Abstract
The CRISPR-Cas9 RNA-guided DNA endonuclease has contributed to an explosion of advances in the life sciences that have grown from the ability to edit genomes within living cells. In this review we summarize CRISPR-based technologies that enable mammalian genome editing and their various applications. We describe recent developments that extend the generality, DNA specificity, product selectivity, and fundamental capabilities of natural CRISPR systems, and some of the remarkable advancements in basic research, biotechnology, and therapeutics development that these developments have facilitated.
Introduction
Genome editing, the introduction of a desired change to the sequence of genomic DNA, is driving a revolution in the biomedical sciences and has the potential to provide future treatments for many human diseases with a genetic component. The ideal genome editing tool would edit any genomic locus with high efficiency, high DNA sequence specificity, and little or no undesired byproducts. Such an ideal agent has not yet been developed and is unlikely to exist in nature, given that naturally occurring forms of genome-editing proteins evolved to achieve only partially related functions such as modulation of gene expression or protection from viral infection. Researchers have therefore recognized the need to develop new tools that increase the scope and effectiveness of genome editing, especially in eukaryotic cells and animal models of human disease. Recent efforts have resulted in remarkable advances towards this goal in a relatively short time period.
Early genome editing efforts were enabled by the discovery that the endogenous cellular repair pathway homologous recombination could be used to replace a small portion of the genome of a living cell with an exogenous donor DNA sequence. To use this strategy for genome editing, the exogenous DNA sequence must have homology to the target genomic DNA site. Following transfection of the donor DNA, incorporation at the desired locus spontaneously occurs very inefficiently, at rates of 1 in every ~103 to 109 cells, depending on the cell type and cell state (Smithies et al., 1985; Thomas et al., 1986). This technique of spontaneous homologous recombination was used in mouse embryo-derived stem cells, allowing researchers to generate mice with a desired genotype (Capecchi, 1989).
In addition to the low efficiency of editing using spontaneous homologous recombination, this approach could also induce undesired genome editing events in which the exogenous DNA sequence was incorporated into the genome at random sites more frequently than at the desired locus (Lin et al., 1985). A key advance in overcoming this limitation was the observation, first in yeast and then in mammalian cells, that the introduction of a double-stranded break (DSB) into the genomic locus using a meganuclease, an endonuclease that recognizes and cleaves a long DNA sequence, would stimulate homology-driven DNA incorporation (Figure 1a) (Rudin et al., 1989; Rouet et al., 1994). This “homology-directed repair” (HDR) strategy enhanced the efficiency of the desired genome editing event by two to three orders of magnitude, resulting in targeted incorporation typically being much more efficient than incorporation at random sites in the genome (Choulika et al., 1995; Jasin, 1996).
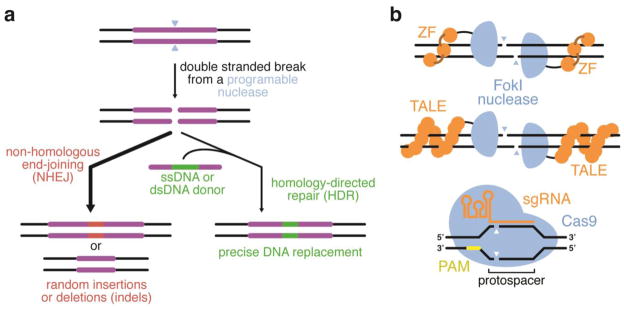
Figure 1.
Genome editing using double-stranded breaks (DSBs). (a) A programmable nuclease incorporates a sequence-specific DSB in genomic DNA. In the absence of a repair template, the cell will process the DSB mostly by NHEJ, resulting in indels at the site of editing. In the presence of a separate DNA template containing sequences homologous to the regions flanking the DSB, HDR can result in incorporation of the repair template into the genomic DNA. (b) ZFNs, TALENs, and CRISPR-based nucleases have also been used to introduce programmable, sequence-specific DSBs. The ability of Cas9 to be reprogrammed to bind a new 23-bp sequence (the protospacer and PAM) by designing a new sgRNA, rather than by engineering a new DNA-binding protein (orange), has transformed the genome editing field.
Despite this breakthrough, genome editing still suffered from two major drawbacks. First, non-homologous end joining (NHEJ) also occurs at the site of DSBs, typically more efficiently than HDR, resulting in stochastic insertions and deletions (indels) of nucleotides at the target locus (Figure 1a) (Jeggo, 1998). While NHEJ-mediated genome editing is useful for gene disruption, when precise genome editing is desired indels are unwanted byproducts. Second, since the probability that a known meganuclease cleaves a particular target locus of interest is extremely small, either a meganuclease recognition site must be incorporated into the genomic locus of interest (Jasin, 1996), or a meganuclease must be engineered to cleave the target locus (Sussman et al., 2004; Rosen et al., 2006; Grizot et al., 2009). Overcoming the first drawback is a focus of current research, and will be discussed later in this review. To address the second drawback, researchers turned to zinc-finger nucleases (ZFNs) (Bibikova et al., 2002; Bibikova et al., 2003; Porteus and Baltimore, 2003; Urnov et al., 2010) and transcription activator-like effector nucleases (TALENs) (Figure 1b) (Li et al., 2011a; Li et al., 2011b; Joung and Sander, 2013), engineered nucleases based on arrays of naturally occurring DNA-binding domains fused to the nonspecific DNA cleavage domain from FokI. Because the amino acid sequences of zinc finger arrays and TALE repeat arrays, unlike most DNA-binding proteins, can be readily designed to bind to virtually any target DNA sequence, ZFNs and TALENs can be engineered to cleave a target genomic loci with fairly high specificity (Carroll, 2008; Boch et al., 2009; Moscou and Bogdanove, 2009; Miller et al., 2011; Zhang et al., 2011; Joung and Sander, 2013). The design of ZFNs is complicated by their extensive protein-DNA contacts, however, and the cloning of TALEN genes is impeded by their highly repetitive nature. In addition, each new target locus requires the design, gene synthesis, expression, and validation of a new ZFN or TALEN protein (Figure 1b) (Urnov et al., 2010; Miller et al., 2011).
This significant barrier to genome editing—that each new target site requires the design and construction of a new nuclease—was substantially lowered by the advent of CRISPR-Cas9 as an RNA-guided DNA endonuclease (Garneau et al., 2010; Jinek et al., 2012; Gasiunas et al., 2012). In this system, a Cas endonuclease protein forms a complex with a “guide RNA” molecule and localizes to a target DNA sequence following simple guide RNA:genomic DNA base pairing rules (Figure 1b) (Doudna and Charpentier, 2014; Hsu et al., 2014). The target DNA sequence (the protospacer) must be both complementary to the guide RNA, and also contain a “protospacer-adjacent motif” (PAM), a short DNA sequence required for compatibility with the particular Cas protein being used (Deveau et al., 2008; Garneau et al., 2010; Sapranauskas et al., 2011; Jinek et al., 2012; Gasiunas et al., 2012). While this new technology places a modest limitation on the number of genomic sites amenable to genome editing due to the PAM requirement, it replaces the complex protein design and engineering tasks associated with ZFNs and TALENs with the much simpler task of designing a new guide RNA for each genomic site of interest using simple Watson-Crick base-pairing (Cong et al., 2013; Mali et al., 2013b; Jinek et al., 2013).
The elucidation of the mechanics of CRISPR-Cas9 (Barrangou et al., 2007; Garneau et al., 2010; Deltcheva et al., 2011; Sapranauskas et al., 2011; Jinek et al., 2012; Gasiunas et al., 2012), and its adaptation for use in eukaryotic genome editing (Cong et al., 2013; Mali et al., 2013b; Wang et al., 2013; Cho et al., 2013; Hwang et al., 2013b; Jinek et al., 2013) has had a transformative impact on the life sciences. The ease with which new DNA sequences can be targeted for genome editing has enabled scientists to rapidly discover new gene functions, develop new cell and animal models of diseases, and make substantial progress towards human therapeutics. In this review we summarize some of the recently developed tools that use CRISPR-Cas9 for the manipulation of mammalian genomes, and their applications in basic science, biotechnology, and medicine.
New natural CRISPR enzymes
Several natural CRISPR nucleases have now been used for mammalian genome editing. Each CRISPR nuclease can vary in size, PAM requirement, and location of the introduced DSB within the protospacer (Table 1). The most commonly used variant is the 1,368-residue Cas9 protein from Streptococcus pyogenes (SpCas9) (Haft et al., 2005). Most known naturally occurring Cas9 nucleases including SpCas9 natively use two different RNA molecules, the CRISPR-RNA (crRNA) and the trans-activating crRNA (tracrRNA) to form a functional guide RNA:Cas9 complex (Deltcheva et al., 2011). The discovery that a single guide RNA (sgRNA) could take the place of the crRNA and the tracrRNA further simplified the use of the CRISPR-Cas9 system such that only one protein and one RNA molecule are needed to achieve RNA-programmed DNA cleavage (Jinek et al., 2012).
Table 1.
Properties of some of the naturally occurring and engineered CRISPR enzymes that have been used for genome editing in mammalian cells.
| Enzyme name | Size (residues) | PAM requirement and cleavage pattern |
|---|---|---|
| SpCas9/FnCas9 | 1368/1629 |
|
| St1Cas9 | 1121 |
|
| St3Cas9 | 1409 |
|
| NmCas9 | 1082 |
|
| SaCas9 | 1053 |
|
| AsCpf1/LbCpf1 | 1307/1228 |
|
| VQR SpCas9 | 1368 |
|
| EQR SpCas9 | 1368 |
|
| VRER SpCas9 | 1368 |
|
| RHA FnCas9 | 1629 |
|
| KKH SaCas9 | 1053 |
|
The relatively simple PAM requirement of NGG contributes to the popularity of SpCas9 for genome editing (Table 1). The Staphylococcus aureus (Sa) Cas9 analog (SaCas9) offers a smaller size (1,053 residues) that facilitates some of the applications described below, but requires a more complex PAM of NNGRRT (Ran et al., 2015; Friedland et al., 2015). Other Cas9 homologs with different PAM requirements have also been used for mammalian genome editing. For example, the Streptococcus thermophilus (St) Cas9 proteins St1Cas9 and St3Cas9 are 1,121 and 1,388 residues and require NNAGAAW and NGGNG PAMs, respectively (Table 1) (Gasiunas et al., 2012; Cong et al., 2013; Gasiunas and Siksnys, 2013; Esvelt et al., 2013; Ran et al., 2013b; Muller et al., 2016). The Neisseria meningitides (Nm) Cas9 protein (NmCas9) is 1,082 residues and requires a NNNNGATT PAM (Table 1) (Gasiunas and Siksnys, 2013; Esvelt et al., 2013; Ran et al., 2013b; Hou et al., 2013; Zhang et al., 2015). Table 1 lists other Cas9 homologs that have been validated for mammalian genome editing. Nevertheless, SpCas9 remains the most widely used homolog as it is the most well characterized, offers a reasonable balance between PAM complexity and construct size, and has been extensively tested in a wide variety of contexts.
Recent progress has uncovered additional nucleases capable of RNA-guided sequence-specific DNA cleavage. For example, both the 1,307-residue Acidaminococcus sp. Cpf1 (AsCpf1) and the 1,228-residue Lachnospiraceae bacterium Cpf1 (LbCpf1) enzymes have been used for mammalian cell genome editing (Zetsche et al., 2015a). In contrast to the known Cas9 homologs, these two enzymes natively require only a crRNA, as opposed to a dual-guide RNA, a TTTN PAM at the 5′ end, rather than the 3′ end, of the protospacer, and cleave the two DNA strands in a staggered, rather than a blunt-ended, configuration (Zetsche et al., 2015a; Fonfara et al., 2016). While these and other RNA-programmed endonucleases already offer researchers a variety of possible genome editing options, the steadily increasing popularity of genome editing, coupled with the development of new precision genome editing techniques such as those described below, suggests the continued importance of discovering additional programmable DNA-binding or DNA-cleaving proteins.
Expanding the targeting scope of Cas9
As genome editing techniques using RNA-guided nucleases become more precise and diverse, the need for agents with different PAM requirements increases. The relatively simple NGG PAM sequence of SpCas9 occurs on average every 8–12 bp in the human genome (Cong et al., 2013; Hsu et al., 2013), a frequency that is not excessively limiting for classical HDR- and NHEJ-based genome editing as multiple DNA cleavage locations can lead to the same desired HDR or NHEJ outcome. The discovery of additional naturally occurring RNA-guided nucleases such as those in Table 1 offer additional targeting flexibility. For other genome-editing techniques such as base editing (see below), or when it is necessary to distinguish between a wild-type and mutant allele, however, precise targeting of a locus with single-nucleotide resolution can be critical. In these cases, the PAM requirements can be a major restriction.
Wild-type SpCas9 has been shown to have some activity on sites with NAG and NGA PAMs, but typically with much lower efficiencies than on sites with canonical NGG PAMs (Jiang et al., 2013; Hsu et al., 2013; Zhang et al., 2014; Kleinstiver et al., 2015b). A recent study used a bacterial selection system to identify three new variants of SpCas9 that can target NGA, NGAG, and NGCG PAMs with high efficiencies and specificities (Table 1) (Kleinstiver et al., 2015b). This study is an exciting example of how a small number of mutations—in these cases, three to four—can substantially alter the PAM specificity of an RNA-guided nuclease.
Researchers have also engineered Cas9 enzymes to exhibit relaxed PAM specificities. In one approach, an unbiased selection system was used to relax the NNGRRT PAM requirement of SaCas9 to NNNRRT (Table 1). The engineered variant had three mutations, and exhibited off-target editing comparable to that of the wild-type enzyme in human cells (Kleinstiver et al., 2015a). In a different study, the crystal structure of FnCas9 was used to guide the rational design of a variant with a relaxed PAM requirement. While the wild-type FnCas9 recognizes a NGG PAM, the engineered variant (which differs from wild-type at three residues) requires only a YG PAM, and can be used to edit mammalian genomes when the protein is pre-complexed with sgRNA and directly injected into zygotes (Table 1) (Hirano et al., 2016). These important advances expand the number of target loci amenable to RNA-guided genome editing.
Improving the DNA specificity of CRISPR-based agents
In addition to expanding the targeting scope of genome editing agents, improving their DNA specificity has also been a major priority. Researchers have revealed the DNA-targeting specificities of CRISPR-based genome editing agents using a variety of approaches. These methods include ChIP-seq (Cencic et al., 2014; Kuscu et al., 2014; Wu et al., 2014; O’Geen et al., 2015), targeted analysis of genomic sites identified through computational predictions (Fu et al., 2013; Hsu et al., 2013), in vitro high-throughput profiling methods (Pattanayak et al., 2013), whole-genome sequencing methods (Smith et al., 2014; Veres et al., 2014; Yang et al., 2014; Kim et al., 2015), the GUIDE-seq method (Tsai et al., 2015), and the BLESS method (Crosetto et al., 2013; Ran et al., 2015). While detailed analyses of these methods are beyond the scope of this review, collectively they have revealed the presence of off-target activity among wild-type Cas9 homologs with certain sgRNAs and established that no simple algorithm or inspection process can accurately and comprehensively predict the off-target substrates of a given Cas9:sgRNA complex (Tsai and Joung, 2016). In many reported cases, off-target sites with more mismatches relative to the on-target site are modified by wild-type CRISPR agents more extensively than sites with fewer mismatches. Indeed, off-target modification in a few studied cases can approach or even exceed the efficiency of on-target modification (Fu et al., 2013; Kuscu et al., 2014; Tsai et al., 2015; Ran et al., 2015). Notably, the inherent specificity of Cpf1 enzymes appears to be higher than that of the SpCas9 variant (Figure 2d) (Kim et al., 2016; Kleinstiver et al., 2016b).
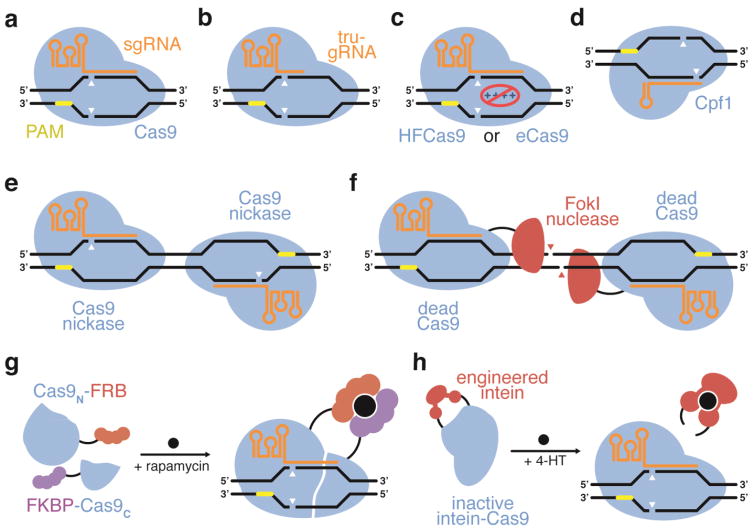
Figure 2.
Strategies for improving the DNA specificity of CRISPR-based agents. (a) Wild-type Cas9 variants have been shown to possess significant off-target activity. (b) DNA specificity can be improved using truncated sgRNAs with wtCas9 (Fu et al., 2014), (c) engineered HFCas9 or eCas9 variants that reduce nonspecific electrostatic interactions between the protein and DNA (Slaymaker et al., 2016; Kleinstiver et al., 2016a), or (d) the Cpf1 CRISPR enzyme (Kim et al., 2016; Kleinstiver et al., 2016b). Alternatively, (e) two Cas9 nickase enzymes (Ran et al., 2013a; Mali et al., 2013a), or (f) dCas9-FokI fusions can be used to require two RNA-programmed binding events to induce a DSB (Guilinger et al., 2014b), increasing specificity. DNA specificity can also be increased by limiting the cellular residence time of wtCas9 using (g) a small molecule-activated split Cas9 (Zetsche et al., 2015b), or (h) a small molecule-activated intein-disrupted Cas9 (Davis et al., 2015).
Researchers have developed several strategies to substantially improve the specificity of SpCas9 (and likely other CRISPR agents) without making any changes to the Cas9 protein sequence. Off-target modification by SpCas9 can be decreased up to several orders of magnitude simply by truncating the sgRNA of SpCas9 to have fewer than 20 nucleotides of complementarity with its target DNA (Figure 2b) (Fu et al., 2014; Tsai et al., 2015). Another strategy that improves the specificity of Cas9 is to decrease its activity or lifetime in cells after it has had sufficient opportunity to modify the target locus. This strategy improves genome-editing specificity as it reduces the amount of time Cas9 can function after its on-target locus has already been modified and only off-target loci are available for modification. For example, the direct delivery of Cas9:sgRNA ribonucleotide protein complexes (RNPs) to cells, which results in transient Cas9 activity, rather than plasmid transfection, which results in long-lasting Cas9 and sgRNA expression, can increase the ratio of on-target genome editing to off-target genome editing by more than an order of magnitude in mammalian cells (Lin et al., 2014b; Kim et al., 2014; Ramakrishna et al., 2014; Zuris et al., 2015; Liu et al., 2015b).
Researchers have also engineered variants of Cas9 that are activated by light or exogenous small molecules. These variants, including an intein-inactivated Cas9 system (Davis et al., 2015) and a small molecule-dimerized split Cas9 system (Zetsche et al., 2015b), have been shown to substantially improve genome editing specificity in mammalian cells compared with wild-type Cas9 by carefully controlling the temporal window within which active Cas9 is generated so that less active Cas9 is present after modification of the on-target loci is complete (Figure 2g,h). Similar systems, such as light-activated Cas9 variants (Nihongaki et al., 2015a; Hemphill et al., 2015; Jain et al., 2016), split Cas9 variants (Truong et al., 2015; Wright et al., 2015), small-molecule induction of Cas9 (Dow et al., 2015), and an engineered allosterically regulated Cas9 (Oakes et al., 2016) could also be used to reduce off-target genome editing following these same principles.
An additional strategy to reduce off-target genome editing through Cas9 engineering is to require that two separate Cas9 binding events take place at the same locus in order to result in DNA cleavage. Cas9 can be converted to a nickase enzyme (Cas9n) by inactivating either of its two catalytic residues (Mali et al., 2013a; Ran et al., 2013b). By designing two sgRNAs that bring separate Cas9n molecules to nick opposite DNA strands, double-stranded breaks only occur with simultaneous binding events (Figure 2e). This strategy reduces the theoretical likelihood of off-target events from 1/n to ~1/n2; in practice paired nicking reduced off-target activity up to several orders of magnitude in mammalian cells while retaining on-target activity (Ran et al., 2013a; Mali et al., 2013a). Inactivation of both catalytic residues results in dCas9, which cannot cleave either DNA strand but retains its ability to bind to a target DNA sequence. Fusion of the nonspecific restriction endonuclease FokI, which requires dimerization to become catalytically competent, to dCas9 results in an engineered variant that requires dual guide RNAs to coordinate FokI-dCas9 dimerization at a specific locus (Figure 2f) (Guilinger et al., 2014b; Tsai et al., 2014; Wyvekens et al., 2015). This approach results in up to two orders of magnitude of improved specificity in mammalian cells although with somewhat reduced activity compared to wild-type Cas9. Because ChIP-seq experiments suggest that dCas9 binding is more promiscuous than Cas9 cleavage (Wu et al., 2014), however, a paired nickase approach may offer additional specificity advantages compared with the use of FokI-dCas9 dimers.
Recently, structure-guided engineering of SpCas9 has yielded variants with improved specificity. Several previous studies have implicated excess DNA-binding energy as a source of off-target genome editing activity among ZFNs, TALENs, and Cas9 (Gupta et al., 2011; Pattanayak et al., 2011; Pattanayak et al., 2013; Guilinger et al., 2014a). By introducing just three to four mutations into SpCas9 that neutralize nonspecific electrostatic interactions between the protein and the sugar-phosphate backbone of its target DNA, its DNA specificity increases dramatically (Figure 2c) (Slaymaker et al., 2016; Kleinstiver et al., 2016a).
Together, these advances in enhancing the specificity of CRISPR-based genome editing agents in mammalian cells are key developments that increase their promise both as research tools, and as potential future therapeutics.
Improving the product selectivity of genome editing agents
Because all of the above genome editing tools are programmable nucleases that create a double-stranded break in DNA, a major limitation to the use of these tools is the introduction of stochastic indels at the site of genome editing due to NHEJ (Figure 1a). While certain applications rely on these indels to disrupt genes, splicing, or regulatory sequences, for many applications the unpredictable insertion or deletion of nucleotides at the target locus is not desired, and precise editing in which the predominant product is the replacement of one allele for another is required. Although HDR is capable of effecting precise allele replacement, HDR efficiency is typically quite low (< 5%), depends on many factors including cell type and cell state, and is competitive with NHEJ. DNA nicks do not commonly result in NHEJ, and thus the usage of a single Cas9 nickase with a donor DNA template usually results in lower indel frequency (Cong et al., 2013). This single nickase strategy, however, also results in substantially decreased HDR-mediated editing efficiencies compared to that of wild-type Cas9 or double nicking strategies, and is not amenable to all cell types.
To improve the product selectivity of genome editing to favor precise allele replacement, researchers have inhibited certain endogenous DNA repair components to favor HDR over NHEJ (Figure 3d). For example, the small molecule Scr7 is known to inhibit DNA ligase IV, a key component of NHEJ (Srivastava et al., 2012; Vartak and Raghavan, 2015). Administration of Scr7 in combination with Cas9, a sgRNA, and a donor DNA template has been shown to enhance HDR:NHEJ ratios substantially in some systems (Vartak and Raghavan, 2015; Chu et al., 2015; Maruyama et al., 2015; Pinder et al., 2015), but not in others (Song et al., 2016). Small molecules targeting DNA-dependent protein kinase (DNA-PKcs), another key element of NHEJ, have also been successfully to enhance HDR outcomes (Robert et al., 2015). Likewise, shRNA-mediated silencing of KU70, KU80, or DNA ligase IV (all involved in NHEJ) substantially suppresses NHEJ-mediated indel formation and increases HDR-mediated genome editing when administered in combination with Cas9, a sgRNA, and a donor DNA template (Chu et al., 2015).
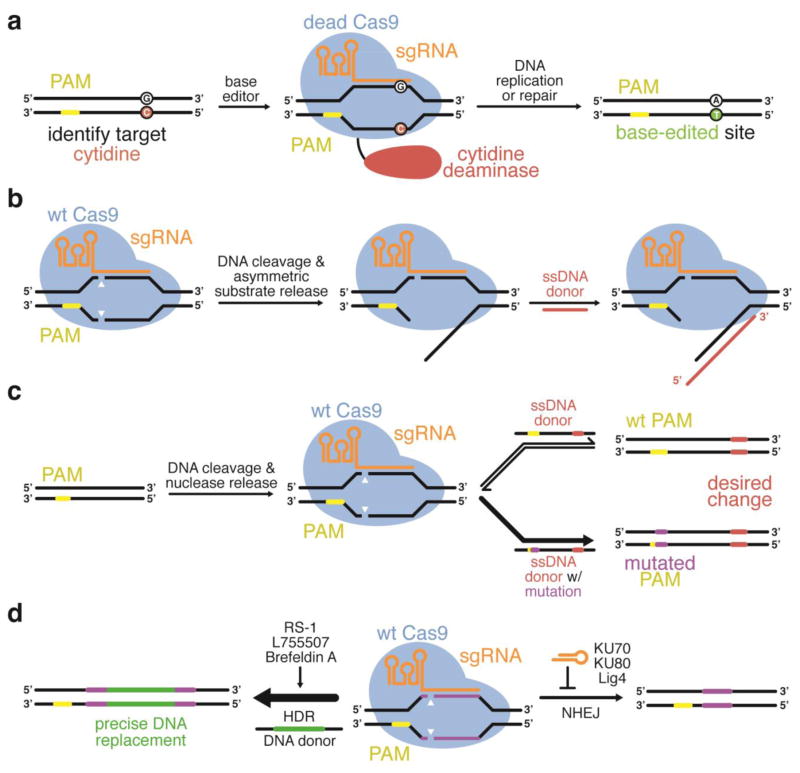
Figure 3.
Approaches that improve the product selectivity of genome editing agents. Wild-type Cas9 will induce undesired indels when the desired product is a precise DNA modification. (a) Base editing is capable of editing G:C base pairs to A:T base pairs with high conversions and very low indel rates (Nishida et al., 2016; Komor et al., 2016). (b) The ssDNA donor used during HDR can be designed such that it anneals with the DNA strand that is initially released by Cas9 following DNA cleavage to enhance HDR efficiency (Richardson et al., 2016). (c) In some cases, HDR strategies can also be designed to install a silent mutation into the PAM in order to prevent re-cutting by Cas9 following HDR (Paquet et al., 2016). (d) Small molecule inhibitors of NHEJ (Srivastava et al., 2012; Robert et al., 2015; Vartak and Raghavan, 2015; Chu et al., 2015; Maruyama et al., 2015), enhancers of HDR (Yu et al., 2015; Pinder et al., 2015; Song et al., 2016), or cell-cycle synchronizers (Lin et al., 2014b) can be used to increase the ratio of HDR:NHEJ genome editing products.
As an alternative strategy to suppressing NHEJ, a small molecule activator of Rad51, a protein involved in HDR, can augment HDR-mediated genome editing when added to cells treated with Cas9, a sgRNA, and a donor DNA template (Figure 3d) (Pinder et al., 2015; Song et al., 2016). A high-throughput screen designed to identify potential small molecules capable of enhancing HDR-mediated genome editing has been used to discover additional compounds that can be administered to cells to increase precision genome editing (Yu et al., 2015). HDR is known to be dependent on many variables, such as the cell- and tissue-type, the location of the target DNA within the chromosome, and the cell cycle (Biswas et al., 1992; Saleh-Gohari and Helleday, 2004; Heyer et al., 2010; Miyaoka et al., 2016). Chemically synchronizing cells to arrest them in G-phase by blocking M-phase before delivering Cas9 RNP and donor DNA template will increase HDR rates (Lin et al., 2014b).
The use of exogenous molecules to manipulate components of the NHEJ or HDR pathways therefore can be used to enhance HDR outcomes (Figure 3d), but such treatments are typically very perturbative to cells, are cell-type specific, and impair the cell’s ability to perform native DNA repair functions. These strategies therefore may have limited relevance in a therapeutic context or in research settings in which preserving native cell states is a priority.
Judicious design of the donor DNA template can also enhance HDR-mediated precision genome editing efficiency. The observation that Cas9 dissociates from its cleaved DNA substrate asymmetrically prompted a study on the effects of donor template geometry on HDR rates. Researchers discovered that if the ssDNA template is designed such that it can anneal to the DNA strand that is released by Cas9 first, HDR-mediated genome editing rates can be improved (Figure 3b) (Richardson et al., 2016). Frequently, the high levels of indels are exacerbated by the ability of an HDR-mediated product to remain a substrate for subsequent cleavage by Cas9 (Figure 3c). In some cases, this reprocessing of desired HDR product can be avoided by installing silent (or acceptable) mutations into the donor template such that the HDR product contains mutations in the PAM or the PAM-proximal region of the protospacer, thereby blocking Cas9 from cutting the product (Figure 3c) (Paquet et al., 2016).
“Base editing” is a new strategy to introduce point mutations in an RNA-programmed manner that does not rely on HDR or double-stranded DNA breaks (Komor et al., 2016). The fusion to dCas9 of a cytidine deaminase enzyme that operates on single-stranded DNA (but not duplex DNA) allows C to U conversion within a small (~3–5-base) window of the protospacer, exploiting the presence of a short segment of accessible single-stranded DNA in the “R-loop” ternary complex (Jiang et al., 2016) between Cas9, the guide RNA, and the target DNA. To make base editing efficient and permanent in mammalian cells, the dCas9–cytidine deaminase fusion protein was further engineered to inhibit base excision repair at the site of the edit, and to induce cellular mismatch repair to replace the original C:G base pair with a T:A base pair (Figure 3a) (Komor et al., 2016). Base editing efficiencies are typically much higher than the efficiency of HDR-mediated point mutation. In addition, because base editing avoids making double-stranded DNA breaks, indel formation is minimized (Nishida et al., 2016; Komor et al., 2016). Current limitations of this new strategy include off-target editing that parallels Cas9 off-target DNA cleavage, the requirement of an NGG PAM a specific distance from the target C, and the inability to distinguish among multiple Cs present within the editing window. Advances described above that expand the targeting scope and improve the DNA specificity of Cas9 may offer solutions to some of these limitations.
Epigenome Editing
In addition to changing the DNA sequence of a given genome, researchers have also begun to edit the epigenome in order to alter the regulation of a target gene, rather than its sequence identity (Thakore et al., 2016). One technique to increase expression of a specific gene is to tether the dCas9:sgRNA complex to a transcriptional activator and program it to bind near the transcriptional start site of a gene of interest. For example, the transcriptional activation domain VP64, which consists of four tandem copies of the Herpes Simplex Viral Protein 16 (VP16), can be fused directly to the C-terminus of dCas9 and used to increase the expression of a wide variety of different genes (Figure 4a) (Mali et al., 2013a; Maeder et al., 2013; Perez-Pinera et al., 2013). This fusion has been shown to be target sequence-specific and capable of opening chromatin (Polstein et al., 2015). A light-activated version of this system has been developed by fusing dCas9 and VP64 or p65, the transactivation domain of NF-KB, to each member of a light-activated heterodimerizing pair of proteins (Figure 4a) (Nihongaki et al., 2015b; Polstein and Gersbach, 2015). In addition, the small molecule-dimerized split Cas9 system described above has been adapted to generate a small molecule-activated dCas9-VP64 activator (Figure 4a) (Zetsche et al., 2015b). These systems allow spatial and temporal dynamic control of gene activation.
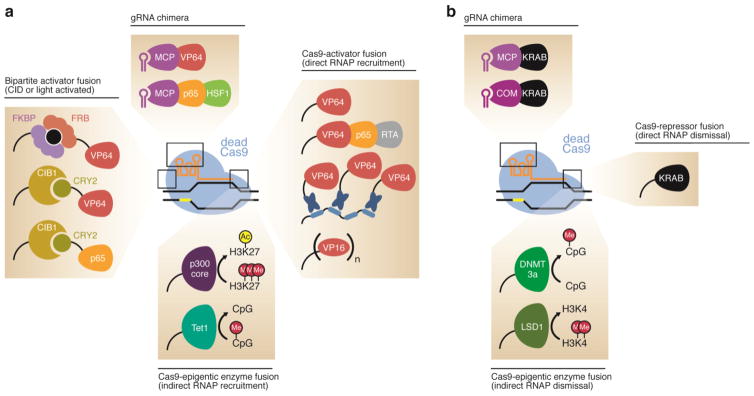
Figure 4.
CRISPR-based epigenome editing. (a) RNA-programmed gene activators can be assembled through the direct fusion of dCas9 with the transcriptional activators VP64 (Mali et al., 2013a; Maeder et al., 2013; Perez-Pinera et al., 2013; Tanenbaum et al., 2014) and VPR (Chavez et al., 2015), the histone acetyltransferase enzyme p300 (Hilton et al., 2015), or the DNA demethylase Tet1 (Xu et al., 2016; Morita et al., 2016; Liu et al., 2016). Alternately, the transcriptional activators VP64 and p65-HSF1 can be attached to the sgRNA (Zalatan et al., 2015; Konermann et al., 2015). Light-activated and small molecule-activated variants can also be used (Nihongaki et al., 2015b; Zetsche et al., 2015b; Polstein and Gersbach, 2015). (b) RNA-programmed gene repressors can be assembled by attaching the transcriptional repressor domain KRAB to dCas9 by either a direct fusion between the two proteins (Gilbert et al., 2013; Lawhorn et al., 2014; Thakore et al., 2015) or via the sgRNA. Alternatively, dCas9 can be fused to the DNA methyltransferase enzyme DNMT3a (McDonald et al., 2016; Liu et al., 2016; Vojta et al., 2016) or the histone demethylase LSD1 (Kearns et al., 2015) to result in a transcriptional repressor.
A variety of second-generation dCas9-activator fusions have been subsequently engineered that incorporate varying copies of VP16 (Cheng et al., 2013; Chakraborty et al., 2014), the tripartite activator VPR (VP64-p65-Rta, where Rta is the transcriptional activation domain of the Epstein-Barr virus) (Chavez et al., 2015), or the repeating peptide array SunTag that subsequently recruits multiple copies of an antibody-VP64 fusion (Tanenbaum et al., 2014) (Figure 4a). These various constructs offer strong gene activation in a variety of mammalian cell types (Chavez et al., 2016). Alternately, transcriptional activators can be attached to the sgRNA through an RNA hairpin that binds with very high affinity to the MS2 bacteriophage coat protein (Peabody, 1993). Coexpression of wild-type dCas9 or dCas9-VP64, the sgRNA-hairpin construct, and a MS2-VP64 (Zalatan et al., 2015; Konermann et al., 2015) or MS2-p65-HSF1 (where HSF1 is the activation domain from the human heat-shock transcription factor 1) (Konermann et al., 2015) fusion protein results in assembly of an RNA-guided transcriptional activator (Figure 4a). The combination of dCas9-VP64 and MS2-p65-HSF1, termed the synergistic activation mediator (SAM), exhibits particularly robust transcriptional activation (Konermann et al., 2015).
Conversely, dCas9 can be used as a transcriptional repressor on its own, simply exploiting its high affinity for target DNA and ability to block components of the transcriptional machinery (Qi et al., 2013), or as a fusion to the Kruppel-associated box (KRAB) transcriptional repressor (Figure 4b) (Gilbert et al., 2013; Lawhorn et al., 2014; Thakore et al., 2015). Together, these techniques of activating (CRISPRa) or repressing (CRISPRi) genes at will can be combined to allow researchers to reversibly modulate gene expression with a dynamic range of several orders of magnitude (Gilbert et al., 2014). A public tool is available to facilitate the design of appropriate sgRNAs to enable efficient CRISPRi and CRISPRa (Liu et al., 2015a). Researchers have also used genome-wide sgRNA libraries in high-throughput CRISPRi and CRISPRa screens to identify genes that result in phenotypes of interest when up- or down-regulated (Gilbert et al., 2014). Orthogonal RNA-protein binding modules have been incorporated into dCas9 transcriptional regulation complexes that allow for simultaneous gene activation and repression (Zalatan et al., 2015).
The packaging of DNA into chromatin is key feature of eukaryotic DNA, and researchers have begun to develop tools that allow chromatin manipulation in a sequence-programmed manner (Keung et al., 2015). The fusion of the DNA methyltransferase enzyme DNMT3A to dCas9 results in an epigenetic modifier that methylates CpG islands within ~100 bp of the sgRNA-programmed genomic loci (Figure 4b) (McDonald et al., 2016; Liu et al., 2016; Vojta et al., 2016). Tethering the catalytic domain of the DNA demethylase Tet1 to dCas9 has been used for targeted DNA demethylation (Figure 4a). These fusions can induce over 90% demethylation of CpG islands within a 200-bp range of the target site (Xu et al., 2016; Morita et al., 2016; Liu et al., 2016). Researchers have also developed a histone-modifying CRISPR-based tool in which the catalytic domain of human acetyltransferase p300 was fused to the C-terminus of dCas9. This fusion catalyzes histone H3 lysine 27 (H3K27) acetylation at loci up to thousands of base pairs from the sgRNA-specified locus and results in transcriptional activation of genes (Figure 4a) (Hilton et al., 2015). Alternately, the histone demethylase LSD1 has been fused to dCas9, allowing for demethylation of dimethylated histone H3 lysine 4 (H3K4me2) at sites >350 bp from the sgRNA (Figure 4b) (Kearns et al., 2015). The much larger activity window of the methylation tools, unlike the small window of base editing, arises from the use of domains that operate on double-stranded DNA in the reported epigenome editing agents, in contrast with the use of single-stranded DNA-specific enzymes in base editors.
Delivery of genome-editing and epigenome-editing agents
Although their substrates are intracellular, the genome-editing and epigenome-editing agents described above are all macromolecules and therefore do not spontaneously enter cells. The delivery of genome-editing agents into cells has therefore been the subject of intense research over the past several decades and remains a significant barrier to some applications of genome editing (Bartus et al., 1998; Gaj et al., 2013). For many research applications, the transfection of plasmid DNA expressing genome editing proteins and guide RNAs is sufficient. In other cases including in vivo therapeutic applications, however, DNA transfection is not possible, and alternative methods to deliver genome editing agents are needed.
A number of effective ex vivo methods have been used to deliver proteins or their encoding genes into cultured mammalian cells. These methods include electroporation or nucleofection, lipid-based transfection, viruses, cationic peptides, and other approaches (Luo and Saltzman, 2000; Maasho et al., 2004; Zeitelhofer et al., 2007; Cockrell and Kafri, 2007; Yin et al., 2014a). For some cell types including many cancer cell lines and certain blood cells, ex vivo delivery methods when applied to genome-editing proteins such as Cas9 can be very effective, resulting in the exposure of the vast majority of treated cells to the genome-editing agent (Heckl et al., 2014). For other cell types of interest, including hematopoietic stem cells and some primary cells (i.e, cells taken directly from tissue, rather than replicated in culture), even ex vivo delivery using a wide variety of methods has proven challenging (Amsellem et al., 2003; Lombardo et al., 2007).
Viral delivery of genome editing agents has been explored using lentivirus, adenovirus, and adeno-associated virus (AAV) (Gori et al., 2015) (Figure 5a). Lentiviruses are able to infect non-dividing cells and have been used in vivo to efficiently transduce a variety of specific target organs (Cockrell and Kafri, 2007). Furthermore, the packaging limit of lentivirus is ~8.5 kb (although inserts larger than ~3 kb are packaged less efficiently), sufficient to package most Cas9 genes, guide RNA expression constructs, and required promoter and regulatory sequences (Kumar et al., 2001; Yacoub et al., 2007). Lentiviruses have been successfully used to deliver Cas9 and sgRNA genes into mice to characterize the contributions of a panel of tumor suppressor genes to the progression of lung cancer (Sanchez-Rivera et al., 2014).
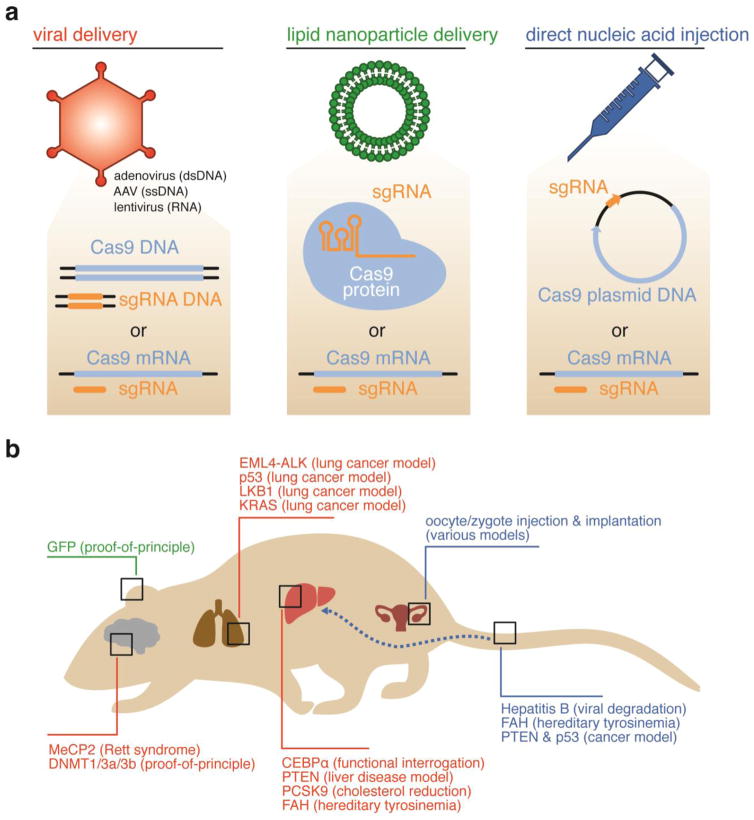
Figure 5.
Strategies for in vivo delivery of CRISPR-based genome editing agents. (a) Viral (orange)-, lipid nanoparticle (green)-, and direct nucleic acid injection (blue)-mediated delivery of CRISPR-based genome editing agents have all been successfully used to achieve in vivo genome editing. (b) These methods have been used to deliver genome editing agents to a variety of mammalian organs shown. The genes that were modified within each organ are shown in a color corresponding to the delivery method used, matching the colors in (a).
Adenoviruses are also capable of infecting both replicating and non-replicating cells, but do not integrate their DNA into the host cell genome, and can elicit a strong immune response in animals (Wang et al., 2004). Adenovirus-mediated delivery of Cas9 has been used to achieve in vivo genome editing in mouse lungs (Maddalo et al., 2014) and livers (Cheng et al., 2014; Ding et al., 2014; Wang et al., 2015a).
Finally, AAV variants engineered for gene therapy can infect both dividing and non-dividing cells, do not integrate its DNA into the host genome, and do not elicit a significant immune response in the host (Wang et al., 2004). A variety of serotypes of AAV are known, offering delivery into different tissue types. However, AAV has a packaging limit of ~4.5 kb of foreign DNA (Wu et al., 2010). Thus, packaging into AAV genes encoding SpCas9 (4.2 kb), a sgRNA, a donor DNA template, and associated promoters and regulatory sequences is generally not possible. The gene encoding SaCas9 (3.2 kb) is significantly smaller than that encoding SpCas9, and can be packaged along with an sgRNA and associated promoters into a single AAV vector (Ran et al., 2015). Alternatively, genes encoding SpCas9 and its sgRNA have been packaged into separate AAV vectors for in vivo genome editing in mouse brains (Swiech et al., 2015) and livers (Yang et al., 2016). An additional option is to use a split-intein Cas9 system and package each half into separate AAV vectors. Upon coinfection, the Cas9-intein halves associate and undergo protein splicing to yield a complete, covalently intact Cas9 protein. This system has successfully mediated genome editing in cultured human and mouse cells (Truong et al., 2015).
To circumvent the challenges associated with delivery of a Cas9-encoding gene, researchers have developed a mouse that expresses Cas9. A Cre recombinase-inducible Cas9 transgene was inserted into the Rosa26 locus of the mouse. The resulting mouse can be crossed with mice expressing Cre recombinase from a tissue-specific promoter to generate mice that express Cas9 in specific tissues. An sgRNA and donor DNA can then be delivered using AAV or lentivirus to initiate genome editing (Platt et al., 2014).
Hydrodynamic injection (HDI) of plasmid DNA is a convenient and efficient non-viral delivery option that is particularly well-suited for DNA delivery into hepatocytes in rodents (Suda and Liu, 2007) (Figure 5a). Researchers have shown that DNA plasmids encoding Cas9 and sgRNA can be delivered to the livers of mice infected with hepatitis B virus (HBV) via a hydrodynamic tail vein injection. The resulting Cas9:sgRNA complexes cleave HBV viral DNA, resulting in a gradual decrease in HBV expression in the animals (Lin et al., 2014a; Zhen et al., 2015; Ramanan et al., 2015). This technique has also been used to generate mouse liver cancer models. In this study, a plasmid encoding Cas9 and sgRNAs was delivered via HDI to disrupt tumor suppressor genes. By co-injecting with a donor template for HDR, precise mutations can be introduced to genes of interest as well (Xue et al., 2014).
Another delivery option that can result in germline genome editing in animals is the direct injection of Cas9 mRNA and purified sgRNA into oocytes or zygotes (Figure 5b). This technique has been used to knock-out specific genes in mice (Wang et al., 2013; Li et al., 2013a; Yang et al., 2013a), rats (Li et al., 2013b), zebrafish (Hruscha et al., 2013; Hwang et al., 2013a; Chang et al., 2013; Hsu et al., 2013), flies (Bassett et al., 2013), frogs (Nakayama et al., 2013), pigs (Hai et al., 2014), and monkeys (Niu et al., 2014). By co-injecting with a donor DNA, precise HDR-mediated genome editing has been achieved in mice (Wang et al., 2013; Yang et al., 2013a; Wu et al., 2013), nematodes (Lo et al., 2013), and zebrafish (Hruscha et al., 2013; Chang et al., 2013; Auer et al., 2014).
A DNA- and mRNA-free in vivo delivery method directly delivers purified RNPs using cationic lipids (Zuris et al., 2015) (Figure 5a). Due to the polyanionic charge of the sgRNA and its tight association with Cas9 protein, commercially available cationic lipid nucleic acid transfection reagents can efficiently deliver Cas9:sgRNA complexes into mammalian cells both in cell culture and in vivo. This strategy has been successfully used to perform genome editing in mouse inner ear hair cells and in neurons (Zuris et al., 2015; Wang et al., 2016). An additional advantage of the direct Cas9:sgRNA protein:RNA complex delivery over mRNA or DNA delivery is the more transient nature of protein delivery, which results in substantially higher DNA specificity and less off-target editing, for the reasons discussed above (Kim et al., 2014; Zuris et al., 2015; Liu et al., 2015b).
Applications of CRISPR-based genome editing
CRISPR components have been used for a variety of creative applications. Here we summarize some recent uses of CRISPR-based genome editing in eukaryotic cells and organisms for basic research, biotechnology, and therapeutics development. Although a major and important application of Cas9 is the generation of animal and cell models of diseases, including target gene knockouts, these have been reviewed recently and are not covered in this review (Sander and Joung, 2014; Mou et al., 2015).
Genome editing in human primary cells
Pluripotent stem cells are undifferentiated primary cells that are able to differentiate into virtually any cell type of the human body. Induced pluripotent stem cells (iPSCs) are pluripotent stem cells that can be directly generated from adult somatic cells, and have proven useful for regenerative medicine research (Takahashi and Yamanaka, 2006; Yu et al., 2007). Genome editing in iPSCs allows the precise study of human genetic variants in a wide variety of tissues in cell culture. The use of Cas9 for mammalian genome editing advanced this capability by allowing scientists to perform genetic manipulations in iPSCs at efficiencies that are difficult to attain with TALENs or ZFNs (Ding et al., 2013; Yang et al., 2013b; Byrne et al., 2014; Hockemeyer and Jaenisch, 2016).
Cells isolated from an individual can be genetically modified in a specific way using Cas9, then differentiated alongside identical but unmodified cells. These otherwise isogenic cells allow researchers to directly determine the impact of a given genetic variation on a disease phenotype, or to simply study gene function. Cas9-mediated, iPSC-derived knockout cell lines for a variety of different genes have been reported for use in loss-of-function studies (Yang et al., 2013b; González et al., 2014; Smith et al., 2015; Liao et al., 2015; Chen et al., 2015b; Liang et al., 2015; Wang et al., 2015b). This technique can also be combined with CRISPRi to specifically, rapidly, and reversibly down-regulate specific genes in iPSCs and iPSC-derived cell types (Mandegar et al., 2016). Similarly, specific mutations have been introduced into iPSCs and iPSC-derived cells using Cas9 and HDR-mediated genome editing for the study of genetic diseases (Yang et al., 2013b; Hou et al., 2013; Rong et al., 2014; Smith et al., 2015).
A central goal of regenerative medicine is to replace unhealthy or diseased cells with heathy ones. One approach to this goal is cell therapy, in which primary cells are genetically manipulated then implanted into patients in order to cure or treat genetic diseases (Mironov et al., 2004). Several studies have used Cas9 to correct genetic mutations in patient-derived primary cells, including Duchene muscular dystrophy (DMD) (Li et al., 2015b; Ousterout et al., 2015), Fanconi anemia (Osborn et al., 2015), hemophilia (Park et al., 2015), cystic fibrosis (Schwank et al., 2013), and beta thalassemia (Xie et al., 2014). Additionally, primary immune cells have been made resistant to HIV infection by Cas9-mediated knockout of CCR5 or CXCR4, receptors for HIV entry (Wang et al., 2014b; Li et al., 2015a; Hou et al., 2015; Schumann et al., 2015).
Together, these studies highlight how CRISPR-based genome editing technologies have accelerated biological studies in primary cells. The development of more precise, specific, and capable genome editing technologies will likely further augment our understanding of diseased cells as these new technologies are applied to primary cell editing.
CRISPR-based treatment of animal models of human genetic disease
Cas9-mediated genome editing in vivo has been used to correct disease-associated alleles in animal models of genetic diseases. The zygotes of mice heterozygous for a dominant-negative cataract-causing mutation in the CRYGC gene were injected with Cas9 mRNA and an sgRNA targeting only the mutant allele. HDR-mediated correction resulted in cataract-free progeny (Wu et al., 2013). This technique was also applied to mdx mice, a model of DMD harboring a mutation in the gene encoding dystrophin. Cas9, sgRNA, and a donor template were injected into mouse zygotes, resulting in genetically mosaic progeny with 2 to 100% gene correction and varying degrees of phenotypic rescue (Long et al., 2014). Disease correction, both genotypically and phenotypically, in post-natal mdx mice has also been reported following AAV-mediated delivery of SaCas9 or SpCas9 and sgRNA to skeletal and cardiac muscle cells to delete the mutated exon from the dystrophin gene (Long et al., 2016; Nelson et al., 2016; Tabebordbar et al., 2016). Finally, a mouse model of hereditary tyrosinemia type I (HT1) was corrected in vivo using Cas9. These mice have a homozygous G to A point mutation in the fumarylacetoacetate hydrolase (FAH) gene, which results in severe liver damage. Plasmid encoding Cas9 and sgRNA and a ssDNA oligo were delivered via HDI into adult mice. Wild-type Fah protein was detected in ~1/250 liver cells, and significantly less liver damage was observed as compared to untreated controls (Yin et al., 2014b). Correction of this point mutation was also achieved using the concomitant lipid-mediated delivery of Cas9 mRNA and AAV-mediated delivery of sgRNA and a repair template. This combined viral and non-viral delivery method yielded point mutation correction in > 6% of hepatocytes (Yin et al., 2016).
These studies collectively demonstrate significant progress towards developing treatments for genetic diseases. Nearly all genetic diseases, including DMD, currently have no cure and in many cases those affected suffer from low quality of life and a shortened life expectancy. These significant advances suggest potential treatments and even potential cures of human genetic diseases.
High-throughput genetic screens
The ease with which CRISPR-based tools can be reprogrammed simply by constructing a new guide RNA has facilitated genome-wide screening. CRISPR-based knockout screens are reported to be more efficient and specific than RNAi, enabling researchers to obtain more reliable and robust results (Sanjana, 2016). In these experiments, sgRNA libraries and Cas9 are delivered into cells. Researchers then screen the treated cells for a phenotype of interest (Sanjana et al., 2014; Shalem et al., 2015). CRISPR-mediated knockout screens have been used to identify genes involved in cancer progression (Shalem et al., 2014; Chen et al., 2015a; Shi et al., 2015) drug resistance (Shalem et al., 2014; Wang et al., 2014a), the immune response (Parnas et al., 2015), vulnerability to bacterial toxins (Zhou et al., 2014), and other biomedically relevant phenotypes.
Alternatively, Cas9-mediated saturation mutagenesis, in which an sgRNA library is targeted to thoroughly mutate a targeted region of the genome, can be used in a high-throughput manner to obtain high-resolution information on genetic loci. For example, a sgRNA library targeting the BCL11A enhancer was used to dissect the importance of each portion of the enhancer on HbF levels. Similarly, a single sgRNA was combined with a donor DNA library to replace a 6-bp portion of the BRCA1 gene with all possible hexamers, and to replace exon 18 of BRCA1 with all possible SNP variations to dissect the mutations’ effects on transcript processing (Kiani et al., 2015).
Together, these studies demonstrate how Cas9 has enhanced the ability to screen the modification of many loci within the human genome in a single experiment. These types of studies provide researchers with powerful tools to dissect complex cellular signaling pathways, determine gene function, identify targets for therapeutic intervention, and predict drug side effects.
Gene Drives
Gene drives are a particularly powerful application of CRISPR technology. Gene drives, genetic elements that insert themselves into target sites lacking that element, convert heterozygous alleles to homozygous alleles within an organism. In organisms that support sexual reproduction, gene drives enable non-Mendelian inheritance of alleles that can spread throughout a population. This spread can be rapid for species with short reproductive generation times. While gene drives can be implemented in other ways, Cas9-based gene drives are very efficient. CRISPR gene drives were first demonstrated by inserting into the Drosophila genome a construct consisting of Cas9 and an sgRNA flanked by two “homology arms” that match the genomic sequence surrounding the sgRNA-programmed target site. Cas9:sgRNA complex expression results in cleavage of the wildtype allele, and incorporation of the Cas9:sgRNA cassette via HDR to produce a homozygous mutant organism (Gantz and Bier, 2015). In mosquitos, researchers developed a Cas9-based gene drive that inserts a gene inducing a parasite-resistance phenotype (Gantz et al., 2015; Hammond et al., 2016). Gene drives have the potential to allow the facile generation of animal models with recessive mutations, as well as the genome editing of whole populations of rapidly reproducing organisms—a remarkably powerful capability that should be considered only with great thoughtfulness and caution (Oye et al., 2014; Esvelt et al., 2014).
Conclusion
The discovery and characterization of CRISPR systems have transformed genome editing and the life sciences. The development of new CRISPR technologies that extend the generality, DNA specificity, product selectivity, and even the fundamental capabilities of natural CRISPR systems has driven and accelerated this transformation. These technologies have armed researchers with powerful new tools to study living systems and human disease. Incredibly, they have also made it possible to imagine a near-term future in which the treatment of genetic diseases is within our reach. As these technologies grow in scope and capability, ethical and regulatory guidelines must also be thoughtfully developed (Baltimore et al., 2015) to ensure a balance between realizing the enormous potential of these tools to benefit mankind and minimizing the risk of their misuse.
Acknowledgments
We are grateful to Professor Jennifer Doudna and her coworkers Mitchell O’Connell and Alexandra Seletsky, Professor Keith Joung, Professor Gerald Joyce, and Professor Feng Zhang for their helpful comments. This work was supported by U.S. National Institutes of Health (NIH) R01 EB022376 (formerly R01 GM065400), NIH R35GM118062, F-Prime Biomedical Research Initiative (A28161), and the Howard Hughes Medical Institute. A.C.K. is a Ruth L. Kirchstein National Research Service Awards Postdoctoral Fellow (F32 GM 112366-2). A.H.B. was supported by the Harvard Chemical Biology Program and a National Science Foundation Graduate Research Fellowship. The authors declare competing financial interests: A.C.K. and D.R.L. have filed patent applications on base editing. D.R.L. is a consultant and co-founder of Editas Medicine, a company that seeks to develop genome-editing therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amsellem S, Pflumio F, Bardinet D, Izac B, Charneau P, Romeo PH, Dubart-Kupperschmitt A, Fichelson S. Ex vivo expansion of human hematopoietic stem cells by direct delivery of the HOXB4 homeoprotein. Nat Med. 2003;9:1423–1427. doi: 10.1038/nm953. [DOI] [PubMed] [Google Scholar]
- Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Research. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, Corn JE, Daley GQ, Doudna JA, Fenner M, et al. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348:36–38. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science. 2007;315:1709. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bartus RT, Tracy MA, Emerich DF, Zale SE. Sustained Delivery of Proteins for Novel Therapeutic Products. Science. 1998;281:1161–1162. doi: 10.1126/science.281.5380.1161. [DOI] [PubMed] [Google Scholar]
- Bassett Andrew R, Tibbit C, Ponting Chris P, Liu JL. Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cell Reports. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing Gene Targeting with Designed Zinc Finger Nucleases. Science. 2003;300:764–764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted Chromosomal Cleavage and Mutagenesis in Drosophila Using Zinc-Finger Nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Vagner V, Ehrlich SD. Efficiency of homologous intermolecular recombination at different locations on the Bacillus subtilis chromosome. Journal of Bacteriology. 1992;174:5593–5596. doi: 10.1128/jb.174.17.5593-5596.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science. 2009;326:1509. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Byrne SM, Mali P, Church GM. Chapter Six - Genome Editing in Human Stem Cells. In: Jennifer AD, Erik JS, editors. Methods in Enzymology. Academic Press; 2014. pp. 119–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- Carroll D. Zinc-finger Nucleases as Gene Therapy Agents. Gene therapy. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Miura H, Malina A, Robert F, Ethier S, Schmeing TM, Dostie J, Pelletier J. Protospacer Adjacent Motif (PAM)-Distal Sequences Engage CRISPR Cas9 DNA Target Cleavage. PLoS ONE. 2014;9:e109213. doi: 10.1371/journal.pone.0109213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Ji H, Kabadi Ami M, Gersbach Charles A, Christoforou N, Leong Kam W. A CRISPR/Cas9-Based System for Reprogramming Cell Lineage Specification. Stem Cell Reports. 2014;3:940–947. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer PRE, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Meth. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, et al. Comparison of Cas9 activators in multiple species. Nat Meth. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana Neville E, Zheng K, Shalem O, Lee K, Shi X, Scott David A, Song J, Pan Jen Q, Weissleder R, et al. Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis. Cell. 2015a;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao J, Xiong M, Petersen Andrew J, Dong Y, Tao Y, Huang, Cindy T-L, Du Z, Zhang SC. Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell Stem Cell. 2015b;17:233–244. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Peng J, Yan Y, Cao P, Wang J, Qiu C, Tang L, Liu D, Tang L, Jin J, et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Letters. 2014;588:3954–3958. doi: 10.1016/j.febslet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotech. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Molecular and Cellular Biology. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotech. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- Cockrell AS, Kafri T. Gene delivery by lentivirus vectors. Molecular Biotechnology. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Meth. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. Small Molecule-Triggered Cas9 Protein with Improved Genome-Editing Specificity. Nature chemical biology. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage Response to CRISPR-Encoded Resistance in Streptococcus thermophilus. Journal of Bacteriology. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell stem cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, Musunuru K. Permanent Alteration of PCSK9 With In Vivo CRISPR-Cas9 Genome Editing. Circulation Research. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346 doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Dow LE, Fisher J, O’Rourke KP, Muley A, Kastenhuber ER, Livshits G, Tschaharganeh DF, Socci ND, Lowe SW. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotech. 2015;33:390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Meth. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 2014;3:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biology. 2015;16:1–10. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotech. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotech. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz VM, Bier E. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science. 2015;348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proceedings of the National Academy of Sciences. 2015;112:E6736–E6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Siksnys V. RNA-dependent DNA endonuclease Cas9 of the CRISPR system: Holy Grail of genome editing? Trends in Microbiology. 2013;21:562–567. doi: 10.1016/j.tim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Gilbert Luke A, Horlbeck Max A, Adamson B, Villalta Jacqueline E, Chen Y, Whitehead Evan H, Guimaraes C, Panning B, Ploegh Hidde L, Bassik Michael C, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert Luke A, Larson Matthew H, Morsut L, Liu Z, Brar Gloria A, Torres Sandra E, Stern-Ginossar N, Brandman O, Whitehead Evan H, Doudna Jennifer A, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González F, Zhu Z, Shi ZD, Lelli K, Verma N, Li Qing V, Huangfu D. An iCRISPR Platform for Rapid, Multiplexable, and Inducible Genome Editing in Human Pluripotent Stem Cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori JL, Hsu PD, Maeder ML, Shen S, Welstead GG, Bumcrot D. Delivery and Specificity of CRISPR-Cas9 Genome Editing Technologies for Human Gene Therapy. Hum Gene Ther. 2015;26:443–451. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Research. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Pattanayak V, Reyon D, Tsai SQ, Sander JD, Joung JK, Liu DR. Broad Specificity Profiling of TALENs Results in Engineered Nucleases With Improved DNA Cleavage Specificity. Nature methods. 2014a;11:429–435. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotech. 2014b;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Research. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A Guild of 45 CRISPR-Associated (Cas) Protein Families and Multiple CRISPR/Cas Subtypes Exist in Prokaryotic Genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Research. 2014;24:372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotech. 2016;34:78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotech. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A. Optical Control of CRISPR/Cas9 Gene Editing. Journal of the American Chemical Society. 2015;137:5642–5645. doi: 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual review of genetics. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotech. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H, Gootenberg Jonathan S, Horii T, Abudayyeh Omar O, Kimura M, Hsu Patrick D, Nakane T, Ishitani R, Hatada I, Zhang F, et al. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Jaenisch R. Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell. 2016;18:573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P, Chen S, Wang S, Yu X, Chen Y, Jiang M, Zhuang K, Ho W, Hou W, Huang J, et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Scientific Reports. 2015;5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of the National Academy of Sciences. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- Hsu Patrick D, Lander Eric S, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotech. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JRJ. Heritable and Precise Zebrafish Genome Editing Using a CRISPR-Cas System. PLoS ONE. 2013a;8:e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JRJ, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotech. 2013b;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain PK, Ramanan V, Schepers AG, Dalvie NS, Panda A, Fleming HE, Bhatia SN. Development of Light-Activated CRISPR Using Guide RNAs with Photocleavable Protectors. Angewandte Chemie International Edition. 2016 doi: 10.1002/anie.201606123. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends in Genetics. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- Jeggo PA. DNA Breakage and Repair. In: Jeffery JCDTF, Hall C, Francesco G, editors. In Advances in Genetics. Academic Press; 1998. pp. 185–218. [DOI] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotech. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Meth. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keung AJ, Joung JK, Khalil AS, Collins JJ. Chromatin regulation at the frontier of synthetic biology. Nat Rev Genet. 2015;16:159–171. doi: 10.1038/nrg3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S, Chavez A, Tuttle M, Hall RN, Chari R, Ter-Ovanesyan D, Qian J, Pruitt BW, Beal J, Vora S, et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat Meth. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Meth. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon S-h, Kim J-S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotech. 2016 doi: 10.1038/nbt.3609. advance online publication. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016a;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotech. 2015a;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh JRJ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015b;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotech. 2016b doi: 10.1038/nbt.3620. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Keller B, Makalou N, Sutton RE. Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther. 2001;12:1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotech. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Lawhorn IEB, Ferreira JP, Wang CL. Evaluation of sgRNA Target Sites for CRISPR-Mediated Repression of TP53. PLoS ONE. 2014;9:e113232. doi: 10.1371/journal.pone.0113232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Guan X, Du T, Jin W, Wu B, Liu Y, Wang P, Hu B, Griffin GE, Shattock RJ, et al. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. Journal of General Virology. 2015a;96:2381–2393. doi: 10.1099/vir.0.000139. [DOI] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotech. 2013a;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- Li Hongmei L, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H, et al. Precise Correction of the Dystrophin Gene in Duchenne Muscular Dystrophy Patient Induced Pluripotent Stem Cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015b;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Research. 2011a;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Research. 2011b doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotech. 2013b;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. Journal of Biotechnology. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FL, Sperle K, Sternberg N. Recombination in mouse L cells between DNA introduced into cells and homologous chromosomal sequences. Proceedings of the National Academy of Sciences. 1985;82:1391–1395. doi: 10.1073/pnas.82.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids. 2014a;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014b;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wei Z, Dominguez A, Li Y, Wang X, Qi LS. CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics. 2015a;31:3676–3678. doi: 10.1093/bioinformatics/btv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gaj T, Yang Y, Wang N, Shui S, Kim S, Kanchiswamy CN, Kim JS, Barbas CF., III Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells. Nat Protocols. 2015b;10:1842–1859. doi: 10.1038/nprot.2015.117. [DOI] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young Richard A, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167:233–247. e217. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo TW, Pickle CS, Lin S, Ralston EJ, Gurling M, Schartner CM, Bian Q, Doudna JA, Meyer BJ. Precise and Heritable Genome Editing in Evolutionarily Diverse Nematodes Using TALENs and CRISPR/Cas9 to Engineer Insertions and Deletions. Genetics. 2013;195:331–348. doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotech. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9–mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotech. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- Maasho K, Marusina A, Reynolds NM, Coligan JE, Borrego F. Efficient gene transfer into the human natural killer cell line, NKL, using the Amaxa nucleofection system. Journal of Immunological Methods. 2004;284:133–140. doi: 10.1016/j.jim.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Meth. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotech. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandegar Mohammad A, Huebsch N, Frolov Ekaterina B, Shin E, Truong A, Olvera Michael P, Chan Amanda H, Miyaoka Y, Holmes K, Spencer CI, et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotech. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JI, Celik H, Rois LE, Fishberger G, Fowler T, Rees R, Kramer A, Martens A, Edwards JR, Challen GA. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biology Open. 2016 doi: 10.1242/bio.019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotech. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Mironov V, Visconti RP, Markwald RR. What is regenerative medicine? Emergence of applied stem cell and developmental biology. Expert Opinion on Biological Therapy. 2004;4:773–781. doi: 10.1517/14712598.4.6.773. [DOI] [PubMed] [Google Scholar]
- Miyaoka Y, Berman JR, Cooper SB, Mayerl SJ, Chan AH, Zhang B, Karlin-Neumann GA, Conklin BR. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Scientific Reports. 2016;6:23549. doi: 10.1038/srep23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, Sakai A, Nakashima H, Hata K, Nakashima K, et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotech. 2016 doi: 10.1038/nbt.3658. advance online publication. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A Simple Cipher Governs DNA Recognition by TAL Effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Mou H, Kennedy Z, Anderson DG, Yin H, Xue W. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Medicine. 2015;7:1–11. doi: 10.1186/s13073-015-0178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24:636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Rivera RMC, Madhavan S, Pan X, Ran FA, Yan WX, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotech. 2015a;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. CRISPR-Cas9-based Photoactivatable Transcription System. Chemistry & Biology. 2015b;22:169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016 doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- O’Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJ. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Research. 2015;43:3389–3404. doi: 10.1093/nar/gkv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes BL, Nadler DC, Flamholz A, Fellmann C, Staahl BT, Doudna JA, Savage DF. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat Biotech. 2016;34:646–651. doi: 10.1038/nbt.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Gabriel R, Webber BR, DeFeo AP, McElroy AN, Jarjour J, Starker CG, Wagner JE, Joung JK, Voytas DF, et al. Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum Gene Ther. 2015;26:114–126. doi: 10.1089/hum.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6 doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oye KA, Esvelt K, Appleton E, Catteruccia F, Church G, Kuiken T, Lightfoot SBY, McNamara J, Smidler A, Collins JP. Regulating gene drives. Science. 2014;345:626. doi: 10.1126/science.1254287. [DOI] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- Park CY, Kim Duk H, Son Jeong S, Sung Jin J, Lee J, Bae S, Kim JH, Kim DW, Kim JS. Functional Correction of Large Factor VIII Gene Chromosomal Inversions in Hemophilia A Patient-Derived iPSCs Using CRISPR-Cas9. Cell Stem Cell. 2015;17:213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure Thomas M, Herbst Rebecca H, Dixit A, Ye Chun J, Przybylski D, Platt Randall J, Tirosh I, Sanjana Neville E, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotech. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Meth. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS. The RNA binding site of bacteriophage MS2 coat protein. The EMBO Journal. 1993;12:595–600. doi: 10.1002/j.1460-2075.1993.tb05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Meth. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder J, Salsman J, Dellaire G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt Randall J, Chen S, Zhou Y, Yim Michael J, Swiech L, Kempton Hannah R, Dahlman James E, Parnas O, Eisenhaure Thomas M, Jovanovic M, et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Perez-Pinera P, Kocak DD, Vockley CM, Bledsoe P, Song L, Safi A, Crawford GE, Reddy TE, Gersbach CA. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Research. 2015;25:1158–1169. doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric Nucleases Stimulate Gene Targeting in Human Cells. Science. 2003;300:763–763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Qi Lei S, Larson Matthew H, Gilbert Luke A, Doudna Jennifer A, Weissman Jonathan S, Arkin Adam P, Lim Wendell A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Research. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V, Shlomai A, Cox DBT, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Scientific Reports. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu Patrick D, Lin CY, Gootenberg Jonathan S, Konermann S, Trevino AE, Scott David A, Inoue A, Matoba S, Zhang Y, et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013a;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protocols. 2013b;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotech. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- Robert F, Barbeau M, Éthier S, Dostie J, Pelletier J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Medicine. 2015;7:1–11. doi: 10.1186/s13073-015-0215-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Z, Zhu S, Xu Y, Fu X. Homologous recombination in human embryonic stem cells using CRISPR/Cas9 nickase and a long DNA donor template. Protein & Cell. 2014;5:258–260. doi: 10.1007/s13238-014-0032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Research. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Molecular and Cellular Biology. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N, Sugarman E, Haber JE. Genetic and Physical Analysis of Double-Strand Break Repair and Recombination in Saccharomyces Cerevisiae. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Research. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotech. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE. Genome-scale CRISPR pooled screens. Analytical Biochemistry. 2016 doi: 10.1016/j.ab.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Meth. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Research. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proceedings of the National Academy of Sciences. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G, Koo BK, Sasselli V, Dekkers Johanna F, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent Cornelis K, et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotech. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Abalde-Atristain L, He C, Brodsky BR, Braunstein EM, Chaudhari P, Jang YY, Cheng L, Ye Z. Efficient and Allele-Specific Genome Editing of Disease Loci in Human iPSCs. Mol Ther. 2015;23:570–577. doi: 10.1038/mt.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, et al. Whole-Genome Sequencing Analysis Reveals High Specificity of CRISPR/Cas9 and TALEN-Based Genome Editing in Human iPSCs. Cell stem cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nature Communications. 2016;7:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Nambiar M, Sharma S, Karki Subhas S, Goldsmith G, Hegde M, Kumar S, Pandey M, Singh Ram K, Ray P, et al. An Inhibitor of Nonhomologous End-Joining Abrogates Double-Strand Break Repair and Impedes Cancer Progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- Suda T, Liu D. Hydrodynamic Gene Delivery: Its Principles and Applications. Mol Ther. 2007;15:2063–2069. doi: 10.1038/sj.mt.6300314. [DOI] [PubMed] [Google Scholar]
- Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R, Jr, Stoddard BL, Seligman LM. Isolation and Characterization of New Homing Endonuclease Specificities at Individual Target Site Positions. Journal of Molecular Biology. 2004;342:31–41. doi: 10.1016/j.jmb.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotech. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanenbaum Marvin E, Gilbert Luke A, Qi Lei S, Weissman Jonathan S, Vale Ronald D. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Meth. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Meth. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Truong DJJ, Kühner K, Kühn R, Werfel S, Engelhardt S, Wurst W, Ortiz O. Development of an intein-mediated split–Cas9 system for gene therapy. Nucleic Acids Research. 2015;43:6450–6458. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotech. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotech. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Vartak SV, Raghavan SC. Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing. FEBS Journal. 2015;282:4289–4294. doi: 10.1111/febs.13416. [DOI] [PubMed] [Google Scholar]
- Veres A, Gosis Bridget S, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski Michael E, Musunuru K. Low Incidence of Off-Target Mutations in Individual CRISPR-Cas9 and TALEN Targeted Human Stem Cell Clones Detected by Whole-Genome Sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, Klasić M, Zoldoš V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Research. 2016 doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AY, Peng PD, Ehrhardt A, Storm TA, Kay MA. Comparison of adenoviral and adeno-associated viral vectors for pancreatic gene delivery in vivo. Human gene therapy. 2004;15:405–413. doi: 10.1089/104303404322959551. [DOI] [PubMed] [Google Scholar]
- Wang D, Mou H, Li S, Li Y, Hough S, Tran K, Li J, Yin H, Anderson DG, Sontheimer EJ, et al. Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Hum Gene Ther. 2015a;26:432–442. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila Chikdu S, Dawlaty Meelad M, Cheng Albert W, Zhang F, Jaenisch R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proceedings of the National Academy of Sciences. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin M, Pedrosa E, Hrabovsky A, Zhang Z, Guo W, Lachman HM, Zheng D. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Molecular Autism. 2015b;6:1–18. doi: 10.1186/s13229-015-0048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science. 2014a;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 Gene Disruption via Lentiviral Vectors Expressing Cas9 and Single Guided RNA Renders Cells Resistant to HIV-1 Infection. PLoS ONE. 2014b;9:e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, Doudna JA. Rational design of a split-Cas9 enzyme complex. Proceedings of the National Academy of Sciences. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotech. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. Correction of a Genetic Disease in Mouse via Use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Wu Z, Yang H, Colosi P. Effect of Genome Size on AAV Vector Packaging. Molecular Therapy. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyvekens N, Topkar VV, Khayter C, Joung JK, Tsai SQ. Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum Gene Ther. 2015;26:425–431. doi: 10.1089/hum.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, Kan YW. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Research. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, Ruan K, Wang F, Xu G-l, Hu R. A CRISPR-based approach for targeted DNA demethylation. Cell Discovery. 2016;2:16009. doi: 10.1038/celldisc.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoub Na, Romanowska M, Haritonova N, Foerster J. Optimized production and concentration of lentiviral vectors containing large inserts. The Journal of Gene Medicine. 2007;9:579–584. doi: 10.1002/jgm.1052. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila Chikdu S, Cheng Albert W, Shi L, Jaenisch R. OneStep Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013a;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Grishin D, Wang G, Aach J, Zhang C-Z, Chari R, Homsy J, Cai X, Zhao Y, Fan J-B, et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;5 doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Research. 2013b;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotech. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014a;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotech. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotech. 2014b;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, et al. Small Molecules Enhance CRISPR Genome Editing in Pluripotent Stem Cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zalatan Jesse G, Lee Michael E, Almeida R, Gilbert Luke A, Whitehead Evan H, La Russa M, Tsai Jordan C, Weissman Jonathan S, Dueber John E, Qi Lei S, et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitelhofer M, Vessey JP, Xie Y, Tubing F, Thomas S, Kiebler M, Dahm R. High-efficiency transfection of mammalian neurons via nucleofection. Nat Protocols. 2007;2:1692–1704. doi: 10.1038/nprot.2007.226. [DOI] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg Jonathan S, Abudayyeh Omar O, Slaymaker Ian M, Makarova Kira S, Essletzbichler P, Volz Sara E, Joung J, van der Oost J, Regev A, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015a;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotech. 2015b;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotech. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ge X, Yang F, Zhang L, Zheng J, Tan X, Jin ZB, Qu J, Gu F. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Scientific Reports. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rajan R, Seifert HS, Mondragón A, Sontheimer Erik J. DNase H Activity of Neisseria meningitidis Cas9. Molecular Cell. 2015;60:242–255. doi: 10.1016/j.molcel.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, Dong LH, Song HF, Gao X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotech. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]