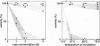Abstract
We have mutated several residues of the first of the two HMG-boxes of mammalian HMG1. Some mutants cannot be produced in Escherichia coli, suggesting that the peptide fold is grossly disrupted. A few others can be produced efficiently and have normal DNA binding affinity and specificity; however, they are more sensitive towards heating and chaotropic agents than the wild type polypeptide. Significantly, the mutation of the single most conserved residue in the rather diverged HMG-box family falls in this 'in vitro temperature-sensitive' category, rather than in the non-folded category. Finally, two other mutants have reduced DNA binding affinity but unchanged binding specificity. Overall, it appears that whenever the HMG-box can fold, it will interact specifically with kinked DNA.
Full text
PDF





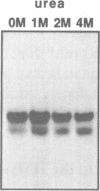
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Falciola L., Ferrari S., Lilley D. M. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992 Mar;11(3):1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M. E. Interaction of a protein from rat liver nuclei with cruciform DNA. EMBO J. 1988 Mar;7(3):843–849. doi: 10.1002/j.1460-2075.1988.tb02883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne C., Sautiere P., Duguet M., de Recondo A. M. Identification of a single-stranded DNA binding protein from rat liver with high mobility group protein 1. J Biol Chem. 1982 Mar 25;257(6):2722–2725. [PubMed] [Google Scholar]
- Bordo D., Argos P. Suggestions for "safe" residue substitutions in site-directed mutagenesis. J Mol Biol. 1991 Feb 20;217(4):721–729. doi: 10.1016/0022-2836(91)90528-e. [DOI] [PubMed] [Google Scholar]
- Brown S. J., Kellett P. J., Lippard S. J. Ixr1, a yeast protein that binds to platinated DNA and confers sensitivity to cisplatin. Science. 1993 Jul 30;261(5121):603–605. doi: 10.1126/science.8342024. [DOI] [PubMed] [Google Scholar]
- Bruhn S. L., Pil P. M., Essigmann J. M., Housman D. E., Lippard S. J. Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2307–2311. doi: 10.1073/pnas.89.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M., Lehn D. A., Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990 Jul 30;1049(3):231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- Carballo M., Puigdomènech P., Palau J. DNA and histone H1 interact with different domains of HMG 1 and 2 proteins. EMBO J. 1983;2(10):1759–1764. doi: 10.1002/j.1460-2075.1983.tb01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Harley V. R., Pontiggia A., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992 Dec;11(12):4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser K. D., Feix G. Isolation and characterization of maize cDNAs encoding a high mobility group protein displaying a HMG-box. Nucleic Acids Res. 1991 May 25;19(10):2573–2577. doi: 10.1093/nar/19.10.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Bustin M. Hierarchy of binding sites for chromosomal proteins HMG 1 and 2 in supercoiled deoxyribonucleic acid. Biochemistry. 1985 Mar 12;24(6):1428–1433. doi: 10.1021/bi00327a022. [DOI] [PubMed] [Google Scholar]
- Harley V. R., Jackson D. I., Hextall P. J., Hawkins J. R., Berkovitz G. D., Sockanathan S., Lovell-Badge R., Goodfellow P. N. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992 Jan 24;255(5043):453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- Isackson P. J., Fishback J. L., Bidney D. L., Reeck G. R. Preferential affinity of high molecular weight high mobility group non-histone chromatin proteins for single-stranded DNA. J Biol Chem. 1979 Jul 10;254(13):5569–5572. [PubMed] [Google Scholar]
- Jantzen H. M., Admon A., Bell S. P., Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990 Apr 26;344(6269):830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- Kelly M., Burke J., Smith M., Klar A., Beach D. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 1988 May;7(5):1537–1547. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodrubetz D., Burgum A. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J Biol Chem. 1990 Feb 25;265(6):3234–3239. [PubMed] [Google Scholar]
- Lilley D. M. DNA--protein interactions. HMG has DNA wrapped up. Nature. 1992 May 28;357(6376):282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- Miller J. B. Myoblast diversity in skeletal myogenesis: how much and to what end? Cell. 1992 Apr 3;69(1):1–3. doi: 10.1016/0092-8674(92)90111-o. [DOI] [PubMed] [Google Scholar]
- Ner S. S. HMGs everywhere. Curr Biol. 1992 Apr;2(4):208–210. doi: 10.1016/0960-9822(92)90541-h. [DOI] [PubMed] [Google Scholar]
- Parisi M. A., Clayton D. A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991 May 17;252(5008):965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- Pil P. M., Lippard S. J. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992 Apr 10;256(5054):234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- Pontiggia A., Negri A., Beltrame M., Bianchi M. E. Protein HU binds specifically to kinked DNA. Mol Microbiol. 1993 Feb;7(3):343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- Read C. M., Cary P. D., Crane-Robinson C., Driscoll P. C., Norman D. G. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993 Jul 25;21(15):3427–3436. doi: 10.1093/nar/21.15.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeck G. R., Isackson P. J., Teller D. C. Domain structure in high molecular weight high mobility group nonhistone chromatin proteins. Nature. 1982 Nov 4;300(5887):76–78. doi: 10.1038/300076a0. [DOI] [PubMed] [Google Scholar]
- Schulman I. G., Wang T., Wu M., Bowen J., Cook R. G., Gorovsky M. A., Allis C. D. Macronuclei and micronuclei in Tetrahymena thermophila contain high-mobility-group-like chromosomal proteins containing a highly conserved eleven-amino-acid putative DNA-binding sequence. Mol Cell Biol. 1991 Jan;11(1):166–174. doi: 10.1128/mcb.11.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman S. E., Gibson D., Wang A. H., Lippard S. J. X-ray structure of the major adduct of the anticancer drug cisplatin with DNA: cis-[Pt(NH3)2(d(pGpG))]. Science. 1985 Oct 25;230(4724):412–417. doi: 10.1126/science.4048939. [DOI] [PubMed] [Google Scholar]
- Sinclair A. H., Berta P., Palmer M. S., Hawkins J. R., Griffiths B. L., Smith M. J., Foster J. W., Frischauf A. M., Lovell-Badge R., Goodfellow P. N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990 Jul 19;346(6281):240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis A., Amsterdam A., Belanger C., Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected]. Genes Dev. 1991 May;5(5):880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- Wagner C. R., Hamana K., Elgin S. C. A high-mobility-group protein and its cDNAs from Drosophila melanogaster. Mol Cell Biol. 1992 May;12(5):1915–1923. doi: 10.1128/mcb.12.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir H. M., Kraulis P. J., Hill C. S., Raine A. R., Laue E. D., Thomas J. O. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993 Apr;12(4):1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992 Aug;11(8):3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kitzing E., Lilley D. M., Diekmann S. The stereochemistry of a four-way DNA junction: a theoretical study. Nucleic Acids Res. 1990 May 11;18(9):2671–2683. doi: 10.1093/nar/18.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]