Abstract
Clustered regularly interspaced short palindromic repeat (CRISPR) loci and their flanking CRISPR-associated (cas) genes make up RNA-guided, adaptive immune systems in prokaryotes whose effector proteins have become powerful tools for basic research and biotechnology. While the Cas effector proteins are remarkably diverse, they commonly rely on protospacer-adjacent motifs (PAMs) as the first step in target recognition. PAM sequences are known to vary considerably between systems and have proven to be difficult to predict, spurring the need for new tools to rapidly identify and communicate these sequences. Recent advances have also shown that Cas proteins can be engineered to alter PAM recognition, opening new opportunities to develop CRISPR-based tools with enhanced targeting capabilities. In this review, we discuss the properties of the CRISPR PAM and the emerging tools for determining, visualizing, and engineering PAM recognition. We also propose a standard means of orienting the PAM to simplify how its location and sequence are communicated.
Keywords: Cas9, Cpf1, CRISPR-Cas systems, PFS, rPAM
Graphical Abstract
Introduction
Over the past 11 years, CRISPR (Clustered Regularly Interspaced Palindromic Repeats) loci and their associated Cas (CRISPR-associated) proteins have transitioned from curious prokaryotic immune systems to revolutionary tools for fundamental biomolecular research, biotechnology, agriculture, and medicine [1–5]. A key driver has been the ease by which the effector proteins of these systems can be utilized as programmable nucleases to specifically bind and/or cleave selected DNA or RNA sequences. Because these proteins and their guide RNAs are easier and cheaper to implement than all comparable technologies, CRISPR technologies have become a standard for applications in genome editing, gene regulation, and DNA imaging and are being explored for gene drives and sequence-specific antimicrobials [6–16]. CRISPR-Cas immune systems have also proven to be remarkably diverse, with new and emerging systems poised to further advance existing applications or drive entirely new ones [17–20]. Despite this diversity, CRISPR-Cas systems rely on a common set of rules for target recognition: complementary between the guide RNA and the target sequence, and a protospacer-adjacent motif (PAM) flanking the target [21–29]. This review details the nature of the PAM and recent efforts to identify, disseminate, and alter the recognized PAM sequences for different CRISPR-Cas systems. We describe the field’s current understanding of what defines a PAM as well as available methods to identify and communicate these sequences. We also discuss efforts to re-engineer PAM recognition and generate CRISPR-Cas systems with altered or improved DNA recognition capabilities.
CRISPR Biology
CRISPR-Cas systems naturally function as adaptive immune systems that protect bacteria and archaea from foreign genetic material such as bacteriophages or plasmids. The ability to uniquely recognize foreign sequences stems from the CRISPR array, a short stretch of DNA composed of alternating conserved repeats and target-specific spacers [1,30–32]. Each spacer is directly acquired from a fragment of the foreign genetic material called the protospacer, allowing the CRISPR array to possess heritable memory of the infection [30]. The CRISPR array is transcribed and subsequently processed into individual units called CRISPR RNAs (crRNAs) [33–35]. These RNAs associate with the system’s Cas effector proteins to form a ribonucleoprotein surveillance complex. Once assembled, the complex scans the cell for PAM sequences. Upon binding, the complex interrogates the extent of base pairing between the downstream sequence and the spacer portion of the crRNA. Extensive base pairing leads to the Cas proteins cleaving or degrading the target sequence, resulting in the clearance of the foreign invader.
CRISPR-Cas systems possess a diverse compilation of genes and are found throughout the prokaryotic world. Current estimates place CRISPR-Cas systems within ~50% of bacterial genomes and ~90% of archaeal genomes [18], although recent metagenomics sequencing analyses have suggested that the frequency of CRISPR-Cas systems in nature could be much lower [36]. Furthermore, only a subset of the identified systems may be active. Roughly 100 protein families have been associated with these systems, where the varying prevalence and co-occurrence of these genes has spurred the development of numerous classification schemes [18,19,37,38]. The most recent scheme breaks the systems into two classes, six types, and 19 subtypes [18,19]. The two classes are differentiated by whether a protein complex (Class 1) or individual protein (Class 2) serves as the effector of immune surveillance. The six types (I – VI) are defined by the presence of a signature gene encoding a protein responsible for nucleic-acid cleavage (e.g. Cas3 for Type I systems, Cas9 for Type II systems). The types also differ in their mechanisms of crRNA processing and target recognition, as well as whether the target is DNA (Types I, II, V), RNA (Type VI), or both (Type III) [39,40]. The subtypes are named by addition of a letter to the type (e.g. I-A, II-C) and are defined based on the specific cas genes and their configuration. As the vast expanse of CRISPR-Cas systems in the prokaryotic world remain poorly characterized, more unique functions and systems are expected to be discovered and will ultimately yield new CRISPR tools and technologies.
PAM Characteristics
The PAM was first observed in 2008, when Horvath and coworkers noticed conserved sequences that flanked protospacers acquired by Streptococcus thermophilus after being challenged with a lytic bacteriophage [21,22]. The following year, Mojica and coworkers uncovered similar motifs for multiple CRISPR-Cas systems through bioinformatics analyses, which established PAMs as generalized features of these systems [23]. Each report coined different names for these motifs—CRISPR motifs or protospacer-adjacent motifs (PAMs), respectively--where the latter became the accepted terminology.
The first insights into the function of these motifs first came from studies of the Type III CRISPR-Cas system in Staphylococcus epidermidis by Marraffini and Sontheimer, who demonstrated that these flanking sequences are essential for self/non-self discrimination (Figure 1) [41]. They specifically showed that the system uses these flanking sequences to differentiate between the CRISPR array (self) and the foreign invader (non-self), which both harbor a sequence perfectly complementary to the CRISPR RNA spacer. While the mechanism of discrimination appeared to rely on base pairing between the flanking regions of the spacer and protospacer [41], it established the theme that flanking sequences such as PAMs are critical for protospacer selection and target recognition. This insight was upheld as others showed that the PAM was an essential element for target recognition and cleavage [9,42].
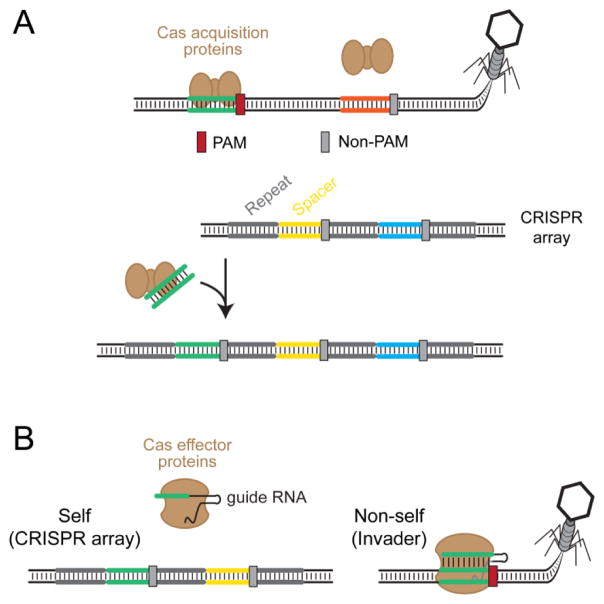
Figure 1.
Function of the CRISPR PAM. CRISPR-Cas systems naturally utilize PAMs to discriminate between self and non-self. The spacer portion of the CRISPR RNA is perfectly complementary to both its own CRISPR array (self) and the protospacer within the foreign invader’s genetic material (non-self). The systems differentiate between these two through the flanking PAM that is absent within the CRISPR array (gray) and present within the invader’s genetic material (red). Because the PAM is essential for target recognition, the CRISPR-Cas systems will target the invader’s genetic material but not the CRISPR array.
Extensive structural and biochemical analyses have helped reveal how the PAM participates in target recognition [43–47]. Cas effector proteins directly bind the PAM sequence through protein-DNA interactions and subsequently unzip the downstream DNA sequence. The effector proteins then interrogate the extent of base pairing between one strand of the DNA target and the spacer portion of the CRISPR RNA. Sufficient complementarity between the two drives target cleavage. Critically, if a PAM sequence is absent, the effector proteins do not interrogate the downstream sequence even if it is perfectly complementary to the spacer. We refer to this unrecognized sequence as a non-PAM. Aside from its function in immune recognition, the PAM also plays an integral role on spacer acquisition. In this case, the acquisition proteins alone or in coordination with effector proteins recognize defined PAM sequences while acquiring new spacers, ensuring that each new spacer can recognize the invading DNA [48–53]. As PAMs for acquisition and interference can be different [29,54,55], the associated sequences have been respectively termed spacer acquisition motifs (SAMs) and target interference motifs (TIMs) [56,57]. While this distinction is important, we refer to both motifs as PAMs given our primary focus on immune defense and the limited adoption of the terms SAMs and TIMs.
Despite the common role of the PAM in target recognition, its characteristics vary between the different types of CRISPR-Cas systems. One major difference is the location of the PAM. Using the non-target strand of the protospacer as a reference, the PAM is located on the 5′ of the protospacer for Type I and V systems and on the 3′ end of the protospacer for Type II systems. Note that the target strand also has been used to specify the PAM [29,43,58,59], creating some confusion about the exact location and sequence of any reported PAM; see Box 1 and Figure 2 for more information on these orientations and why we recommend the guide-centric orientation used in this review. Figure 3 illustrates the location of the PAM for different CRISPR-Cas system types given this orientation. In the case of Type III and Type VI systems, limited evidence suggests that the PAM is located within the target RNA [39,40]. Because of this unique location, the PAM for these systems was renamed the RNA PAM (rPAM) or the protospacer-flanking sequence (PFS), respectively. Given that Type III systems are thought to rely on base pairing between the crRNA 5′ handle and the region flanking the target DNA sequence [41], more work is needed to determine whether this mechanism or the rPAM is normally implemented and whether they occur separately or together.
Box 1.
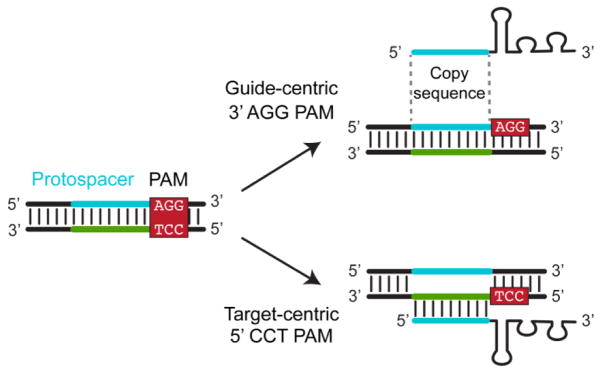
Standardizing the orientation of the CRISPR PAM. The PAM sequence flanks the protospacer within the target DNA. Because of the double-stranded nature of DNA, only one strand needs to be reported along with its location relative to the protospacer. To date, both strands have been used to report PAM sequences, where the selected strand often trends with the particular type of CRISPR-Cas system. The problems are that consensus PAM sequences are reported without the orientation, and the use of either strand creates confusion about the exact location of the PAMs. Here, we describe both orientations, which we term target-centric and guide-centric, and argue for the guide-centric orientation to be universally adopted. Both orientations are illustrated in Figure 2. Under the target-centric orientation, the PAM is located on the same strand that base pairs with the guide RNA. In many cases, the PAM on this strand is specifically recognized by the Cas effector proteins [43,58], lending a mechanistic argument to this orientation. The target-centric orientation is regularly employed for Type I systems [29,43,58,59]. Under the guide-centric orientation, the PAM is located on the strand that matches the sequence guide RNA. This match lends to guide-RNA design, where the sequence flanking the identified PAM is used to create the guide portion of the RNA. This orientation is used for Type II and V systems [21–23,61,69]. While the two orientations are equivalent, we propose the universal adoption of the guide-centric orientation: the Cas9 effector proteins from Type II systems are the most widely recognized and published, and the PAMs for these proteins are always reported in the guide-centric orientation. Adopting this orientation would therefore have a smaller impact on the existing body of CRISPR literature.
Figure 2.
Two orientations for reporting PAM sequences. The PAM sequence is located within double-stranded DNA, where either strand of the PAM can be reported along with its location relative to the protospacer. Under the target-centric orientation, the PAM is reported from the strand that base pairs with the guide RNA. Under the guide-centric orientation, the PAM is reported from the strand that matches the guide RNA and is used for guide-RNA design.
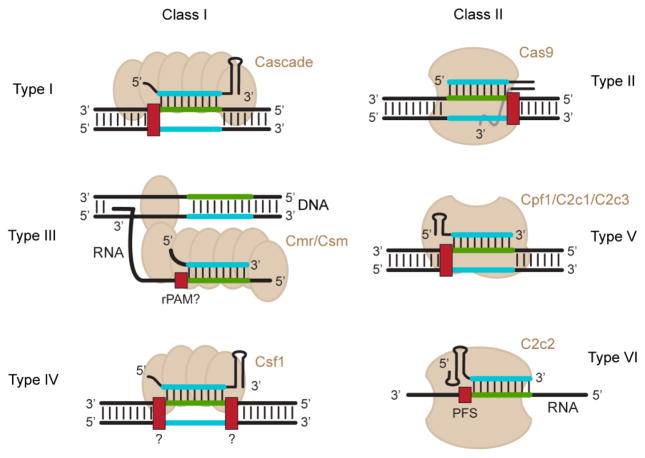
Figure 3.
PAM orientation and targets for different types of CRISPR-Cas systems. CRISPR-Cas systems are subdivided into two classes and six types. Representative illustrations of the effector proteins for each type are shown. The dsDNA orientation has been flipped from Figure 2 to accommodate the guide-centric PAM orientation. Note that the effector proteins for the V-B and V-C subtypes require a tracrRNA similar to Type II systems. For Type III and VI systems, the crRNA binds target RNAs. Type III systems cleave transcribed dsDNA due to its proximity to the RNA target [40]. Note that the mechanism by which Type III and VI systems recognize their nucleic-acid targets are still under investigation. PAM orientations are presented for all systems except for Type IV systems, which remain uncharacterized.
Aside from location, the composition of the PAM can vary widely. The composition includes the sequences comprising the PAM, the length of the linker (represented by N’s, where N any one of the four possible bases) separating the protospacer and the sequence-specific portion of the PAM, and the promiscuity in deviating from a defined consensus sequence. As one example, the widely used Type II-A system from Streptococcus pyogenes recognizes a NGG PAM, and to a lesser extent, an NAG PAM [9,23,60]. Separately, one Type II-A system from Streptococcus thermophilus recognizes an NNAGAA PAM but has the ability to recognize other sequences such as NNGGAAA and may accommodate changes in its linker length of 2 nucleotides [21,22,61,62]. Finally, the Type I-E system from Escherichia coli has one of the most promiscuous PAM recognition capabilities, with at least nine recognized PAM sequences (NAAG, NAGG, NATG, NGAG, NTAG, NAAC, NAAA, NAAT, NATA) and a strong nucleotide preference at the N position for some of these PAMs [29,58,63,64]. Table 1 contains representative consensus sequences for the most active PAMs for each characterized subtype. Given that PAMs can vary widely even within a given subtype [21–23,65], more work is needed to fully interrogate the diversity of PAMs in nature.
Table 1.
Consensus PAMs for representative CRISPR-Cas systems. The consensus PAM sequences represent the most active PAM sequences and are reported using the guide-centric orientation (see Box 1). Dashes indicate a subtype lacking any system with a predicted or characterized PAM. rPAM: RNA PAM. PFS: protospacer-flanking sequence.
| Classification
|
Source organism | PAM location | Consensus sequence* | Reference | |
|---|---|---|---|---|---|
| Class | Type/subtype | ||||
| 1 | I-A | P. furiosus | 5′ | YCN | [59] |
| 1 | I-B | C. difficile | 5′ | CCW | [87] |
| 1 | I-C | B. halodurans | 5′ | YYC | [64] |
| 1 | I-D | Cyanothece sp. PCC 8802 | - | - | [18] |
| 1 | I-E | E. coli | 5′ | AWG | [63,64] |
| 1 | I-F | P. aeruginosa | 5′ | CC | [88] |
| 1 | I-U | G. sulfurreducens | - | - | [18] |
|
| |||||
| 1 | III-A | S. epidermidis | None | None | [18] |
| 1 | III-B | P. furiosus | 3′ | MMA (rPAM) | [40] |
| 1 | III-C | M. thermautotrophicus str. Delta H | - | - | [18] |
| 1 | III-D | Roseiflexus sp. RS-1 | - | - | [18] |
|
| |||||
| 1 | IV | A. ferrooxidans | - | - | [18] |
|
| |||||
| 2 | II-A | S. pyogenes | 3′ | NGG | [9,23,60] |
| 2 | II-A | S. thermophilus (CRISPR1) | 3′ | NNAGAA | [21,22,61] |
| 2 | II-A | S. aureus | 3′ | NNGRRT | [65,76] |
| 2 | II-B | L. pneumophila str. Paris | - | - | [18] |
| 2 | II-C | N. meningitidis | 3′ | NNNNGWWT | [61] |
|
| |||||
| 2 | V-A | F. novicida | 5′ | TTN | [64,69,73] |
| 2 | V-B | A. acidoterrestris | 5′ | TTN | [19] |
| 2 | V-C | Metagenomic datasets | - | - | [18] |
|
| |||||
| 2 | VI | L. shahii | 3′ | D (PFS) | [39] |
D = A, G, T; M = A, C; N = A, C, G, T; W = A, T; Y = C, T.
PAM Determination
The PAM is an essential feature of CRISPR-Cas systems, whether for the biological function of the system or for harnessing the system as a biomolecular tool. Determining the full set of functional PAM sequences, however, has proven difficult. This challenge has spurred the development of multiple methods to determine PAMs (Figure 4). Below we describe the available methods along with their particular advantages and disadvantages. While all of these methods reproduce the same highly active PAM sequences, they can often identify differing sup-optimal PAMs, which can impact target selection and off-target predictions. Thus, the best option for PAM determination will likely depend on the particular CRISPR-Cas system and its end use.
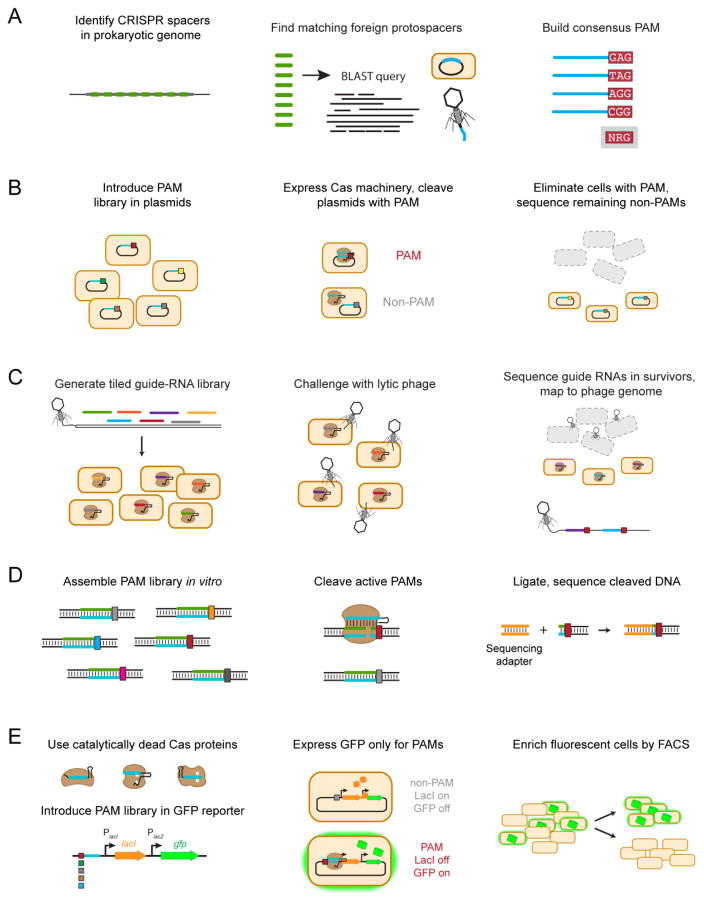
Figure 4.
Methods for PAM determination. (A) In silico PAM determination. A BLAST search of metagenomic databases identifies potential protospacers. Matches found on foreign nucleic acid elements from bacteriophages and plasmids are aligned to elucidate a single consensus sequence. (B) PAM determination through plasmid clearance. Plasmids harboring a library of potential PAM sequences are transformed into cells expressing the Cas proteins and the targeting guide RNA, and functional PAM sequences are identified based on their depletion from the library. (C) PAM determination through bacteriophage clearance. A library of guide RNAs are designed so their targets tile along a lytic bacteriophage genome. Guide RNAs targeting a site flanked by a functional PAM protect the bacteriophage attack, as revealed by sequencing the enriched guide RNAs and mapping their locations and flanking sequences within the bacteriophage genome. (D) In vitro PAM determination through DNA cleavage. A large PAM library is incubated with purified Cas proteins and transcribed crRNAs in a reaction buffer. Functional PAMs within this library are cleaved, and a sequencing adapter (shown in orange) can then be ligated for high-throughput sequencing. Alternatively, intact DNA can be sequenced, revealing PAM sequences that were depleted because of DNA cleavage. (E) In vivo PAM determination through DNA binding. Catalytically-dead effector proteins are targeted to a PAM library upstream of the lac operon regulating GFP expression. If a functional PAM sequence is present, binding by the catalytically-dead effector proteins represses expression of LacI, resulting in de-repression of GFP. Fluorescence cells are then isolated by fluorescent-activated cell sorting and sequenced.
Protospacer identification in silico
Mojica and coworkers introduced the first means of identifying PAMs as part of their original observation of this motif [23]. Under this method, each spacer sequence from a natural CRISPR array is subjected to a nucleotide BLAST search for homologous sequences [66]. Strong matches that appear to be derived from bacteriophages or plasmids are compiled, and the flanking sequences are aligned to discern a general motif. This method offers a rapid means of identifying potential functional PAM sequences that can be evaluated experimentally. With the recent development of automated online tools such as CRISPRTarget [67], the analysis can be completed in less than a day. One disadvantage is a strong dependence on available sequencing information, which may not contain the associated invader sequences. Another is the challenge of deciding whether a BLAST hit represents the true source of the spacer, particularly when mismatches are present between the spacer and the putative protospacer sequence. Third, even if the protospacer is the source of the spacer, some of the protospacers may have accumulated mutations in the PAM. Finally, the PAMs associated with acquisition can represent a subset of those that elicit targeting [48,49,68], giving a narrow impression of the PAMs that elicit interference. For these many reasons, the in silico method represents a convenient starting point but often lacks sufficient sequence information and is less suited to obtain a comprehensive picture of the PAM for a given system.
Plasmid clearance in vivo
The second and most common method screens for functional PAM sequencing using the natural ability of CRISPR-Cas systems to clear foreign genetic material. This method utilizes an effector protein that is either already present in its native host [59] or is imported into a convenient host such as Escherichia coli [60,61,69]. To generate potential PAM sequences, a randomized nucleotide library is inserted next to a target sequence within a plasmid. Plasmids harboring the PAM library are transformed into the host and then subjected to next-generation sequencing. Any plasmids harboring functional PAM sequences would be cleared by the Cas effector proteins, resulting in a substantially lower frequency in the library. This method has been the most widely used because it recapitulates the natural function of CRISPR-Cas systems and has the potential to comprehensively determine all functional PAM sequences. One disadvantage is that the method employs a negative selection, which requires extensive library coverage to identify the missing sequences. Furthermore, because the screen is in vivo, there is a general limit on the library size, and the Cas proteins must be functionally expressed in a non-native host. Finally, the readout of this method is the frequency of PAM escape, whether by mutating the target or by promoting DNA repair. This is problematic particularly for less-active PAM sequences that may translate poorly to other CRISPR-based applications, such as genome editing or gene regulation [54,64,70].
One recent variation on this method generates a library of guide RNAs that tile along the genome of a lytic bacteriophage [39]. The host harboring the CRISPR-Cas system is transformed with the plasmids harboring the guide-RNA library and is then infected with the bacteriophage. Cells survive if the target location on the bacteriophage genome is flanked by a PAM, resulting in the enrichment of guide RNAs targeting functional PAM sequences. This strategy was applied to elucidate the PFS for the RNA-targeting Type VI effector protein C2c2 using the single-stranded RNA bacteriophage MS2 [39]. By selecting for cells targeting protospacers flanked by a functional PFS, the screen provided a positive selection. The major limitations of this variation are that the guide-RNA library is much smaller than the equivalent PAM library, not all possible PAMs may be sufficiently represented within the bacteriophage genome, and guide-RNA libraries will be much more expensive to generate than PAM libraries. Therefore, this strategy offers some unique advantages over the traditional method if a sufficiently large library can be generated and the associated costs are acceptable.
DNA cleavage in vitro
In contrast to the in vivo plasmid-clearance methods described above, methods have been developed to determine PAMs under in vitro conditions. These methods involve an in vitro cleavage reaction that combines purified Cas effector proteins, the in vitro-transcribed guide RNAs, and a target DNA library of potential PAM sequences. Following the cleavage reaction, the PAM library is subjected to next-generation sequencing. The reaction can be conducted as a positive screen by ligating adapters to the ends of cleaved library members [70,71], or as a negative screen by sequencing the intact library members [69]. In vitro methods generally offer numerous advantages particularly over in vivo methods. For instance, the screened library can be multiple orders-of-magnitude larger for in vitro screens than for in vivo screens because there are no limitations from transformation efficiencies or cloning in vitro. Furthermore, in vitro reactions grant exquisite control over the assay conditions, such as component concentrations, reaction temperature, and the reaction time. Finally, the ability to sequence the cleavage products represents a positive screen that can reliably identify functional PAM sequences. One downside is that reconstituting the complete system requires complete knowledge of the required components as well as a protein purification. Another is that the ligation step requires a double-stranded break, which excludes Type I systems that cleave and degrade the target. Finally, assay conditions can deviate from the cellular environment, whether it is the buffer conditions, the relative stoichiometry of Cas effector proteins and targets, or the reaction times. This deviation can yield artificial PAM assignments as recently highlighted when Karvelis and coworkers varied the stoichiometry of Cas9 and the target DNA [71]. These in vitro methods thus provide powerful screens to comprehensively determine PAMs for many CRISPR-Cas systems, although the resulting PAMs may not translate well in vivo.
DNA binding in vivo
The high-throughput experimental methods described above all relied on target cleavage. However, the ability to generate catalytically-dead Cas effector proteins affords the development of PAM determination methods based on DNA binding [8–10]. We recently reported an in vivo DNA-binding method that utilizes catalytically-dead effector proteins to regulate the expression of GFP in E. coli [64]. As part of the screen, binding by the catalytically-dead effector proteins blocks expression of the LacI repressor, which would otherwise block expression of GFP. Based on this configuration, cells harboring a functional PAM sequence fluorescence, allowing these cells to be isolated by fluorescence-activated cell sorting and subjected to next-generation sequencing. The unique benefits of this method include a positive screen based on the expression of GFP only in the presence of a functional PAM, and the ability to tune the assay stringency by titrating in the LacI inhibitor IPTG. One limitation of the method is the need to identify and inactivate the catalytic domains of the desired CRISPR-Cas systems, particularly if the systems have not undergone initial characterization. Another is the size of the limited library size that can be screened akin to any in vivo screen. Finally, the absence of nuclease activity can yield PAMs that promote efficient binding but not efficient cleavage, even though the PAMs elucidated to-date using this method closely aligned with those determined by cleavage-based methods [61,63,64,69,71–73]. The in vivo DNA binding method thus may be best suited for better understanding the biophysics of DNA recognition or in applications centered on DNA binding or gene regulation. PAM determination methods that rely on DNA binding in vitro are also under development that could introduce the advantages of in vitro screens [74].
PAM Reporting
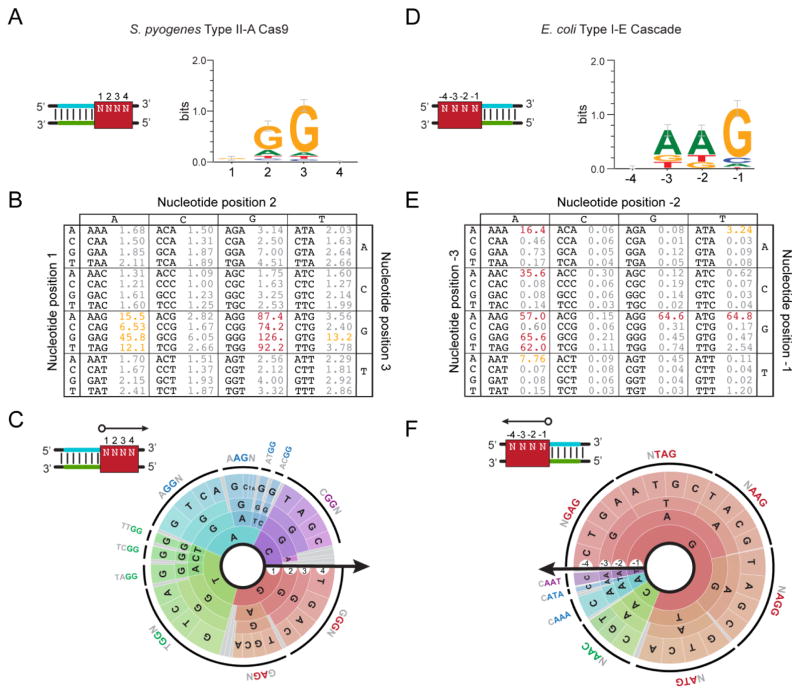
PAM determination methods often generate a large collection of functional PAM sequences that vary in their extent of enrichment (for positive screens) or depletion (for negative screens). The question is how to best report this information without providing the complete list of all sequences and their associated enrichment or depletion scores. A number of reporting schemes have been described, where each is illustrated in Figure 5 using published PAM determination data for the widely used Type II-A system from S. pyogenes and the well-characterized Type I-E system from Escherichia coli [64,72]. As illustrated in this figure, each reporting scheme manages a trade-off between simplicity and information content.
Figure 5.
Reporting the CRISPR PAM. (A–C) Visualizing the PAM for the S. pyogenes Type II-A system. The reported PAMs are based on data from the plasmid-clearance method conducted by Kleinstiver and coworkers [72]. (D–F) Visualizing the PAM for the E. coli Type I-E system. The reported PAMs are based on data from the DNA-binding method conducted by Leenay and coworkers [64]. (A) The consensus sequence and sequence logo. The consensus sequence represents a compilation of all highly-active functional PAM sequences. The sequence logo displays the sequence conservation of each base at each position in functional PAM sequences [89]. The red box demonstrates the PAM location assuming a guide-centric orientation (see Figure 2). (B) The PAM table. The table displays the enrichment or depletion scores from the conducted screen for each possible sequence. Higher values represent more active PAMs. PAMs with similar activities are colored. (C) The PAM wheel. Individual sequences are read following a radius of the circle. The arrow shows the orientation of each base in relation to the protospacer. The larger the radian occupied by the sequence, the greater its enrichment or depletion score. The outer ring depicts a common functional PAM sequence. PAM wheels are also available as interactive .html files [64].
Sequence logos and consensus sequences have been the most common reporting schemes to date starting with the original discovery of the PAM [21,22]. Sequence logos display the conservation of each base within each position: more conserved or highly active bases appear as larger letters, while less conserved or less active bases appear as smaller letters. Consensus sequences report a single sequence that captures the most dominant set of functional PAM sequences. For instance, the consensus sequence for the S. pyogenes Cas9 is NGG, while the sequence logo shows a small but notable A in the middle position reflecting partial activity of an NAG PAM [23,60]. Conversely, the consensus sequence for the promiscuous E. coli I-E system has been reported as AWG (where W is A or T) while the sequence logo shows other, smaller letters at each of the three positions [23,58]. Both schemes are easy to interpret and can be expanded to any sequence length. However, they also sacrifice individual functional PAM sequences and their relative activity. This loss of information is manageable if the PAMs are simple or only the most active functional PAM sequences are required for the final use of the interrogated CRISPR-Cas system, such as for designing highly active guide RNAs. However, these reporting schemes can obscure the true number of target sites that can be targeted by the system or the prediction of off-target effects by discarding or masking less active functional PAMs. One workaround is reporting multiple consensus sequences that are classified as more or less active [61], although this scheme still misses the full range of functional PAM sequences and activities. In cases where the PAM is relatively simple (e.g. NGG for the S. pyogenes Cas9) or the user is only interested in the most active PAMs, then sequence logos and consensus sequences are sufficient.
A separate scheme that better captures individual functional PAM sequences and activities can be described as a PAM table (Figure 5B,E). The table is similar to a codon table in which each base of the codon appears along the edges and each of the 64 cells represents a distinct sequence. In the case of the PAM table, each cell conveys the activity or enrichment/depletion score for that specific PAM sequence. Cells with similar activities or scores can be colored, although the groupings are somewhat arbitrary. The upside of the table is that it displays all PAM sequences and activities in a relatively compact format, as illustrated for PAMs recognized by the S. pyogenes Cas9 and by multiple CRISPR-Cas systems present in Pyrococcus furiosus [59,60]. The major downside is that the table is difficult to expand beyond three bases, greatly limiting the size of a PAM library that can be reported.
We recently developed a reporting scheme called the PAM wheel that also captures individual sequences and activities that can convey larger PAM libraries [64] (Figure 5C,F). The PAM wheel are derived from interactive Krona plots [75]. Each PAM sequence is read by moving along a radius of the circle, where the accompanying arrow specifies the location of each base in relation to the protospacer. The relative activity of each PAM sequence scales with the size of the radial arc. The outer, black rings designate functional PAM sequences. As a Krona plot, any sector of the wheel can be expanded to better view a defined subset of sequences, such as PAMs that begin with a G. The ability to fully interrogate all possible sequences grants a comprehensive picture of the PAM landscape that is otherwise obscured by the other available reporting schemes. For instance, the PAM wheel revealed a potential two-base linker for the S. pyogenes Cas9 or strong base preferences at the −4 position for some functional PAM sequences for the E. coli Type I-E system (Figure 5). The most notable disadvantage is that the PAM wheel can be difficult to interpret, particularly in comparison to the other described methods. There is still room for additional reporting schemes that effectively capture and convey the higher-order information content of the PAM, and it will be up to the CRISPR research community to settle on a common scheme or set of schemes to convey PAM sequences and activities.
PAM Engineering
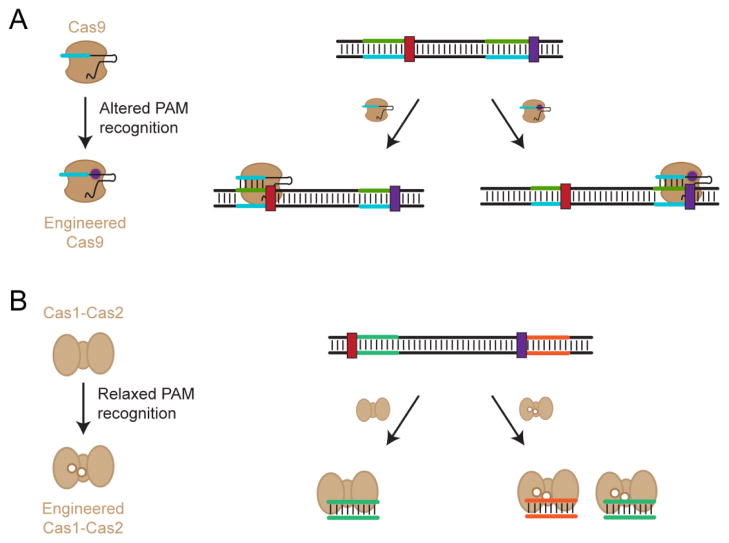
While the PAM remains an essential feature for self/non-self discrimination, it also restricts which sequences can be targeted by a given CRISPR-Cas system and impacts the likelihood of off-target effects. Accordingly, there has been intense interest in modifying Cas proteins—particularly the widely used Cas9 effector proteins—to change the recognized PAM [68,72,76,77]. This interest has been fueled by the growing number of structural studies that have pinpointed the PAM-interacting domains and how these domains interact with the PAM as part of target recognition [78–80]. The PAM-interacting domains are relatively modular, allowing Nishimasu and coworkers to swap these domains between the closely related S. pyogenes Cas9 and S. thermophilus CRISPR3 Cas9 proteins, thereby changing each Cas9’s PAM recognition (Figure 6) [45]. Rationally mutating the protein residues involved in PAM binding has proven more difficult as illustrated for the S. pyogenes Cas9 [9], although Anders and coworkers successfully changed PAM recognition using a variant of this protein [77].
Figure 6.
Engineering Cas proteins with altered or relaxed PAM recognition. (A) Changing PAM specificity for the Cas9 effector protein. This protein was engineered to recognizes an alternative PAM but not the original PAM, thereby changing which sequences can be targeted by this Cas9. (B) Relaxing PAM specificity for the Cas1 and Cas2 acquisition proteins. These proteins still recognize the original PAM as well as additional PAMs, thereby broadening the sequences that can be acquired with these proteins.
The most successful PAM engineering efforts to-date combined random mutagenesis of the DNA binding residues or the entire protein with a high-throughput dual selection. Kleinstiver and coworkers applied this combination to change PAM recognition for the S. pyogenes Cas9 and the compact Staphylococcus aureus Cas9. These efforts generated a variant of the S. pyogenes Cas9 with only three point mutations (Cas9 VQR) that changed the recognized PAM from NGG to NGA. The new Cas9 exhibited a lower propensity for off-target effects for the selected targets [72] despite more promiscuous PAM recognition [64]. These efforts also generated a variant of the S. aureus Cas9 that relaxed its recognized PAM consensus sequence from NNGRRT (where R is A or G) to NNNRRT [76]. Relaxing the requirement for a G did not impact the propensity for off-target effects despite the expectation that a more flexible PAM would reduce its contribution to targeting specificity [76,81]. Applying similar strategies to other Cas effector proteins such as Cpf1 could generate an expanded set of proteins that are no longer limited to their natural PAM sequences, thereby expanding the types of sequences that can be targeted for genome editing and other applications.
Shipman and coworkers recently extended PAM engineering to the acquisition proteins Cas1 and Cas2 from the E. coli Type I-E system [68]. These proteins function together to integrate new spacers into the CRISPR array [82–84]. While the work by Shipman and coworkers sought to utilize these proteins to incorporate synthetic spacers for long-term memory storage, the two proteins were also engineered to relax their PAM recognition requirements. The engineering was performed using error-prone PCR, followed by integrating spacers from non-canonical PAMs [68]. The resulting variants of Cas1 and Cas2 still slightly preferred an AAG PAM, although to a lesser extent than the wild-type versions. Taken together, these accomplishments highlight the potential for engineering diverse Cas effector proteins and acquisition proteins for tightened, relaxed, or non-native PAM recognition.
PERSPECTIVES
Our knowledge of what comprises a PAM has been overwhelmingly shaped by a few well-characterized Cas effector proteins. However, these few proteins stand in contrast to the large diversity of systems currently spanning 19 subtypes and thousands of individual proteins [18,19] (Table 1). Furthermore, characterization of different Cas9 proteins from II-A systems revealed large variation in the PAM: for instance, those for Cas9 proteins from S. pyogenes (NGG PAM), S. thermophilus (NNAGAA PAM), and S. aureus (NNGRRT PAM). We predict that characterizing multiple systems within other subtypes has the potential to reveal similarly diverse PAMs. By performing bioinformatics and structural analyses of these effector proteins, we could learn how PAM recognition domains uniquely recognize different PAM sequences, informing future efforts to engineer PAM recognition. Elucidating PAMs within different subtypes could also reveal the full range extent of PAM sequences and lengths present in nature, highlighting which sequences may be more readily accessible through PAM engineering.
High-throughput methods have been invaluable tools to determine PAMs for varying CRISPR-Cas systems. One consistent insight from these methods is the plasticity of the PAM. Rather than a static sequence that is either correct or incorrect, the PAM comprises a range of sequences with varying activities. This is highlighted by the Type V-A Cpf1 effector protein from Francisella novicida, which has a reported PAM consensus of NTTN but exhibits clear bias within the first T and both N’s [64,69,73]. Another unique insight is the co-dependency between the PAM and the protospacer. Despite the general assumption that these entities operate independently, separate studies with the Type I-E system from E. coli have shown that the relative activity of different PAMs depended on the specific target sequence [54,64]. Whether this co-dependency extends to other systems remains to be explored and may need to become a standard part of any attempt at high-throughput PAM determination. Finally, there is evidence that PAM determination methods can predict slightly different PAMs for the same CRISPR-Cas system. For instance, characterization of the Streptococcus thermophilus CRISPR1 Cas9 using an in vivo plasmid-clearance assay, an in vitro cleavage assay, and an in vivo binding assay revealed different suboptimal PAMs [64,71,72]. While these differences may be attributed to the selected PAM reporting schemes, they highlight how the available PAM determination methods can yield disparate results. These differences can be important, as elucidated PAMs may not be universally relevant across all applications. As a result, it may be best to perform PAM determination methods that best align with the final application (i.e. in vitro cleavage assays for genome editing; binding assays for transcriptional regulation).
The ability to engineer PAM recognition holds great potential for CRISPR technologies. One potential outcome is revising the process of guide-RNA design. The current process involves identifying a PAM within a genetic locus and then selecting the flanking sequence as the target. However, PAM engineering could be used to generate a collection of effector proteins that together recognize all possible PAM sequences. For instance, the collection could include 15 derivatives of the S. pyogenes Cas9 that each recognize a variant of the NGG consensus PAM. Separately, the PAM could be lengthened to recognize a more specific sequence that is likely to be found across a genome. These mutations could be combined with those known to better reject mismatches between the guide RNA and the target [85,86], potentially yielding highly specific Cas effector proteins with negligible off-target effects.
Highlights.
The PAM is an essential feature for target recognition
PAMs are associated with all characterized types of CRISPR-Cas systems
Multiple methods are available for PAM determination and visualization
Cas proteins can be engineered to alter, tighten, or relax PAM recognition
We propose a standard means of orienting the PAM for all CRISPR-Cas types
Acknowledgments
We thank Michelle Luo, Matthew Waller, and Rodolphe Barrangou for constructive feedback on the manuscript. The work was supported by funding from the National Science Foundation (CBET-1403135, MCB-1452902 to C.L.B.), the National Institutes of Health (1R35GM119561-01 to C.L.B., 5T32GM008776-15 to R.T.L.), and Agilent Technologies (to C.L.B.). C.L.B. and R.T.L. have submitted provisional patent applications on CRISPR technologies. C.L.B. is a co-founder of Locus Biosciences and a member of its scientific advisory board.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 6.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033.RNA-Guided. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bikard D, Marraffini LA. Control of gene expression by CRISPR-Cas systems. F1000Prime Rep. 2013;5 doi: 10.12703/p5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–822. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo ML, Mullis AS, Leenay RT, Beisel CL. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2014;43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. Elife. 2014:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S, Burt A, Windbichler N, Crisanti A, Nolan T. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34:78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel C. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio. 2014;5 doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anton T, Bultmann S, Leonhardt H, Markaki Y. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus. 2014;5:163–172. doi: 10.4161/nucl.28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B, Gilbert La, Cimini Ba, Schnitzbauer J, Zhang W, Li G, Park J, Blackburn EH, Weissman JS, Lei S. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001.Dynamic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, Van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 18.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:1–13. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo ML, Leenay RT, Beisel CL. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol Bioeng. 2016;113:930–943. doi: 10.1002/bit.25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath P, Romero DA, Coute-Monvoisin A-C, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 24.Garneau JE, Dupuis M-È, Villion M, Romero Da, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 25.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2013:1–9. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul Ü, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders APL, Dickman MJ, Doudna JA, Boekema EJ, Heck AJR, van der Oost J, Brouns SJJ. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 29.Westra ER, van Erp PBG, Künne T, Wong SP, Staals RHJ, Seegers CLC, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJJ, van der Oost J. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 31.Jansen R, Van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 32.Bolotin A, Quinquis B, Sorokin A, Dusko Ehrlich S. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 33.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deltcheva E, Chylinski K, Sharma CM, Gonzales K. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886.CRISPR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burstein D, Sun CL, Brown CT, Sharon I, Anantharaman K, Probst AJ, Thomas BC, Banfield JF. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat Commun. 2016;7:10613. doi: 10.1038/ncomms10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1101/054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, Terns MP. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR–Cas system. Genes Dev. 2016;30:447–459. doi: 10.1101/gad.272153.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhoua K, Barendregt A, Westphal W, Heck AJR, Boekema EJ, Dickman MJ, Douda J. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci. 2011;108:15010–15010. doi: 10.1073/pnas.1111854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulepati S, Héroux A, Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna J. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yosef I, Shitrit D, Goren MG, Burstein D, Pupko T, Qimron U. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc Natl Acad Sci U S A. 2013;110:14396–401. doi: 10.1073/pnas.1300108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savitskaya E, Semenova E, Dedkov V, Metlitskaya A, Severinov K. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 2013;10:716–725. doi: 10.4161/rna.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:1–16. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Y, Terns RM, Terns MP, Terns MP, Terns MP. Cas9 function and host genome sampling in type II-A CRISPR–cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kunne T, Kieper SN, Bannenberg JW, Depken M, Suarez-diez M, Brouns SJJ, Ku T, Kieper SN, Bannenberg JW, Vogel AIM, Miellet WR, Klein M. Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol Cell. 2016;63:852–864. doi: 10.1016/j.molcel.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Xue C, Seetharam AS, Musharova O, Severinov K, Brouns SJJ, Severin AJ, Sashital DG. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 2015;43:10831–10847. doi: 10.1093/nar/gkv1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue C, Whitis NR, Sashital DG, Xue C, Whitis NR, Sashital DG. Conformational control of Cascade interference and priming activities in CRISPR immunity. Mol Cell. 2016;64:1–9. doi: 10.1016/j.molcel.2016.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mojica FJM, Díez-Villaseñor C. Right of admission reserved, no matter the path. Trends Microbiol. 2013;21:446–448. doi: 10.1016/j.tim.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 57.Shah S, Erdmann S, Mojica F, Garrett R. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elmore J, Deighan T, Westpheling J, Terns RM, Terns MP. DNA targeting by the type I-G and type I-A CRISPR–Cas systems of Pyrococcus furiosus. Nucleic Acids Res. 2015;43:gkv1140. doi: 10.1093/nar/gkv1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 63.Westra ER, Semenova E, Datsenko KA, Jackson RN, Wiedenheft B, Severinov K, Brouns SJJ. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol Cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ran FA, Cong L, Yan WX, Scott Da, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp Pa, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–190. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 67.Biswas A, Gagnon JN, Brouns SJJ, Fineran PC, Brown CM. CRISPRTarget: Bioinformatic prediction and analysis of crRNA targets. RNA Biol. 2013;10:817–827. doi: 10.4161/rna.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shipman SL, Shipman SL, Nivala J, Macklis JD, Church GM. Molecular recordings by directed CRISPR spacer acquisition. Science. 2016;1175:1–16. doi: 10.1126/science.aaf1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 Is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:1–13. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karvelis T, Gasiunas G, Young J, Bigelyte G, Silanskas A, Cigan M, Siksnys V. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16:253. doi: 10.1186/s13059-015-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, Aryee MJ, Joung JK. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016:1–19. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 74.Boyle EA, Andreasson JOL, Lauren M, Sternberg SH, Wu MJ, Chantal K, Doudna JA, Greenleaf WJ. High-throughput biochemical profiling reveals Cas9 off- target binding and unbinding heterogeneity. bioRxiv. 2016 doi: 10.1101/059782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33:1–7. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anders C, Bargsten K, Jinek M. Structural plasticity of PAM recognition by engineered variants of the RNA-guided endonuclease Cas9. Mol Cell. 2016;61:895–902. doi: 10.1016/j.molcel.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, Liu H, Li N, Zhang B, Yang D, Ma C, Wang S, Wu D, Ma Y, Fan S, Wang J, Gao N, Huang Z. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016:1–16. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- 79.Nishimasu H, Cong L, Winston X, Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y. Crystal structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jackson RN, Golden SM, van Erp PBG, Carter J, Westra ER, Brouns SJJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Datsenko Ka, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 85.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Keith Joung J. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2015;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boudry P, Semenova E, Monot M, Datsenko KA, Lopatina A, Sekulovic O, Ospina-Bedoya M, Severinov L-CFK, Dupuy B, Soutourinaa O. Function of the CRISPR-Cas System of the human pathogen Clostridium difficile. MBio. 2015;6:1–16. doi: 10.1128/mBio.01112-15.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O’Toole GA. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crooks G, Hon G, Chandonia J, Brenner S. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004.1. [DOI] [PMC free article] [PubMed] [Google Scholar]