, Won H, Peterson S, et al. Reverse transcription-PCR-electrospray ionization mass spectrometry for rapid detection of biothreat and common respiratory pathogens. J Clin Microbiol. 2013;51: 3300–7. 10.1128/JCM.01443-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huttner A, Emonet S, Harbarth S, Renzi G, Kaiser L, Schrenzel J. Polymerase-chain reaction/electrospray ionization-mass spectrometry for the detection of bacteria and fungi in bronchoalveolar lavage fluids: a prospective observational study. Clin Microbiol Infect. 2014;20: O1059–66. 10.1111/1469-0691.12749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent JL, Brealey D, Libert N, Abidi NE, O'Dwyer M, Zacharowski K, et al. Rapid diagnosis of infection in the critically ill, a multicenter study of molecular detection in bloodstream infections, pneumonia, and sterile site infections. Crit Care Med. 2015;43: 2283–91. 10.1097/CCM.0000000000001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedberg ST, Olcen P, Fredlund H, Molling P. Real-time PCR detection of five prevalent bacteria causing acute meningitis. APMIS. 2009;117: 856–60. 10.1111/j.1600-0463.2009.02539.x [DOI] [PubMed] [Google Scholar]
- 19.Berglund C, Molling P, Sjoberg L, Soderquist B. Predominance of staphylococcal cassette chromosome mec (SCCmec) type IV among methicillin-resistant Staphylococcus aureus (MRSA) in a Swedish county and presence of unknown SCCmec types with Panton-Valentine leukocidin genes. Clin Microbiol Infect. 2005;11: 447–56. 10.1111/j.1469-0691.2005.01150.x [DOI] [PubMed] [Google Scholar]
- 20.Arbique JC, Poyart C, Trieu-Cuot P, Quesne G, Carvalho Mda G, Steigerwalt AG, et al. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J Clin Microbiol. 2004;42: 4686–96. 10.1128/JCM.42.10.4686-4696.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho Mda G, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, et al. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol. 2007;45: 2460–6. 10.1128/JCM.02498-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson P, Ripa T. Staphylococcus aureus throat colonization is more frequent than colonization in the anterior nares. J Clin Microbiol. 2006;44: 3334–9. 10.1128/JCM.00880-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nurjadi D, Lependu J, Kremsner PG, Zanger P. Staphylococcus aureus throat carriage is associated with ABO-/secretor status. J Infect. 2012;65: 310–7. 10.1016/j.jinf.2012.05.011 [DOI] [PubMed] [Google Scholar]
- 24.Johnson AP. Methicillin-resistant Staphylococcus aureus: the European landscape. J Antimicrob Chemother. 2011;66 Suppl 4: iv43–iv8. [DOI] [PubMed] [Google Scholar]
- 25.Laupland KB, Lyytikainen O, Sogaard M, Kennedy KJ, Knudsen JD, Ostergaard C, et al. The changing epidemiology of Staphylococcus aureus bloodstream infection: a multinational population-based surveillance study. Clin Microbiol Infect. 2013;19: 465–71. 10.1111/j.1469-0691.2012.03903.x [DOI] [PubMed] [Google Scholar]
- 26.Strålin K, Ehn F, Giske CG, Ullberg M, Hedlund J, Petersson J, et al. The IRIDICA PCR/Electrospray Ionization-Mass Spectrometry assay on bronchoalveolar lavage for bacterial etiology in mechanically ventilated patients with suspected pneumonia. PLoS One. 2016;11: e0159694 10.1371/journal.pone.0159694 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Culture results for S. pneumoniae in relation to CT-values for the lytA gene. (B) Relation between semi-quantitative PCR/ESI-MS levels and CT-values for the lytA gene.
(TIF)
In the sample indicated by a triangle, heavy growth of K. pneumoniae was detected.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.


