Abstract
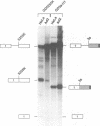
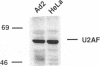
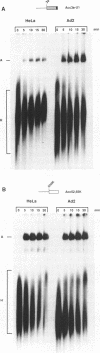
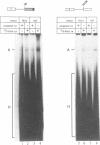
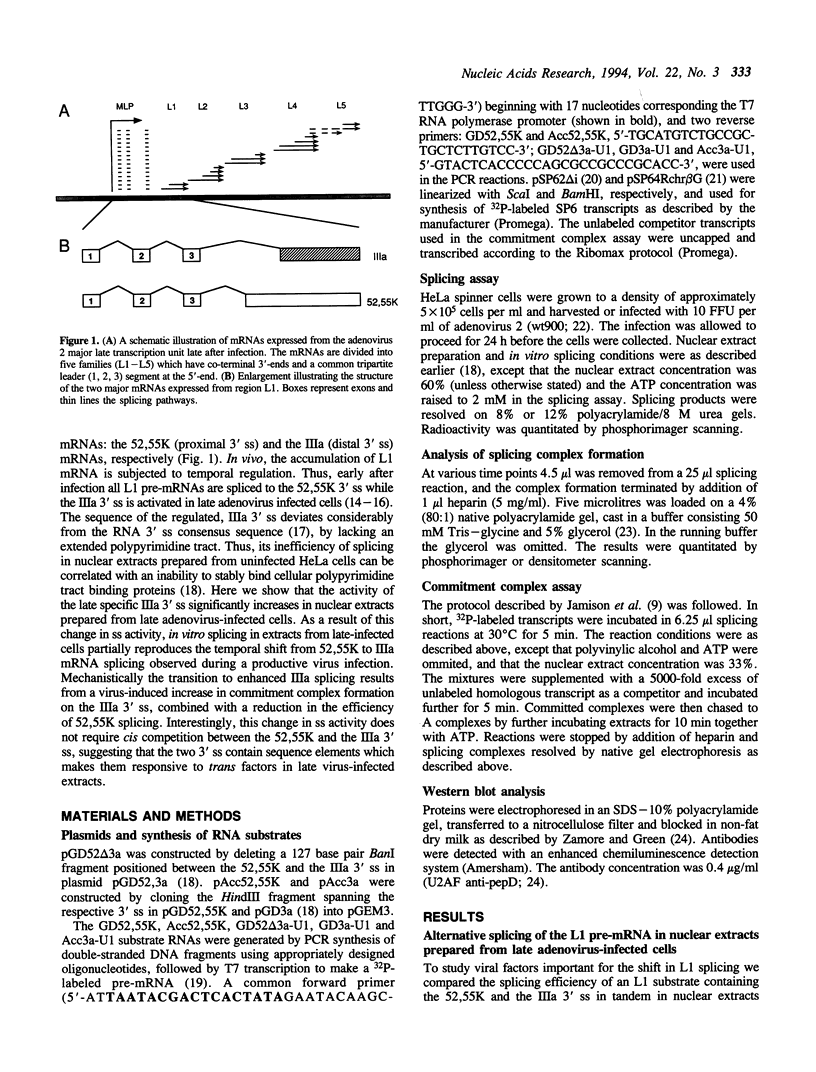
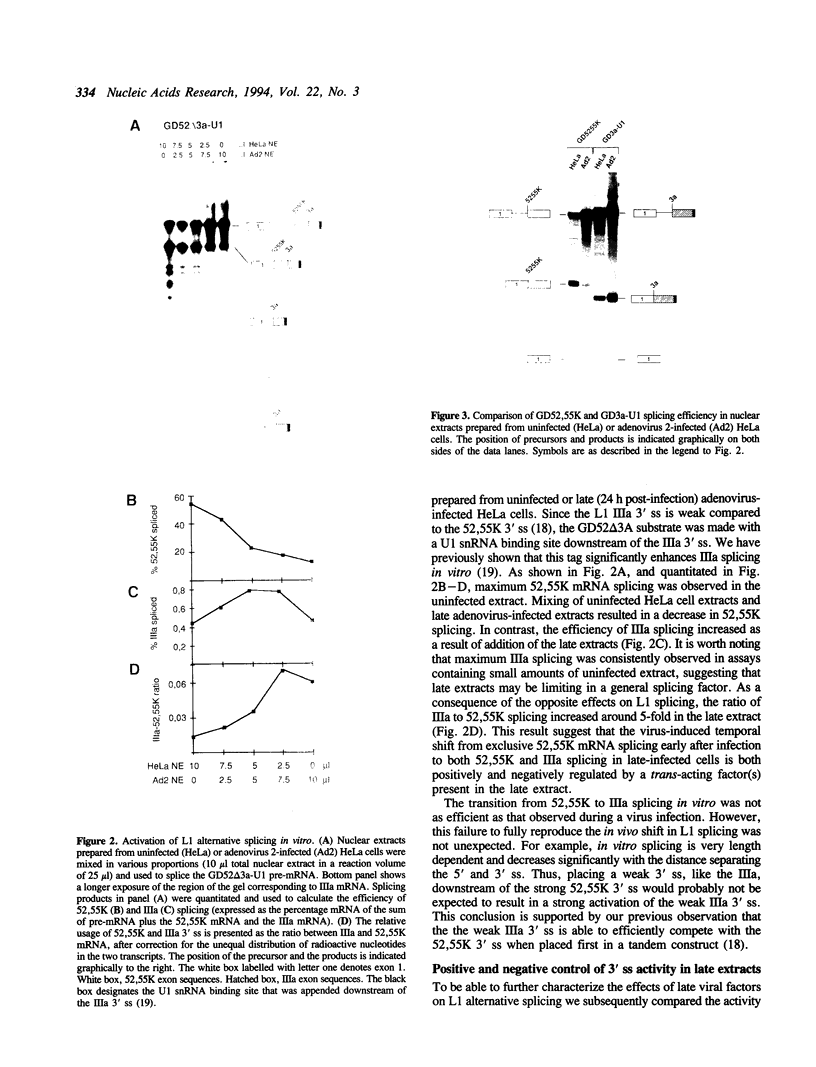
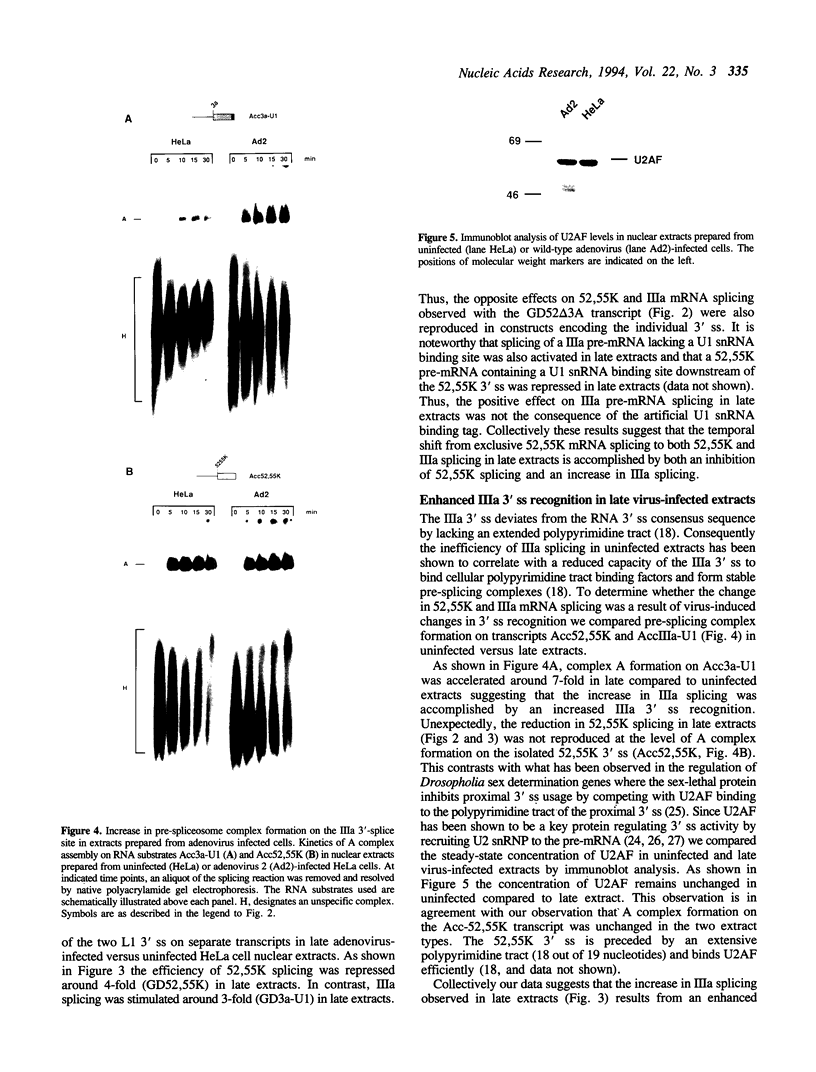
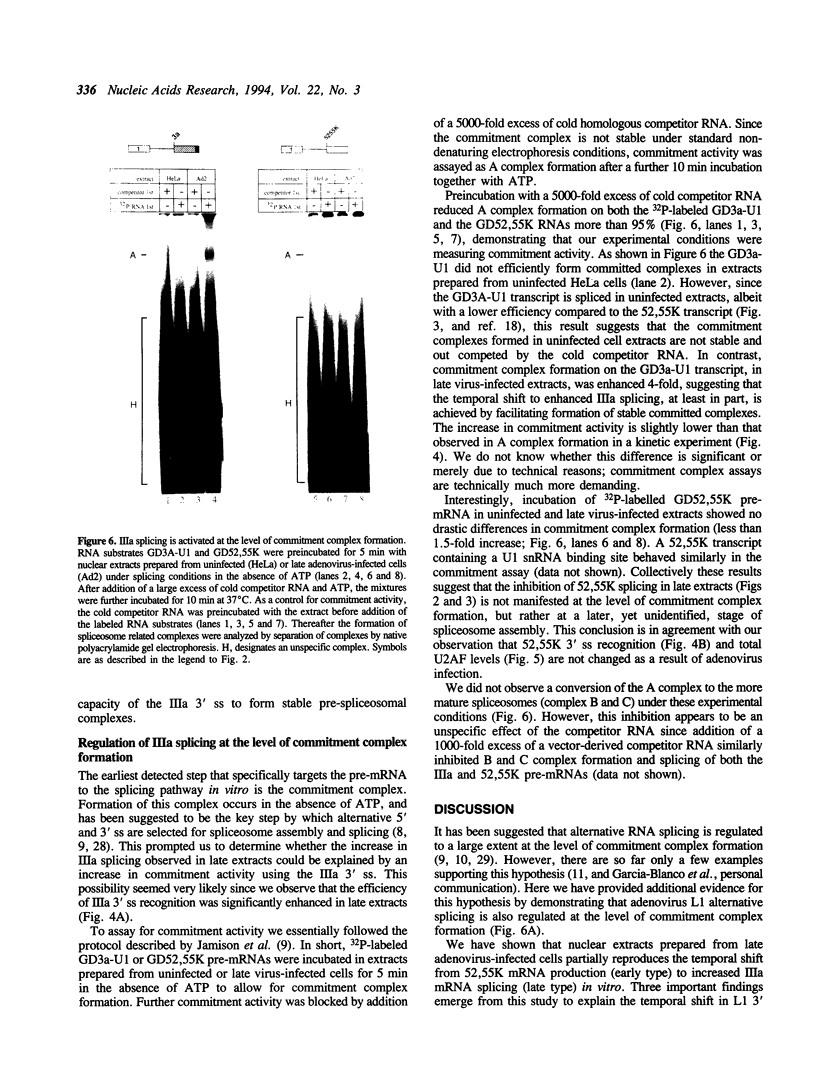
The adenovirus late region 1 (L1) represents an example of an alternatively spliced gene where one 5' splice site is spliced to two alternative 3' splice sites, to produce two mRNAs; the 52,55K and IIIa mRNAs, respectively. Accumulation of the L1 mRNAs is temporally regulated during the infectious cycle. Thus, the proximal 3' splice site (52,55K mRNA) is used at all times during the infectious cycle whereas the distal 3' splice site (IIIa mRNA) is used exclusively late in infection. Here we show that in vitro splicing extracts prepared from late adenovirus-infected cells reproduces the virus-induced temporal shift from proximal to distal 3' splice site selection in L1 pre-mRNA splicing. Two stable intermediates in spliceosome assembly have been identified; the commitment complex and the pre-spliceosome (or A complex). We show that the transition in splice site activity in L1 alternative splicing results from an increase in the efficiency of commitment complex formation using the distal 3' splice site in extracts prepared from late virus-infected cells combined with a reduction of the efficiency of proximal 3' splice site splicing. The increase in commitment activity on the distal 3' splice site is paralleled by a virus-induced increase in A complex formation on the distal 3' splice site. Importantly, the virus-induced shift from proximal to distal L1 3' splice site usage does not require cis competition between the 52,55K and the IIIa 3' splice sites, but rather results from the intrinsic property of the two 3' splice sites which make them respond differently to factors in extracts prepared from virus-infected cells.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akusjärvi G., Persson H. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature. 1981 Jul 30;292(5822):420–426. doi: 10.1038/292420a0. [DOI] [PubMed] [Google Scholar]
- Beltz G. A., Flint S. J. Inhibition of HeLa cell protein synthesis during adenovirus infection. Restriction of cellular messenger RNA sequences to the nucleus. J Mol Biol. 1979 Jun 25;131(2):353–373. doi: 10.1016/0022-2836(79)90081-0. [DOI] [PubMed] [Google Scholar]
- Bennett M., Michaud S., Kingston J., Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992 Oct;6(10):1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., Lewis J. B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979 Oct 25;134(2):265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Fu X. D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993 Sep 2;365(6441):82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- Hasson T. B., Soloway P. D., Ornelles D. A., Doerfler W., Shenk T. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J Virol. 1989 Sep;63(9):3612–3621. doi: 10.1128/jvi.63.9.3612-3621.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Jamison S. F., Crow A., Garcia-Blanco M. A. The spliceosome assembly pathway in mammalian extracts. Mol Cell Biol. 1992 Oct;12(10):4279–4287. doi: 10.1128/mcb.12.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Kreivi J. P., Zerivitz K., Akusjärvi G. Sequences involved in the control of adenovirus L1 alternative RNA splicing. Nucleic Acids Res. 1991 May 11;19(9):2379–2386. doi: 10.1093/nar/19.9.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreivi J. P., Zerivitz K., Zefrivitz K., Akusjärvi G. A U1 snRNA binding site improves the efficiency of in vitro pre-mRNA splicing. Nucleic Acids Res. 1991 Dec 25;19(24):6956–6956. doi: 10.1093/nar/19.24.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Utans U. Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J. 1991 Jun;10(6):1503–1509. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. RNA splicing. Question of commitment. Nature. 1993 Sep 2;365(6441):14–14. doi: 10.1038/365014a0. [DOI] [PubMed] [Google Scholar]
- Michaud S., Reed R. A functional association between the 5' and 3' splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 1993 Jun;7(6):1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- Michaud S., Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991 Dec;5(12B):2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 1989 Oct;3(10):1562–1571. doi: 10.1101/gad.3.10.1562. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Wilson M. C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981 Mar 12;290(5802):113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Svensson C., Akusjärvi G. Defective RNA splicing in the absence of adenovirus-associated RNAI. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4690–4694. doi: 10.1073/pnas.83.13.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993 Jul 16;74(1):105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- Ulfendahl P. J., Linder S., Kreivi J. P., Nordqvist K., Sevensson C., Hultberg H., Akusjärvi G. A novel adenovirus-2 E1A mRNA encoding a protein with transcription activation properties. EMBO J. 1987 Jul;6(7):2037–2044. doi: 10.1002/j.1460-2075.1987.tb02468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcárcel J., Singh R., Zamore P. D., Green M. R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993 Mar 11;362(6416):171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991 Jan;10(1):207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerivitz K., Kreivi J. P., Akusjärvi G. Evidence for a HeLa cell splicing activity that is necessary for activation of a regulated adenovirus 3' splice site. Nucleic Acids Res. 1992 Aug 11;20(15):3955–3961. doi: 10.1093/nar/20.15.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Leung H., Weiner A. M. The natural 5' splice site of simian virus 40 large T antigen can be improved by increasing the base complementarity to U1 RNA. Mol Cell Biol. 1987 Aug;7(8):3018–3020. doi: 10.1128/mcb.7.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]