Abstract
Trachoma is caused by Chlamydia trachomatis. Clinical grading with the WHO simplified system can be highly repeatable provided graders are adequately trained and standardized. At the community level, rapid assessments are useful for confirming the absence of trachoma but do not determine the magnitude of the problem in communities where trachoma is present. New rapid assessment protocols incorporating techniques for obtaining representative population samples (without census preparation) may give better estimates of the prevalence of clinical trachoma. Clinical findings do not necessarily indicate the presence or absence of C. trachomatis infection, particularly as disease prevalence falls. The prevalence of ocular C. trachomatis infection (at the community level) is important because it is infection that is targeted when antibiotics are distributed in trachoma control campaigns. Methods to estimate infection prevalence are required. While culture is a sensitive test for the presence of viable organisms and nucleic acid amplification tests are sensitive and specific tools for the presence of chlamydial nucleic acids, the commercial assays presently available are all too expensive, too complex, or too unreliable for use in national programs. There is an urgent need for a rapid, reliable test for C. trachomatis to assist in measuring progress towards the elimination of trachoma.
INTRODUCTION
The World Health Organization (WHO) defines blindness as visual acuity in the better eye of less than 3/60 with available refractive correction, which predicts the inability to walk safely without assistance. The best published estimate suggests that 5.9 million people in the world fulfill this criterion because of trachoma, which makes it responsible for about 15% of all cases of blindness (227). In addition to those already blind, an estimated 600 million people live in areas of Africa, the Middle East, and Central and South America, Asia, Australia, and the Pacific Islands where trachoma is endemic (225, 227). The accuracy of these estimates is questionable, however (6). There is a pressing need for further research on the distribution and prevalence of disease (116).
EPIDEMIOLOGY
Patterns of Distribution
Trachoma is, first and foremost, a disease of poverty. It thrives in remote, marginalized, and displaced populations. Within areas where it is endemic, the distribution of disease is heterogeneous. Some communities are badly affected, while others with seemingly similar community-level risk factors (such as poor access to water and sanitation) are not. In affected communities, clustering of disease by subvillage (241), compound (11), and bedroom (11) has been noted. This clustering at different scales is reminiscent of fractal geometry (169) and probably reflects the importance of transmission of infection between members of the same family (11, 18) and (in some settings) transmission between families with close social ties (172a).
CAUSATIVE ORGANISM
Historical Perspective
Trachoma has been recognized for millennia as a blinding disease. It has been known in Egypt for more than 3,500 years (62, 109, 130). Its contagious nature was recognized in Syria in the thirteenth century (1), but upon first coming to the attention of European surgeons during the Napoleonic campaigns in Egypt in 1798 to 1799, the French determined that it was due either to sand or to noxious night vapors. The British, on the other hand, believed that it was caused by a virus and took appropriate measures; their infantry suffered a lower incidence of blindness (62). In the late nineteenth and early twentieth centuries, the discovery of clinical trachoma in would-be U.S. immigrants disembarking at Ellis Island, New York, N.Y., was responsible for more than half of all medical detentions there and resulted for many in deportation back to the port of origin (137; http://www.americanparknetwork.com/parkinfo/sl/history/journey.html; http://www.infectiousdiseasenews.com/200201/immigrants.asp).
The causative agent of trachoma was not visualized until 1907, when Halberstaedter and von Prowazek described the presence of inclusion bodies (Halberstaedter-Prowazek bodies) inside infected cells. They believed the organism to be a protozoon (90). The transmissibility of trachoma was by then already firmly established in the minds of the public. Hundreds of Russian and Austro-Hungarian First World War conscripts, for example, evaded military service by infecting their own eyes with discharges wiped from the eyes of trachoma patients (62). Meanwhile, unconvinced by the findings of Halberstaedter and von Prowazek, researchers nominated a variety of bacteria, fungi, and viruses as the underlying pathogen (62). It was not until 1957 in Peking that T'ang et al. completed the first successful isolation, using chicken embryos whose yolk sacs had been inoculated with material from infected human eyes (212). They were able to serially passage the organism in eggs and to use this material to infect the eyes of rhesus monkeys, producing characteristic clinical signs of trachoma and, on one occasion, inclusion bodies. Based on filtration experiments, they believed the trachoma agent to be a virus (212). T'ang et al.'s methods were successfully replicated by Collier and Sowa in the Gambia in 1958 (45). The isolates obtained were noted both to have the same antigen as and to physically resemble the agents of psittacosis and lymphogranuloma venereum (45) and the agent of some cases of cervicitis and mucopurulent conjunctivitis of the newborn (110).
As knowledge of the nature of these organisms accumulated, there was considerable debate over whether they represented a transitional remnant on the degenerate evolutionary pathway that Green had hypothesized (82) for the descent of viruses from bacteria, or whether they should be placed wholly within one or another of these classes. In 1966, Moulder published a comprehensive review of the growth, division, structure, chemical composition, and metabolism of the group, taking into account the definitions of viruses and bacteria that had recently been proposed by Lwoff and Stanier, respectively. He concluded fairly unequivocally that chlamydiae were intracellular bacteria, with a distinctive developmental cycle and unique structure (151). The Taxonomy Committee of the American Society for Microbiology unified these organisms in the genus Chlamydia and supported their status as bacteria (166). Today, some 15 major bacterial groupings are recognized, and the chlamydiae are the only ones whose members are all exclusively intracellular parasites of eukaryotes (66).
Developmental Cycle
Chlamydiae lack cytochromes and so cannot synthesize their own ATP. They are therefore obligate intracellular organisms, requiring energy-rich metabolic intermediates from host cells in order to complete their replication cycle (30). To permit egress from infected cells and entry of new ones, a metabolically inert, extracellular infectious form known as the elementary body alternates with the metabolically active, dividing, intracellular form, the reticulate body.
Elementary bodies of chlamydiae are spherical (or, rarely, pear-shaped) and 0.2 to 0.3 μm in diameter (42). They appear to bind to susceptible host cells via heparin bridges. There is considerable interest in identifying the chlamydial ligands involved in heparin binding: candidates include the major outer membrane protein and the cysteine-rich protein OmcB, both of which are found in the chlamydial outer membrane complex (203, 209). Successful attachment of the elementary body is followed by its entry into the host cell. Although hosts are nonprofessional phagocytes, their oxidative and glycolytic pathways must be intact for entry to occur, suggesting that they participate actively in elementary body ingestion (150). Tyrosine phosphorylation of host cell proteins and actin cytoskeletal rearrangement may be involved (69, 179). Although elementary bodies are taken up 10 to 100 times more efficiently than latex particles of the same size or Escherichia coli (34), the active participation of the elementary body appears to be minimal; elementary body envelopes are internalized just as efficiently as whole elementary bodies (65). The processes involved in attachment and uptake may differ between species of chlamydiae and even between variants of the same species (179, 196).
Once inside a host cell, the elementary body reorganizes into a 0.5- to 1.0-μm-diameter reticulate body within a membrane-bound vacuole known as an inclusion. The reticulate body grows and replicates by binary fission, remaining within the inclusion membrane (derived from the cytoplasmic membrane of the host) for the entire duration of the organism's intracellular phase. After a period of exponential growth, progeny differentiate back into elementary bodies. A number of late-phase proteins are synthesized during the reticulate body-to-elementary body transformation, including chlamydial outer membrane complex proteins OmcB and OmcA and two histone H1-like proteins, Hc1 and Hc2, which are involved in compaction of the chlamydial chromosome (42, 85). Elementary bodies are released into the extracellular environment by fusion of the membrane of the inclusion with that of the host cell or upon host cell lysis (30, 150). The specific processes involved in cellular exit are poorly defined. In tissue culture, the entire developmental cycle from attachment to exit takes between 48 and 72 h.
Structure
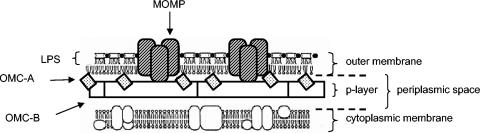
With an electron microscope, an elementary body is seen to have a granular cytoplasm, reflecting the presence of 70S ribosomes, and an eccentrically placed nucleoid containing condensed DNA (140). The cell envelope is double layered, resembling the cell envelope of gram-negative bacteria (30). The cell wall (the portion of the cell envelope lying external to the cytoplasmic membrane) can itself be resolved into two layers: an inner (p) layer composed of hexagonally arrayed structures, and a granular outer layer containing the outer membrane (42). The inner layer therefore lies within the periplasmic space (Fig. 1).
FIG. 1.
Model of the elementary body cell wall, after Everett and Hatch (67). LPS, lipopolysaccharide.
Cylindrical projections radiate from the outer membrane of the elementary body. Each projection has its inner end at the cytoplasmic (inner) membrane, extending outwards to penetrate the outer membrane through the center of a membrane-bound rosette. A rosette is made up of eight or nine protein subunits: the number of subunits varies between species of chlamydiae (141). DNA strands can be seen connecting the nucleoid with the cytoplasmic membrane subjacent to the projections (249).
Reticulate bodies are larger than elementary bodies and contain diffuse, fibrillar DNA plus a high concentration of ribosomes. The cell envelope appears less complex than that of the elementary body, lacking the hexagonally packed structures of the elementary body periplasmic space. The surface of the reticulate body outer membrane, however, contains projections and rosettes at even higher densities than are seen on elementary bodies (139). The outer end of the projections appear to contact the inclusion membrane, leading to the hypothesis that projection-rosette complexes have a secretory function analogous to the type III secretory system found in other bacterial species (42, 104).
Cellular components important for diagnostic assays: MOMP and lipopolysaccharide.
New knowledge of the biology of chlamydiae has been accruing very quickly since the chlamydial genome sequence was published in 1998 (202; R. S. Stephens, S. Kalman, C. Fenner, and R. David, 1998, Chlamydia genome project [http://chlamydia-www.berkeley.edu:4231], accessed 15 January 2003). In this review, discussion of chlamydial structural antigens will be limited to two components of the chlamydial outer membrane complex that are of relevance to the diagnosis of ocular chlamydial infection, the major outer membrane protein (MOMP) and lipopolysaccharide. First, however, a brief comment about the likely function of the chlamydial outer membrane complex may be helpful.
Most bacteria have peptidoglycan, a complex cross-linked polymer, in their cell envelope. In gram-negative organisms, peptidoglycan is found in the periplasmic space, while in gram-positive organisms it lies immediately outside the cytoplasmic membrane and may constitute up to 50% of total cell wall material (30). Its function is to help maintain cell shape and integrity despite the relatively high internal osmotic pressure of the bacterium. Penicillin and other β-lactam antibiotics inhibit the growth of susceptible organisms by preventing the formation of peptide cross-links in peptidoglycan. This effect is mediated through bacterial penicillin-binding proteins. Chlamydiae produce penicillin-binding proteins, and attempts to grow them in the presence of penicillin result in the formation of aberrant inclusions, but, surprisingly, they do not appear to contain appreciable amounts of peptidoglycan (17, 35). This paradox raises interesting questions about the biology of the organism (41, 76) and demands an alternative explanation for the rigidity and osmotic stability of the elementary body. These properties are presently thought to be conferred by the chlamydial outer membrane complex.
The chlamydial outer membrane complex was first defined in 1981, when Caldwell et al. (35) reported the outcome of their experiments with the detergent Sarkosyl (sodium N-lauroyl sarcosine), which had been shown to selectively solubilize the cytoplasm and cytoplasmic membranes of gram-negative bacteria (71). Transmission electron microscopy of Sarkosyl-treated elementary bodies showed empty elementary body particles with an apparently intact outer membrane (35). Caldwell et al.'s method has since been used as the standard method for purification of the chlamydial outer membrane complex. Using sodium dodecyl sulfate-polyacrylamide gel electrophoresis of elementary body lysates, the same group identified a protein of about 40 kDa that was found in a number of chlamydial strains. They went on to show that this protein, which soon became known as the major outer membrane protein (MOMP), was one of the elements of the Sarkosyl-insoluble chlamydial outer membrane complex and that it constituted about 60% of the total protein mass of the elementary body cell wall. 125I labeling of the protein indicated that it was surface exposed (35, 95, 183).
Hatch et al., trying to extract MOMP from other components of the cell wall, noted that it could not be dissolved in sodium dodecyl sulfate or mercaptoethanol alone but was soluble in a solution containing both of these agents. This implied that disulfide bonds were important in binding MOMP to the chlamydial outer membrane complex and perhaps in maintaining the overall structural stability of the cell wall (95).
It was quickly recognized that the importance of MOMP was not confined to its structural role. Salari and Ward were able to extract MOMP from 14 of the 15 then-known serovars (see below) of the chlamydial species Chlamydia trachomatis and noted minor serovar-specific variations in its molecular weight (183). The existence of species- and subspecies-specific epitopes within the protein was noted (37, 206), before it became clear that MOMP also contained serovar-specific epitopes (138, 162). Even more important, polyclonal (36) and then monoclonal (168, 251) antibodies to MOMP were shown to neutralize infectivity of the live organism. These discoveries raised hopes that the protein would be useful in the development of a protective subunit vaccine. Unfortunately, such hopes have so far remained unfulfilled (129).
Although multiple strands of evidence suggest that MOMP is surface exposed, other data have localized parts of the molecule to the periplasmic space (10). These two conclusions are consistent with MOMP's being an integral membrane protein. It now seems likely that MOMP's physiological function is as a membrane channel or porin permeable to ATP. Prevention of uptake of host cell ATP could potentially be a mechanism by which antibodies to MOMP block cellular infection (248). Although the protein is thought to form trimeric aggregates within the outer membrane (23), its actual conformation within intact chlamydiae is unknown (94).
A model of the cell envelope structure has been suggested by Everett and Hatch (67) (Fig. 1). It has been hypothesized that the structural stability of elementary bodies is maintained by disulfide cross-linking between cysteine residues of MOMP and other membrane proteins (67, 160). A similar supramolecular structure is absent from reticulate bodies, which are osmotically fragile. Protein cross-linking appears to occur during the last 24 h of the intracellular phase of the life cycle (161).
All species of chlamydiae identified to date have a common lipopolysaccharide that differs from the lipopolysaccharide of other bacteria. The molecule is present in the outer membrane of the cell envelope throughout the life cycle (24, 145) and contains polysaccharide epitopes recognized by the human humoral immune system (30, 59).
Taxonomy
In 1980, when the Approved Lists of Bacterial Names were first published, the chlamydiae had two species, Chlamydia trachomatis and Chlamydia psittaci (165). In 1989, isolates previously identified as the TWAR strain of C. psittaci (80) were proposed as a third species, Chlamydia pneumoniae; it was differentiated from other chlamydiae on the basis of the shape of the elementary body, serology, and DNA analysis (79). Another group of strains originally classified as C. psittaci were subsequently reassigned to Chlamydia pecorum following further DNA and serological analyses (75).
In 1999, a paper by Everett et al. attempted to reclassify the family into two genera, Chlamydia and Chlamydophila, which together contain a total of nine species (66). This new nomenclature has proven controversial (J. Schachter, R. S. Stephens, P. Timms, C. Kuo, P. M. Bavoil, S. Birkelund, J. Boman, H. Caldwell, L. A. Campbell, M. Chernesky, G. Christiansen, I. N. Clarke, C. Gaydos, J. T. Grayston, T. Hackstadt, R. Hsia, B. Kaltenboeck, M. Leinonnen, D. Ocjius, G. McClarty, J. Orfila, R. Peeling, M. Puolakkainen, T. C. Quinn, R. G. Rank, J. Raulston, G. L. Ridgeway, P. Saikku, W. E. Stamm, D. Taylor-Robinson, S. P. Wang, and P. B. Wyrick, letter, Int. J. Syst. Evol. Microbiol. 51:249, 2001; K. Everett and A. Andersen, authors' reply to letter, Int. J. Syst. Evol. Microbiol. 51:251-253, 2001). Whether or not the proposed changes enter general use, the designation of the pathogen responsible for human trachoma will remain Chlamydia trachomatis.
Classification and Tropism of C. trachomatis Strains
Three biovars (groups of strains distinguishable from others of the same species on the basis of physiological characteristics) of C. trachomatis are recognized: mouse pneumonitis, lymphogranuloma venereum, and trachoma.
The mouse pneumonitis biovar (which gains species status in the Everett et al. classification) includes two strains: MoPn, which is found in the respiratory tract of mice, and SFPD, which has been isolated from the intestines of hamsters (66). Neither strain is known to infect humans.
The other two C. trachomatis biovars preferentially infect humans. They are closely related. Four serotypes or serovars are currently included in the lymphogranuloma venereum biovar, and 15 are currently included in the trachoma biovar. Each of the 19 can be distinguished from the others on the basis of binding affinity for monoclonal antibodies. They can also be differentiated by polymorphisms in the sequence of MOMP or in the sequence of the gene omp1, which codes for MOMP. There is very limited variation in these sequences between isolates of any given serovar (66). Separation of the lymphogranuloma venereum and trachoma strains into two biovars is based on tissue tropism: lymphogranuloma venereum strains can invade lymphatic tissue, while trachoma strains are restricted to mucosal epithelial cells.
The lymphogranuloma venereum serovars (L1, L2, L2a, and L3) are rare. All are sexually transmitted, although the eye may also act as the portal of entry. Infection is associated with a suppurative adenitis, usually of the inguinal or perirectal nodes, as well as systemic symptoms. The disease is most commonly seen in tropical and subtropical areas (30).
The trachoma serovars of C. trachomatis are designated by the letters A through K, plus Ba, Da, Ia, and Ja (66, 207). Different serovars have different tissue preferences. Serovars A, B, Ba, and C are the usual ocular isolates from patients with clinical trachoma in regions where trachoma is endemic, while D to K, Da, Ia, and Ja are typically associated with genital tract disease. The latter are the commonest causes of urethritis and mucopurulent cervicitis in females and nongonococcal urethritis in males. They have also been linked to female pelvic inflammatory disease, infertility, ectopic pregnancy, and chronic pelvic pain; male epididymitis, prostatitis, and infertility; neonatal conjunctivitis and pneumonia; and various arthritides.
Strains are tissue selective rather than specific. Even where trachoma is endemic, genital serovars are occasionally found in the eye. Serovars D (15, 31, 127), E (31), F (31, 97, 127), J (93), K (127), and L2 (with serovar A coinfection) (31) have all been isolated from conjunctival swabs taken from individuals with typical clinical signs of active trachoma. Similarly, ocular C. trachomatis strains are sometimes isolated from the genital tract. Frost et al. determined the serovars of 435 isolates taken from male and female attendees at sexually transmitted disease, perinatal, and family planning clinics in Canada and found that 5% were serovar Ba and 2% were serovar C strains (73).
Genome of C. trachomatis
Chromosome.
Chlamydia trachomatis contains a single ≈1,043,000-bp chromosome (202). The first gene to be analyzed was that coding for MOMP, which was designated omp1. In 1986, Stephens et al. sequenced omp1 from a C. trachomatis L2 strain after cloning and expressing the gene in an E. coli λ bacteriophage (205). Comparison of this gene with that from C. trachomatis serovars that were subsequently sequenced revealed extensive omp1 sequence variation. Most of the polymorphisms were localized to four 40- to 90-bp-long variable domains (VDs), designated VD1, VD2, VD3, and VD4, regularly distributed among the relatively conserved constant domains (CDs). Examination of the accessibility of MOMP segments to digestion by proteolytic enzymes suggests surface exposure of variable domain-encoded peptide sequences, with localization of the protein's amino and carboxy termini inside the periplasmic space (10). Serovar specificity of C. trachomatis appears to be determined by particular residues within VD1, VD2, and VD4 (10, 19). Yuan et al. found that for each serovar, the variable domain coding for the most hydrophilic and charged amino acid sequence contained the serovar-specific epitope (250). Later studies, however, indicated that omp1 of a given serovar can incorporate multiple distinct serovar-specific epitopes, each of which may be found in a different VD (19). Collectively, these findings (plus the demonstration that anti-MOMP antibodies neutralize the organism, as discussed above) indicate that the omp1 gene product, MOMP, spans the outer membrane of the cell envelope and presents immunologically important epitopes, coded for by one or more VDs, at the cell surface.
Heterogeneity in omp1 constant domains between urogenital and trachoma isolates of the same Ba and C serovars has been identified. The altered nucleotide sequences produce changes in the amino acid sequences of MOMP and could potentially play a role in determining the tissue tropism or virulence of the organism (74). More extensive analysis by Stothard et al. of 69 strains representing 17 serovars has not, however, supported an association between omp1 sequence and tissue tropism, disease presentation, or epidemiologic success (207).
The first complete C. trachomatis genome sequence (a serovar D isolate) was published by Stephens et al. in 1998 (202; R. S. Stephens, S. Kalman, C. Fenner, and R. David 1998, Chlamydia genome project [http://chlamydia-www.berkeley.edu:4231],accessed 15 January 2002). Notable findings included the localization of an entire set of genes required for peptidoglycan synthesis (despite the lack of demonstrable peptidoglycan in the organism, as discussed above) and genes encoding ATP biosynthetic pathways (despite C. trachomatis's apparent inability to make its own ATP). The genome sequence of a C. trachomatis mouse pneumonitis strain was subsequently also analyzed, which highlighted the presence of a plasticity zone near the chlamydial chromosome's origin of replication (176). This zone includes genes coding for enzymes involved in tryptophan synthesis. Ocular but not genital serovars of the C. trachomatis trachoma biovar have recently been found to carry a deletion or frameshift mutation at this locus. Ocular strains are therefore unable to synthesize tryptophan from exogenous indole (38, 70). This finding is the first known point of difference in the biosynthetic abilities of ocular and genital strains (70).
Plasmid.
In addition to the chromosome, chlamydiae commonly possess an extrachromosomal genetic element. The 7.4-kb plasmid pCT was first isolated from a C. trachomatis L2 strain by Palmer and Falkow in 1986 (167). Their studies identified pCT DNA in laboratory strains of all C. trachomatis serovars that cause human infection as well as in 200 separate clinical isolates. The plasmid is very highly conserved, with less than 1% variation in nucleotide sequence (222; M. E. Ward, 2002, Chlamydial plasmids [http://www.chlamydiae.com/chlamydiae/docs/biology/genome_plasmid.htm], accessed 26 June 2002). Because of this sequence conservation, and because maintenance of superfluous extrachromosomal DNA seems unlikely in a bacterium with a genome one quarter the size of that of E. coli, it was suggested that the plasmid might be essential for chlamydial growth or replication (46, 167). However, several naturally occurring C. trachomatis strains lacking the plasmid have since been isolated, including an L2 cultured from a patient with proctocolitis (171), a genotype B variant cultured from a male urethral swab (68), and a serovar E cultured from a male urethral swab (208). Such strains are thought to be rare (M. E. Ward, website cited above, accessed 26 June 2002), and no plasmid-free ocular isolates have been reported to date. Estimates of the mean number of plasmids per elementary body include 10 (167) (determined with a C. trachomatis L2 strain), 7 to 10 (211) (C. trachomatis L2), and 4 (172) (C. trachomatis L1). This estimate and the possibility of chlamydial infection without the presence of plasmid DNA both have implications for determining the likely sensitivity of some laboratory assays for C. trachomatis, as will be discussed later.
NATURAL HISTORY AND CLINICAL FEATURES
Clinically, trachoma can be divided into its acute (active) and chronic or late-stage manifestations, but acute and chronic signs can occur at the same time in the same individual. In areas where it is endemic, repeated episodes of active disease occur, particularly during childhood, and are probably required for later development of the chronic sequelae (81).
The degree of distress caused by ocular infection with C. trachomatis ranges from minimal to severe. Many infections are asymptomatic. In other cases, following an incubation period of 5 to 10 days, conjunctival infection produces an irritated, red eye and scanty mucopurulent discharge. Involvement of the cornea in the acute inflammatory process can cause pain and photophobia (184). In general, symptoms are milder than would be expected from the appearance of the eye (53).
The first sign of infection is a nonspecific vasodilation of conjunctival blood vessels (184). Specific changes may be noted after infection of several weeks duration (44), with the development of follicles subjacent to the conjunctivae of the fornices, the tarsal plates, and the limbus. Follicles are lymphoid germinal centers and are found immediately below the epithelial cell layer. They are grey or creamy masses 0.2 to 3.0 mm or more in diameter (47). It is not uncommon to find one or two follicles in normal healthy eyes, usually towards the lateral or medial canthi. Because the superficial layer of the conjunctival stroma lacks lymphoid tissue until about 3 months after birth (111), newborns are unable to mount a follicular response to ocular chlamydial infection (223). Papillae may also be noted at this stage: in mild cases, a few isolated, small red dots can be seen with the naked eye. With the aid of a slit lamp, papillae appear as small swellings of the conjunctiva, each with a central vascular core. When inflammation is severe, an intense papillary reaction on the tarsal conjunctiva is associated with a diffuse thickening of the conjunctiva, obscuration of the deep tarsal vessels, and, sometimes, eyelid edema. If the cornea is involved in the inflammatory process, a superficial punctate keratitis may be noted upon instillation of fluorescein into the conjunctival sac. Superficial infiltrates or pannus (subepithelial infiltration of fibrovascular tissue into the peripheral cornea, once thought to be found to at least some degree in every case of trachoma) (62) also indicate corneal inflammation. Follicles, papillae, and these corneal signs are features of active disease; the signs discussed below are all manifestations of late-stage trachoma. Note, however, that pannus may persist long after active disease has subsided.
Resolution of follicles may be accompanied by scarring of the subepithelial conjunctiva. Scar deposition is most prominent in the upper tarsal plate, although the conjunctival fornices, the bulbar conjunctiva, and the upper part of the cornea may also be involved. In areas where trachoma is endemic, upper tarsal plate scars derived from repeated episodes of infection can eventually accumulate to such an extent that they becomes visible macroscopically after eversion of the upper lid, appearing as white bands against the erythematous background of the conjunctiva. At the limbus, replacement of follicles by scar results in the formation of translucent depressions in the corneoscleral junction called Herbert's pits.
If sufficient tarsoconjunctival scarring accumulates, contraction of it over the years will cause the upper eyelid to turn inward so that the lashes rub against the globe. This is known as trichiasis. When the whole lid margin is turned in, the condition is known as entropion. Scars around the bases of hair follicles can pull individual eyelashes into contact with the cornea, even without entropion (184). Trichiasis is intensely irritating. Sufferers may use homemade forceps to remove their own lashes or attempt to keep their lids elevated with strips of cloth tied around their heads.
Besides being painful, trichiasis injures the cornea. In addition to the direct abrasive effect of the in-turned lashes, secondary bacterial and fungal infections of the cornea and corneal drying due to scarring of forniceal-mucous, lacrimal, and Meibomian glands accelerate epithelial damage. Collagenous scar is laid down as part of the repair process. Because scars are opaque, vision can be affected by scarring that involves the central part of the cornea.
CLINICAL DIAGNOSIS
History
Because active trachoma is usually found in children, is an almost universal experience of residents of communities where it is hyperendemic, and seems to cause little discomfort, there are generally few reported symptoms. Individuals with trichiasis can be symptomatic. The degree of distress depends on the number of lashes touching the globe, whether or not the cornea is abraded, and whether there is associated blepharospasm. The symptoms have been described above.
Examination
Examination of the eye for clinical signs of trachoma involves careful inspection of the lashes, cornea, and limbus, then eversion of the upper lid, and inspection of the tarsal conjunctiva. Binocular magnifying loupes (×2.5) and adequate lighting are required; if available, a slit lamp can be used, but it is not essential. Signs of interest are described above.
Differential Diagnosis
Follicles are not pathognomonic for trachoma but are reasonably predictive of it when seen in individuals living in communities where trachoma is endemic. The differential diagnosis of follicular conjunctivitis includes adult inclusion conjunctivitis (caused by infection with urogenital strains of C. trachomatis); other bacterial infections, particularly Moraxella spp. and Streptococcus pneumoniae; viral infections, including adenovirus, molluscum contagiosum, and herpes simplex virus; pediculosis palpebrarum; and toxic conjunctivitis secondary to topical drugs or eye cosmetics. The giant cobblestone papillae of vernal keratoconjunctivitis (spring catarrh) may be mistaken for follicles at first glance but are both clinically and histologically distinct (72, 111, 223).
Papillae are poorly specific for trachoma, particularly if there are no accompanying follicles. They form part of the conjunctival tissue's response to many acute and chronic inflammatory disorders and are therefore seen in bacterial, viral, and allergic conjunctivitides.
In areas where trachoma is endemic, pannus, conjunctival scarring, and trichiasis are nearly always attributable to trachoma. Herbert's pits are pathognomonic of past trachomatous inflammation. Corneal opacity, however, has many possible etiologies, including previous traumatic injury, viral, bacterial, and fungal infections, and a variety of other conditions. The prevalence of corneal scar is therefore not necessarily a good estimate of the contribution of trachoma to the overall burden of blindness and visual impairment.
Grading Systems
Grading systems are used in an effort to standardize diagnosis in field surveys and research studies. In the English language literature since 1900, at least 10 different systems or variations on systems have been published: by MacCallan in 1908 (131) and 1931 (130); by the WHO Expert Committee on Trachoma in 1962 (3); by the Fourth WHO Scientific Group on Trachoma Research in 1966 (4); by Tarizzo in 1973 (213); by Dawson, Jones, and Darougar in 1975 (52) and 1981 (53); by Darougar and Jones (47); by Tielsch, West, Johnson, Tizazu, Schwab, Chirambo, and Taylor in 1987 (226, 228); and by Thylefors, Dawson, Jones, West, and Taylor in 1987 (226). In addition, Sarkies in 1965 (185) and Melese, Alemayehu, Bejiga, Adamu, and Worku in 2003 (143) contributed subclassifications of trachomatous trichiasis and entropion.
The several variants of the MacCallan trachoma classification (130, 131) were probably derived from a description of the four stages of trachoma by Aetus of Amida in the sixth century (245). The MacCallan systems were durably popular in the ophthalmological literature of the first half of the twentieth century and still occasionally appear in papers in peer-reviewed journals (147) and consulting room reference texts (177), despite (i) implying that the clinical course is linear, ignoring trachoma's multicyclic nature; (ii) not mentioning corneal opacity or visual impairment and therefore having little prognostic value; and (iii) lacking clear definitions that would allow one stage to be reliably differentiated from the next (47, 52, 53, 109). Some of the later schemes are exceptionally complex: the 1966 proposal by the Fourth WHO Scientific Group on Trachoma Research, for example, took more than four pages to outline, recommending the grading of up to 19 signs, each of which had its own scale (4). Two classifications are in current general use and will be discussed here. The others have largely been superseded.
Dawson, Jones, and Tarizzo 1981 (modified WHO system or FPC system).
This grading system was developed “to describe more precisely the intensity of active trachoma” (52; p. 279) than did the MacCallan classification. An embryonic form of the FPC (follicles, papillae, cicatrices) system can be found in the 1962 WHO Expert Committee on Trachoma Third Report (3). Development of the system can be traced through a number of subsequent publications (52, 213), before its appearance in final (for WHO) form in the 1981 WHO manual “Guide to Trachoma Control in Programmes for the Prevention of Blindness” (53). The modified system includes five signs, each of which is graded independently in the right and left eye, as outlined here.
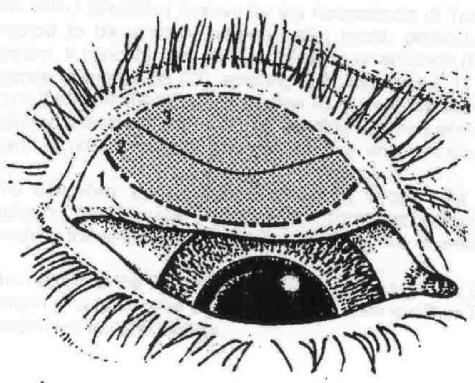
In the modified WHO system (53), the upper tarsal follicles (F) are graded F0 for no follicles, F1 for follicles present but no more than five in zones 2 and 3 together (Fig. 2), F2 for more than five follicles in zones 2 and 3 together but less than five in zone 3, and F3 for five or more follicles in each of the three zones. Upper tarsal papillary hypertrophy and diffuse infiltration (P) are graded P0 for absent, normal appearance; P1 for minimal, individual vascular tufts (papillae) prominent but deep subconjunctival vessels on the tarsus not obscured; P2 for moderate, more prominent papillae and normal vessels appear hazy even when seen by the naked eye; and P3 for pronounced, conjunctiva thickened and opaque, normal vessels on the tarsus are hidden over more than half of the surface. Conjunctival scarring (C) is graded C0 for no scarring on the conjunctiva; C1 for mild, fine, scattered scars on the upper tarsal conjunctiva or scars on the other parts of the conjunctiva; C2 for moderate, more severe scarring but without shortening or distortion of the upper tarsus; and C3 for severe scarring with distortion of the upper tarsus. Trichiasis/entropion (T/E) is scored as T/E0 for no trichiasis or entropion, T/E1 for lashes deviated towards the eye but not touching the globe, T/E2 for lashes touching the globe but not rubbing on the cornea, and T/E3 for lashes constantly rubbing on the cornea. Corneal scarring (CC) is scored CC0 for absent, CC1 for minimal scarring or opacity not involving the visual axis (determining whether or not corneal opacity “involves the visual axis” [the line between the fovea and the target] is impossible to determine by inspection [231, 236]: the term “over the pupil” might have been more practical, but the definitions here are reproduced as they appeared in the original publication) and with clear central cornea, CC2 for moderate scarring or opacity involving the visual axis, with the pupillary margin visible through the opacity, and CC3 for severe central scarring or opacity, with the pupillary margin not visible through the opacity.
FIG. 2.
Upper tarsal conjunctival zones used for scoring F in the FPC system. “Zones are defined by two imaginary lines which, as viewed on the everted tarsal surface, are approximately parallel with the upper tarsal border and curve upward towards their lateral extremities… Zone 1 includes the entire upper tarsal border and adjacent lateral tarsal surface; zone 2 occupies the area between zones 1 and 3 and extends to the lateral quarters of the lid margin; zone 3 includes the tarsal conjunctiva adjacent to the central half of the lid margin and, at its center, covers just less than half the vertical extent of the tarsal surface” (reference 53, p. 14). Modified by Matthew Burton and reproduced with the permission of M. Burton and the World Health Organization.
The system selects the upper tarsal conjunctiva to provide an “index of trachomatous inflammation in the eye as a whole” (reference 53, p. 14). The intensity of inflammation is classified as trivial, mild, moderate, or severe with Table 1, which is reproduced here as it appears in the WHO guide. (In that document, the meaning of “key sign” is not explained.)
TABLE 1.
Intensity of inflammation classification scheme proposed by Dawson et al.
| Intensity | Grade
|
||
|---|---|---|---|
| Follicles | Papillae | Key sign | |
| Severe | F3 (or F2 or F1)a | P3 | P3 |
| Moderate | F3 | P2 | F3 |
| Mild | F2 | P0, P1, or P2 | F2 |
| Trivial (insignificant or absent) | F0 or F1 | P0, P1, or P2 | F0 or F1 |
“The follicles may be obscured by severe papillary hypertrophy and diffuse infiltration (P3)” (reference 53, p. 14).
With this classification system (with a few minor alterations), Tielsch et al. (228) found the intra- and interobserver agreement of four well-trained, experienced ophthalmologists working in the field to be variable and often poor. For nonspecialist health personnel, the modified WHO system is thought to be too complex (226). However, it still enjoys a degree of popularity with some experts (13, 32, 48).
Thylefors, Dawson, Jones, West, and Taylor, 1987 (WHO simplified system).
The WHO simplified system (226) was designed as a cut-down version of the FPC system, with which it was intended to coexist. Thylefors et al. considered the simplified scheme suitable for use by “less experienced observers” in “population based surveys or for the simple assessment of the disease at the community level” (reference 226, p. 480). It provides considerably less information than the FPC scale. However, the simplified system has enjoyed broad acceptance and is now widely used in research, community assessment, and program monitoring by both nonspecialists and ophthalmologists. The system requires the examiner to assess an individual for the presence or absence of each of five signs (Fig. 3).
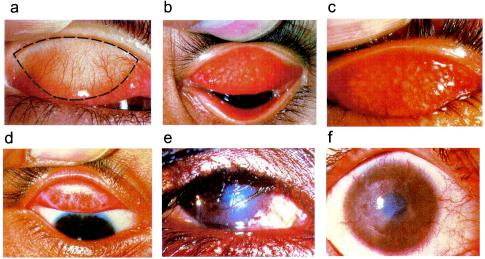
FIG. 3.
WHO simplified system. (a) Normal conjunctiva, showing area to be examined. (b) Follicular trachomatous inflammation (TF). (c) Intense trachomatous inflammation (TI) (and follicular trachomatous inflammation). (d) Conjunctival scarring (TS). (e) Trichiasis (TT). (f) Corneal opacity (CO). Reproduced with the permission of the World Health Organization.
The WHO simplified system (226) uses the following criteria: TF, trachomatous inflammation, follicular, the presence of five or more follicles at least 0.5 mm in diameter in the central part of the upper tarsal conjunctiva; TI, trachomatous inflammation, intense, pronounced inflammatory thickening of the upper tarsal conjunctiva obscuring more than half the normal deep tarsal vessels; TS, trachomatous conjunctival scarring, the presence of easily visible scars in the tarsal conjunctiva; TT, trachomatous trichiasis, at least one eyelash rubs on the eyeball or evidence of recent removal of in-turned eyelashes; and CO, corneal opacity, easily visible corneal opacity over the pupil so dense that at least part of the pupil margin is blurred when viewed through the opacity. In this system, the presence of follicular or intense trachomatous inflammation represents active disease.
Preliminary testing of this system by its developers (after testing and modifying a prototype) involved four observers each examining 179 cases (226). The interobserver agreement measurements found in this study are presented in Table 2. The degree of agreement is indicated with the kappa statistic, a measure of observer reliability for categorical data that estimates the extent of agreement not due to chance between two sets of observations of the same variable. Kappa has possible values between −1 and +1, with −1 indicating complete disagreement, and +1 complete agreement. Landis and Koch (118) set arbitrary divisions for describing the relative strength of agreement associated with this measurement, as follows: <0.00, poor; 0.00 to 0.20, slight; 0.21 to 0.40, fair; 0.41 to 0.60, moderate; 0.61 to 0.80, substantial; and 0.81 to 1.00, almost perfect.
TABLE 2.
Interobserver agreement for the WHO simplified system: second study from Thylefors et al. (226)
| Clinical signa | Avg kappa statistic | Kappa statistic range |
|---|---|---|
| TF | 0.70 | 0.66-0.74 |
| TI | 0.45 | 0.38-0.49 |
| TS | 0.75 | 0.67-0.79 |
| TT | 0.76 | 0.68-0.81 |
| CO | 0.68 | 0.48-0.71 |
TF, inflammation, follicular; TI, inflammation, intense; TS, conjunctival scarring; TT, trichiasis; CO, corneal opacity.
Improved interobserver agreement was reported from further assessment of the system in Tanzania (219). These studies involved comparisons between an experienced ophthalmologist who had participated in the original development of the system and two ophthalmic nurses and an ophthalmologist trained by that individual. Two separate trials were performed to assess interobserver agreement. In the first, 25 eyes were examined by each of four observers, and the scores of the three others were compared to those of the instructor. In the second, a single ophthalmic nurse and the instructor evaluated follicular trachomatous inflammation, intense trachomatous inflammation, and trachomatous conjunctival scarring only in 20 eyes. The results are shown in Table 3 and Table 4.
TABLE 3.
Interobserver agreement for the WHO simplified system: first trial of 25 eyes by Taylor et al. (219) following 3 h of training
| Clinical signa | Kappa statistic for each of three trainees compared to the experienced opthalmologist
|
Average kappa statistic | ||
|---|---|---|---|---|
| 2 | 3 | 4 | ||
| TF | 0.80 | 0.91 | 0.60 | 0.77 |
| TI | 0.66 | 0.71 | 0.88 | 0.78 |
| TS | 0.73 | 0.39 | 0.27 | 0.46 |
| TT | 1.00 | 1.00 | 1.00 | 1.00 |
| CO | 1.00 | 0.63 | 0.83 | 0.82 |
See Table 2, footnote a.
TABLE 4.
Interobserver agreement for the WHO simplified system: second trial of 20 eyes by Taylor et al. (219) following several hours of further training
| Clinical signa | Kappa statistic |
|---|---|
| TF | 0.79 |
| TI | 0.95 |
| TS | 0.87 |
See Table 2, footnote a.
It is notable from these data that good agreement is not guaranteed even when the observers under comparison are all qualified, experienced personnel trained by the same teacher. The reliability of examiners with various amounts of training, experience, and enthusiasm operating under different conditions and at different times, or even of the same observer over time has not been determined. This is not unique to the WHO simplified system (7) and has been a long-standing problem for evaluating trachoma control interventions (54). In fact, diagnostic reproducibility has been rigorously proven for few signs in clinical medicine.
Comparability of Grading Schemes
The WHO simplified and FPC systems are often said to be directly comparable, allowing derivation of simplified system grades from FPC grades without separate assessment of patients. In the original paper describing the simplified system, Thylefors et al. included a table comparing it with Dawson et al.'s FPC scheme (Table 5). This comparison is not strictly correct: the systems are not directly comparable, although the discrepancies are relatively minor. A diagnosis of follicular trachomatous inflammation requires five or more follicles in the central part of the upper tarsal plate, while F2 is defined as more than five follicles (i.e., six or more) in zones 2 and 3 together: the boundaries for follicular trachomatous inflammation absent versus follicular trachomatous inflammation present and F1 versus F2 do not coincide. Additionally, the woolliness of the definitions of conjunctival scarring in both systems make comparison of grades for this sign problematic.
TABLE 5.
Comparison of the simple grading of trachoma with the grading used in a more detailed systema
| Simplified systemb | Detailed system | Implication |
|---|---|---|
| TF | F2 or F3 | Presence of inflammatory trachoma |
| TI | P3 | Severe intensity of inflammation |
| TS | C1, C2, or C3 | Presence of cicatricial trachoma |
| TT | T/E2 or T/E3 | Potentially disabling lesion |
| CO | CC2 or CC3 | Visually disabling lesion |
LABORATORY DIAGNOSIS
In general, the diagnosis of trachoma is made on clinical grounds. This is appropriate. Laboratory testing is typically unavailable or unaffordable for clinical care in areas where trachoma is endemic, antibiotics used against active disease can usually be provided at low or no cost to the patient, and those antibiotics are well tolerated by both children and adults, making presumptive treatment for suspected chlamydial infection a logical therapeutic approach. It should also be noted here that, even if it were to be detected, a single bout of ocular C. trachomatis infection would not constitute trachoma. The clincal signs which accompany repeated reinfection over a period of months to years (81) are required for the disease entity to be considered.
However, as will be discussed in detail later, clinical signs of active disease do not necessarily mean that the conjunctiva is currently infected with C. trachomatis. Detection of the presence or absence of C. trachomatis is often desirable for research purposes. (Later in this paper, we will present an argument that there may be an additional role for laboratory assays in community assessment, particularly for certifying the elimination of trachoma as a public health problem.) The available assays include microscopy of conjunctival scrapings, isolation in cell culture, direct fluorescent antibody, enzyme immunoassay, serology, nucleic acid hybridization probes, and nucleic acid amplification tests. The characteristics of all of these assays are compared in Table 6. The sections below present the basic principles of the assays and their advantages and disadvantages for detecting ocular C. trachomatis. Laboratory diagnosis of urogenital C. trachomatis infections has been comprehensively reviewed by Black (26).
TABLE 6.
Comparison of assays available for diagnosis of ocular C. trachomatis infection
| Test | Detection target | Specimens | Check for specimen adequacy? | Transport conditions | Technical complexity | Processing time | Performance characteristicsa
|
Cost | |
|---|---|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | ||||||||
| Microscopy | |||||||||
| Giemsa/iodine | Inclusions | Conjunctival scraping | Yes | No refrigeration required | ++ | 30 min | 30-60 | 50-70 | + |
| Fluorescein | Antigen | Conjunctival swab | ++ | 30 min | 60-80 | 80-95 | ++ | ||
| Culture | Infectious organisms | Conjunctival swab | No | Cold chain in transport medium | +++ | 48-72 h | 50-70 | 100 | ++++ |
| Enzyme immunoassay | |||||||||
| Lab based | Antigen | Conjunctival swab | No | No refrigeration required | ++ | 4-6 h | 60-80 | 80-95 | + |
| Rapid | + | 30 min | 50-70 | 80-95 | ++ | ||||
| Nucleic acid hybridization | DNA | Conjunctival swab | No | No refrigeration requiredb | ++++ | 4-6 h | 60-80 | 95-100 | +++ |
| Nucleic acid amplification | DNA or RNA | Conjunctival swab | No | No refrigeration requiredb | ++++ | 4-6 h | 90-100 | 95-100 | ++++ |
| 1-2 h for real-time PCR | |||||||||
| Serology | Human antichlamydial antibody (IgA, IgG, or IgM) | Serum, tears | Yes | No refrigeration required | +++ | 4-6 h | ? | ? | +++ |
Performance compared against a reference standard of culture and/or nucleic acid amplification test. The ranges given represent best current estimates; the difficulties involved in assigning numerical values to the sensitivity and specificity of diagnostic tests for ocular C. trachomatis are discussed in the text.
Requirements for transport medium and transport conditions vary for different assays.
Microscopy
Examination of stained conjunctival scrapings for C. trachomatis inclusion bodies is the oldest method for detection of ocular infection. With Giemsa, the stain used by Halberstaedter and von Prowazek (90) and the one still most commonly used (9, 149, 190, 214, 224) until microscopy was superseded by superior diagnostic techniques, mature inclusions appear as dark purple masses in the cytoplasm of epithelial cells. Acridine orange and iodine are alternative stains (213); the latter is quicker than Giemsa (108) and was preferred by some (92, 200). Gram staining, however, is unreliable; the reaction is negative or variable (30). Microscopy requires trained technicians (224), is time-consuming, and is probably the least sensitive method for diagnosis (189). Additionally, collection of conjunctival scrapings with a metal blade is painful and unpopular (126). Tests that are less traumatic, more rapid, and more sensitive have displaced microscopy of scrapings to the sidelines of Chlamydia diagnostics.
Cell Culture
Chlamydiae are fastidious organisms. Successful culture relies on the use of enriched sucrose phosphate transport medium and strict maintenance of the cold chain during transport. In the laboratory, clinical specimens are inoculated onto McCoy cells (11, 31, 57, 78, 100, 102, 125, 126, 134-136, 189, 193, 238, 242), HeLa 229 cells (31, 37, 43, 123, 168, 183), or L434 mouse fibroblasts (M. E. Ward, 2002, Classic diagnostic methods: cell culture [http://www.chlamydiae.com/chlamydiae/restricted/docs/labtests/diag_cellcult.htm], accessed 26 June 2002). Usually the cell layer is irradiated (78, 213) or pretreated with DEAE-dextran (27, 43, 102, 114, 168, 183) or mitomycin C (Ward, website cited above, accessed 26 June 2002) to enhance uptake of the organism. The specimen is centrifuged onto the monolayer to aid cellular infection, and the culture is incubated for 2 to 3 days in the presence of cycloheximide, which inhibits host protein synthesis. Determining whether the culture is positive or negative requires staining with iodine or Giemsa or the use of labeled poly- or monoclonal antibody (204). One or more blind passages (in which apparently negative cultures are homogenized and inoculated onto fresh monolayers) are sometimes performed to ensure that low-level infection is not overlooked. Identification of one inclusion is sufficient to record a positive result.
Despite the high specificity of isolation, a number of problems are associated with use of it as a diagnostic test. Inhibition of chlamydial growth in culture can, in theory, be caused by cytokines or antibodies produced by infected tissues and introduced into the culture medium with the clinical sample (180, 243). Even when using purified elementary body stock, which (by definition) should be free of inhibitors, the data suggest that only about one elementary body in several hundred (or more) is capable of successfully infecting tissue culture cells (105); the reasons for this are unclear. Because of the organism's stringent requirements for special transport medium, the need for strict maintenance of the cold chain, and the complicated nature of the culture protocol, there are multiple opportunities for variation in factors that impact isolation efficiency and, therefore, sensitivity (186). This makes it difficult to compare results between laboratories or even to compare one run to the next in the same laboratory. Additionally, chlamydial culture is expensive and time-consuming and requires special expertise.
Although cell culture is considered the gold standard for laboratory diagnosis, it is now accepted that isolation of C. trachomatis in cell culture is less than 100% sensitive (14, 43, 186, 189, 193).
Direct Fluorescent Antibody
Immunofluorescence is a technique for detecting cellular molecules. Reagents labeled with fluorescent dye that bind specifically to target proteins are used. A number of different immunofluorescence techniques are possible, depending on whether sample antibody or antigen is the target molecule, and on whether the fluorescent dye is attached to the reagent that binds to the sample (direct immunofluorescence) or attached to the reagent that binds to an intermediate reagent that binds to the sample (indirect immunofluorescence). A direct fluorescent antibody test, the Syva MicroTrak (Syva, Palo Alto, Calif.), was the first diagnostic reagent that used a monoclonal antibody against C. trachomatis and began the move away from culture to techniques that do not rely on chlamydial viability (M. E. Ward, 2002, Classic diagnostics: immunofluorescence [http://www.chlamydiae.com/chlamydiae/restricted/docs/labtests/diag_IF.htm], accessed 25 June 2002). The MicroTrak uses labeled antibody to detect a species-specific epitope in MOMP.
For the clinician, specimen collection for the direct fluorescent antibody test is straightforward: conjunctival cells and exudates from a swab are smeared onto a slide in the field or clinic, fixed with methanol (189), and air dried. The slides are easy to transport and store (126). In the laboratory, the fixed sample is stained with fluorescein isothiocyanate-conjugated monoclonal antibody and examined under a fluorescence microscope. Performing an epithelial cell count provides a straightforward method for determining the adequacy of the sample (216). Samples taken from the upper tarsal conjunctiva yield a greater concentration of infected cells than those from the lower fornix (91).
The MicroTrak performed extremely well in initial sensitivity and specificity trials (232), and the fact that specimens could be transported to the laboratory at ambient temperature gave it a considerable advantage over tissue culture. As a result, the MicroTrak has been widely used in trachoma research studies (28, 31, 55, 57, 100, 135, 189, 210, 217, 240, 242, 244). Comparison between studies is made difficult by the fact that the threshold for defining a positive slide varies from one group of investigators to the next. The major practical disadvantages with the direct fluorescent antibody test are that the average time required to screen each slide is more than 20 min (216), and, because the criteria for positivity are so subjective, specificity is highly dependent on the competence of the microscopist (108).
Enzyme Immunoassay
Enzyme-linked immunosorbent assays, also known as enzyme immunoassays, are designed to detect antigens or antibodies by producing an enzyme-triggered color change. For C. trachomatis, enzyme immunoassay usually refers to an antigen detection test, with antibody used to detect chlamydial antigen contained in the specimen. There are many C. trachomatis enzyme immunoassays on the market, each with slightly different configurations, but almost all detect chlamydial lipopolysaccharide with the same sandwich immunoassay principle.
Chlamydial antigens are eluted from collected swabs in specimen dilution buffer. An aliquot of sample eluate is placed onto a solid-phase support (such as a microtiter plate well or a polystyrene bead) to which antibodies that bind chlamydial antigens have been attached. Bound chlamydial antigen is then detected by the addition of a second antibody conjugated to a developer, such as horseradish peroxidase (103). Following a washing step to remove unbound components, a colorless substrate that is transformed by peroxidase to a colored product is added to the well. The presence of chlamydial antigen-antibody complexes is demonstrated by detecting the color change with a spectrophotometer.
Enzyme immunoassays do not require immediate refrigeration of clinical specimens following collection (128), and specimen processing can be completed in 4 h. High throughput can be achieved by batching. If processing is delayed, however, prolonged sample storage can reduce sensitivity (216). Specificity is also a problem. Chlamydial lipopolysaccharide shares epitopes with a number of other bacterial species (148). As a result, Staphylococcus aureus, Haemophilus aegyptius, Klebsiella pneumoniae, Gardnerella vaginalis, Neisseria gonorrhoeae, Escherichia coli, Streptococcus agalactiae, Moraxella lacunata, Chlamydia psittaci, the Salmonella enterica serovar Minnesota Re mutant, Acinetobacter lwoffii and Acinetobacter calcoaceticus var. anitratus can all react in this type of assay (182, 189, 220; M. M. Rothburn, H. Mallinson, and K. J. Mutton, Letter, Lancet ii:982-983, 1986). Conjunctival infection or sample contamination with any of these organisms could therefore produce a false-positive result. A confirmatory assay that selectively blocks binding of the chlamydia-specific epitope can be used to separate true positives from false positives and thereby increase the specificity of the test (148).
The Boots CellTech (later Dako) IDEIA (Boots CellTech, Slough, England) (12, 125, 128, 164, 238) incorporated a detection system with the potential for improved specificity, with murine monoclonal antibody to chlamydial lipopolysaccharide in place of the polyclonal antibody used in most other tests (133). The detection principles were later further altered by attaching multiple copies of an antilipopolysaccharide monoclonal antibody-alkaline phosphatase complex to a dextran backbone. In this format, designated polymer conjugate enhancement, each copy of lipopolysaccharide in the sample is able to capture multiple copies of the enzyme, resulting in dual amplification of the signal (39). There are insufficient data, however, to conclude that this test has better specificity or sensitivity than other enzyme immunoassays (113).
A number of rapid point-of-care tests with the enzyme immunoassay format are also available. Results are available 30 min after sample collection, but the consensus on these tests seems to be that they sacrifice sensitivity for speed (174, 246).
Serology
The first serological test used for diagnosis was a complement fixation test that detected serum antibodies against the polysaccharide antigens of lipopolysaccharide. Because these epitopes are common to all chlamydial species (59), the specificity of the test was low (230). Additionally, it had low sensitivity for ocular infections (108, 214).
The microimmunofluorescence technique developed by Wang and Grayston (237) was the first method used to classify strains of C. trachomatis into serovars. The serovar-specific antigens delineated by this test can be used in an indirect fluorescent antibody assay to detect antichlamydial antibodies in serum or tears, with greater sensitivity than achieved with complement fixation (230). Serial dilutions of the sample are placed on glass slides to which antigens of different C. trachomatis serovars have been fixed (Individual Antigen Serovar Kit; Washington Research Foundation, Seattle, Wash.). Following incubation, the slides are probed with fluorescein-labeled anti-human immunoglobulin. Testing for the presence of immunoglobulin A, immunoglobulin G, and immunoglobulin M can be performed separately (132, 216).
Detecting antichlamydial antibody in serum is difficult, subjective, and tedious and has poor specificity and poor reproducibility. The potential advantage of distinguishing between acute, subacute, and chronic infection is not borne out, even with the use of paired acute- and convalescent-phase sera, because production of immunoglobulin M antibody is not stimulated by ocular reinfection with a previously encountered C. trachomatis serotype (230). Its uses are limited. Tear microimmunofluorescence has better correlation with clinical trachoma but suffers from low sensitivity and the same practical disadvantages (216).
Direct Hybridization Probe Tests
Early attempts to use direct nucleic acid hybridization for the diagnosis of chlamydial infection used radiolabeled C. trachomatis DNA and autoradiography, which required an exposure time of 36 h or more. It was successfully used to detect infected cells from tissue cultures, ocular swabs, and cervical smears (57, 102). Unfortunately, the sensitivity was thought to be lower than that of culture (102). Commercial applications of the technique incorporate significant improvements.
The Gen-Probe PACE 2 (Gen-Probe, San Diego, Calif.) is a nucleic acid probe or hybridization probe test. The probe is a synthetic single-stranded DNA molecule complementary to a region of chlamydial rRNA. The sample is heated so that cells are lysed and rRNA is released. The probe, labeled with an acridinium ester, is added; it forms a stable DNA-RNA hybrid with its target sequence. Detection is performed with a hybridization protection assay: following binding, selection reagent is added, which hydrolyses the acridinium ester on unhybridized probes and thereby deactivates it; the acridinium ester on hybridized probes is protected within the double helix of the DNA-RNA complex. With subsequent addition of hydrogen peroxide, bound acridinium ester releases a pulse of light that can be detected with a luminometer. Because there are about 2,000 copies of rRNA per chlamydial cell, sensitivity is slightly better than that achieved by enzyme immunoassay (119, 247), without the need for amplification of nucleic acid.
A second hybridization probe test, the Hybrid Capture II (HCII; Digene, Gaithersburg, Md.), uses an enzyme immunoassay to achieve signal amplification and is therefore classed as a nucleic acid probe-signal amplification assay. Alkali is added to the clinical sample to lyse cells and denature double-stranded DNA (separate it into its two component strands). An RNA hybridization probe with nucleotide sequence complementary to chlamydial DNA is introduced; because the DNA has been denatured, the probe can anneal to its target region. The RNA-DNA complex is then captured by antibody bound to microwell plates. Antibodies labeled with alkaline phosphatase are added to bind to the bound complex. After washing, the presence or absence of bound DNA can be determined by adding a substrate for alkaline phosphatase: cleavage of the substrate results in emission of light (77; M. E. Ward, 2002, Molecular diagnostics: direct hybridization probe tests [http://www.chlamydiae.com/chlamydiae/restricted/docs/labtests/diag_hybridization.htm], accessed 1 July 2002). The test is rapid and reproducible (77), but, for the diagnosis of ocular chlamydial infection, direct hybridization assays have been largely overlooked in favor of the nucleic acid amplification tests discussed below.
PCR
PCR is a technique for amplifying DNA, and assays based on it are part of the group of nucleic acid amplification tests. PCR uses the enzyme DNA polymerase. A variant of this enzyme is found in the nucleus of all replicating cells. In vivo, its function is to duplicate DNA during the cell's preparation for its own division. In 1983, Mullis realized that exponential growth in the number of copies of a target DNA sequence could be achieved in vitro if repeated rounds of DNA polymerase-catalyzed duplication were made to occur back to back with a thermostable DNA polymerase (153, 181).
There are three phases of the reaction. All take place in the same vessel but at different temperatures. The reaction mixture contains an aliquot of the sample, an excess of deoxyribonucleotide triphosphates, DNA polymerase, and two primers (short, synthetic oligonucleotides that flank the region to be amplified); one primer is complementary to the sense strand, and one to the antisense strand at the opposite end of the target sequence. First the mixture is heated to between 90 and 95°C to break apart the two strands of target DNA, allowing the primers access to complementary sequences on the target. At a reduced temperature, the primers anneal to these binding regions. The mixture is then heated again to enhance the activity of the DNA polymerase, which extends each chain from the 3′ end of its annealed primer to produce two double-stranded copies of the target sequence. Both of the two newly synthesized primer extension products contain the appropriate primer-binding regions and, after heat-induced separation from the target, can themselves function as templates alongside the original templates in the next round of duplication. Multiple repetitions of the denaturation-annealing-extension process therefore result in exponential accumulation of the target. PCR is now usually automated in a thermal cycler, which rapidly and reliably changes the temperature of the reaction vessel to provide appropriate conditions for each stage of the amplification process.
PCR is ideally suited to the detection of DNA of fastidious and noncultivatable infectious agents because it does not rely on the presence of viable organisms in the sample. The first bacterium for which a PCR-based detection method was published was C. trachomatis (63).
A number of different nucleic acid sequences have been used as targets in PCRs for the detection of C. trachomatis. These include the chlamydial cryptic plasmid (pCT) (43, 97, 134, 163, 164), omp1, coding for MOMP (27, 63, 96, 134, 163), the gene coding for 16S rRNA (2, 43, 134), and omp2, coding for OmcB (239). With the exception of pCT, all of these targets are sequences found on the C. trachomatis chromosome, which includes two complete rRNA operons and single copies of omp1 and omp2.
PCR directed at plasmid genes (164) or omp1 (27) is thought to be both sensitive and specific for the diagnosis of C. trachomatis infection. Primers designed for the chlamydial rRNA gene amplify this DNA sequence in C. trachomatis, C. psittaci, and C. pneumoniae (43), which reduces the specificity of the assay. The omp2-based PCR (detecting the gene coding for OMC-B) has also been found to return positive results on samples containing any of these three species; subsequent restriction endonuclease digestion and gel electrophoresis permit species and strain identification of isolates (239), but this assay has not been used extensively in published studies to date.
Mahoney et al. (134) estimated that plasmid-based PCRs are between 10 and 10,000 times more sensitive than PCRs directed against C. trachomatis chromosomal genes. This is probably at least partly attributable to the presence of multiple copies of the plasmid per chlamydial cell (134, 163, 164). Bailey et al. (13) also suggested that using a plasmid target gives greater sensitivity. Using serial dilutions of DNA standards, they calculated the lower detection limit of their plasmid-based PCR to be 1 to 10 elementary bodies, compared with 10 to 100 elementary bodies for a PCR against omp1.
A commercial PCR kit, Amplicor Chlamydia (Roche Diagnostic Systems, Branchburg, N.J.) targets a 207-bp sequence within pCT. An aliquot of prepared sample is added to the PCR master mix, which contains heat-stable polymerase, an internal control (see below), two biotinylated primers, cofactors, deoxynucleotide triphosphates, and the enzyme AmpErase (uracil-N-glycosylase). The deoxynucleotide triphosphate dUTP is included in place of deoxythymidine triphosphate. Together, the substitution of thymidine by deoxyuridine in the reaction mixture (and therefore, ultimately, in the amplified product) and the addition of AmpErase help to prevent DNA produced during PCR from contaminating subsequent runs (122). AmpErase degrades DNA containing deoxyuridine by breaking the deoxyribose chain at the C-1 position. Naturally occurring DNA does not contain deoxyuridine and is therefore not a substrate for the enzyme. As the reaction mixture is heated in the first thermal cycling step, any contaminating product DNA molecules are cut into short oligonucleotides by breakages induced at each deoxyuridine residue, rendering them nonamplifiable. AmpErase becomes inactive above 55°C, and, because the reaction vessel is held above this temperature for the duration of the amplification process, the target amplicon is not affected. Following amplification, before the reaction mixture has a chance to cool, AmpErase is destroyed by immediate addition of denaturation solution (178).
Denaturation after cycling also separates the double-stranded amplified product into single strands of DNA. An aliquot of the reaction solution is placed in a microwell to which oligonucleotide probes complementary to the pCT target sequence have been bound. Specific amplified product hybridizes to the probes, and the microwell plate is washed. Avidin-horseradish peroxidase conjugate is then added, which binds to any amplicon that has been captured in the microwell. After another washing step, and hydrogen peroxide and 3,3′,5,5′-tetramethylbenzidine are added; bound horseradish peroxidase catalyzes the formation of a colored compound from the 3,3′,5,5′-tetramethylbenzidine. Optical density can be read with a photometer (178).
Reproducibility problems have been reported in a very small proportion of samples tested with the Amplicor assay, with results obtained from multiple aliquots of the same specimen yielding optical densities ranging from negative (<0.200) to the upper positive limit of the photometer (>3) even when tested in the manufacturer's own laboratories (170). Similarly, the result obtained from a PCR with primers for pCT does not invariably correspond to that from a PCR with primers for omp1 (170). Possible explanations include contamination of PCR tubes, contamination of the microwell plates used in the detection step, nonspecific binding of primers during amplification, nonspecific hybridization during detection, a very low concentration of organisms, plasmid-free strains, technical errors, or the presence of inhibitors that undergo time-dependent inactivation (20, 57, 170). Investigation of the reasons for these inconsistencies is ongoing (188).
The possible presence of inhibitors of DNA amplification within samples needs to be kept in mind. They can be found in specimens obtained from the conjunctiva and from the urogenital tract (28, 135, 234). Inhibition may in some instances be overcome by prolonged storage prior to processing, which presumably allows time for degradation of the inhibitory factor (20, 112, 135), sample dilution (28, 57, 135, 234), freezing at −70°C (57), or heat treatment at 95°C for 10 min (234). The Amplicor assay gives the user the additional option of monitoring each individual test for inhibition by including an internal control in each amplification reaction. The internal control is a synthetic nucleic acid sequence with primer binding regions identical to those of the test-specific target and a randomized internal sequence that is similar in length and base composition to the target. As a result, the internal control and the target region of the plasmid from the clinical specimen are coamplified with equivalent efficiencies. The probe binding region of the internal control is unique, allowing reliable differentiation from plasmid DNA (178). Postamplification, detection of the target and of the internal control is undertaken separately. A positive test for amplification of the internal control indicates that PCR was not inhibited and therefore that inhibition was unlikely to have produced a false-negative result.
The nature of PCR, in which small quantities of DNA are multiplied exponentially, makes it inherently vulnerable to spuriously positive results following contamination of samples during collection, preparation, or processing. Air-borne target DNA in the laboratory is a particular danger (164). The use of separate laboratory zones for sample preparation, amplification, and detection; restriction of sample handling to negative-pressure safety cabinets; stringent adherence to procedures for decontamination; and inclusion of negative controls all help to protect (and demonstrate protection of) the integrity of results. Similar precautions must also be taken to safeguard the reliability of the other nucleic acid amplification tests described below.
Roche also produces an automated version of the Amplicor system, known as the Cobas Amplicor. For urogenital swabs and urine, the two formats appear to have comparable sensitivity and specificity (233).
Ligase Chain Reaction
DNA ligase is an enzyme that links fragments of DNA by inducing the formation of phosphodiester bonds between the 5′ phosphate of the first fragment and the 3′ hydroxyl of the second. Its function is to repair nicks in the phosphodiester backbone of double-stranded DNA and thus will only catalyze the linkage of DNA fragments that are annealed adjacent to each other opposite their complementary sequences. The ligase chain reaction uses a thermostable form of this enzyme as part of an iterative process of denaturation, annealing, and ligation.
In the gapped ligase chain reaction, DNA ligase functions alongside DNA polymerase in a process that combines the ligase chain reaction with PCR. Two pairs of oligonucleotide probes are used. Each pair is designed to anneal to adjacent regions on the same strand, leaving a gap of a few nucleotides. This gap is filled by DNA polymerase. DNA ligase then joins the 3′ probe of each pair to the intervening nucleotide sequence (25).
The Abbott LCx (Abbott Laboratories, Abbott Park, Ill.) targeted a sequence of pCT (146) in a gapped ligase chain reaction that was widely used for laboratory diagnosis of genital and ocular infection. However, because of reproducibility problems, this product has been withdrawn.
Strand Displacement Assay
The strand displacement assay was developed as an alternative to PCR and ligase chain reaction assays for amplifying target DNA sequences. Fundamentally, the strand displacement assay is a variant of the PCR assay. It uses a restriction endonuclease to create a nick in one strand of double-stranded DNA and an exonuclease-deficient form of E. coli DNA polymerase I (or equivalent) to extend from the site of the nick while displacing intact the downstream complement of the target (235). Because repeated denaturation is not required, the reaction can proceed isothermally. A commercially available strand displacement assay, the BDProbeTecET (Becton Dickinson, Franklin Lakes, N.J.), targets C. trachomatis plasmid DNA and incorporates a real-time fluorescence detection system, allowing amplification and detection to occur in the same sealed microwell (121, 201). No published studies have used strand displacement assay for identifying chlamydiae in ocular swabs.
Transcription-Mediated Amplification
Transcription-mediated amplification mimics the RNA replication strategy of retroviruses, producing an RNA amplicon by means of cDNA intermediates (84). It can be used against any type of nucleic acid target. The commercial assay developed for detection of C. trachomatis, the Gen-Probe AMP-CT (Gen-Probe, San Diego, Calif.), is directed against rRNA. Reverse transcriptase and RNA polymerase are used to achieve amplification. Like the strand displacement assay, the ranscription-mediated amplification assay can be carried out without thermal cycling. Transcription-mediated amplification has not yet been evaluated for use with ocular swabs.
Quantitative PCR
Culture, the direct fluorescent antibody test, and the first generation of enzyme immunoassays can be optimized to function as semiquantitative techniques. Unfortunately, these tests lack sensitivity. The nucleic acid amplification tests discussed so far (which are highly sensitive) are purely qualitative. They are difficult to use for quantification because of the exponential nature of DNA reproduction: small changes in amplification efficiency cause a large change in the amount of DNA product (195). In other words, the amount of DNA present at the end of 40 PCR cycles may depend as much on minor variations in reagent concentrations, the properties of any contaminating DNA, or the presence of inhibitors as it does on the amount of DNA present in the original sample. Small between-tube variations in amplification efficiency on the same run may be caused by temperature differences along the heating block and other factors that are difficult to characterize. For these reasons, reliable quantification of the number of copies of a DNA sequence in a sample cannot be achieved by comparing the end PCR yield with the end PCR yield of an external control (195).
Kinetic quantitative PCR assumes that the efficiency of DNA synthesis (E, a number between 0 and 1) remains constant from one cycle to the next for at least part of the amplification process and that during the time in which E is constant, the strength of the signal from the detection system (often, for example, fluorescence) is linearly dependent on the amount of accumulated product in the reaction vessel. These assumptions are only valid during the exponential phase of PCR (Fig. 4); in the background phase, the background signal is greater than the signal generated by the amplified product; in the saturation or plateau phase, amplification efficiency falls because inhibitory reaction products accumulate, the concentration of polymerase becomes limiting, and product renaturation competes with primer binding during annealing (175). With these assumptions, however, the amount of DNA present prior to the beginning of the reaction can be determined as follows.
FIG. 4.
PCR profile. The signal (shown on the vertical axis) is related to the amount of amplified product and has been acquired once per cycle. 1, background phase; 2, exponential phase; 3, plateau phase.
For any period of a cycles during which E is constant, the number of copies of the target (Pi) present at the end of the final cycle of that period is given by Pi = Pi-a × (1 + E)a (equation 1) (173), where Pi-a is the number of copies of the target present in the reaction vessel at the beginning of the period (the end of cycle i − a). Therefore, E = −1 + (Pi/Pi-a)1/a (173).
To ensure that E has remained constant up to the point at which the last measurement of product is made, E must be estimated at least twice. As a result, the number of copies of target present in the reaction vessel must be determined three or more times during amplification. Once the value of E has been calculated, the number of copies of the target present before amplification commenced (P0) can be derived from the measured amount of product after n cycles by rearranging equation 1 (173): P0 = Pn/(1 + E)n.
True kinetic (or real-time) PCR can be achieved by having the reactions take place in closed, optically clear glass vessels and analyzing product generation by detection and quantification of a fluorescent reporter after each extension step. This eliminates the need for repeated tube opening and post-PCR processing, improves accuracy, reduces the risk of sample contamination, and enhances automation (21, 173). A number of different approaches to fluorescence monitoring have been developed; the details of these are probably beyond the scope of this review. To calibrate the signal produced by the detection system, a standard curve should be generated for each run by amplifying serial dilutions of a known concentration of the target sequence (173).
Eastick et al. (64) presented the development of kinetic quantitative PCRs for both omp1 and plasmid sequences of C. trachomatis. Testing 501 urine specimens from women attending a genitourinary clinic, they found that their quantitative PCR was about as sensitive as the Roche Cobas Amplicor PCR kit. Seven specimens that were discrepant by Cobas and by quantitative PCR were thought to contain chlamydial loads around the detection limit of both assays. In their abstract, the authors mention that their assay was able to quantify DNA over a 10,000-fold range, but no data on actual chlamydial loads are included.
Huang et al. (105) were able to detect as few as one copy of their target omp1 sequence. Using serial dilutions of C. psittaci B577 stock, they found an approximately linear relationship between the number of copies of chlamydial DNA target and the number of inclusion-forming units in culture. Recently, an assay similar to the assay of Huang et al. has been used to quantify ocular C. trachomatis infection in all members of three communities where trachoma is endemic, before (199) and at intervals after (198) community-wide antibiotic treatment. These studies will provide further clues about the epidemiology of trachoma and help inform rational strategies for trachoma control programs.
Other groups have developed quantitative PCR assays and applied them to the study of nonocular chlamydial infections (115, 158, 229) and chlamydial infections in mammals other than humans (58, 98). There appears to be a general consensus that the sensitivity, specificity, dynamic range, rapidity, and automation of quantitative PCR make it a useful research tool.
Sensitivity and Specificity of Laboratory Tests
Cell culture has long been regarded as the gold standard of Chlamydia diagnosis because its specificity is thought to be nearly perfect. Its sensitivity is known to be imperfect. True infection status has therefore been impossible to determine, and numerical values assigned to the specificity or sensitivity of diagnostic tests vary significantly from one estimate to the next. Despite this, such values are routinely quoted.
Newer tests, such as the nucleic acid amplification tests, are believed (for biological reasons) to be more sensitive than cell culture. Investigators comparing the performance of these tests against that of culture assume that at least some of the apparent false positives (by the new test) are actually true positives that have been missed by culture. To estimate the performance characteristics of the new test, these apparent false positives are often evaluated with discrepant analysis. In this procedure, gold-standard-negative, new-test-positive samples are further tested by one or more other appeal assays; the return of one or more positive results from these assays labels the sample a true positive. In evaluating the nucleic acid amplification tests for C. trachomatis, the appeal tests used have typically been other nucleic acid amplification tests, often with a different method of amplification or a different target nucleic acid sequence (83, 87, 144).
Hadgu has argued that discrepant analysis is fundamentally flawed, because the restriction of retesting to the apparent false positives must upwardly bias sensitivity and specificity estimates for the new test regardless of the accuracy of the appeal tests used for resolution (86, 87). In fact, any test used for resolution, no matter how ridiculous (McAdam [142], for example, suggests tossing a coin), can only improve or leave unaltered the calculated sensitivity and specificity of the new test (142); the magnitude of the bias depends on the underlying prevalence of disease and the independence of the appeal and new tests (83, 142). This criticism has provoked vigorous and often heated debate (88, 89, 99, 144, 187, 191, 192).
Despite the rancor, there is broad acceptance that nucleic acid amplification tests represent a significant advance in the diagnosis of chlamydial infections. They appear to be significantly more sensitive than assays available previously (40), with the potential to detect chlamydiae “beyond clinically relevant levels” (reference 61, p. 731). Kuipers et al. directly compared the sensitivities of a number of assays with dilutions of purified C. trachomatis elementary bodies spiked into urine and found that the lower limits of detection were 2 elementary bodies for an omp1-based in-house PCR, 2 × 103 for both direct fluorescent antibody (MicroTrak) and enzyme immunoassay (ChlamydiaEIA, Syva, San Jose, Calif.), and 2 × 104 for the PACE 2 hybridization probe and IDEIA (113). With a similar dilution series, and assuming the presence of 10 plasmids per organism, Shattock et al. estimated the detection limits of the Cobas Amplicor, Amplicor plate kit, and LCx assays to be approximately 1 to 2, 2 to 4, and 2 elementary bodies (per tested aliquot), respectively (197). Additionally, the nucleic acid amplification tests seem to be highly specific (107). Quality control in manufacturing and the skill of the user are probably more likely to affect the accuracy of nucleic acid amplification tests than the underlying biological characteristics of the assay (188).
Correlation of Laboratory Tests with Clinical Signs of Trachoma
Advances in techniques for the laboratory diagnosis of C. trachomatis have generally been driven by the requirement for improved detection of urogenital infections. As a result, the bulk of the literature examines their utility in that setting. This analysis considers only the relationship of test positivity with clinical signs of ocular disease and only uses the results of studies obtained with either the FPC or WHO simplified grading schemes. Where information from trachoma treatment trials has been included, only baseline (pretreatment) data are presented. The data are shown in Table 7.
TABLE 7.
Published data on the correlation of qualitative laboratory tests for ocular C. trachomatis infection with clinical signs of trachoma, where signs were graded using either the modified WHO system (53) or the WHO simplified system (226)
| Country | Subjects | Prevalence of trachoma in study subjects or in population from which subjects were selected | Testa | Prevalence of positive testsb | Reference(s) |
|---|---|---|---|---|---|
| Egypt | Children 1-10 years of age with at least moderately severe active trachoma, screened at a medical clinic | “Holoendemic” | Giemsa | 13 (14%) of 91 examined | 189 |
| Nepal | All children aged 1-10 years from 6 villages examined; Giemsa staining performed on 15 children arbitrarily selected from those with active disease | 46 (6%) of 726 examined had active disease | Giemsa | 0 of 15 swabbed | 16 |
| Tunisia | Children aged 6-8 years attending schools of 4 villages screened; conjunctival scrapings collected from those with active trachoma of moderate or severe intensity (n = 162; results given for 161) | Not stated; all children from whom samples were collected had active disease of moderate or severe intensity | Giemsa | 51 (32%) of 161 | 50 |
| Mexico | All available residents of 2 villages examined; scrapings obtained from all children aged 1-10 years in every fourth family | Approximately 25% of those under 10 years had P3 and F1 OR F3 OR F2 | Giemsa | 19 (24%) of 81 children with P3 and F1, OR F3, OR F2 | 218 |
| Egypt | Children 1-10 years of age with at least moderately severe active trachoma, screened at a medical clinic | “Holoendemic” | Culture (cycloheximide-treated McCoy cells); serial passage after 96 h if negative on initial inoculation | 34 (37%) of 91 examined | 189 |
| The Gambia | All available residents (911 of 950) of 1 village examined; swabs taken from everyone with active disease and a random sample of those without active disease | 111 (12.2%) of 911 examined had active disease | Culture (cycloheximide-treated McCoy cells) | 1 (1%) of 97 without active disease; 31 (43%) of 72 with active disease (19% of those with mild active disease; 63% of those with moderate active disease; 50% of those with severe active disease) | 11, 238 |
| The Gambia | In one village (population ≈1,000), all available residents examined; in the second (population ≈600), residents of 5 randomly selected compounds examined; swabs collected from the more severely affected eye of 63 cases (31 with clinical evidence of active trachoma and 32 without) | In the first village, the prevalence of active trachoma in children under 15 yr was 22%; in the second village it was 13% | Culture (cycloheximide-treated McCoy cells) | 2 (6%) of 32 with insignificant or no active disease; 4 (40%) of 10 with mild active disease; 5 (63%) of 8 with moderate active disease; 5 (38%) of 13 with severe active disease | 126 |
| The Gambia | All residents of one village (n ≈ 900) examined twice, in 1985 and 1986; in 1985 swabs collected from a randomized, age-stratified sample of the population; in 1986 only from clinically active cases | 22% of those aged ≤15 yrs had active disease | Culture (cycloheximide-treated McCoy cells) | 3 (3%) of 90 with insignificant or no active disease, 30 (18%) of 169 with active disease | 128 |
| Nepal | All children aged ≤5 yr in three villages of Lumbini Zone, western Nepal (n = 430); specimens for 400 subjects reported | Moderate to severe intensity of inflammation in 85 (21%) of 430 examined | Culture (cycloheximide-treated McCoy cells); serial passage after 96 h on 2 duplicate plates if negative on initial inoculation | 44 (11%) of 400, ≈5% of 267 with no active disease, ≈13% of 54 with mild active disase, ≈11% of 18 with moderate active disease, ≈31% of 61 with severe active disease (read from graph) | 57 |
| Tanzania | Stratified random sample of 20 villages drawn; within each village, a cluster sample of children aged 1-7 yr and their mothers or female caretakers examined; swabs taken from all those examined in the first 9 villages (n = 1,671) | 589 (54%) of 1,090 children aged 1-7 yrs examined had active trachoma; 52 (9%) of 581 women examined had active trachoma | Culture considered inadequate if cell monolayer completely destroyed in both 1st and 2nd passages (n = 23) or if it had been completely destroyed in one and partially destroyed in the other (n = 12) | 34 (4%) of 927 with no sign of trachoma, 150 (31%) of 481 with TF only, 73 (51%) of 142 with TI, 7 (9%) of 77 with TS only, 1 (11%) of 9 with TT without TF or TI | 215 |
| USA | Students aged 12 to 21 attending the Stewart Indian School near Carson City, Nev.; all were American Indians from reservations in the southwestern U.S. | “Highly endemic” | In-house immunofluorescence of conjunctival scrapings with polyclonal fluorescein-labeled serum from a rabbit immunized with a yolk sac-grown strain of C. trachomatisb agent | 9 (53%) of 17 with clinical signs of active disease (based on severity of purulent discharge, follicular or papillary hypertrophy, pannus, and conjunctival and corneal scarring) in 1965, 8 (53%) of 15 with clinical signs of active disease in 1966, 3 (60%) of 5 with clinical signs of active disease in 1967, 5 (17%) of 29 without clinical signs of active disease in 1965, 11 (35%) of 31 without clinical signs of active disease in 1966, 15 (37%) of 41 without clinical signs of active disease in 1967 | 51, 91 |
| Egypt | Children 1-10 yr of age with at least moderately severe active trachoma screened at a medical clinic | “Holoendemic” | DFA (MicroTrak) slides considered positive if ≥10 EBs seen | 35 (38%) of 91 examined | 189 |
| The Gambia | In one village (population ≈1000), all available residents examined; in the second (population≈600), residents of 5 randomly selected compounds examined; swabs collected from the more severely affected eye of 63 cases (31 with clinical evidence of active trachoma and 32 without) | In the first village, the prevalence of active trachoma in children under 15 yr was 22%; in the second village it was 13% | DFA (MicroTrak) slides considered positive if ≥10 EBs seen | 1 (3%) of 32 with insignificant or no active disease, 2 (20%) of 10 with mild active disease, 3 (38%) of 8 with moderate active disease, 4 (31%) of 13 with severe active disease | 126 |
| Nepal | All children aged ≤5 years in three villages of Lumbini Zone, western Nepal (n = 430); specimens for 400 subjects reported | Moderate to severe intensity of inflammation in 85 (21%) of 430 examined | DFA (MicroTrak) slides considered positive if ≥10 EBs seen | 33 (8%) of 400, ≈15% of 267 with no active disease, ≈28% of 54 with mild active disease, ≈22% of 18 with moderate active disease, ≈34% of 61 with severe active disease (read from graph) | 57 |
| Tanzania | One index child from each of 234 households with children aged 1-7 yr randomly selected from a village with no previous trachoma control program | 137 (59%) of 234 examined had active disease | DFA (MicroTrak) slides considered positive if ≥5 EBs seen | 1 (1%) of 97 without TF or TI, 28 (28%) of 100 with TF but not TI, 22 (60%) of 37 with TI ± TF, | 28 |
| Tanzania | Stratified random sample of 20 villages drawn; within each village, cluster sample of children aged 1-7 yr and their mothers or female caretakers examined; swabs taken from all those examined in the first 9 villages (n = 1,671) | 589 (54%) of 1,090 children aged 1-7 yr examined had active trachoma; 52 (9%) of 581 women examined had active trachoma | DFA (MicroTrak) slides considered positive if ≥5 EBs seen; slides considered inadequate if <200 epithelial cells seen; 188 slides were inadequate but 2 of these had >10 EBs and were included | 48 (6%) of 813 with no sign of trachoma, 221 (49%) of 447 with TF only, 100 (71%) of 140 with TI, 16 (21%) of 76 with TS only, 1 (11%) of 9 with TT without TF or TI | 215 |
| The Gambia | All residents of two trachoma-endemic villages (n = 1363) examined; swabs taken from 1,348 subjects for EIA | 201 (15%) of 1,348 swabbed for EIA | EIA (Novo Nordisk) | 66 (52%) of 126 with mild active disease, 37 (79%) of 47 with moderate active disease, 22 (79%) of 28 with severe active disease, 75 (7%) of 1147 without active disease | 13 |
| The Gambia | All residents of one village (n ≈ 900) examined twice, 1985 and 1986; in 1985 swabs collected from a randomized, age-stratified sample of the population, in 1986 from the whole population | 22% of those aged ≤15 yr had active disease | EIA-PCE (Boots Celltech) | 56 (25%) of 228 with active disease, 49 (4.9%) of 997 with insignificant or no disease, 10 (20%) of 49 with severe scarring (C3) | 128 |
| Egypt | Children 1-10 yr of age with at least moderately severe active trachoma | “Holoendemic” | EIA (Chlamydiazyme) | 36 (40%) of 91 | 189 |
| The Gambia | All inhabitants of one village (n ≈ 950) examined in 1990; swabs taken from all those with active disease and from all members of 2 households, one containing substantial numbers of trachoma cases, the other free of clinical trachoma | 96 (11%) of 844 inhabitants of the village had clinically active trachoma | EIA (Novobiolabs) | 0 of 37 without active disease, 10 (83%) of 12 with severe active disease, 8 (29%) of 28 with moderate disease, 6 (11%) of 56 with mild disease | 14, 96 |
| Algeria | Children aged 3-17 yr living in 9 schools and nurseries in Saharan refugee camps (n = 527) | 13 (2.5%) of 527 examined had TF | Rapid EIA (Clearview Chlamydia MF) | 0 of 13 with TF, 2/514 (0.4%) without TF | 106 |
| The Gambia | All residents of one village (n ≈ 900) examined in 1984; swabs taken from everyone with active disease and a random sample of those without active disease | Same population as in reference 128; prevalence data provided in this paper only as a graph of prevalence against age for those 0-18 yr old (using pooled data from three studies in the village); peak prevalence of active disease (in 2-year-olds) of ≈37% | Tear MIF-ELISA for slgA | 38 (56%) of 68 with active disease, 10 (43%) of 23 without active disease but with scarring | 238 |
| Kenya | Children with abnormal ocular discharge visiting a health center after an invitation issued for families with eye problems to attend | 90% of persons in the district had “past or current findings suggestive of trachoma” in a survey performed 7-9 yr before the study | Serology (serum MIF for IgM and IgG) positive result defined as an antibody titer of ≥1:8 | 32 (82%) of 39 with 3 or more follicles, 18 (38%) of 47 with papillary hypertrophy sufficient to obscure underlying blood vessels, but 2 or fewer follicles | 31 |
| Nepal | All children aged ≤5 yr in three villages of Lumbini Zone, western Nepal (n = 430); specimens for 400 subjects reported | Moderate to severe intensity of inflammation in 85 (21%) of 430 examined | In-house DNA probe with radiolabeled cloned plasmid DNA from serovar C strain TW3 | 70 (18%) of 400, ≈10% of 267 with no active disease, ≈22% of 54 with mild active disease, ≈22% of 18 with moderate active disease, ≈46% of 61 with severe active disease (read from graph) | 57 |
| Australia | Children aged <15 yr from one Aboriginal community (n = 221) screened for trachoma; of those with follicular trachoma (n = ?), 48 had conjunctival swabs taken | Not specified; in “many Aboriginal communities in central Australia. . .20% of children (have) characteristic eyelid inflammation” | Hybridization probe (Gen-Probe PACE 2) | 0 (0%) of 48 with TF | 117, 120 |
| The Gambia | All residents of two trachoma-endemic villages (n = 1,363) examined; swabs taken from 1,332 subjects for PCR | 200 (15%) of 1,332 swabbed for PCR had active disease | In-house PCR against pCT | 83 (65%) of 128 with mild active disease, 38 (86%) of 44 with moderate active disease, 23 (82%) of 28 with severe active disease, 85 (7.5%) of 1132 without active disease | 13 |
| Algeria | Children aged 3-17 yr living in 9 schools and nurseries in Saharan refugee camps (n = 527) | 13 (2%) of 527 examined had TF | In-house PCR against pCT | 2 (15%) of 13 with TF, 10 (2%) of 514 without TF | 106 |
| The Gambia | All inhabitants of one village (n ≈ 950) examined in 1990; swabs taken from all those with active disease and from all members of two households, one containing substantial numbers of trachoma cases, the other free of clinical trachoma | 96 (11%) of 844 inhabitants of the village had clinically active trachoma | In-house PCR against MOMP | 2 (5%) of 37 without active disease, 8 (67%) of 12 with severe active disease, 16 (57%) of 28 with moderate disease, 25 (45%) of 56 with mild disease | 14, 96 |
| Tanzania | Swabs obtained from 129 individuals aged between 1 and 68 yr living in 2 trachoma-endemic villages; correlation between infection and disease reported for 50 swabs chosen by randomly selecting 24 PCR-positive swabs and 26 PCR-negative swabs and retrieving the clinical (field) information | Not stated | In-house PCR-EIA | 12 (63%) of 19 with TI, 3 (28%) of 8 with TF but not TI | 29 |
| Tanzania | One index child from each of 234 households with children aged 1-7 yr randomly selected from a village with no previous trachoma control program | 137 (59%) of 234 examined had active disease | In-house PCR-EIA against MOMP | 23 (24%) of 97 without TF or TI, 54 (54%) of 100 with TF but not TI, 35 (95%) of 37 with TI ± TF | 28 |
| Tanzania | 4,932 women aged 18 yr or older from 11 villages examined in 1989; follow-up examination planned for all women with scars living in 6 of 11 villages and a random sample of women without scars from the same villages; 523 of 745 with scars examined; 503 of 749 without scars examined | 147 (5%) of 1,014 examined had active disease | In-house PCR-EIA against MOMP | 17 (4%) of 453 without scars at baseline or active disease at F/U, 5 (31%) of 16 without scars at baseline, with TF (but not TI?) at F/U, 14 (44%) of 32 without scars at baseline, with TI at F/U, 24 (5.8%) of 414 with scars at baseline, without active disease at F/U, 2 (33%) of 6 with scars at baseline, with TF (but not TI?) at F/U, 34 (37%) of 93 with scars at baseline, with TI at F/U, 36 (7.2%) of 499 without scars at baseline, without TT at F/U, 0 (0%) of 2 without scars at baseline, with TT at F/U, 51 (11%) of 466 with scars at baseline, without TT at F/U, 9 (19%) of 47 with scars at baseline, with TT at F/U | 154 |
| Australia | Children aged <15 yr from one Aboriginal community (n = 221) screened for trachoma; of those with follicular trachoma (n = ?), 48 had conjunctival swabs taken | Not specified; in “many Aboriginal communities in central Australia. . .20% of children (have) characteristic eyelid inflammation” | PCR (in-house; target gene not specified) | 7 (15%) of 48 with TF | 117, 120 |
| Tanzania | All residents of one subvillage in Rombo district invited (956 of 978 residents examined) | 174 (18%) of 956 examined had active disease | PCR (Amplicor) | 17 (27%) of 63 with TF but not TI, 41 (37%) of 111 with TI with or without TF, 2 (2%) of 86 with TS but neither TF nor TI, 31 (4%) of 696 with no sign of trachoma | 199 |
| Tanzania | All residents of one subvillage in Kongwa district invited (874 of 1,017 residents examined; results available for 871) | 312 (36%) of 871 examined had active disease | PCR (Amplicor) | 161 (74%) of 271 with TF but not TI, 80 (84%) of 95 with TI with or without TF, 48 (52%) of 93 with TS but neither TF nor TI, 207 (44%) of 466 with no sign of trachoma | 199 |
| The Gambia | All residents of 14 trachoma-endemic villages (1,319 residents examined) | 103 (8%) of 1319 examined had active disease | PCR (Amplicor) | 14 (17%) of 84 with TF but not TI, 9 (47%) of 19 with TI with or without TF, 8 (10%) of 81 with TS but neither TF nor TI, 64 (6%) of 1135 with no sign of trachoma | 33, 199 |
| The Gambia | All residents of 14 trachoma-endemic villages (1,319 residents examined) | 103 (8%) of 1319 examined had active disease | PCR (Amplicor) | 65 (5.5%) of 1182 with P0, 10 (14%) of 69 with P1, 11 (22%) of 49 with P2, 9 (47%) of 19 with P3, 69 (6%) of 1146 with F0, 4 (5%) of 74 with F1, 6 (10%) of 62 with F2, 16 (43%) of 37 with F3 | 33 |
| Australia | Children aged <15 yr from one Aboriginal community (n = 221) screened for trachoma; of those with follicular trachoma (n = ?), 48 had conjunctival swabs taken | Not specified; in “many Aboriginal communities in central Australia. . .20% of children (have) characteristic eyelid inflammation” | PCR (Cobas Amplicor) | 3 (6%) of 48 with TF | 117, 120 |
| Egypt | All inhabitants of two trachoma-endemic villages | 46.3% of those 0-10 yr old had active disease; 3.6% of those >10 yr old had active disease | LCR (LCx) | 382 (29%) of 1,330 without active trachoma, 84 (25%) of 331 with mild follicular trachoma (F1, P1, P2), 176 (61%) of 288 with follicular trachoma only (F2, F3), 97 (81%) of 120 with severe inflammatory trachoma (P3) | 194 |
| Egypt | All children aged 1-5 yr living in two trachoma-endemic villages (an age-defined subset of the population of 194 immediately above) | 224 (62%) of 360 examined had active disease | LCR (LCx) | 45 (33%) of 136 without TF or TI, 155 (69%) of 224 with TF and/or TI | 22 |
| Egypt | All children aged 6-10 yr living in two trachoma-endemic villages (an age-defined subset of the population of 194 above) | 129 (35%) of 371 examined had active disease | LCR (LCx) | 65 (27%) of 242 without TF or TI, 90 (70%) of 129 with TF and/or TI | 22 |
| Egypt | All children aged 11-15 yr living in two trachoma-endemic villages (an age-defined subset of the population of 194 above) | 24 (7.3%) of 328 examined had active disease | LCR (LCx) | 70 (23%) of 304 without TF or TI, 15 (63%) of 24 with TF and/or TI | 22 |
| The Gambia | All inhabitants of several trachoma-endemic villages | Active disease in 34% of 0-10-yr-olds; active disease in 4.1% of >10-yr-olds | LCR (LCx) | 241 (24%) of 990 without active trachoma, 221 (46%) of 480 with mild follicular trachoma (F1, P1, P2), 81 (52%) of 155 with follicular trachoma only (F2, F3), 85 (70%) of 122 with severe inflammatory trachoma (P3) | 194 |
| Tanzania | All inhabitants of two trachoma-endemic villages | Active disease in 60.1% of 0-10-yr-olds; active disease in 17.2% of >10 yr-olds | LCR (LCx) | 22 (2%) of without active trachoma, 61 (11%) of 574 with mild follicular trachoma (F1, P1, P2), 169 (41%) of 408 with follicular trachoma only (F2, F3), 239 (55%) of 436 with severe inflammatory trachoma (P3) | 194 |
| Nepal | 17 of 18 wards in two village development committees were separated into randomization units of 1, 2, or 3 wards; 50 randomly chosen children aged 1-7 yr were chosen from each unit; LCR performed on all sampled clinically active cases (n = 117) and on a randomly chosen 118 normals | Pretreatment survey of 1,597 children aged 1-10 yr in 5 arbitrarily chosen wards showed a prevalence of active disease of 28.5% | LCR (LCx) | 29 (25%) of 117 with active disease, 5 (4%) of 118 clinically normal cases | 101 |
| Nepal | All children aged 1-10 yr from 6 villages examined; swabs taken from all those with active disease (TF or TI) and one eighth of those without active disease (selected at random) | 46 (6%) of 726 examined had active disease | LCR (LCx) | 0 of 46 with active disease (TF and/or TI in the swabbed eye), 0 of 44 without active disease | 16 |
| Egypt | All inhabitants of two villages (n = 2,067) | 408 (20%) of 2,067 examined had active disease | LCR (LCx) | 465 (28%) of 1659 without active disease, 176 (61%) of 288 with TF but not TI, 97 (81%) of 120 with TI | 48 |
| Australia | Children aged <15 yr from one Aboriginal community (n = 221) screened for trachoma; of those with follicular trachoma (n = ?), 48 had conjunctival swabs taken | Not specified; in “many Aboriginal communities in central Australia. . .20% of children (have) characteristic eyelid inflammation” | LCR (LCx) | 8 (17%) of 48 with TF | 117, 120 |
| China | All individuals (any age) with clinically active trachoma (TF or TI) in 7 villages randomly selected from Dongfang District, Hainan Province; no two cases were from the same household | Prevalence in 1-7-yr-old children 2%; sample age range 2-75 yr was though 21 of 25 swabs were from children <10 yr | LCR (LCx) | 2 (8%) of 25 swabbed (all had TF or TI) | 221 |
| Nepal | 55 children aged 1-10 yr were randomly selected from each of 4 villages in which the prevalence of active trachoma was found to be <10%; swabs taken from selected children who were clinically active | 7% in 1-10-yr-old children | LCR (LCx) | 2 (8%) of 24 swabbed (all had TF or TI) | 221 |
| Nepal | 55 children aged 1-10 yr were randomly selected from a village in which the prevalence of active trachoma was found to be >30%; swabs taken from selected children who were clinically active | 39% in 1-10-yr-old children | LCR (LCx) | 15 (63%) of 24 swabbed (all had TF or TI) | 221 |
DFA, direct fluorescent antibody; F/U, follow-up; EB, elementary body; EIA, enzyme immunoassay; sIgA, secretory immunoglobulin A; MIF, microimmunofluorescence; LCR, ligase chain reaction.
See Table 2, footnote a.
The data suggest that, although individuals with the most severe inflammation have the highest proportion of positive assays (194), not all individuals with active disease test positive for ocular C. trachomatis, even in areas where trachoma is hyperendemic, and regardless of the assay used. Equally, the absence of signs is no guarantee of a negative result. Hypotheses for the poor correlation of laboratory tests with clinical findings fall into three categories: wrong test result; wrong clinical diagnosis; and true finding. Consideration of the last of these possibilities may help us further elucidate the natural history of human ocular chlamydial infection.
Factors influencing the accuracy of the test.
Collecting a conjunctival swab or scrape involves sampling cells. If the intensity of infection is low, the proportion of conjunctival cells that are infected will be low, so the number of chlamydiae collected in a fixed number of sampled cells will have a Poisson distribution. Chance (sampling variation) will determine the likelihood that one swab contains sufficient cells with sufficient numbers of elementary bodies and reticulate bodies to exceed the test's threshold for returning a positive result. With the direct fluorescent antibody test, Taylor et al., defining a positive slide as one in which five or more elementary bodies were seen, found a discordance of 10% within pairs of replicate ocular specimens taken from the same individuals 5 min apart with the direct fluorescent antibody test (216). This was attributed to sampling variation. It was believed unlikely that this result was due to inadequate specimen collection in one or both of the samples, since all the samples comprising the discordant pairs had satisfactory conjunctival epithelial cell counts. Rather, discordance seemed to occur when a low chlamydial load was present.
Unfortunately, it is impossible to collect a fixed number of conjunctival cells. Factors affecting cell yield include the intensity and maturity of infection; the abrasiveness and absorptive ability of the swab; the force with which the swab is applied; the speed, distance traveled, and rotation of the swab head; the subject's reaction to the procedure; and the presence of any pathology rendering the conjunctival tissues more friable.
The relative contribution of antigen or nucleic acid from free elementary bodies within extracellular fluid is equally difficult to quantify and would probably vary quite markedly during the natural course of infection. Taylor et al., for example, found direct fluorescent antibody discordance within pairs of samples taken 2 to 8 days apart to be 22%, suggesting a degree of short-term biological variation in addition to the difference attributable to sampling (216).
Trachoma is usually found in more remote communities of poor countries. Suboptimal storage conditions and delays in shipping or processing can be expected. In these circumstances, deterioration of the sample between collection and testing is possible and may affect test performance (117, 194).
Occasional technical errors and minor variations in protocol are inevitable and may affect the reliability of any assay. Inadequate sampling of conjunctival epithelium (particularly in children unwilling to submit to examination) and cross-contamination of samples at the point of collection are both potential problems. In the laboratory, several tests are particularly operator dependent: microscopy, culture, and direct fluorescent antibody tests all demand expertise, patience, and concentration; microscopy and microimmunofluorescence are by nature somewhat subjective.
The billionfold amplification of chlamydial nucleic acid in the nucleic acid amplification tests raises the possibility of cross contamination of samples that are subsequently tested in the same laboratory.
Some substances present in ocular specimens might inhibit chlamydial growth in tissue culture. Some substances present in ocular specimens inhibit DNA or RNA polymerase, DNA ligase, or reverse transcriptase. The Amplicor kit gives the user the option of specifically checking each assay for successful amplification, and strategies (such as preheating the specimen to 95°C) that inactivate some inhibitors of nucleic acid amplification have been empirically determined. Equivalent steps to exclude the presence of growth inhibitors in culture would be more expensive and time-consuming.
Some assays target epitopes (enzyme immunoassay for lipopolysaccharide) or genes (PCR for rRNA or omp2) that react with similar molecules in organisms other than C. trachomatis. Even when the target is specific, nonspecific nucleic acid amplification or nonspecific hybridization during detection can theoretically occur, producing false-positive results.
The existence of plasmid-free strains of C. trachomatis has already been mentioned. The usefulness of Amplicor and LCx as diagnostic tools depends critically on the fact that these strains are extremely rare.
Factors influencing the accuracy of clinical diagnosis.
Currently accepted thresholds for clinical diagnosis of trachoma exclude some individuals with good evidence of disease (13). For example, with the WHO simplified system (226), active trachoma is considered present when five follicles are seen in the central part of the upper tarsal conjunctiva, while the presence of four follicles (assuming there are no other signs of trachoma) is considered normal. In both the WHO simplified system and the FPC system (53), pannus (once considered a sine qua non of diagnosis (8, 62, 245), and follicles seen at the limbus or in the fornices are ignored.
Conversely, both grading systems force a diagnosis of trachoma on individuals who have inflammatory conjunctival disease of other etiology. As already mentioned, papillae are poorly predictive of chlamydial infection when seen in the absence of follicles or in an area where trachoma is not highly endemic. It has also been postulated that the characteristic trachomatous follicular reaction may be elicited by nonchlamydial stimuli in individuals with a history of active trachoma (16).
In brief, then, the clinical grading schemes in current use have imperfect sensitivity and specificity. Final judgment of diagnostic tests on the basis of a comparison with the presence or absence of the standard signs is inappropriate.
Factors relating to the natural history of infection.
It is conceivable that, in the absence of any clinical signs of disease, some positive tests represent transient contamination of the eye with an inoculum too small to establish infection of epithelial cells (Fig. 5a).
FIG. 5.
Hypothesized models for the relationship between infection, disease, and test positivity following introduction of C. trachomatis into the conjunctival sac (↓ = C. trachomatis inoculum). (a) Two inocula fail to establish productive conjunctival infection. (b) Inoculum establishes productive infection, which results in the appearance of clinical signs of active trachoma. (c) Repeated reinfection causing sustained active disease, but resulting in shortened duration of infection and nucleic acid amplification test positivity through development of protective immunity. (d) Persistent infection (marked *). (e) Cryptic infection (marked **). NAAT, nucleic acid amplification test.
There is good evidence that the development and resolution of signs lags behind the start and finish of the period of laboratory positivity by direct fluorescent antibody and enzyme immunoassay tests (217) (Fig. 5b). A similar offset between laboratory evidence of infection and clinical status has been noted with PCR (13). In other words, it is possible that active disease may become clinically apparent weeks to months after infection and that a similar period may elapse between clearance of infection (or cessation of bacterial shedding) and the disappearance of clinical disease (13). Individuals in areas where trachoma is hyperendemic may be so frequently exposed to infectious inocula that the clinical signs of active disease are always present (Fig. 5c).
Conversely, assay-detectable infection sometimes persists in human conjunctival tissue after the resolution of (or without) clinical signs (28, 194) (Fig. 5d) This can be termed persistent infection; unfortunately, there is no consistency in the literature in the use of this term. The mechanism for the occurrence of persistent infection is unclear.
Additionally, interrupted chlamydial development with resultant chronic, laboratory test-negative, cryptic infection has been documented in vitro (152, 238) (Fig. 5e). If such a phenomenon occurrs in vivo, it is possible that a clinically quiet, assay-negative human conjunctiva could nevertheless have a chronic C. trachomatis infection.
Which Laboratory Tests Are Useful?
By virtue of their straightforward sampling requirements, high sensitivity, high specificity, and (manufacturing quality control issues aside) high level of interbatch and interobserver repeatability, the nucleic acid amplification tests are superior to the other assays described in this review. They are extremely useful for detecting, quantifying, and genotyping C. trachomatis DNA (124). The nucleic acid amplification tests will therefore continue to improve our understanding of the pathogenesis of infection, the role that persistent infection plays in the development of the blinding complications of trachoma, the epidemiology of ocular infection, and the impact of interventions designed to control trachoma blindness. Wherever possible, research studies that address these questions should employ nucleic acid amplification tests as the gold standard for assessing infection status.
However, the nucleic acid amplification tests only detect chlamydial nucleic acid, which does not prove that replicating or even viable organisms were present in the eye at the time of sample collection. Residual C. trachomatis DNA from a resolved ocular infection (or contamination of eye, swab, transport tube, microwell, or capillary with target DNA) could lead to a false-positive result. Currently, culture is the only method by which we can determine infectiousness and the only method by which we can assess the susceptibility of C. trachomatis isolates to antichlamydial antibiotics. (Assays for C. trachomatis-specific mRNA may eventually prove to be an alternative means for demonstrating the presence of replicating organism.)
Usually, however, neither culture nor the nucleic acid amplification tests are available or affordable for community-level assessment of populations where trachoma is endemic. Why this presents a problem for trachoma control programs will be discussed below.
COMMUNITY ASSESSMENT
Measures to control trachoma are currently being stepped up in an effort to meet the WHO-supported target of global elimination of trachoma as a public health problem by the year 2020 (5). It is clear that control cannot be achieved through management of symptomatic individuals presenting to health care facilities; a package of interventions addressing individual and community risk factors for trachomatous blindness needs to be instituted at the village level. Areas where trachoma is endemic are often extensive, but the extent to which neighboring communities are affected by the disease can vary considerably. It is therefore necessary to assess the needs of each community in turn. Because trachoma typically affects the most medically underserved populations, existing data are rarely available for program staff. There is a need for reliable techniques that can quickly identify communities in which priority interventions are required.
WHO Simplified Grading Scheme at the Community Level
Two observations will be made here about the use of the WHO simplified system in large surveys.
Follicular trachomatous inflammation is thought to provide reliable evidence of active trachoma (at least in places where trachoma is moderately to highly endemic; see below); intense trachomatous inflammation has lower specificity. Follicular trachomatous inflammation is therefore the most important sign for determining whether or not a given community requires interventions to treat disease and reduce transmission. It is important to maximize the reliability of the follicular trachomatous inflammation grade in surveys undertaken with multiple examiners. Anecdotal evidence suggests that when a few follicles are found in the central part of the tarsal conjunctiva, insufficient to meet the criteria for the diagnosis of follicular trachomatous inflammation, some examiners are reluctant to ignore them, and therefore they assign a grade of follicular trachomatous inflammation. This leads to inaccuracies. To overcome this problem, in our own research programs, follicular trachomatous inflammation is graded as absent (0), present (1), or mild (M, one to four follicles in the central part of the upper tarsal conjunctiva). The M grade has no value in itself; its sole purpose is to protect against overdiagnosis of follicular trachomatous inflammation.
The presence of trichiasis is taken to indicate a need for urgent intervention to prevent blindness. However, the urgency and nature of the intervention actually recommended (and the patient's likelihood of consenting to it) may depend on the number of lashes touching the globe, the number of lashes touching the cornea, and whether or not there is existing corneal opacity. If such distinctions are made when collecting survey data, it can help program staff to determine probable requirements for surgical services.
Trachoma Rapid Assessment
In 2001, WHO published a method for rapid, low-cost identification of communities “likely to have significant problem of blinding trachoma” (reference 159; p. 3). This method for community assessment is known as the trachoma rapid assessment.
The protocol has two phases. First, existing information on trachoma prevalence is sought from available datasets and written reports and, more subjectively, through interviewing individuals with local experience. In this way, an attempt is made to classify regions, districts, and villages as likely or unlikely to be areas where trachoma is endemic. If no information on trachoma endemicity is available to classify an area, it is recommended that its socioeconomic, geographic, and ethnic characteristics be defined and compared with the corresponding characteristics of areas that have been classified. Areas where trachoma is considered likely to be endemic are considered for further evaluation in phase two, field work.
Priority villages for field visits in the trachoma rapid assessment are the most socioeconomically disadvantaged of those identified in phase one. This deliberately selects communities in which clinical evidence of trachoma is most likely to be seen. The principal objective of such an optimally biased approach is to try and find trachoma. The implication is that if, with this method, trachoma is not found somewhere thought to be at high risk, attention may reasonably be shifted to other locations where intervention may be more urgent.
In the field, the assessment team first convenes a group discussion with community members in order to determine whether or not there is local awareness of trichiasis and to gather help in identifying local people who may suffer from it. The eyes of these nominated individuals are examined. A tour of the village is then undertaken in order to locate its most disadvantaged quarters. From at least 15 households or compounds in those areas, 50 children between 1 and 9 years of age are selected for examination. At least half the children should be preschool children, and if households are widely spaced, selection of “scattered” households is recommended. Clinical findings (with the WHO simplified system) (226) are recorded for the worst eye of each child examined. The final part of field work involves the collection of observations on the percentage of children with an unclean face (defined as the presence of “sleep” or ocular discharge around the eyes, or the presence of nasal discharge on the upper lip or cheeks), the percentage of households more than half an hour's walk from a functioning water source, the percentage of households without a functioning latrine, and the percentage of households situated less than 20 m from solid waste, garbage, or animal pens (159).
The major objections that can be raised against the trachoma rapid assessment methodology up to this point are the hit-and-miss nature of trichiasis case identification, the biased nature of sampling for active disease in children, and the lack of evidence showing a relationship between the risk of trachoma in a household and its distance to garbage or animal enclosures. However, short of examining everyone in the village, there is no sure way of identifying all cases of trichiasis, and the sampling method for follicular and intense trachomatous inflammation aims only to obtain a “worst-case estimate” of the prevalence of trachoma in the most disadvantaged households. The real shortfalls in trachoma rapid assessment are its recommendations for data analysis and the way in which these data are subsequently overinterpreted by users.
The trachoma rapid assessment guide suggests that a “rough prevalence estimate” of trichiasis be obtained by adding the number of confirmed cases of trichiasis to the number of suspected (but unexamined) cases and dividing the total by the estimated number of persons in the village. The number of children seen to have follicular and/or intense trachomatous inflammation is divided by the number of children examined to generate “the percentage of active trachoma among children,” Although the guide acknowledges that the children examined “may not be representative of the village children” and specifically warns that the protocol does not generate accurate, population-based data, enable quantification of the size of the trachoma problem, or provide a baseline for evaluation of interventions, it nevertheless recommends that these indicators be used for ranking purposes to determine which communities should be prioritized for trachoma control interventions. Furthermore, because the rough prevalence estimate of trichiasis and the percentage of active trachoma in children are expressed as percentages, they are often taken to be true estimates of disease prevalence and used to make detailed comparisons between communities. The trachoma rapid assessment does not provide and was not intended to provide data reliable enough to be used in this manner.
Required Indicators from Community Assessment
The key variables from assessment are as follows. First, how many people need trichiasis surgery? This is a number, not a prevalence, because the number of people who need surgery is used by the program manager to determine whether the program's resources are adequate to meet the need, and every person with trichiasis needs urgent treatment. It should be noted, however, that the use of the estimated backlog alone (without periodic revisions to account for recurrence and new incident cases) is a serious oversight both for monitoring program impact and for the purpose of planning future services.
Second, which are the worst communities for active disease? These are the communities that need priority implementation of measures to interrupt infection transmission. This indicator is not an exact prevalence but a categorization, which the trachoma rapid assessment can supply with an acceptable degree of uncertainty. If prevalence data are required for monitoring or research purposes, a different survey method needs to be employed.
Sequential Sampling
Myatt et al. have developed a new approach for the rapid trachoma survey, using sequential sampling (156, 157). Used widely in manufacturing and agriculture, sequential sampling contrasts with sample selection in classical epidemiology in that the sample size is not fixed in advance and data collection and analysis occur simultaneously. After the collection of each observation, accumulated data are assessed to determine whether or not the information that has been gathered is sufficient to make a decision (156, 157). The acccumulated observations must be representative of the population under study, however, or serious (and unknown) bias can be introduced. Random sampling of individuals from a census list of the entire village or district population would therefore be useful, but complete census lists are usually unavailable from areas where trachoma is endemic in advance and are time-consuming to collect. Novel techniques for approximating random sampling in rural communities have therefore also been suggested (155). This combination of a sequential sampling framework and practical proxies for random sampling may prove useful for assessing the prevalence of trachoma (and other diseases) in large or poorly defined populations.
Place of Laboratory Tests in Community Assessment
In the Surket district of Nepal in 1997, Baral et al. examined all children aged 1 to 10 years from six arbitrarily chosen villages. Forty-six (6%) of 726 children had clinical signs of active disease (follicular and/or intense trachomatous inflammation) in the right eye. Swabs were taken from the same eye of all those with active disease and a random selection of one in eight of those without active disease. All samples were negative by a ligase chain reaction assay directed against pCT and by a ligase chain reaction assay directed against omp1. Microscopy disclosed the presence of nonchlamydial bacteria on 3 of 15 swabs, gram-negative bacilli consistent with Moraxella on two slides, and gram-positive cocci consistent with Streptococcus pneumoniae on one. It is possible that these and other organisms are able to reproduce the clinical picture of active trachoma, particularly in those who have previously had a follicular reaction to ocular chlamydial infection (16).
In a separate study in low-prevalence areas of Nepal and China, only 8% of clinically active cases were ligase chain reaction positive (221). Additionally, even in areas where trachoma is hyperendemic, the correlation between disease and infection appears to worsen following antibiotic treatment. In the Egyptian arm of the trial testing azithromycin in the control of trachoma, 1,039 residents in one village were each offered a total of three doses of oral azithromycin; swabs were taken before treatment and 13 to 14 months afterwards. The positive predictive value of active trachoma for ligase chain reaction positivity was 67% before azithromycin and only 8% at follow-up (49). It could be inferred that the prevalence of active disease provides a poor indicator for assessing the success of antibiotic treatment, or the need for retreatment.
The poor positive predictive value of active trachoma for infection in populations with declining prevalence of signs of active trachoma raises the question of how elimination of trachoma from an area will be certified. The absence or a very low prevalence of C. trachomatis conjunctival infection in a representative sample of the population, proven by a nucleic acid amplification test or other sensitive assay, may ultimately be required. The expense of this method of assessment could be reduced by pooling specimens (60). Better still, the development of an inexpensive, simple, reliable assay that could be used in the field would greatly assist the process of community assessment.
Acknowledgments
We thank Rosa Arques for excellent administrative support and the International Trachoma Initiative, the Wellcome Trust/Burroughs Wellcome Fund, the Edna McConnell Clark Foundation, and the Christoffel Blinden Mission for financial assistance.
REFERENCES
- 1.al-Rifai, K. M. 1988. Trachoma through history. Int. Ophthalmol. 12:9-14. [DOI] [PubMed] [Google Scholar]
- 2.An, Q., G. Radcliffe, R. Vassallo, D. Buxton, W. J. O'Brien, D. A. Pelletier, W. G. Weisburg, J. D. Klinger, and D. M. Olive. 1992. Infection with a plasmid-free variant chlamydia related to Chlamydia trachomatis identified by using multiple assays for nucleic acid detection. J. Clin. Microbiol. 30:2814-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 1962. Expert committee on trachoma: third report. W.H.O. Tech. Rep. Ser. 234:1-48. [Google Scholar]
- 4.Anonymous. 1966. Fourth W.H.O. scientific group on trachoma research: report. W.H.O. Tech. Rep. Ser. 330:1-24. [PubMed] [Google Scholar]
- 5.Anonymous. 1997. Planning for the global elimination of trachoma (GET): report of a W.H.O. consultation (W.H.O./PBL/97.60). World Health Organization, Geneva, Switzerland.
- 6.Anonymous. 2004. Report of the seventh meeting of the W.H.O. Alliance for the Global Elimination of Trachoma (W.H.O./PBD/GET/04.1). World Health Organization, Geneva, Switzerland.
- 7.Assaad, F. A., and F. Maxwell-Lyons. 1967. Systematic observer variation in trachoma studies. Bull W.H.O. 36:885-900. [PMC free article] [PubMed] [Google Scholar]
- 8.Assaad, F. A., and F. Maxwell-Lyons. 1966. The use of catalytic models as tools for elucidating the clinical and epidemiological features of trachoma. Bull W.H.O. 34:341-355. [PMC free article] [PubMed] [Google Scholar]
- 9.Babalola, O. E., and S. D. Bage. 1992. The persistence of chlamydial inclusions in clinically quiescent trachoma. West Afr. J. Med. 11:55-61. [PubMed] [Google Scholar]
- 10.Baehr, W., Y. X. Zhang, T. Joseph, H. Su, F. E. Nano, K. D. Everett, and H. D. Caldwell. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. USA 85:4000-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey, R., C. Osmond, D. C. Mabey, H. C. Whittle, and M. E. Ward. 1989. Analysis of the household distribution of trachoma in a Gambian village using a Monte Carlo simulation procedure. Int. J. Epidemiol. 18:944-951. [DOI] [PubMed] [Google Scholar]
- 12.Bailey, R. L., P. Arullendran, H. C. Whittle, and D. C. Mabey. 1993. Randomised controlled trial of single-dose azithromycin in treatment of trachoma. Lancet 342:453-456. [DOI] [PubMed] [Google Scholar]
- 13.Bailey, R. L., T. J. Hampton, L. J. Hayes, M. E. Ward, H. C. Whittle, and D. C. Mabey. 1994. Polymerase chain reaction for the detection of ocular chlamydial infection in trachoma-endemic communities. J. Infect. Dis. 170:709-712. [DOI] [PubMed] [Google Scholar]
- 14.Bailey, R. L., L. Hayes, M. Pickett, H. C. Whittle, M. E. Ward, and D. C. Mabey. 1994. Molecular epidemiology of trachoma in a Gambian village. Br. J. Ophthalmol. 78:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballard, R. C., H. G. Fehler, P. Fotheringham, E. E. Sutter, and J. D. Treharne. 1983. Trachoma in South Africa. Soc. Sci. Med. 17:1755-1765. [DOI] [PubMed] [Google Scholar]
- 16.Baral, K., S. Osaki, B. Shreshta, C. R. Panta, A. Boulter, F. Pang, V. Cevallos, J. Schachter, and T. Lietman. 1999. Reliability of clinical diagnosis in identifying infectious trachoma in a low-prevalence area of Nepal. Bull. W.H.O. 77:461-466. [PMC free article] [PubMed] [Google Scholar]
- 17.Barbour, A. G., K. Amano, T. Hackstadt, L. Perry, and H. D. Caldwell. 1982. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J. Bacteriol. 151:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barenfanger, J. 1975. Studies on the role of the famiy unit in the transmission of trachoma. Am. J. Trop. Med. Hyg. 24:509-515. [DOI] [PubMed] [Google Scholar]
- 19.Batteiger, B. E. 1996. The major outer membrane protein of a single Chlamydia trachomatis serovar can possess more than one serovar-specific epitope. Infect. Immun. 64:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauwens, J. E., A. M. Clark, and W. E. Stamm. 1993. Diagnosis of Chlamydia trachomatis endocervical infections by a commercial polymerase chain reaction assay. J. Clin. Microbiol. 31:3023-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bieche, I., M. Olivi, M. H. Champeme, D. Vidaud, R. Lidereau, and M. Vidaud. 1998. Novel approach to quantitative polymerase chain reaction using real-time detection: application to the detection of gene amplification in breast cancer. Int. J. Cancer 78:661-666. [DOI] [PubMed] [Google Scholar]
- 22.Bird, M., C. R. Dawson, J. S. Schachter, Y. Miao, A. Shama, A. Osman, A. Bassem, and T. M. Lietman. 2003. Does the diagnosis of trachoma adequately identify ocular chlamydial infection in trachoma-endemic areas? J Infect. Dis. 187:1669-1673. [DOI] [PubMed] [Google Scholar]
- 23.Birkelund, S., A. G. Lundemose, and G. Christiansen. 1988. Chemical cross-linking of Chlamydia trachomatis. Infect. Immun. 56:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birkelund, S., A. G. Lundemose, and G. Christiansen. 1989. Immunoelectron microscopy of lipopolysaccharide in Chlamydia trachomatis. Infect. Immun. 57:3250-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birkenmeyer, L. G., and I. K. Mushahwar. 1991. DNA probe amplification methods. J. Virol. Methods 35:117-126. [DOI] [PubMed] [Google Scholar]
- 26.Black, C. M. 1997. Current methods of laboratory diagnosis of Chlamydia trachomatis infections. Clin. Microbiol. Rev. 10:160-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bobo, L., F. Coutlee, R. H. Yolken, T. Quinn, and R. P. Viscidi. 1990. Diagnosis of Chlamydia trachomatis cervical infection by detection of amplified DNA with an enzyme immunoassay. J. Clin. Microbiol. 28:1968-1973. (Erratum, J. Clin. Microbiol. 29:2912.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bobo, L., B. Munoz, R. Viscidi, T. Quinn, H. Mkocha, and S. West. 1991. Diagnosis of Chlamydia trachomatis eye infection in Tanzania by polymerase chain reaction/enzyme immunoassay. Lancet 338:847-850. [DOI] [PubMed] [Google Scholar]
- 29.Bobo, L., N. Novak, H. Mkocha, S. Vitale, S. West, and T. C. Quinn. 1996. Evidence for a predominant proinflammatory conjunctival cytokine response in individuals with trachoma. Infect. Immun. 64:3273-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooks, G. F., J. S. Butel, L. N. Ornston, E. Jawetz, J. L. Melnick, and E. A. Adelberg. 1991. Medical microbiology, 19th ed. Appleton & Lange, Norwalk, Conn.
- 31.Brunham, R. C., M. Laga, J. N. Simonsen, D. W. Cameron, R. Peeling, J. McDowell, H. Pamba, J. O. Ndinya-Achola, G. Maitha, and F. A. Plummer. 1990. The prevalence of Chlamydia trachomatis infection among mothers of children with trachoma. Am. J. Epidemiol. 132:946-952. [DOI] [PubMed] [Google Scholar]
- 32.Burton, M., M. Holland, N. Faal, A. Aguirre, E. Aryee, G. Johnson, D. Mabey, S. West, and R. Bailey. 2002. Ocular Chlamydia trachomatis load in relation to clinical signs of trachoma in the Gambia. In J. Schachter, G. Christiansen, I. N. Clarke, M. R. Hammerschlag, B. Kaltenboeck, C. C. Kuo, R. G. Rank, G. L. Ridgway, P. Saikku, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Chlamydial infections: proceedings of the 10th International Symposium on Human Chlamydial Infections, June 16-21, 2002, Antalya, Turkey. International Chlamydia Symposium, San Francisco, Calif.
- 33.Burton, M. J., M. J. Holland, N. Faal, E. A. Aryee, N. D. Alexander, M. Bah, H. Faal, S. K. West, A. Foster, G. J. Johnson, D. C. Mabey, and R. L. Bailey. 2003. Which members of a community need antibiotics to control trachoma? Conjunctival Chlamydia trachomatis infection load in Gambian villages. Investig. Ophthalmol. Vis. Sci. 44:4215-4222. [DOI] [PubMed] [Google Scholar]
- 34.Byrne, G. I., and J. W. Moulder. 1978. Parasite-specified phagocytosis of Chlamydia psittaci and Chlamydia trachomatis by L and HeLa cells. Infect. Immun. 19:598-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldwell, H. D., and L. J. Perry. 1982. Neutralization of Chlamydia trachomatis infectivity with antibodies to the major outer membrane protein. Infect. Immun. 38:745-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caldwell, H. D., and J. Schachter. 1982. Antigenic analysis of the major outer membrane protein of Chlamydia spp. Infect. Immun. 35:1024-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Investig. 111:1757-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chernesky, M., D. Jang, D. Copes, J. Patel, A. Petrich, K. Biers, A. Sproston, and J. Kapala. 2001. Comparison of a polymer conjugate-enhanced enzyme immunoassay to ligase chain reaction for diagnosis of Chlamydia trachomatis in endocervical swabs. J. Clin. Microbiol. 39:2306-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chernesky, M. A. 2002. Chlamydia trachomatis diagnostics. Sex. Transm. Infect. 78:232-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chopra, I., C. Storey, T. J. Falla, and J. H. Pearce. 1998. Antibiotics, peptidoglycan synthesis and genomics: the chlamydial anomaly revisited. Microbiology 144:2673-2678. [DOI] [PubMed] [Google Scholar]
- 42.Christiansen, G., and S. Birkelund. 2002. Chlamydia structure-a molecular approach to understand the structure of Chlamydia. In J. Schachter, G. Christiansen, I. N. Clarke, M. R. Hammerschlag, B. Kaltenboeck, C. C. Kuo, R. G. Rank, G. L. Ridgway, P. Saikku, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Chlamydial infections: proceedings of the 10th International Symposium on Human Chlamydial Infections, June 16-21, 2002, Antalya, Turkey. International Chlamydia Symposium, San Francisco, Calif.
- 43.Claas, H. C., W. J. Melchers, I. H. de Bruijn, M. de Graaf, W. C. van Dijk, J. Lindeman, and W. G. Quint. 1990. Detection of Chlamydia trachomatis in clinical specimens by the polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 9:864-868. [DOI] [PubMed] [Google Scholar]
- 44.Collier, L. H., S. Duke-Elder, and B. R. Jones. 1958. Experimental trachoma produced by cultured virus. Br. J. Ophthalmol. 42:705-720. [DOI] [PMC free article] [PubMed]
- 45.Collier, L. H., and J. Sowa. 1958. Isolation of trachoma virus in embryonate eggs. Lancet i:993-996. [DOI] [PubMed] [Google Scholar]
- 46.Comanducci, M., S. Ricci, R. Cevenini, and G. Ratti. 1990. Diversity of the Chlamydia trachomatis common plasmid in biovars with different pathogenicity. Plasmid 23:149-154. [DOI] [PubMed] [Google Scholar]
- 47.Darougar, S., and B. R. Jones. 1983.Trachoma. Br. Med. Bull. 39:117-122. [DOI] [PubMed] [Google Scholar]
- 48.Dawson, C., B. Munoz, A. Shama, A. Osman, A. Sheta, S. Sallam, S. West, and J. Schachter. 2002. Antibiotic treatment of trachoma in the Egyptian arm of the ACT trial: laboratory testing and clinical examinations. In J. Schachter, G. Christiansen, I. N. Clarke, M. R. Hammerschlag, B. Kaltenboeck, C. C. Kuo, R. G. Rank, G. L. Ridgway, P. Saikku, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Chlamydial infections: Proceedings of the 10th International Symposium on Human Chlamydial Infections, June 16-21, 2002, Antalya, Turkey. International Chlamydia Symposium, San Francisco, Calif.
- 49.Dawson, C., B. Munoz, A. Sheta, S. Sallam, S. West, and J. Schachter. 2000. Clinical and laboratory assessment of azithromycin or topical tetracycline for trachoma in the ACT trial in Egypt, p. 392. In P. Saikku (ed.), Proceedings of the 4th Meeting of the European Society for Chlamydia Research. Universitas Helsingiensis, Helsinki, Finland.
- 50.Dawson, C. R., T. Daghfous, I. Hoshiwara, K. Ramdhane, M. Kamoun, C. Yoneda, and J. Schachter. 1982. Trachoma therapy with topical tetracycline and oral erythromycin: a comparative trial. Bull. W.H.O. 60:347-355. [PMC free article] [PubMed] [Google Scholar]
- 51.Dawson, C. R., L. Hanna, and E. Jawetz. 1967. Controlled treatment trials of trachoma in American Indian children. Lancet ii:961-964. [DOI] [PubMed] [Google Scholar]
- 52.Dawson, C. R., B. R. Jones, and S. Darougar. 1975. Blinding and non-blinding trachoma: assessment of intensity of upper tarsal inflammatory disease and disabling lesions. Bull W.H.O. 52:279-282. [PMC free article] [PubMed] [Google Scholar]
- 53.Dawson, C. R., B. R. Jones, and M. L. Tarizzo. 1981. Guide to trachoma control in programs for the prevention of blindness. World Health Organization, Geneva, Switzerland.
- 54.Dawson, C. R., and J. Schachter. 2002. Should trachoma be treated with antibiotics? Lancet 359:184-185. [DOI] [PubMed] [Google Scholar]
- 55.Dawson, C. R., J. Schachter, S. Sallam, A. Sheta, R. A. Rubinstein, and H. Washton. 1997. A comparison of oral azithromycin with topical oxytetracycline/polymyxin for the treatment of trachoma in children. Clin. Infect. Dis. 24:363-368. [DOI] [PubMed] [Google Scholar]
- 56.Dean, D., L. Palmer, C. R. Pant, P. Courtright, S. Falkow, and P. O'Hanley. 1989. Use of a Chlamydia trachomatis DNA probe for detection of ocular chlamydiae. J. Clin. Microbiol. 27:1062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Barbeyrac, B., P. Rodriguez, B. Dutilh, P. Le Roux, and C. Bebear. 1995. Detection of Chlamydia trachomatis by ligase chain reaction compared with polymerase chain reaction and cell culture in urogenital specimens. Genitourin. Med. 71:382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeGraves, F. J., D. Gao, and B. Kaltenboeck. 2002. Frequent natural chlamydial infection in cattle found with high-sensitivity high-throughput quantitative PCR platform. In J. Schachter, G. Christiansen, I. N. Clarke, M. R. Hammerschlag, B. Kaltenboeck, C. C. Kuo, R. G. Rank, G. L. Ridgway, P. Saikku, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Chlamydial infections: Proceedings of the 10th International Symposium on Human Chlamydial Infections, June 16-21, 2002, Antalya, Turkey. International Chlamydia Symposium, San Francisco, Calif.
- 59.Dhir, S. P., S. Hakomori, G. E. Kenny, and J. T. Grayston. 1972. Immunochemical studies on chlamydial group antigen (presence of a 2-keto-3-deoxycarbohydrate as immunodominant group). J. Immunol. 109:116-122. [PubMed] [Google Scholar]
- 60.Diamant, J., R. Benis, J. Schachter, J. Moncada, F. Pang, H. C. Jha, R. C. Bhatta, T. Porco, and T. Lietman. 2001. Pooling of Chlamydia laboratory tests to determine the prevalence of ocular Chlamydia trachomatis infection. Ophthal. Epidemiol. 8:109-117. [DOI] [PubMed] [Google Scholar]
- 61.Dille, B. J., C. C. Butzen, and L. G. Birkenmeyer. 1993. Amplification of Chlamydia trachomatis DNA by ligase chain reaction. J. Clin. Microbiol. 31:729-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duke-Elder, W. S. 1937. Textbook of ophthalmology. Volume II: Clinical methods of examination, congenital and developmental anomalies, general pathological and therapeutic considerations, diseases of the outer eye. Henry Kimpton, London, England.
- 63.Dutilh, B., C. Bebear, P. Rodriguez, A. Vekris, J. Bonnet, and M. Garret. 1989. Specific amplification of a DNA sequence common to all Chlamydia trachomatis serovars using the polymerase chain reaction. Res. Microbiol. 140:7-16. [DOI] [PubMed] [Google Scholar]
- 64.Eastick, K., D. Longhurst, I. D. Paul, J. Leece, E. O. Caul, P. J. Horner, and A. J. Herring. 2000. Sensitive detection of Chlamydia trachomatis using the LightCycler -a new reference test, p. 93. In P. Saikku (ed.), Proceedings of the 4th meeting of the European Society for Chlamydia Research. Universitas Helsingiensis, Helsinki, Finland.
- 65.Eissenberg, L. G., P. B. Wyrick, C. H. Davis, and J. W. Rumpp. 1983. Chlamydia psittaci elementary body envelopes: ingestion and inhibition of phagolysosome fusion. Infect. Immun. 40:741-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Everett, K., R. Bush, and A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 67.Everett, K. D., and T. P. Hatch. 1995. Architecture of the cell envelope of Chlamydia psittaci 6BC. J. Bacteriol. 177:877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farencena, A., M. Comanducci, M. Donati, G. Ratti, and R. Cevenini. 1997. Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect. Immun. 65:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fawaz, F. S., C. van Ooij, E. Homola, S. C. Mutka, and J. N. Engel. 1997. Infection with Chlamydia trachomatis alters the tyrosine phosphorylation and/or localization of several host cell proteins including cortactin. Infect. Immun. 65:5301-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fehlner-Gardiner, C., C. Roshick, J. H. Carlson, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-26903. [DOI] [PubMed] [Google Scholar]
- 71.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedlaender, M. H., and J. Cameron. 1988. Vernal keratoconjunctivitis and trachoma. Int. Ophthalmol. 12:47-51. [DOI] [PubMed] [Google Scholar]
- 73.Frost, E. H., S. Deslandes, and D. Bourgaux-Ramoisy. 1993. Chlamydia trachomatis serovars in 435 urogenital specimens typed by restriction endonuclease analysis of amplified DNA. J. Infect. Dis. 168:497-501. [DOI] [PubMed] [Google Scholar]
- 74.Frost, E. H., S. Deslandes, D. Gendron, D. Bourgaux-Ramoisy, and P. Bourgaux. 1995. Variation outside variable segments of the major outer membrane protein distinguishes trachoma from urogenital isolates of the same serovar of Chlamydia trachomatis. Genitourin. Med. 71:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fukushi, H., and K. Hirai. 1992. Proposal of Chlamydia pecorum sp. nov. for Chlamydia strains derived from ruminants. Int. J. Syst. Bacteriol. 42:306-308. [DOI] [PubMed] [Google Scholar]
- 76.Ghuysen, J. M., and C. Goffin. 1999. Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob. Agents Chemother. 43:2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Girdner, J. L., A. P. Cullen, T. G. Salama, L. He, A. Lorincz, and T. C. Quinn. 1999. Evaluation of the digene hybrid capture II CT-ID test for detection of Chlamydia trachomatis in endocervical specimens. J. Clin. Microbiol. 37:1579-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gordon, F. B., and A. L. Quan. 1965. Isolation of the trachoma agent in cell culture. Proc. Soc. Exp. Biol. Med. 118:354-359. [DOI] [PubMed] [Google Scholar]
- 79.Grayston, J. T., C. C. Kuo, L. A. Campbell, and S. P. Wang. 1989. Chlamydia pneumoniae sp. nov. for Chlamydia sp. strain TWAR. Int. J. Syst. Bacteriol. 39:88-90. [Google Scholar]
- 80.Grayston, J. T., C. C. Kuo, S. P. Wang, and J. Altman. 1986. A new Chlamydia psittaci strain, TWAR, isolated in acute respiratory tract infections. N. Engl. J. Med. 315:161-168. [DOI] [PubMed] [Google Scholar]
- 81.Grayston, J. T., S. P. Wang, L. J. Yeh, and C. C. Kuo. 1985. Importance of reinfection in the pathogenesis of trachoma. Rev. Infect. Dis. 7:717-725. [DOI] [PubMed] [Google Scholar]
- 82.Green, R. G. 1935. On nature of filterable viruses. Science 82:443-445. [DOI] [PubMed] [Google Scholar]
- 83.Green, T. A., C. M. Black, and R. E. Johnson. 1998. Evaluation of bias in diagnostic-test sensitivity and specificity estimates computed by discrepant analysis. J. Clin. Microbiol. 36:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guatelli, J. C., K. M. Whitfield, D. Y. Kwoh, K. J. Barringer, D. D. Richman, and T. R. Gingeras. 1990. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc. Natl. Acad. Sci. USA 87:1874-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hackstadt, T., W. Baehr, and Y. Ying. 1991. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc. Natl. Acad. Sci. USA 88:3937-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hadgu, A. 1997. Bias in the evaluation of DNA-amplification tests for detecting Chlamydia trachomatis. Stat. Med. 16:1391-1399. [DOI] [PubMed] [Google Scholar]
- 87.Hadgu, A. 1996. The discrepancy in discrepant analysis. Lancet 348:592-593. [DOI] [PubMed] [Google Scholar]
- 88.Hadgu, A. 2000. Discrepant analysis is an inappropriate and unscientific method. J. Clin. Microbiol. 38:4301-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hadgu, A. 1999. Discrepant analysis: a biased and an unscientific method for estimating test sensitivity and specificity. J. Clin. Epidemiol. 52:1231-1237. [DOI] [PubMed] [Google Scholar]
- 90.Halberstaedter, L., and S. von Prowazek. 1907. Über zelleinschlüsse parasitärer nature beim trachom. Arb. Gesundh. Amte. 26:44-47. [Google Scholar]
- 91.Hanna, L., C. R. Dawson, O. Briones, P. Thygeson, and E. Jawetz. 1968. Latency in human infections with TRIC agents. J. Immunol. 101:43-50. [PubMed] [Google Scholar]
- 92.Harper, I. A. 1973. Laboratory problems in the diagnosis of trachoma. Trans. Ophthalmol. Soc. U. K. 93:641-646. [PubMed] [Google Scholar]
- 93.Harrison, H. R., W. T. Boyce, S. P. Wang, G. N. Gibb, J. E. Cox, and E. R. Alexander. 1985. Infection with Chlamydia trachomatis immunotype J associated with trachoma in children in an area previously endemic for trachoma. J. Infect. Dis. 151:1034-1036. [DOI] [PubMed] [Google Scholar]
- 94.Hatch, T. P. 1996. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 178:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hatch, T. P., D. W. Vance, Jr., and E. Al-Hossainy. 1981. Identification of a major envelope protein in Chlamydia spp. J. Bacteriol. 146:426-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hayes, L. J., R. L. Bailey, D. C. Mabey, I. N. Clarke, M. A. Pickett, P. J. Watt, and M. E. Ward. 1992. Genotyping of Chlamydia trachomatis from a trachoma-endemic village in the Gambia by a nested polymerase chain reaction: identification of strain variants. J. Infect. Dis. 166:1173-1177. [DOI] [PubMed] [Google Scholar]
- 97.Hayes, L. J., S. Pecharatana, R. L. Bailey, T. J. Hampton, M. A. Pickett, D. C. Mabey, P. J. Watt, and M. E. Ward. 1995. Extent and kinetics of genetic change in the omp1 gene of Chlamydia trachomatis in two villages with endemic trachoma. J. Infect. Dis. 172:268-272. [DOI] [PubMed] [Google Scholar]
- 98.Helps, C., N. Reeves, S. Tasker, and D. Harbour. 2001. Use of real-time quantitative PCR to detect Chlamydophila felis infection. J. Clin. Microbiol. 39:2675-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hilden, J. 1997. Discrepant analysis—or behaviour? Lancet 350:902. [DOI] [PubMed] [Google Scholar]
- 100.Holland, S. M., A. P. Hudson, L. Bobo, J. A. Whittum-Hudson, R. P. Viscidi, T. C. Quinn, and H. R. Taylor. 1992. Demonstration of chlamydial RNA and DNA during a culture-negative state. Infect. Immun. 60:2040-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holm, S. O., H. C. Jha, R. C. Bhatta, J. S. Chaudhary, B. B. Thapa, D. Davis, R. P. Pokhrel, M. Yinghui, M. Zegans, J. Schachter, K. D. Frick, L. Tapert, and T. M. Lietman. 2001. Comparison of two azithromycin distribution strategies for controlling trachoma in Nepal. Bull W.H.O. 79:194-200. [PMC free article] [PubMed] [Google Scholar]
- 102.Horn, J. E., M. L. Hammer, S. Falkow, and T. C. Quinn. 1986. Detection of Chlamydia trachomatis in tissue culture and cervical scrapings by in situ DNA hybridization. J. Infect. Dis. 153:1155-1159. [DOI] [PubMed] [Google Scholar]
- 103.Howard, L. V., P. F. Coleman, B. J. England, and J. E. Herrmann. 1986. Evaluation of chlamydiazyme for the detection of genital infections caused by Chlamydia trachomatis. J. Clin. Microbiol. 23:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hsia, R. C., Y. Pannekoek, E. Ingerowski, and P. M. Bavoil. 1997. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 25:351-359. [DOI] [PubMed] [Google Scholar]
- 105.Huang, J., F. J. DeGraves, D. Gao, P. Feng, T. Schlapp, and B. Kaltenboeck. 2001. Quantitative detection of Chlamydia spp. by fluorescent PCR in the LightCycler. BioTechniques 30:150-157. [DOI] [PubMed] [Google Scholar]
- 106.Javaloy, J., C. Ferrer, M. T. Vidal, and J. L. Alio. 2003. Follicular conjunctivitis caused by Chlamydia trachomatis in an infant Saharan population: molecular and clinical diagnosis. Br. J. Ophthalmol. 87:142-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Johnson, R. E., T. A. Green, J. Schachter, R. B. Jones, E. W. Hook, 3rd, C. M. Black, D. H. Martin, M. E. St Louis, and W. E. Stamm. 2000. Evaluation of nucleic acid amplification tests as reference tests for Chlamydia trachomatis infections in asymptomatic men. J. Clin. Microbiol. 38:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jones, B. R. 1974. Laboratory tests for chlamydial infection. Their role in epidemiological studies of trachoma and its control. Br. J. Ophthalmol. 58:438-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jones, B. R. 1975. The prevention of blindness from trachoma. Trans. Ophthalmol. Soc. U. K. 95:16-33. [PubMed] [Google Scholar]
- 110.Jones, B. R., L. H. Collier, and C. H. Smith. 1959. Isolation of virus from inclusion blennorrhoea. Lancet i:902-905. [DOI] [PubMed] [Google Scholar]
- 111.Kanski, J. J. 1999. Clinical ophthalmology: a systematic approach, 4th ed. Butterworth-Heinemann, Oxford, England.
- 112.Kellogg, J. A., J. W. Seiple, J. L. Klinedinst, E. S. Stroll, and S. H. Cavanaugh. 1995. Improved PCR detection of Chlamydia trachomatis by using an altered method of specimen transport and high-quality endocervical specimens. J. Clin. Microbiol. 33:2765-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuipers, J., K. Scharmann, J. Wollenhaupt, E. Nettelnbreker, S. Hopf, and H. Zeidler. 1995. Sensitivities of PCR, MicroTrak, ChlamydiaEIA, IDEIA, and PACE 2 for purified Chlamydia trachomatis elementary bodies in urine, peripheral blood, peripheral blood leukocytes, and synovial fluid. J. Clin. Microbiol. 33:3186-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuo, C., S. Wang, B. B. Wentworth, and J. T. Grayston. 1972. Primary isolation of TRIC organisms in HeLa 229 cells treated with DEAE-dextran. J. Infect. Dis. 125:665-668. [DOI] [PubMed] [Google Scholar]
- 115.Kuoppa, Y., J. Boman, L. Scott, U. Kumlin, I. Eriksson, and A. Allard. 2002. Quantitative detection of respiratory Chlamydia pneumoniae infection by real-time PCR. J. Clin. Microbiol. 40:2273-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kuper, H., A. W. Solomon, J. Buchan, M. Zondervan, A. Foster, and D. Mabey. 2003. A critical review of the SAFE strategy for the prevention of blinding trachoma. Lancet Infect. Dis. 3:372-381. [DOI] [PubMed] [Google Scholar]
- 117.Laming, A. C., and P. J. Hallsworth. 1999. Detection of chlamydial nucleic acid in follicular trachoma. Med. J. Aust. 170:190. [DOI] [PubMed] [Google Scholar]
- 118.Landis, J. R., and G. G. Koch. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159-174. [PubMed] [Google Scholar]
- 119.Lauderdale, T. L., L. Landers, I. Thorneycroft, and K. Chapin. 1999. Comparison of the PACE 2 assay, two amplification assays, and Clearview EIA for detection of Chlamydia trachomatis in female endocervical and urine specimens. J. Clin. Microbiol. 37:2223-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leach, A. J., T. M. Shelby-James, M. Mayo, M. Gratten, A. C. Laming, B. J. Currie, and J. D. Mathews. 1997. A prospective study of the impact of community-based azithromycin treatment of trachoma on carriage and resistance of Streptococcus pneumoniae. Clin. Infect. Dis. 24:356-362. [DOI] [PubMed] [Google Scholar]
- 121.Little, M. C., J. Andrews, R. Moore, S. Bustos, L. Jones, C. Embres, G. Durmowicz, J. Harris, D. Berger, K. Yanson, C. Rostkowski, D. Yursis, J. Price, T. Fort, A. Walters, M. Collis, O. Llorin, J. Wood, F. Failing, C. O'Keefe, B. Scrivens, B. Pope, T. Hansen, K. Marino, K. Williams, et al. 1999. Strand displacement amplification and homogeneous real-time detection incorporated in a second-generation DNA probe system, BDProbeTecET. Clin. Chem 45:777-784. [PubMed] [Google Scholar]
- 122.Longo, M. C., M. S. Berninger, and J. L. Hartley. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93:125-128. [DOI] [PubMed] [Google Scholar]
- 123.Lucero, M. E., and C. C. Kuo. 1985. Neutralization of Chlamydia trachomatis cell culture infection by serovar-specific monoclonal antibodies. Infect. Immun. 50:595-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mabey, D., and A. W. Solomon. 2003. Application of molecular tools in the control of blinding trachoma. Am. J. Trop. Med. Hyg. 69:11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mabey, D. C., R. L. Bailey, M. E. Ward, and H. C. Whittle. 1992. A longitudinal study of trachoma in a Gambian village: implications concerning the pathogenesis of chlamydial infection. Epidemiol. Infect. 108:343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mabey, D. C., and S. Booth-Mason. 1986. The detection of Chlamydia trachomatis by direct immunofluorescence in conjunctival smears from patients with trachoma and patients with ophthalmia neonatorum using a conjugated monoclonal antibody. J. Hyg. (London) 96:83-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mabey, D. C., T. Forsey, and J. D. Treharne. 1987. Serotypes of Chlamydia trachomatis in the Gambia. Lancet ii:452. [DOI] [PubMed] [Google Scholar]
- 128.Mabey, D. C., J. N. Robertson, and M. E. Ward. 1987. Detection of Chlamydia trachomatis by enzyme immunoassay in patients with trachoma. Lancet ii:1491-1492. [DOI] [PubMed] [Google Scholar]
- 129.Mabey, D. C., A. W. Solomon, and A. Foster. 2003. Trachoma. Lancet 362:223-229. [DOI] [PubMed] [Google Scholar]
- 130.MacCallan, A. F. 1931. The epidemiology of trachoma. Br. J. Ophthalmol. 15:369-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.MacCallan, A. F. 1908. Ophthalmic conditions in the government schools in Egypt and their amelioration. Ophthalmoscope vi:856-863, 947-952. [Google Scholar]
- 132.Mahmoud, E. A., A. E. Elhassan, H. E. Babikir, G. Froman, and P. A. Mardh. 1994. Antichlamydial activity of lacrimal fluid in patients with trachoma. Int. Ophthalmol. 18:87-91. [DOI] [PubMed] [Google Scholar]
- 133.Mahony, J., S. Castriciano, J. Sellors, I. Stewart, I. Cunningham, S. Landis, W. Seidelman, L. Grant, C. Devlin, and M. Chernesky. 1989. Diagnosis of Chlamydia trachomatis genital infections by cell culture and two enzyme immunoassays detecting different chlamydial antigens. J. Clin. Microbiol. 27:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mahony, J. B., K. E. Luinstra, J. W. Sellors, and M. A. Chernesky. 1993. Comparison of plasmid- and chromosome-based polymerase chain reaction assays for detecting Chlamydia trachomatis nucleic acids. J. Clin. Microbiol. 31:1753-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mahony, J. B., K. E. Luinstra, J. W. Sellors, L. Pickard, S. Chong, D. Jang, and M. A. Chernesky. 1994. Role of confirmatory PCRs in determining performance of Chlamydia Amplicor PCR with endocervical specimens from women with a low prevalence of infection. J. Clin. Microbiol. 32:2490-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Malaty, R., S. Zaki, M. E. Said, D. W. Vastine, D. W. Dawson, and J. Schachter. 1981. Extraocular infections in children in areas with endemic trachoma. J. Infect. Dis. 143:853. [DOI] [PubMed] [Google Scholar]
- 137.Markel, H. 2000. “The eyes have it”: trachoma, the perception of disease, the United States Public Health Service, and the American Jewish immigration experience, 1897-1924. Bull. Hist. Med. 74:525-560. [DOI] [PubMed] [Google Scholar]
- 138.Matikainen, M. T., and P. Terho. 1983. Immunochemical analysis of antigenic determinants of Chlamydia trachomatis by monoclonal antibodies. J. Gen. Microbiol. 129:2343-2350. [DOI] [PubMed] [Google Scholar]
- 139.Matsumoto, A. 1982. Electron microscopic observations of surface projections on Chlamydia psittaci reticulate bodies. J. Bacteriol. 150:358-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Matsumoto, A. 1988. Structural characteristics of chlamydial bodies, p. 21-45. In A. L. Baron (ed.), Microbiology of chlamydia. CRC Press, Boca Raton, Fla.
- 141.Matsumoto, A. 1982. Surface projections of Chlamydia psittaci elementary bodies as revealed by freeze-deep-etching. J. Bacteriol. 151:1040-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McAdam, A. J. 2000. Discrepant analysis: how can we test a test? J. Clin. Microbiol. 38:2027-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Melese, M., W. Alemayehu, A. Bejiga, Y. Adamu, and A. Worku. 2003. Modified grading system for upper eyelid trachomatous trichiasis. Ophthalmic Epidemiol. 10:75-80. [DOI] [PubMed] [Google Scholar]
- 144.Miller, W. C. 1998. Can we do better than discrepant analysis for new diagnostic test evaluation? Clin. Infect. Dis. 27:1186-1193. [DOI] [PubMed] [Google Scholar]
- 145.Miyashita, N., and A. Matsumoto. 1992. Establishment of a particle-counting method for purified elementary bodies of chlamydiae and evaluation of sensitivities of the IDEIA Chlamydia kit and DNA probe by using the purified elementary bodies. J. Clin. Microbiol. 30:2911-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Miyashita, N., A. Matsumoto, Y. Niki, and T. Matsushima. 1996. Evaluation of the sensitivity and specificity of a ligase chain reaction test kit for the detection of Chlamydia trachomatis. J. Clin. Pathol. 49:515-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moalic, E., J. M. Dueymes, R. Baron, and A. M. Le Flohic. 2000. Cross-sectional survey of trachoma in school age children in the region of Thies (Senegal). Pediatr. Infect. Dis. J. 19:979-983. [DOI] [PubMed] [Google Scholar]
- 148.Moncada, J., J. Schachter, G. Bolan, J. Engelman, L. Howard, I. Mushahwar, G. Ridgway, G. Mumtaz, W. Stamm, and A. Clark. 1990. Confirmatory assay increases specificity of the chlamydiazyme test for Chlamydia trachomatis infection of the cervix. J. Clin. Microbiol. 28:1770-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mordhorst, C. H., and N. Hegazy. 1974. Laboratory study of trachoma in Egyptian rural schoolchildren. Bull. W.H.O. 51:167-171. [PMC free article] [PubMed] [Google Scholar]
- 150.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Moulder, J. W. 1966. The relation of the psittacosis group (Chlamydiae) to bacteria and viruses. Annu. Rev. Microbiol. 20:107-130. [DOI] [PubMed] [Google Scholar]
- 152.Moulder, J. W., N. J. Levy, and L. P. Schulman. 1980. Persistent infection of mouse fibroblasts (L cells) with Chlamydia psittaci: evidence for a cryptic chlamydial form. Infect. Immun. 30:874-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mullis, K. B. 1990. The unusual origin of the polymerase chain reaction. Sci. Am. 61:64-65. [DOI] [PubMed] [Google Scholar]
- 154.Munoz, B., L. Bobo, H. Mkocha, M. Lynch, Y. H. Hsieh, and S. West. 1999. Incidence of trichiasis in a cohort of women with and without scarring. Int. J. Epidemiol. 28:1167-1171. [DOI] [PubMed] [Google Scholar]
- 155.Myatt, M. 2004. Acceptance sampling TRA (ASTRA) resource CD v1.01. International Trachoma Initiative, New York, N.Y.
- 156.Myatt, M., H. Limburg, D. Minassian, and D. Katyola. 2003. Field trial of applicability of lot quality assurance sampling survey method for rapid assessment of prevalence of active trachoma. Bull. W.H.O. 81:877-885. [PMC free article] [PubMed] [Google Scholar]
- 157.Myatt, M., D. Minassian, and H. Limburg. 2002. Preliminary report on work towards the development of a new survey method for rapid assessment of trachoma prevalence in rural communities. The Institute of Ophthalmology, London, United Kingdom.
- 158.Mygind, T., S. Birkelund, N. Birkebaek, L. Oestergaard, J. Jensen, and G. Christiansen. 2002. Determination of PCR efficiency in chelex-100 purified clinical samples and comparison of real-time quantitative PCR and conventional PCR for detection of Chlamydia pneumoniae. BMC Microbiol. 2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Negrel, A. D., H. R. Taylor, and S. West. 2001. Guidelines for the rapid assessment for blinding trachoma. World Health Organization, Geneva, Switzerland.
- 160.Newhall, W. J., and R. B. Jones. 1983. Disulfide-linked oligomers of the major outer membrane protein of chlamydiae. J. Bacteriol. 154:998-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Newhall, W. J. T. 1987. Biosynthesis and disulfide cross-linking of outer membrane components during the growth cycle of Chlamydia trachomatis. Infect. Immun 55:162-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Newhall, W. J. t., P. Terho, C. E. Wilde, 3rd, B. E. Batteiger, and R. B. Jones. 1986. Serovar determination of Chlamydia trachomatis isolates by using type-specific monoclonal antibodies. J. Clin. Microbiol. 23:333-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ossewaarde, J. M., M. Rieffe, M. Rozenberg-Arska, P. M. Ossenkoppele, R. P. Nawrocki, and A. M. van Loon. 1992. Development and clinical evaluation of a polymerase chain reaction test for detection of Chlamydia trachomatis. J. Clin. Microbiol. 30:2122-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ostergaard, L., S. Birkelund, and G. Christiansen. 1990. Use of polymerase chain reaction for detection of Chlamydia trachomatis. J. Clin. Microbiol. 28:1254-1260. (Erratum, 31:3081.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Page, L. A. 1968. Proposal for the recognition of two species in the genus Chlamydia Jones, Rake, and Stearns, 1945. Int. J. Syst. Bacteriol. 18:51-66. [Google Scholar]
- 166.Page, L. A. 1966. Revision of the family Chlamydiaceae rake (Rickettsiales): unification of the psittacosis-lymphogranuloma venereum-trachoma group of organisms in the genus Chlamydia Jones, Rake and Stearns, 1945. Int. J. Syst. Bacteriol. 16:223-252. [Google Scholar]
- 167.Palmer, L., and S. Falkow. 1986. A common plasmid of Chlamydia trachomatis. Plasmid 16:52-62. [DOI] [PubMed] [Google Scholar]
- 168.Peeling, R., I. W. Maclean, and R. C. Brunham. 1984. In vitro neutralization of Chlamydia trachomatis with monoclonal antibody to an epitope on the major outer membrane protein. Infect. Immun. 46:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pennington, T. H. 1992. Fractals and medical microbiology. Rev. Med. Microbiol. 3:120-128. [Google Scholar]
- 170.Peterson, E. M., V. Darrow, J. Blanding, S. Aarnaes, and L. M. de la Maza. 1997. Reproducibility problems with the AMPLICOR PCR Chlamydia trachomatis test. J. Clin. Microbiol. 35:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Peterson, E. M., B. A. Markoff, J. Schachter, and L. M. de la Maza. 1990. The 7.5-kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid 23:144-148. [DOI] [PubMed] [Google Scholar]
- 172.Pickett, M. A., J. S. Everson, and I. N. Clarke. 2000. Determination of chlamydial plasmid copy number using a fluorescent, 5′-exonuclease (TAQMAN) assay. In P. Saikku (ed.), Proceedings of the 4th meeting of the European Society for Chlamydia Research. Universitas Helsingiensis, Helsinki, Finland.
- 172a.Polack, S., A. W. Solomon, N. D. E. Alexander, P. A. Massae, S. Safari, J. F. Shao, A. Foster, and D. Mabey. Trans. R. Soc. Trop. Med. Hyg., in press. [DOI] [PMC free article] [PubMed]
- 173.Raeymaekers, L. 2000. Basic principles of quantitative PCR. Mol. Biotechnol. 15:115-122. [DOI] [PubMed] [Google Scholar]
- 174.Rani, R., G. Corbitt, R. Killough, and E. Curless. 2002. Is there any role for rapid tests for Chlamydia trachomatis? Int. J. Sex. Transm. Dis. AIDS 13:22-24. [DOI] [PubMed] [Google Scholar]
- 175.Rasmussen, R. 2001. (posting date). Quantification on the LightCycler instrument. Idaho Technology. http://www.idahotech.com/lightcycler_u/lectures/quantification_on_lc.htm. accessed 27 January 2003. (Online.)
- 176.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rhee, D. J., M. F. Pyfer, M. A. Friedberg, and C. J. Rapuano (ed.). 1999. Wills eye manual: office and emergency room diagnosis and treatment of eye disease (3rd ed.). Lippincott Williams & Wilkins, Philadelphia, Pa.
- 178.Roche Diagnostic Systems, Inc. 1997. Amplicor Chlamydia trachomatis/Neisseria gonorrhoeae (CT/NG) test. Package insert. Roche Diagnostic Systems, Inc., Branchburg, N.J.
- 179.Rockey, D. D. 2002. Chlamydial interactions with host cells: recent progress and remaining issues. In J. Schachter, G. Christiansen, I. N. Clarke, M. R. Hammerschlag, B. Kaltenboeck, C. C. Kuo, R. G. Rank, G. L. Ridgway, P. Saikku, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Chlamydial infections: Proceedings of the 10th International Symposium on Human Chlamydial Infections, June 16-21, 2002, Antalya, Turkey. International Chlamydia Symposium, San Francisco, Calif.
- 180.Rothermel, C. D., J. Schachter, P. Lavrich, E. C. Lipsitz, and T. Francus. 1989. Chlamydia trachomatis -induced production of interleukin-1 by human monocytes. Infect. Immun. 57:2705-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Saiki, R. K., S. Scharf, F. Faloona, K. B. Mullis, G. T. Horn, H. A. Erlich, and N. Arnheim. 1985. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350-1354. [DOI] [PubMed] [Google Scholar]
- 182.Saikku, P., M. Puolakkainen, M. Leinonen, M. Nurminen, and A. Nissinen. 1986. Cross-reactivity between Chlamydiazyme and Acinetobacter strains. N. Engl. J. Med. 314:922-923. [DOI] [PubMed] [Google Scholar]
- 183.Salari, S. H., and M. E. Ward. 1981. Polypeptide composition of Chlamydia trachomatis. J. Gen. Microbiol. 123:197-207. [DOI] [PubMed] [Google Scholar]
- 184.Sandford-Smith, J. 1997. Eye diseases in hot climates, 3rd ed. Butterworth Heinemann, London, United Kingdom.
- 185.Sarkies, J. W. 1965. Early changes in margin of upper eyelid in entropion complicating trachoma. Br. J. Ophthalmol. 49:538-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schachter, J. 1998. Chlamydia trachomatis: the more you look, the more you find-how much is there? Sex. Transm. Dis. 25:229-231. [DOI] [PubMed] [Google Scholar]
- 187.Schachter, J. 1998. Two different worlds we live in. Clin. Infect. Dis. 27:1181-1185. [DOI] [PubMed] [Google Scholar]
- 188.Schachter, J., and J. Moncada. 2002. Nucleic acid amplification tests to diagnose Chlamydia trachomatis genital infection -the glass is more than half full. In J. Schachter, G. Christiansen, I. N. Clarke, M. R. Hammerschlag, B. Kaltenboeck, C. C. Kuo, R. G. Rank, G. L. Ridgway, P. Saikku, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Chlamydial infections: Proceedings of the 10th International Symposium on Human Chlamydial Infections, June 16-21, 2002, Antalya, Turkey. International Chlamydia Symposium, San Francisco, Calif.
- 189.Schachter, J., J. Moncada, C. R. Dawson, J. Sheppard, P. Courtright, M. E. Said, S. Zaki, S. F. Hafez, and A. Lorincz. 1988. Nonculture methods for diagnosing chlamydial infection in patients with trachoma: a clue to the pathogenesis of the disease? J. Infect. Dis. 158:1347-1352. [DOI] [PubMed] [Google Scholar]
- 190.Schachter, J., C. H. Mordhorst, B. W. Moore, and M. L. Tarizzo. 1973. Laboratory diagnosis of trachoma: a collaborative study. Bull. W.H.O. 48:509-515. [PMC free article] [PubMed] [Google Scholar]
- 191.Schachter, J., W. E. Stamm, and T. C. Quinn. 1996. Discrepant analysis and screening for Chlamydia trachomatis. Lancet 348:1308-1309. [DOI] [PubMed] [Google Scholar]
- 192.Schachter, J., W. E. Stamm, and T. C. Quinn. 1998. Discrepant analysis and screening for Chlamydia trachomatis. Lancet 351:217-218. [DOI] [PubMed] [Google Scholar]
- 193.Schachter, J., W. E. Stamm, T. C. Quinn, W. W. Andrews, J. D. Burczak, and H. H. Lee. 1994. Ligase chain reaction to detect Chlamydia trachomatis infection of the cervix. J. Clin. Microbiol. 32:2540-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Schachter, J., S. K. West, D. Mabey, C. R. Dawson, L. Bobo, R. Bailey, S. Vitale, T. C. Quinn, A. Sheta, S. Sallam, H. Mkocha, and H. Faal. 1999. Azithromycin in control of trachoma. Lancet 354:630-635. [DOI] [PubMed] [Google Scholar]
- 195.Schnell, S., and C. Mendoza. 1997. Enzymological considerations for a theoretical description of the quantitative competitive polymerase chain reaction (QC-PCR). J. Theor. Biol. 184:433-440. [DOI] [PubMed] [Google Scholar]
- 196.Schramm, N., and P. B. Wyrick. 1995. Cytoskeletal requirements in Chlamydia trachomatis infection of host cells. Infect. Immun. 63:324-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shattock, R. M., C. Patrizio, P. Simmonds, and S. Sutherland. 1998. Detection of Chlamydia trachomatis in genital swabs: comparison of commercial and in house amplification methods with culture. Sex. Transm. Infect. 74:289-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Solomon, A. W., N. D. Alexander, M. J. Holland, A. Aguirre, P. A. Massae, A. Natividad-Sancho, S. Safari, R. W. Peeling, R. L. Bailey, S. K. West, A. Foster, and D. C. Mabey. 2003. Longitudinal study of the effect of azithromycin on ocular Chlamydia trachomatis load in a trachoma-endemic community of Tanzania. Trans. R. Soc. Trop. Med. Hyg. 97:270.
- 199.Solomon, A. W., M. J. Holland, M. J. Burton, S. K. West, N. D. Alexander, A. Aguirre, P. A. Massae, H. Mkocha, B. Munoz, G. J. Johnson, R. W. Peeling, R. L. Bailey, A. Foster, and D. C. Mabey. 2003. Strategies for control of trachoma: observational study with quantitative PCR. Lancet 362:198-204. [DOI] [PubMed] [Google Scholar]
- 200.Sowa, S., J. Sowa, L. H. Collier, and W. Blyth. 1965. Trachoma and allied infections in a Gambian village. Medical Research Council Special Report 308:1-88. [PubMed] [Google Scholar]
- 201.Spears, P. A., C. P. Linn, D. L. Woodard, and G. T. Walker. 1997. Simultaneous strand displacement amplification and fluorescence polarization detection of Chlamydia trachomatis DNA. Anal. Biochem. 247:130-137. [DOI] [PubMed] [Google Scholar]
- 202.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 203.Stephens, R. S., K. Koshiyama, E. Lewis, and A. Kubo. 2001. Heparin-binding outer membrane protein of chlamydiae. Mol. Microbiol. 40:691-699. [DOI] [PubMed] [Google Scholar]
- 204.Stephens, R. S., C. C. Kuo, and M. R. Tam. 1982. Sensitivity of immunofluorescence with monoclonal antibodies for detection of Chlamydia trachomatis inclusions in cell culture. J. Clin. Microbiol. 16:4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Stephens, R. S., G. Mullenbach, R. Sanchez-Pescador, and N. Agabian. 1986. Sequence analysis of the major outer membrane protein gene from Chlamydia trachomatis serovar L2. J. Bacteriol. 168:1277-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Stephens, R. S., M. R. Tam, C. C. Kuo, and R. C. Nowinski. 1982. Monoclonal antibodies to Chlamydia trachomatis: antibody specificities and antigen characterization. J. Immunol. 128:1083-1089. [PubMed] [Google Scholar]
- 207.Stothard, D. R., G. Boguslawski, and R. B. Jones. 1998. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 66:3618-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Stothard, D. R., J. A. Williams, B. Van Der Pol, and R. B. Jones. 1998. Identification of a Chlamydia trachomatis serovar E urogenital isolate which lacks the cryptic plasmid. Infect. Immun. 66:6010-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Su, H., L. Raymond, D. D. Rockey, E. Fischer, T. Hackstadt, and H. D. Caldwell. 1996. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc. Natl. Acad. Sci. USA 93:11143-11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tabbara, K. F., A. Abu-el-Asrar, O. al-Omar, A. H. Choudhury, and Z. al-Faisal. 1996. Single-dose azithromycin in the treatment of trachoma. A randomized, controlled study. Ophthalmology 103:842-846. [DOI] [PubMed] [Google Scholar]
- 211.Tam, J. E., C. H. Davis, R. J. Thresher, and P. B. Wyrick. 1992. Location of the origin of replication for the 7.5-kb Chlamydia trachomatis plasmid. Plasmid 27:231-236. [DOI] [PubMed] [Google Scholar]
- 212.T’ang, F. F., H. L. Chang, Y. T. Huang, and K. C. Wang. 1957. Studies on the etiology of trachoma with special reference to isolation of the virus in chick embryo. Chin. Med. J. (Engl.) 75:429-447. [PubMed] [Google Scholar]
- 213.Tarizzo, M. L. 1973. Field methods for the control of trachoma. World Health Organization, Geneva, Switzerland.
- 214.Tarizzo, M. L., B. Nabli, and J. Labonne. 1968. Studies on trachoma. 11. Evaluation of laboratory diagnostic methods under field conditions. Bull. W.H.O. 38:897-905. [PMC free article] [PubMed] [Google Scholar]
- 215.Taylor, H. R., P. A. Rapoza, S. West, S. Johnson, B. Munoz, S. Katala, and B. B. Mmbaga. 1989. The epidemiology of infection in trachoma. Investig. Ophthalmol. Vis. Sci. 30:1823-1833. [PubMed] [Google Scholar]
- 216.Taylor, H. R., J. A. Siler, H. A. Mkocha, B. Munoz, V. Velez, L. Dejong, and S. West. 1991. Longitudinal study of the microbiology of endemic trachoma. J. Clin. Microbiol. 29:1593-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Taylor, H. R., J. A. Siler, H. A. Mkocha, B. Munoz, and S. West. 1992. The natural history of endemic trachoma: a longitudinal study. Am. J. Trop. Med. Hyg. 46:552-559. [DOI] [PubMed] [Google Scholar]
- 218.Taylor, H. R., F. M. Velasco, and A. Sommer. 1985. The ecology of trachoma: an epidemiological study in southern Mexico. Bull. W.H.O. 63:559-567. [PMC free article] [PubMed] [Google Scholar]
- 219.Taylor, H. R., S. K. West, S. Katala, and A. Foster. 1987. Trachoma: evaluation of a new grading scheme in the United Republic of Tanzania. Bull. W.H.O. 65:485-488. [PMC free article] [PubMed] [Google Scholar]
- 220.Taylor-Robinson, D., B. J. Thomas, and M. F. Osborn. 1987. Evaluation of enzyme immunoassay (Chlamydiazyme) for detecting Chlamydia trachomatis in genital tract specimens. J. Clin. Pathol. 40:194-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Thein, J., P. Zhao, H. Liu, J. Xu, H. Jha, Y. Miao, L. Pizzarello, L. Tapert, J. Schachter, M. Mabon, S. Osaki-Holm, T. Lietman, and A. Paxton. 2002. Does clinical diagnosis indicate ocular chlamydial infection in areas with a low prevalence of trachoma? Ophthalmic Epidemiol. 9:263-269. [DOI] [PubMed] [Google Scholar]
- 222.Thomas, N. S., M. Lusher, C. C. Storey, and I. N. Clarke. 1997. Plasmid diversity in Chlamydia. Microbiology 143:1847-1854. [DOI] [PubMed] [Google Scholar]
- 223.Thygeson, P. 1957. Etiology and differential diagnosis of non-trachomatous follicular conjunctivitis. Bull. W.H.O. 16:995-1011. [PMC free article] [PubMed] [Google Scholar]
- 224.Thygeson, P. 1958. The present status of laboratory research in trachoma. Bull. W.H.O. 19:129-152. [PMC free article] [PubMed] [Google Scholar]
- 225.Thylefors, B. 1998. Prevention of blindness—W.H.O.'s mission for vision. World Health Forum 19:53-59. [PubMed] [Google Scholar]
- 226.Thylefors, B., C. R. Dawson, B. R. Jones, S. K. West, and H. R. Taylor. 1987. A simple system for the assessment of trachoma and its complications. Bull. W.H.O. 65:477-483. [PMC free article] [PubMed] [Google Scholar]
- 227.Thylefors, B., A. D. Negrel, R. Pararajasegaram, and K. Y. Dadzie. 1995. Global data on blindness. Bull. W.H.O. 73:115-121. [PMC free article] [PubMed] [Google Scholar]
- 228.Tielsch, J. M., K. P. West, Jr., G. J. Johnson, T. Tizazu, L. Schwab, M. C. Chirambo, and H. R. Taylor. 1987. Trachoma grading: observer trials conducted in southern Malawi. Br. J. Ophthalmol. 71:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Tondella, M. L., D. F. Talkington, B. P. Holloway, S. F. Dowell, K. Cowley, M. Soriano-Gabarro, M. S. Elkind, and B. S. Fields. 2002. Development and evaluation of real-time PCR-based fluorescence assays for detection of Chlamydia pneumoniae. J. Clin. Microbiol. 40:575-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Treharne, J. D., T. Forsey, and B. J. Thomas. 1983. Chlamydial serology. Br. Med. Bull. 39:194-200. [DOI] [PubMed] [Google Scholar]
- 231.Uozato, H., and D. L. Guyton. 1987. Centering corneal surgical procedures. Am. J. Ophthalmol. 103:264-275. [PubMed] [Google Scholar]
- 232.Uyeda, C. T., P. Welborn, N. Ellison-Birang, K. Shunk, and B. Tsaouse. 1984. Rapid diagnosis of chlamydial infections with the MicroTrak direct test. J. Clin. Microbiol. 20:948-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Van Der Pol, B., T. C. Quinn, C. A. Gaydos, K. Crotchfelt, J. Schachter, J. Moncada, D. Jungkind, D. H. Martin, B. Turner, C. Peyton, and R. B. Jones. 2000. Multicenter evaluation of the AMPLICOR and automated COBAS AMPLICOR CT/NG tests for detection of Chlamydia trachomatis. J. Clin. Microbiol. 38:1105-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Verkooyen, R. P., A. Luijendijk, W. M. Huisman, W. H. Goessens, J. A. Kluytmans, J. H. van Rijsoort-Vos, and H. A. Verbrugh. 1996. Detection of PCR inhibitors in cervical specimens by using the AMPLICOR Chlamydia trachomatis assay. J. Clin. Microbiol. 34:3072-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Walker, G. T., M. C. Little, J. G. Nadeau, and D. D. Shank. 1992. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc. Natl. Acad. Sci. USA 89:392-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Walsh, P. M., and D. L. Guyton. 1984. Comparison of two methods of marking the visual axis on the cornea during radial keratotomy. Am. J. Ophthalmol. 97:660-661. [DOI] [PubMed] [Google Scholar]
- 237.Wang, S. P., and J. T. Grayston. 1970. Immunologic relationship between genital TRIC, lymphogranuloma venereum, and related organisms in a new microtiter indirect immunofluorescence test. Am. J. Ophthalmol. 70:367-374. [DOI] [PubMed] [Google Scholar]
- 238.Ward, M., R. Bailey, A. Lesley, M. Kajbaf, J. Robertson, and D. Mabey. 1990. Persisting inapparent chlamydial infection in a trachoma endemic community in the Gambia. Scand. J. Infect. Dis. Suppl. 69:137-148. [PubMed] [Google Scholar]
- 239.Watson, M. W., P. R. Lambden, and I. N. Clarke. 1991. Genetic diversity and identification of human infection by amplification of the chlamydial 60-kilodalton cysteine-rich outer membrane protein gene. J. Clin. Microbiol. 29:1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.West, S., B. Munoz, L. Bobo, T. C. Quinn, H. Mkocha, M. Lynch, B. B. Mmbaga, and R. Viscidi. 1993. Nonocular Chlamydia infection and risk of ocular reinfection after mass treatment in a trachoma hyperendemic area. Investig. Ophthalmol. Vis. Sci. 34:3194-3198. [PubMed] [Google Scholar]
- 241.West, S. K., B. Munoz, V. M. Turner, B. B. Mmbaga, and H. R. Taylor. 1991. The epidemiology of trachoma in central Tanzania. Int. J. Epidemiol. 20:1088-1092. [DOI] [PubMed] [Google Scholar]
- 242.West, S. K., P. Rapoza, B. Munoz, S. Katala, and H. R. Taylor. 1991. Epidemiology of ocular chlamydial infection in a trachoma-hyperendemic area. J. Infect. Dis. 163:752-756. [DOI] [PubMed] [Google Scholar]
- 243.Williams, D. M., D. M. Magee, L. F. Bonewald, J. G. Smith, C. A. Bleicker, G. I. Byrne, and J. Schachter. 1990. A role in vivo for tumor necrosis factor alpha in host defense against Chlamydia trachomatis. Infect. Immun. 58:1572-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wilson, M. C., F. Millan-Velasco, J. M. Tielsch, and H. R. Taylor. 1986. Direct-smear fluorescent antibody cytology as a field diagnostic tool for trachoma. Arch. Ophthalmol. 104:688-690. [DOI] [PubMed] [Google Scholar]
- 245.Woodhouse, D. F. 1973. Some problems of trachoma diagnosis. Trans. Ophthalmol. Soc. U. K. 93:635-640. [PubMed] [Google Scholar]
- 246.Woolley, P. D., and J. Pumphrey. 1997. Application of ‘Clearview Chlamydia’ for the rapid detection of cervical chlamydial antigen. Int. J. STD AIDS 8:257-258. [DOI] [PubMed] [Google Scholar]
- 247.Wylie, J. L., S. Moses, R. Babcock, A. Jolly, S. Giercke, and G. Hammond. 1998. Comparative evaluation of chlamydiazyme, PACE 2, and AMP-CT assays for detection of Chlamydia trachomatis in endocervical specimens. J. Clin. Microbiol. 36:3488-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wyllie, S., R. H. Ashley, D. Longbottom, and A. J. Herring. 1998. The major outer membrane protein of Chlamydia psittaci functions as a porin-like ion channel. Infect. Immun. 66:5202-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wyrick, P. B. 2000. Intracellular survival by Chlamydia. Cell. Microbiol. 2:275-282. [DOI] [PubMed] [Google Scholar]
- 250.Yuan, Y., Y. X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Zhang, Y. X., S. Stewart, T. Joseph, H. R. Taylor, and H. D. Caldwell. 1987. Protective monoclonal antibodies recognize epitopes located on the major outer membrane protein of Chlamydia trachomatis. J. Immunol. 138:575-581. [PubMed] [Google Scholar]