Abstract
The mechanism underlying transcriptional coactivation by the co-repressor CtBP is not established. We previously found that CtBP co-occupies several actively transcribed endocrine genes with the transcription factor NeuroD1 to paradoxically increase transcription by recruiting KDM1A and CoREST. While the importance of the oligomeric form of CtBP for corepression is well established, the role of oligomerization in transcriptional coactivation has received little attention. Here, we examined the importance of the oligomeric state of CtBP for coactivation of NeuroD1-dependent transcription by expressing a CtBP dimerization mutant in cells depleted of endogenous CtBP. Dimerization mutants failed to increase transcription or to associate with KDM1A and CoREST, suggesting that oligomeric, but not monomeric CtBP is required to recruit other proteins needed to activate transcription.
Keywords: CtBP, NeuroD1
Introduction
C-terminal binding protein (CtBP) was originally identified as a cellular protein bound to the C-terminal region of the adenoviral oncoprotein E1A through a conserved PXDLS motif to reduce oncogenic function of E1A [1]. Subsequent studies showed that CtBP associated with a number of DNA binding proteins as part of a large multiprotein complex to function as a transcriptional corepressor. CtBP associated proteins include DNA binding proteins along with a number of histone modifying enzymes [2-4], suggesting that CtBP regulates transcription through changes in chromatin structure. CtBP exists in equilibrium between different forms as monomers, dimers and possibly higher ordered oligomeric forms, which impact its transcriptional function [5-8]. The oligomeric form appears to be necessary for transcriptional repression while the function of monomeric CtBP is not well characterized [2]. In Drosophila melanogaster, CtBP exhibits dual functions in the Wnt pathway, activating some Wnt target genes by its monomeric and oligomeric forms. Repression of other Wnt targets by CtBP is mediated by the oligomeric form [9,10].
CtBP has been occasionally linked to transcriptional activation in mammalian cells as well [9-16]. Some of these studies were done prior to the recognition that CtBP was part of a complex with multiple chromatin modifying proteins. As a result, the contribution of other CtBP associated proteins to transcriptional activation has received limited attention.
The basic helix loop helix transcription factor, NeuroD1 is important for differentiation of endocrine cells in the gastrointestinal tract. We recently found that CtBP paradoxically occupied actively transcribed NeuroD1-bound endocrine genes to promote transcription by a new mechanism involving several proteins in the CtBP co-repressor complex [15]. Transcriptional activation by CtBP required the histone demethylase, KDM1A, which can either activate or repress transcription [17,18]. However, KDM1A, which cannot directly bind to CtBP, requires another CtBP associated co-repressor protein, CoREST, to associate with CtBP. The CtBP binding protein, RREB1 (Ras responsive element binding protein), recruits CtBP to DNA close to NeuroD1 occupied sites. Transcriptional activation resulted from demethylation of H3K9Me1/2 by KDM1A and subsequent acetylation to H3K9Ac by the histone acetyl transferase activity of KAT2B. In the present work we examined the role of CtBP oligomerization in transcriptional activation by CtBP. Our results suggest that in the context examined, oligomeric CtBP is required for transcriptional activation.
Materials and Methods
Plasmids
The CtBP dimerization mutant (DM) contains mutations in the dimerization interface (C134Y, N138R, R141A and R142A) of CtBP that were introduced into Flag tagged CtBP (Fl-CtBP1) [19] by site-directed mutagenesis. The Fl-CtBP1 or the generated Fl-CtBP1-DM mutants were used as templates to construct CtBP shRNA-resistant (R) mutants, Fl-CtBP1-WTR or Fl-CtBP1-DMR. The shRNA resistant mutants were generated by introducing codon neutral nucleotide changes between AA 83 to 91 of CtBP1. Primers used for generating mutants are listed in Supplemental Table S1. The reporter genes pREP4-Secretin-Luc [15], pREP7-RSV-Renilla-luc [20], TOPflash [21], and E-cadherin [22] were previously described. An shRNA that targeted sequences common to CtBP1 and CtBP2 was used to deplete both forms of endogenous CtBP [23]. Stable cell lines expressing this CtBP shRNA, were previously described [15].
Cell Lines
The human duodenal cell line, HuTu 80 was obtained from ATCC. CtBP knockdown (KD) cells were transfected with the CtBP shRNA resistant wild-type CtBP1 expression plasmid (pcDNA-Fl-CtBP1-WTR) or its dimerization mutant (pcDNA-Fl-CtBP1-DMR) and selected with G418 (250 μg/ml) and Hygromycin (500 μg/ml) for one week. Cells were maintained by the same antibiotics.
Antibodies
Antibodies used included: CtBP1 (sc-11390 or sc-17759; Santa Cruz Biotech), LSD1/KDM1 (ab17721; Abcam), CoREST (ab24166; Abcam), NeuroD1 (sc-1084; Santa Cruz Biotech), Sp1(sc-59; Santa Cruz Biotech), GAPDH (sc-32233, Santa Cruz Biotech) and FLAG M2 (3165; Sigma-Aldrich), and RREB1 [24].
Chromatin immunoprecipitation assays
ChIP for Flag on HuTu 80 cells used methods described before [15].
Statistical Methods
Statistical analyses were performed using both the Mann-Whitney test and two-tailed unpaired Student’s t-test. Equivalent results were obtained; t-test results were reported in all figures. P values of < 0.05 were considered significant as noted in figure legends.
qRT-PCR analyses
RNA, isolated using the Qiagen RNeasy Plus Mini kit, was reverse transcribed using the SuperScript II first-strand synthesis system (Invitrogen) with levels determined by quantitative PCR using SYBR green incorporation with ACTB as an internal control. Primer sequences used for cDNA amplification are available upon request.
Results
Characterization of CtBP oligomerization mutants.
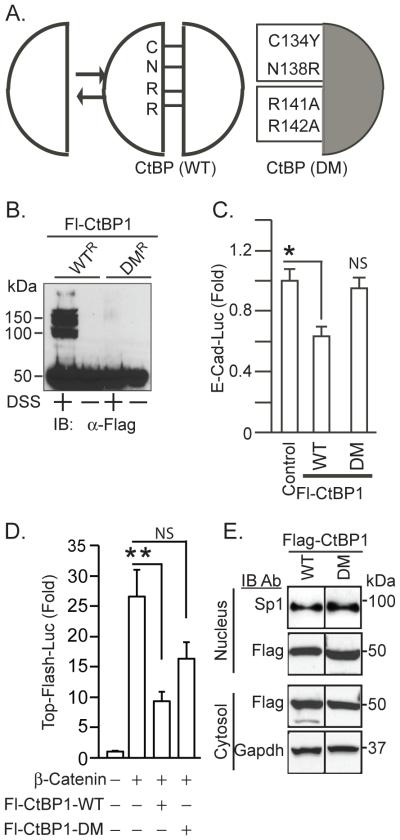
To determine whether potentiation of NeuroD1-dependent transcription by CtBP is mediated by its monomeric or oligomeric form, we generated a dimerization mutant (DM) of human CtBP1 by substitutions of amino acids in the predicted dimerization interface that were conserved in flies and humans (Fig. 1A). These mutations were previously shown to prevent self-association with minimal structural disruption of CtBP [8,9]. In order to confirm that the described substitutions (Fig. 1A) prevented oligomerization, CtBP depleted HuTu 80 cells stably expressing Fl-CtBP1-WTR or its dimerization mutant Fl-CtBP1-DMR were treated with a chemical cross-linker (DSS). The expressed Flag-CtBP1 in cell lysates was then analyzed by immunoblotting for Flag. Cross-linked Fl-CtBP1-WTR gave rise to a mixture of monomeric and oligomeric forms whereas Fl-CtBP1-DMR gave rise only to monomers (Fig. 1B).
Fig. 1.
Mutations in the CtBP dimerization interface prevent oligomerization. (A) amino acid substitutions in the CtBP dimerization mutants. (B) Inability of the mutants to oligomerize. HuTu 80 cells depleted of endogenous CtBP stably expressing Fl-CtBP1-WTR or Fl-CtBP1-DMR were cross-linked with (+) or without (−) 5 mM disuccinimidyl suberate (DSS) for 30 mins. The expressed proteins were detected by immunoblotting with anti-Flag. (C) Monomeric CtBP does not repress E-Cadherin gene transcription. HuTu 80 cells were co-transfected with an E-Cadherin-Luciferase reporter, plus expression plasmids for wild type or mutant Fl-CtBP1. Luciferase activity normalized for transfection efficiency was measured 24 h after transfection. (D) Oligomeric but not monomeric CtBP represses ß catenin dependent transcription. HEK293 cells were co-transfected with TOPflash reporter in the absence (−) or the presence (+) of the indicated expression plasmids. (C, D) Results shown as the mean ± SEM (n=5) *P<0.02, ** P <0.01, NS, not significant. (E) Distribution of wild type and dimerization mutants of Fl-CtBP1, between the cytosol and nucleus. Cytoplasmic and nuclear extracts were prepared from HuTu 80 cells transfected pcDNA-Fl-CtBP1 (WT or DM) and were analyzed for expression of Flag proteins by immunoblotting. Immunoblotting for GAPDH and Sp1 served as loading controls for cytoplasmic and nuclear proteins respectively.
We next examined the ability of our dimerization mutant to repress transcription of the E-Cadherin gene, by co-transfecting HuTu 80 cells with a E-Cadherin-Luciferase reporter with either Fl-CtBP1-WT or the Fl-CtBP1-DM expression plasmids followed by measurement of the reporter gene expression. The promoter for the cellular adhesion molecule E-cadherin is an established target for repression by oligomeric CtBP [2,22,25]. Unlike wild-type CtBP, the dimerization mutant, Fl-CtBP1-DM, did not repress E-Cad expression (Fig. 1 C).
In flies, a number of ß catenin dependent genes associated with Wnt signaling are repressed by oligomeric CtBP while expression of other Wnt targets is increased by either monomeric or wild type oligomeric CtBP [9]. To determine if CtBP similarly affected Wnt signaling in mammalian cells, we used transient expression assays in HEK 293 cells with TOPflash, a Wnt reporter with multiple TCF4 binding sites. Wild type CtBP1 inhibited beta-catenin-dependent transcription of the reporter. While the CtBP dimerization mutant appeared to inhibit reporter expression, the inhibition was not statistically significant despite a trend towards reduced expression (Fig. 1 D). These observations suggest that oligomeric CtBP mediates inhibition of Wnt related transcription by CtBP. Both the wild type and dimerization mutant were similarly distributed between the cytoplasm and the nucleus, suggesting that the inability of monomeric CtBP to inhibit TOPflash reporter activity did not result from changes in the intracellular localization of CtBP (Fig. 1E).
Transcriptional coactivation and the oligomerization state of CtBP
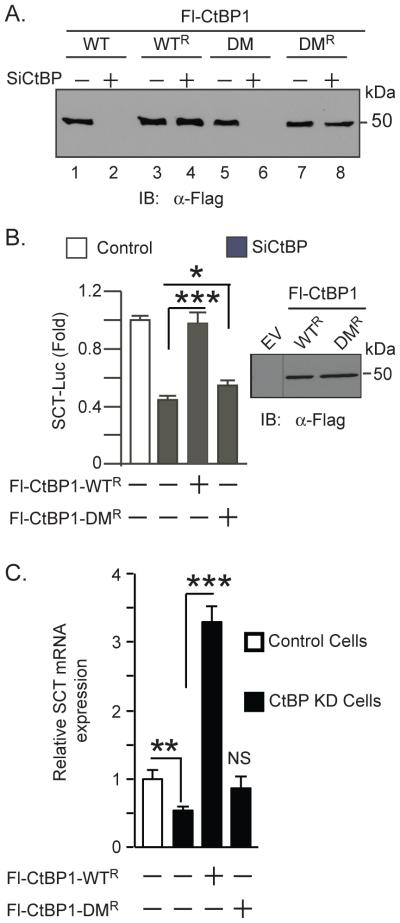
The role of CtBP oligomerization in transcriptional activation has not been characterized. Likewise, it is not known if coactivation of NeuroD1-dependent transcription of the SCT gene described previously requires oligomeric or monomeric forms of CtBP [15]. We depleted expression of both endogenous CtBP genes by RNAi to minimize interference with the CtBP mutants. CtBP was depleted in HuTu 80 cells by stable expression of a CtBP shRNA directed against identical nucleotide sequences present in both CtBP1 and CtBP2. The sequences of this shRNA were previously shown to knockdown both proteins [15,23]. To compare the effects of Fl-CtBP1-WT and Fl-CtBP1-DM mutants on NeuroD1 dependent transcription in a CtBP knockdown background, we constructed wild type and dimerization mutants that were resistant to the CtBP RNAi described above by introducing codon silent mutations to prevent binding to the shRNA. The Fl-CtBP1-WTR and Fl-CtBP1-DMR shRNA resistant mutants were readily expressed in CtBP knockdown cells (Fig. 2A, lanes 4, 8) whereas expression of Fl-CtBP1-WT and Fl-CtBP1-DM was undetectable (Fig. 2A, lanes 2, 6).
Fig. 2.
Oligomeric CtBP increases transcription. (A) Expression of shRNA resistant Flag-CtBP mutants. HuTu 80 cells depleted of endogenous CtBP by an shRNA were examined for Flag protein expression by immunoblotting. (B) Effect of Fl-CtBP1-DMR on SCT-Luciferase reporter gene expression (column 4), in CtBP depleted HuTu 80 cells in the same experiment previously describing the effect of wild type Flag-CtBP1 (columns 1-3) [15]. The shRNA resistant mutants used in the reporter assays were expressed at comparable levels in the transfection experiments (right panel). Results are shown as the mean ± SEM (n>5). (C) Effect of CtBP on expression of endogenous SCT transcripts determined by real time PCR using RNA extracted under the indicated conditions. Relative mRNA expression was normalized to the housekeeping gene, ß-actin. Results expressed as the mean ± SEM (n=7). *P < 0.05, **P < 0.02, ***P < 0.0001. NS, not significantly different from CtBP KD cells.
To determine if monomeric CtBP mediates coactivation of transcription, we cotransfected CtBP depleted HuTu 80 cells with a NeuroD1-dependent SCT-luciferase reporter gene, and Fl-CtBP1-DMR. Monomeric CtBP minimally rescued transcription of the reporter gene (Fig. 2B, column 4) unlike the oligomeric wild type CtBP shown previously in Fig. 1E of reference [15], and included here for comparison (Fig. 2B, columns 1-3). Monomeric CtBP appears to retain the ability to minimally increase the reporter gene expression by approximately 20% over the knockdown level, suggesting that oligomeric CtBP accounts for most of the transcriptional activation of the SCT reporter (Fig. 2B). Wild type and mutant CtBP were expressed at comparable levels in cell lysates, suggesting that the relative inability of Fl-CtBP1-DMR to rescue transcription was not due to differences in expression (Fig. 2B, right panel).
To determine if the endogenous SCT gene was similarly regulated by CtBP, we measured the levels of endogenous SCT transcripts by qRT-PCR in control and CtBP depleted HuTu80 cells. As shown previously[15], we observed a significant reduction of SCT transcript levels in CtBP KD cells (Fig. 2C). Moreover, Fl-CtBP1-WTR but not Fl-CtBP1-DMR significantly increased expression of the SCT transcripts in CtBP depleted cells.
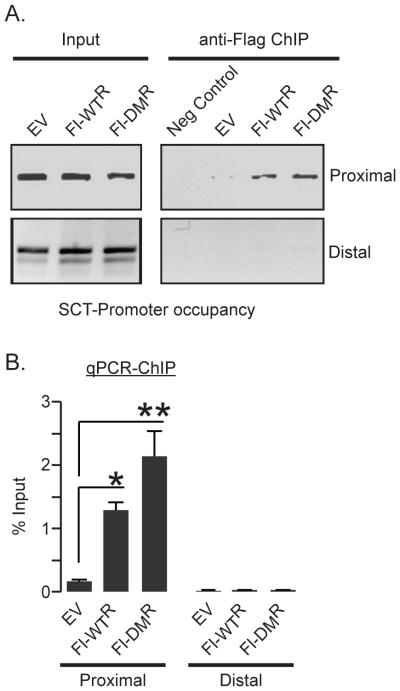
Promoter occupancy represents another potential mechanism for transcriptional regulation. We therefore examined the ability of Fl-CtBP1-WTR or Fl-CtBP1-DMR to occupy the SCT-gene promoter by ChIP with anti-Flag antibody. Both the wild type and the mutant similarly occupied this promoter, making it unlikely that the observed differences in gene expression seen with the monomeric CtBP resulted from the inability to gain promoter access (Fig. 3 A, B). The observed occupancy was specific for the region of the SCT promoter that associates with NeuroD1 and CtBP. Neither form of CtBP occupied distal upstream sequences of the same gene, indicating the specificity of CtBP occupancy (Fig. 3 A, B).
Fig. 3.
Promoter occupancy by oligomeric and monomeric CtBP. (A) ChIP for Fl-CtBP1 at proximal (−469 to −335) (top) and distal sequences (−3811 to −3610) (bottom) of the SCT gene in CtBP depleted HuTu 80 cells expressing Fl-CtBP1 WTR or Fl-CtBP1-DMR. Neg is template free negative control for PCR (B) ChIP-qPCR analyses showing significant enrichment of both monomeric and oligomeric Fl-CtBP1 at the proximal but not at the distal site of the SCT gene. Results shown as the mean ± SEM (n>5), *P< 0.005, **P< 0.001) compared to empty vector (EV) controls.
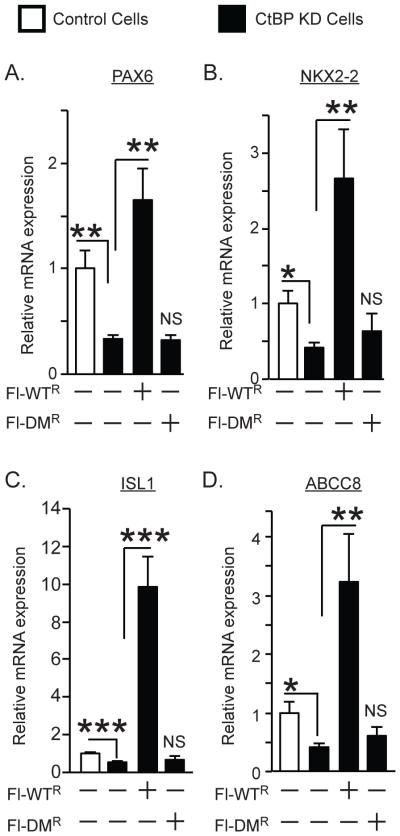
Limited information is available about NeuroD1 targets in human intestinal cells. To determine if oligomeric CtBP regulates other NeuroD1 targets in the intestine, we identified four additional candidate NeuroD1 target genes from a recently published ChIP-seq study that examined NeuroD1 occupied genes in a murine islet cell line [26]. These included three transcription factors PAX6, NKX2-2, and ISL1, all associated with endocrine differentiation in the GI tract. A fourth gene, encodes ABCC8, an ATP-activated K channel involved in regulated insulin secretion from ß cells. All of the selected genes were expressed at high levels in HuTu 80 cells and contained at least one NeuroD1 binding site in the human gene corresponding to a NeuroD1 binding site in the mouse gene. Depletion of CtBP significantly reduced expression of endogenous transcripts for each of these genes, and Fl-CtBP1-WTR but not Fl-CtBP1-DMR significantly increased expression of their messages under CtBP KD background (Fig. 4A-D). These results indicate that CtBP dimerization may have a broader role in transcriptional activation, including additional NeuroD1 targets in the intestine.
Fig. 4.
Regulation of other NeuroD1-targets by CtBP. Effect of wild type and mutant CtBP on expression of endogenous transcripts determined by real time PCR for PAX6 (A), NKX2-2 (B), ISL1 (C) and ABCC8 (D), normalized to ß-actin expression. Results are the mean ± SEM (n≥5). *P < 0.05; **P < 0.02; ***P <0.005.
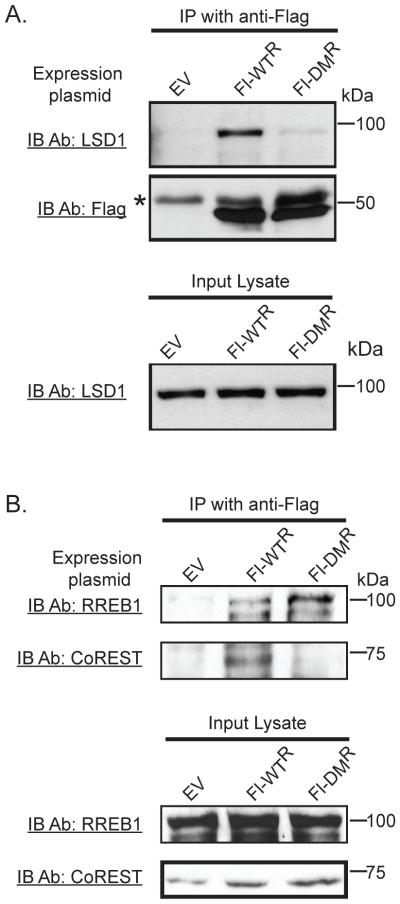
We compared the ability of oligomeric and monomeric CtBP to associate with other proteins that we previously implicated in CtBP stimulated transcription including the histone demethylase, KDM1A (LSD1), which was required for increased NeuroD1 dependent transcription by CtBP [15]. KDM1A was identified in Flag immunoprecipitates of wild type Fl-CtBP1-WTR but not Fl-CtBP1-DMR in lysates from CtBP depleted cells despite the nearly equal expression level of each mutant (Fig. 5A). Our observations suggest that the monomeric form of CtBP cannot associate with KDM1A.
Fig. 5.
KDM1A and CoREST associate with oligomeric but not monomeric CtBP. (A) Coimmunoprecipitation of CtBP and KDM1A. Presence of KDM1A in Flag immunoprecipitates of CtBP depleted HuTu 80 cells expressing Flag-CtBP1 proteins (WTR or DMR) in nuclear extracts. Precipitated proteins were detected by immunoblotting with the indicated antibody. Flag and KDM1A input shown by immunoblotting (lower panels). * denotes a nonspecific band. (B) Association of RREB1 and CoREST with oligomeric and monomeric forms of CtBP. Flag immunoprecipitates were examined for the expression of RREB1 and CoREST by immunoblotting.
Association of KDM1A with CtBP is indirect [2,27,28], requiring another corepressor protein, CoREST, to increase transcription [15]. CoREST co-immunoprecipitated with the wild type CtBP but not with the dimerization mutant, indicating that their association required oligomeric CtBP (Fig. 5B). The inability of monomeric CtBP to associate with CoREST may prevent its association with KDM1A, resulting in reduced transcriptional activation by CtBP.
The CtBP-associated protein, RREB1, binds to DNA sequences close to NeuroD1 occupied sites and directly interacts with NeuroD1 [24]. To determine whether association of CtBP with RREB1 was dependent on CtBP dimerization, we performed coimmunoprecipitation experiments in cells expressing Fl-CtBP1 mutants depleted of endogenous CtBP by shRNA expression. In contrast to KDM1A and CoREST, RREB1 was coimmunoprecipitated by both Fl-CtBP1-WTR and Fl-CtBP1-DMR indicating that both forms of CtBP can bind to RREB1 and that reduced transcriptional activity did not result from the inability of CtBP to associate with RREB1 (Fig. 5B). The ability of monomeric CtBP to bind to RREB1 suggests that the dimerization mutant retains sufficient structure to interact with this protein with two canonical PXDLS CtBP binding motifs [15].
Discussion
CtBP has long been established as a transcriptional corepressor. A few studies suggested that CtBP also increased transcription of a small number of genes although the underlying mechanisms of gene activation or the role of CtBP associated proteins were not characterized [12,13]. Recent ChIP-Seq studies for CtBP suggested that CtBP might be involved in increased transcription much more frequently than was previously recognized. More than 1800 CtBP bound sites across the genome were located within 2.5 kb upstream of transcriptional start sites. When occupied sites were matched to microarray expression profiling data, approximately 10% of CtBP bound sites were associated with genes whose expression increased in CtBP depleted cells, suggesting that CtBP functioned to repress them. Approximately 5% of occupied sites were associated with genes whose expression was reduced in CtBP knockdown cells, implying that their expression was dependent on CtBP [29]. Gene ontology analyses found that genes upregulated by CtBP were associated with a number of pathways involved in cellular homeostasis and cancer. Information about the mechanism of transcriptional activation by CtBP for these genes is limited.
The gene encoding the nucleotide exchange protein, TIAM1 (T-cell lymphoma invasion and metastasis1), is another example of a direct transcriptional target of CtBP whose expression is increased by CtBP and reduced by CtBP RNAi. CtBP associates with the transcription factor KLF8 and binds to multiple KLF8 sites in the TIAM1 promoter region [13].
Although it is well established that dimerization is required for transcriptional co-repression by CtBP [2], the importance of oligomerization for transcriptional activation is not known. Wnt target genes in flies may show increased or reduced expression depending on the oligomeric state of CtBP. Expression of the Wnt target gene, CG6234, was reduced in CtBP knockdown cells, suggesting that CtBP may act to increase its expression. Both monomeric and oligomeric CtBP rescued the reduced expression suggesting that monomeric CtBP may function as a transcriptional activator. Further studies revealed that the transcriptional activation of CG6234 by monomeric CtBP involved association with Pygopus [9]. Another Wnt target gene, nkd, was repressed by oligomeric CtBP, suggesting that transcriptional activation by CtBP may be gene specific. In the present work we found that monomeric CtBP did not increase expression of any of the five NeuroD genes examined in mammalian cells, in contrast to the CG6234 gene in the fly.
Proteomic analysis of CtBP associated proteins revealed that CtBP was part of a large multi-protein complex containing histone-modifying enzymes predominantly associated with transcriptional repression. A common mechanism for CtBP activation of transcription has not been identified from the limited number of genes whose expression is directly increased by CtBP. The function of CtBP associated proteins in transcriptional activation has not been examined extensively.
We previously showed that CtBP occupies several actively transcribed endocrine genes bound to the transcription factor NeuroD1 to increase transcription of neuroendocrine specific genes in the gastrointestinal tract. The increased transcriptional activity was dependent on the presence of multiple histone modifying proteins commonly associated with CtBP containing complexes [15]. The histone demethylase, KDM1A was essential for the effects of CtBP on increasing transcription and its presence at NeuroD1 occupied sites was dependent on CtBP and CoREST. We found that removal of the repressive histone marks at the H3K9 by KDM1A and subsequent acetylation by the NeuroD1-associated protein, KAT2B were necessary for increased transcription.
The present study extends our earlier findings showing that oligomeric but not monomeric CtBP is required to increase transcriptional activity of NeuroD1. The lack of association of KDM1A or CoREST with CtBP monomers accounts for the reduction of transcriptional activity since both of these proteins were required to increase NeuroD1 mediated transcription [15]. Oligomeric CtBP may increase the number of interacting protein interfaces available to recruit other chromatin modifying proteins that may function to either coactivate or co-repress transcription.
Recent studies predict the presence of many additional genes that are potentially transcriptionally activated by CtBP [29]. The mechanisms underlying transcriptional activation of a given CtBP target gene may be gene specific, depending on the specific histone modifying proteins recruited by oligomeric CtBP.
Supplementary Material
Acknowledgements
This work was supported in part by National Institutes of Health grants DK110614, DK100223 and DK90000 to (A.B.L.)
References
- [1].Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–78. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kuppuswamy M, Vijayalingam S, Zhao LJ, Zhou Y, Subramanian T, Ryerse J, Chinnadurai G. Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol Cell Biol. 2008;28:269–81. doi: 10.1128/MCB.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shi Y, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- [4].Zhao LJ, Kuppuswamy M, Vijayalingam S, Chinnadurai G. Interaction of ZEB and histone deacetylase with the PLDLS-binding cleft region of monomeric C-terminal binding protein 2. BMC Mol Biol. 2009;10:89. doi: 10.1186/1471-2199-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Balasubramanian P, Zhao LJ, Chinnadurai G. Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 2003;537:157–60. doi: 10.1016/s0014-5793(03)00119-4. [DOI] [PubMed] [Google Scholar]
- [6].Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- [7].Fjeld CC, Birdsong WT, Goodman RH. Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc Natl Acad Sci U S A. 2003;100:9202–7. doi: 10.1073/pnas.1633591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol Cell. 2002;10:857–69. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- [9].Bhambhani C, Chang JL, Akey DL, Cadigan KM. The oligomeric state of CtBP determines its role as a transcriptional co-activator and co-repressor of Wingless targets. EMBO J. 2011;30:2031–43. doi: 10.1038/emboj.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J. 2006;25:2735–45. doi: 10.1038/sj.emboj.7601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22:5296–307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Itoh TQ, Matsumoto A, Tanimura T. C-terminal binding protein (CtBP) activates the expression of E-box clock genes with CLOCK/CYCLE in Drosophila. PLoS One. 2013;8:e63113. doi: 10.1371/journal.pone.0063113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Paliwal S, Ho N, Parker D, Grossman SR. CtBP2 Promotes Human Cancer Cell Migration by Transcriptional Activation of Tiam1. Genes Cancer. 2012;3:481–90. doi: 10.1177/1947601912463695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–78. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ray SK, Li HJ, Metzger E, Schule R, Leiter AB. CtBP and associated LSD1 are required for transcriptional activation by NeuroD1 in gastrointestinal endocrine cells. Mol Cell Biol. 2014;34:2308–17. doi: 10.1128/MCB.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sundqvist A, Bajak E, Kurup SD, Sollerbrant K, Svensson C. Functional knockout of the corepressor CtBP by the second exon of adenovirus E1a relieves repression of transcription. Exp Cell Res. 2001;268:284–93. doi: 10.1006/excr.2001.5280. [DOI] [PubMed] [Google Scholar]
- [17].Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- [18].Wang J, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–7. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- [19].Zhang Q, Nottke A, Goodman RH. Homeodomain-interacting protein kinase-2 mediates CtBP phosphorylation and degradation in UV-triggered apoptosis. Proc Natl Acad Sci U S A. 2005;102:2802–7. doi: 10.1073/pnas.0409373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol Cell Biol. 2004;24:1188–99. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- [22].Alpatov R, Munguba GC, Caton P, Joo JH, Shi Y, Shi Y, Hunt ME, Sugrue SP. Nuclear speckle-associated protein Pnn/DRS binds to the transcriptional corepressor CtBP and relieves CtBP-mediated repression of the E-cadherin gene. Mol Cell Biol. 2004;24:10223–35. doi: 10.1128/MCB.24.23.10223-10235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bergman LM, Birts CN, Darley M, Gabrielli B, Blaydes JP. CtBPs promote cell survival through the maintenance of mitotic fidelity. Mol Cell Biol. 2009;29:4539–51. doi: 10.1128/MCB.00439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ray SK, Nishitani J, Petry MW, Fessing MY, Leiter AB. Novel transcriptional potentiation of BETA2/NeuroD on the secretin gene promoter by the DNA-binding protein Finb/RREB-1. Mol Cell Biol. 2003;23:259–71. doi: 10.1128/MCB.23.1.259-271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–8. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- [26].Jia S, et al. Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic beta-cell function. EMBO J. 2015;34:1417–33. doi: 10.15252/embj.201490819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–64. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- [28].Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell. 2006;23:377–87. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- [29].Di LJ, et al. Genome-wide profiles of CtBP link metabolism with genome stability and epithelial reprogramming in breast cancer. Nat Commun. 2013;4:1449. doi: 10.1038/ncomms2438. doi:10.1038/ncomms2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.