Abstract
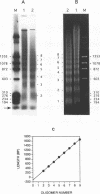
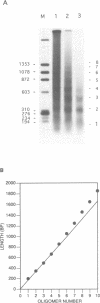
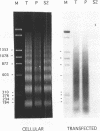
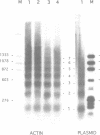
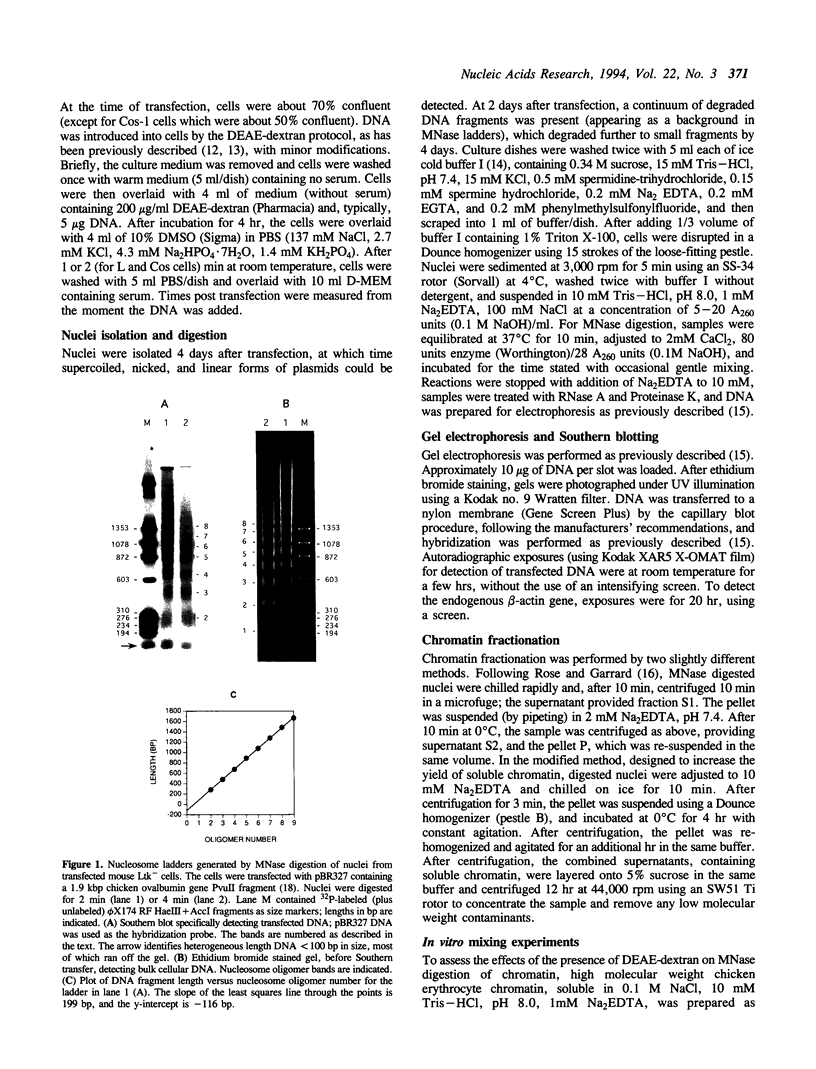
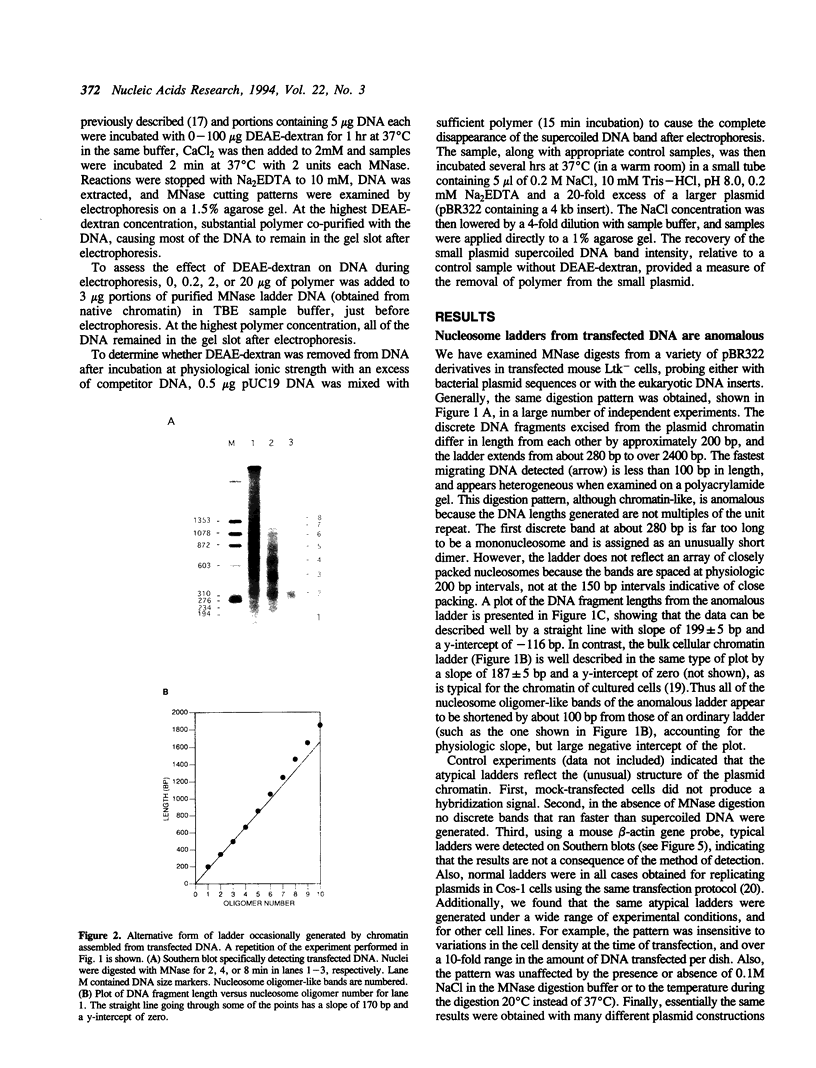
The chromatin structures of a variety of plasmids and plasmid constructions, transiently transfected into mouse Ltk- cells using the DEAE-dextran procedure, were studied by micrococcal nuclease digestion of nuclei and Southern hybridization. Although regularly arranged nucleosome-like particles clearly were formed on the transfected DNA, the nucleosome ladders, in some cases with 13-14 bands, were anomalous. Most often, a ladder of DNA fragments with lengths of approximately 300, 500, 700, 900 bp, etc. was generated. In contrast, typical 180-190 bp multiples were generated from bulk cellular or endogenous beta-actin gene chromatin. Very similar results were obtained with all DNA's transfected, and in a variety of cell lines, provided that plasmid replication did not occur. Additionally, after digestion of nuclei, about 90% of the chromatin fragments that contained transfected DNA sequences could not be solubilized at low ionic strength, in contrast with bulk cellular chromatin, suggesting association with nuclear structures or nuclear matrix. The remaining 10% of transfected DNA sequences, arising from soluble chromatin fragments, generated a typical nucleosome ladder. These results are consistent with the idea that assembly of atypical chromatin structures might be induced by proximity to elements of the nuclear pore complex or by nuclear compartmentalization.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreeva M., Markova D., Loidl P., Djondjurov L. Intranuclear compartmentalization of transcribed and nontranscribed c-myc sequences in Namalva-S cells. Eur J Biochem. 1992 Aug 1;207(3):887–894. doi: 10.1111/j.1432-1033.1992.tb17121.x. [DOI] [PubMed] [Google Scholar]
- Archer T. K., Lefebvre P., Wolford R. G., Hager G. L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992 Mar 20;255(5051):1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghini S., Yaniv M. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J. 1984 Jun;3(6):1243–1253. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croston G. E., Kadonaga J. T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Curr Opin Cell Biol. 1993 Jun;5(3):417–423. doi: 10.1016/0955-0674(93)90006-c. [DOI] [PubMed] [Google Scholar]
- Davis A. H., Reudelhuber T. L., Garrard W. T. Varigated chromatin structures of mouse ribosomal RNA genes. J Mol Biol. 1983 Jun 15;167(1):133–155. doi: 10.1016/s0022-2836(83)80038-2. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hutchison N., Weintraub H. Localization of DNAase I-sensitive sequences to specific regions of interphase nuclei. Cell. 1985 Dec;43(2 Pt 1):471–482. doi: 10.1016/0092-8674(85)90177-1. [DOI] [PubMed] [Google Scholar]
- Jackson D. A. Structure-function relationships in eukaryotic nuclei. Bioessays. 1991 Jan;13(1):1–10. doi: 10.1002/bies.950130102. [DOI] [PubMed] [Google Scholar]
- Jeong S. W., Lauderdale J. D., Stein A. Chromatin assembly on plasmid DNA in vitro. Apparent spreading of nucleosome alignment from one region of pBR327 by histone H5. J Mol Biol. 1991 Dec 20;222(4):1131–1147. doi: 10.1016/0022-2836(91)90597-y. [DOI] [PubMed] [Google Scholar]
- Kaufmann S. H., Coffey D. S., Shaper J. H. Considerations in the isolation of rat liver nuclear matrix, nuclear envelope, and pore complex lamina. Exp Cell Res. 1981 Mar;132(1):105–123. doi: 10.1016/0014-4827(81)90088-4. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- Landsman D., Bustin M. A signature for the HMG-1 box DNA-binding proteins. Bioessays. 1993 Aug;15(8):539–546. doi: 10.1002/bies.950150807. [DOI] [PubMed] [Google Scholar]
- Lauderdale J. D., Stein A. Introns of the chicken ovalbumin gene promote nucleosome alignment in vitro. Nucleic Acids Res. 1992 Dec 25;20(24):6589–6596. doi: 10.1093/nar/20.24.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A., Noll M. Chromatin fine structure of active and repressed genes. Nature. 1981 Jan 15;289(5794):198–203. doi: 10.1038/289198a0. [DOI] [PubMed] [Google Scholar]
- Lois R., Freeman L., Villeponteau B., Martinson H. G. Active beta-globin gene transcription occurs in methylated, DNase I-resistant chromatin of nonerythroid chicken cells. Mol Cell Biol. 1990 Jan;10(1):16–27. doi: 10.1128/mcb.10.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long B. H., Ochs R. L. Nuclear matrix, hnRNA, and snRNA in friend erythroleukemia nuclei depleted of chromatin by low ionic strength EDTA. Biol Cell. 1983;48(2-3):89–98. doi: 10.1111/j.1768-322x.1984.tb00204.x. [DOI] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R. Expression of the herpes thymidine kinase gene in Xenopus laevis oocytes: an assay for the study of deletion mutants constructed in vitro. Nucleic Acids Res. 1980 Dec 20;8(24):5931–5948. doi: 10.1093/nar/8.24.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. P., Albright S. C., Garrard W. T. Nucleosome arrangement with regard to DNA base composition. J Biol Chem. 1979 Sep 25;254(18):9194–9199. [PubMed] [Google Scholar]
- Ohlenbusch H. H., Olivera B. M., Tuan D., Davidson N. Selective dissociation of histones from calf thymus nucleoprotein. J Mol Biol. 1967 Apr 28;25(2):299–315. doi: 10.1016/0022-2836(67)90143-x. [DOI] [PubMed] [Google Scholar]
- Pagano J. S., McCutchan J. H., Vaheri A. Factors influencing the enhancement of the infectivity of poliovirus ribonucleic acid by diethylaminoethyl-dextran. J Virol. 1967 Oct;1(5):891–897. doi: 10.1128/jvi.1.5.891-897.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucho M., Hanahan D., Wigler M. Genetic and physical linkage of exogenous sequences in transformed cells. Cell. 1980 Nov;22(1 Pt 1):309–317. doi: 10.1016/0092-8674(80)90178-6. [DOI] [PubMed] [Google Scholar]
- Reeves R., Gorman C. M., Howard B. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 1985 May 24;13(10):3599–3615. doi: 10.1093/nar/13.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins D. M., Ripley S., Henderson A. S., Axel R. Transforming DNA integrates into the host chromosome. Cell. 1981 Jan;23(1):29–39. doi: 10.1016/0092-8674(81)90267-1. [DOI] [PubMed] [Google Scholar]
- Rose S. M., Garrard W. T. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984 Jul 10;259(13):8534–8544. [PubMed] [Google Scholar]
- Stein A., Künzler P. Histone H5 can correctly align randomly arranged nucleosomes in a defined in vitro system. Nature. 1983 Apr 7;302(5908):548–550. doi: 10.1038/302548a0. [DOI] [PubMed] [Google Scholar]
- Sukegawa J., Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993 Jan 15;72(1):29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Sun Y. L., Xu Y. Z., Bellard M., Chambon P. Digestion of the chicken beta-globin gene chromatin with micrococcal nuclease reveals the presence of an altered nucleosomal array characterized by an atypical ladder of DNA fragments. EMBO J. 1986 Feb;5(2):293–300. doi: 10.1002/j.1460-2075.1986.tb04212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman D. J., Milman G. Short-term, high-efficiency expression of transfected DNA. Mol Cell Biol. 1984 Aug;4(8):1641–1643. doi: 10.1128/mcb.4.8.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Thompson R. J. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977 Apr;10(4):633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Cheng P. F., Conrad K. Expression of transfected DNA depends on DNA topology. Cell. 1986 Jul 4;46(1):115–122. doi: 10.1016/0092-8674(86)90865-2. [DOI] [PubMed] [Google Scholar]