Abstract
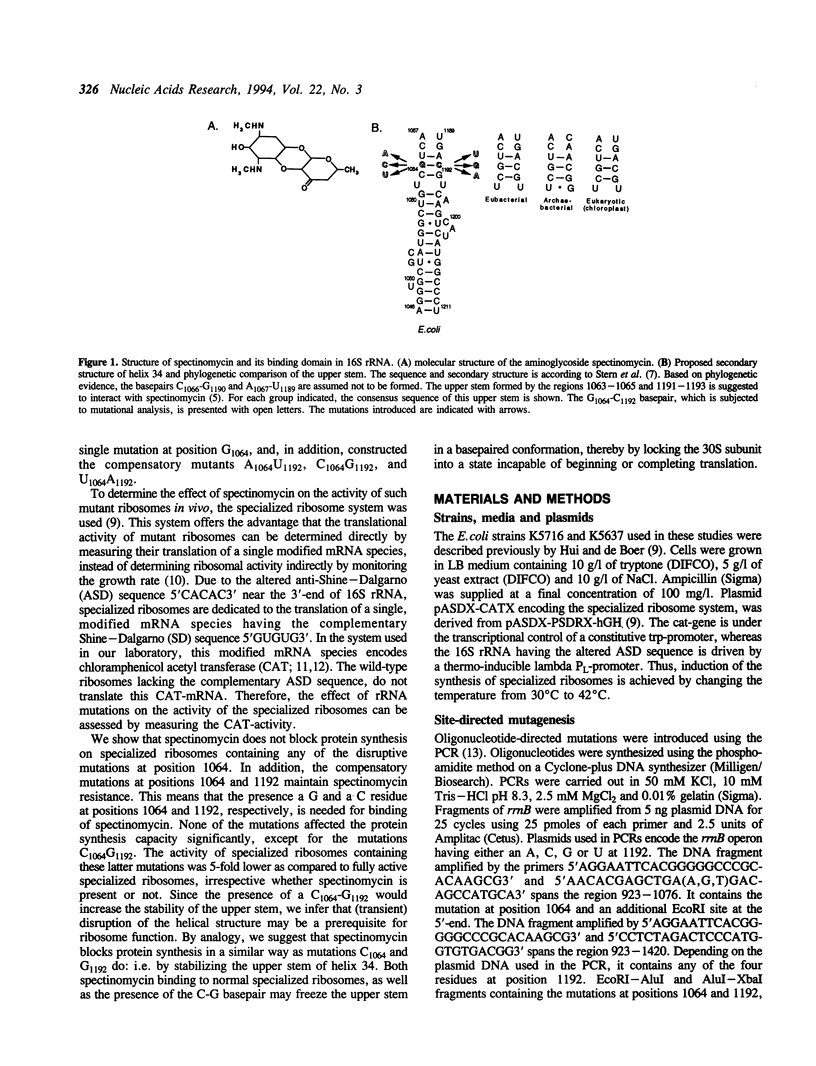
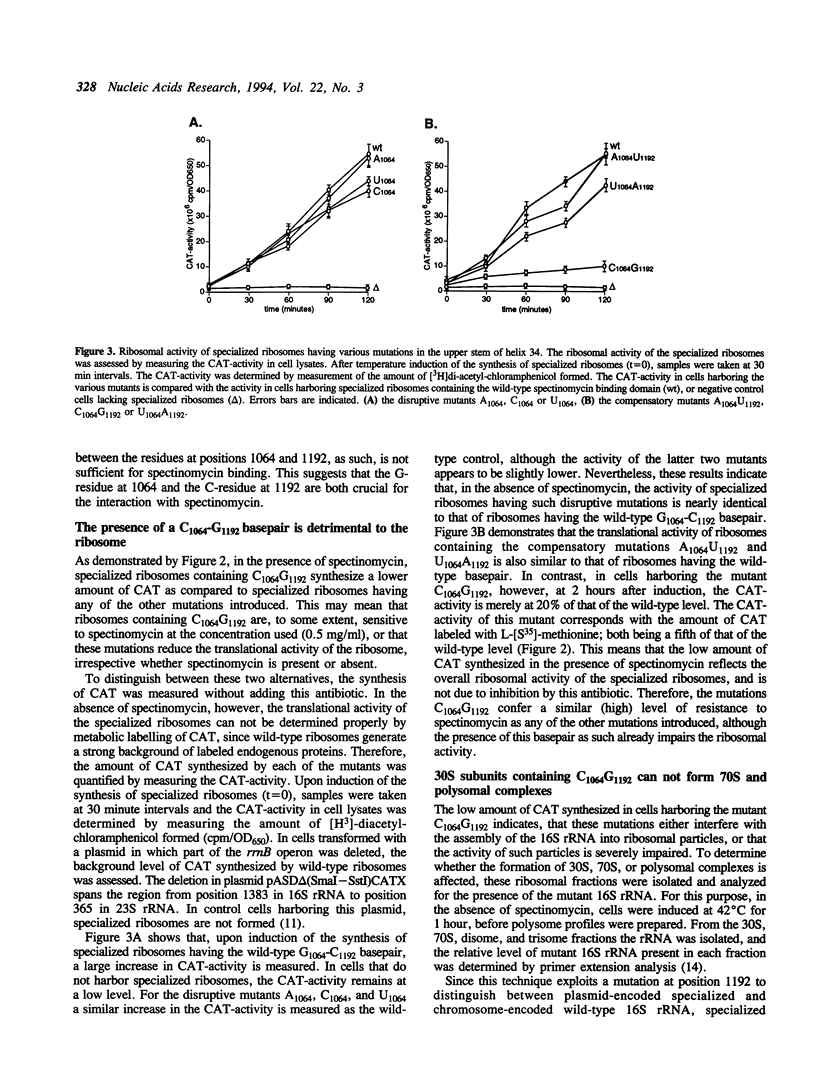
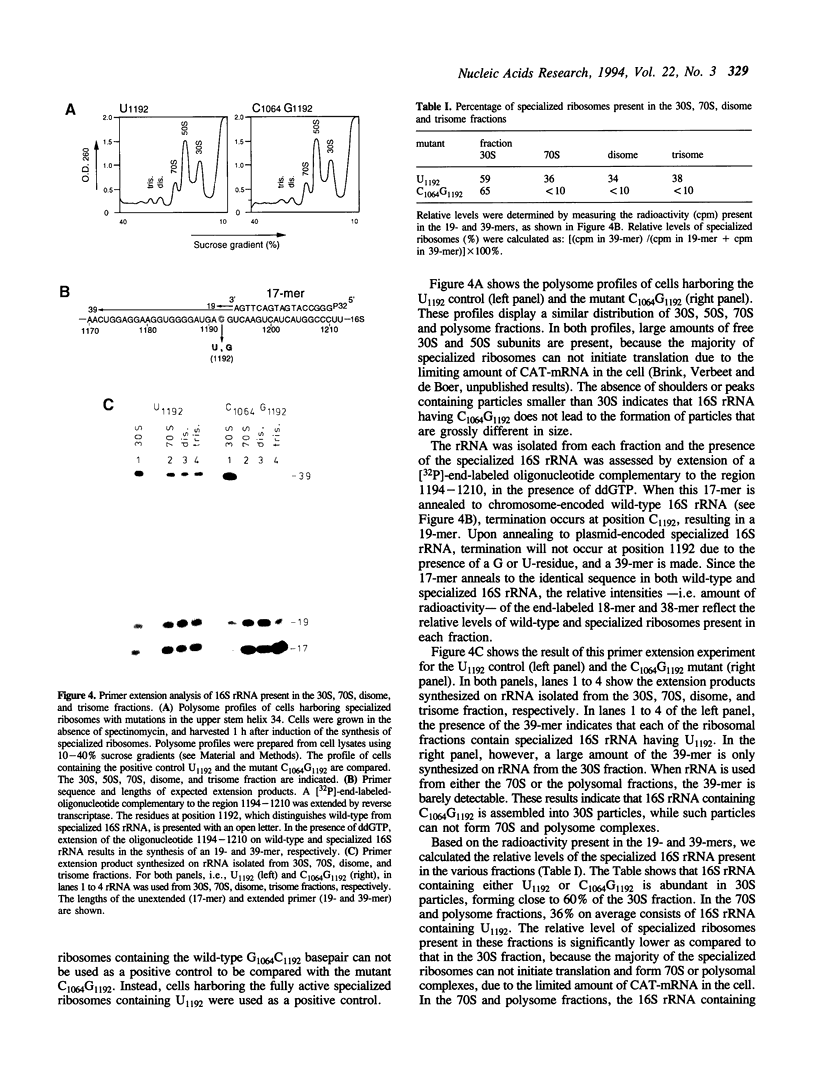
The upper stem of helix 34, consisting of the base-paired sequences C1063G1064U1065 and A1191C1192G1193, is suggested to be involved in the binding of spectinomycin. In E. coli 16S rRNA, each of the three mutations at position C1192 confers resistance to spectinomycin. In chloroplast ribosomes from tobacco plants and algae, resistance is conferred by single mutations at positions 1064, 1191, and 1193 (E. coli numbering). Since each of these mutations disrupt any of the three basepairs in the upper stem of helix 34, it has been postulated that spectinomycin can bind to this region and inhibit protein synthesis, only if its nucleotides are basepaired. We have tested this hypothesis by introducing disruptive and compensatory mutations that alter the basepair G1064-C1192. Using the specialized ribosome system, the translational activity of such mutants was determined, in the absence and presence of spectinomycin. We show that any of the three disruptive mutations A1064, C1064, and U1064 confer resistance, in accordance with the model for spectinomycin binding. Compensatory mutations A1064U1192, C1064G1192, and U1064A1192, however, maintained the resistance. This indicates that a basepaired conformation as such is not sufficient for spectinomycin binding, but rather that a G-C pair at positions 1064 and 1192 is required. In addition, we find that the translational activity of specialized ribosomes containing the mutations C1064G1192 is 5-fold lower compared to that of ribosomes containing any of the other mutations introduced, regardless whether spectinomycin is present or not. Since the introduction of C1064G1192 is expected to increase the stability of the upper stem of helix 34, we suggest that these mutations impair ribosome function by preventing the (transient) disruption of the upper stem. By analogy, we speculate that spectinomycin blocks protein synthesis by stabilizing the upper stem. In both cases, the 30S subunit would be frozen into an inactive conformation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baudin F., Ehresmann C., Romby P., Mougel M., Colin J., Lempereur L., Bachellerie J. P., Ebel J. P., Ehresmann B. Higher-order structure of domain III in Escherichia coli 16S ribosomal RNA, 30S subunit and 70S ribosome. Biochimie. 1987 Oct;69(10):1081–1096. doi: 10.1016/0300-9084(87)90008-3. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Richter A. A., Ehrenberg M., Dahlberg A. E., Kurland C. G. Ribosomal RNA and protein mutants resistant to spectinomycin. EMBO J. 1990 Mar;9(3):735–739. doi: 10.1002/j.1460-2075.1990.tb08167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen A., Davies J., Ozaki M., Mizushima S. Ribosomal protein conferring sensitivity to the antibiotic spectinomycin in Escherichia coli. Science. 1968 Jul 4;165(3888):85–86. [PubMed] [Google Scholar]
- Brimacombe R., Atmadja J., Stiege W., Schüler D. A detailed model of the three-dimensional structure of Escherichia coli 16 S ribosomal RNA in situ in the 30 S subunit. J Mol Biol. 1988 Jan 5;199(1):115–136. doi: 10.1016/0022-2836(88)90383-x. [DOI] [PubMed] [Google Scholar]
- Brink M. F., Verbeet M. P., de Boer H. A. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993 Oct;12(10):3987–3996. doi: 10.1002/j.1460-2075.1993.tb06076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns D. J., Cundliffe E. Bacterial-protein synthesis. A novel system for studying antibiotic action in vivo. Eur J Biochem. 1973 Sep 3;37(3):570–574. doi: 10.1111/j.1432-1033.1973.tb03020.x. [DOI] [PubMed] [Google Scholar]
- De Stasio E. A., Moazed D., Noller H. F., Dahlberg A. E. Mutations in 16S ribosomal RNA disrupt antibiotic--RNA interactions. EMBO J. 1989 Apr;8(4):1213–1216. doi: 10.1002/j.1460-2075.1989.tb03494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm H., Edelman M., Aviv D., Galun E. The molecular basis for rRNA-dependent spectinomycin resistance in Nicotiana chloroplasts. EMBO J. 1987 Nov;6(11):3233–3237. doi: 10.1002/j.1460-2075.1987.tb02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Stark M. J., Dahlberg A. E. Site-directed mutagenesis of ribosomal RNA. Construction and characterization of deletion mutants. J Mol Biol. 1982 Aug 15;159(3):397–416. doi: 10.1016/0022-2836(82)90291-1. [DOI] [PubMed] [Google Scholar]
- Hui A., de Boer H. A. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughrea M., Tam J. In vivo chemical footprinting of the Escherichia coli ribosome. Biochemistry. 1992 Dec 8;31(48):12035–12041. doi: 10.1021/bi00163a011. [DOI] [PubMed] [Google Scholar]
- Leclerc D., Brakier-Gingras L. A conformational switch involving the 915 region of Escherichia coli 16 S ribosomal RNA. FEBS Lett. 1991 Feb 25;279(2):171–174. doi: 10.1016/0014-5793(91)80141-o. [DOI] [PubMed] [Google Scholar]
- Makosky P. C., Dahlberg A. E. Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: an analysis of three mutants. Biochimie. 1987 Aug;69(8):885–889. doi: 10.1016/0300-9084(87)90216-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987 Jun 4;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Pinard R., Payant C., Melançon P., Brakier-Gingras L. The 5' proximal helix of 16S rRNA is involved in the binding of streptomycin to the ribosome. FASEB J. 1993 Jan;7(1):173–176. doi: 10.1096/fasebj.7.1.7678560. [DOI] [PubMed] [Google Scholar]
- Prince J. B., Taylor B. H., Thurlow D. L., Ofengand J., Zimmermann R. A. Covalent crosslinking of tRNA1Val to 16S RNA at the ribosomal P site: identification of crosslinked residues. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5450–5454. doi: 10.1073/pnas.79.18.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V., White S. W. The structure of ribosomal protein S5 reveals sites of interaction with 16S rRNA. Nature. 1992 Aug 27;358(6389):768–771. doi: 10.1038/358768a0. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Borden A., Morgan E. A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Morgan E. A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1984 Jun 11;12(11):4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Triman K., Becker E., Dammel C., Katz J., Mori H., Douthwaite S., Yapijakis C., Yoast S., Noller H. F. Isolation of temperature-sensitive mutants of 16 S rRNA in Escherichia coli. J Mol Biol. 1989 Oct 20;209(4):645–653. doi: 10.1016/0022-2836(89)92000-7. [DOI] [PubMed] [Google Scholar]