SUMMARY
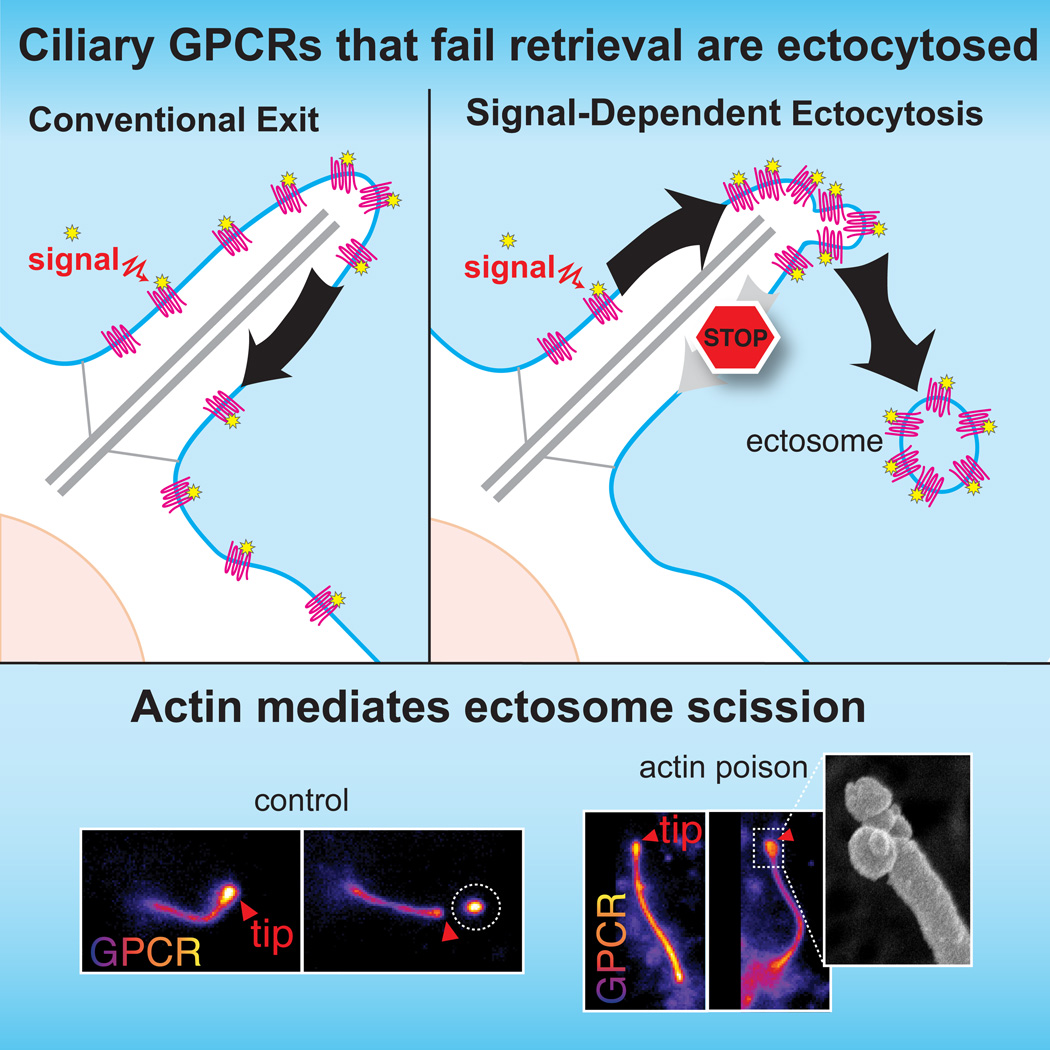
Signaling receptors dynamically exit cilia upon activation of signaling pathways such as Hedgehog. Here we find that when activated G protein coupled receptors (GPCRs) fail to undergo BBSome-mediated retrieval from cilia back into the cell, they concentrate into membranous buds at the tips of cilia before release into extracellular vesicles named ectosomes. Unexpectedly, actin and the actin regulators drebrin and myosin 6 mediate ectosome release from the tip of cilia. Mirroring signal-dependent retrieval, signal-dependent ectocytosis is a selective and effective process that removes activated signaling molecules from cilia. Congruently, ectocytosis compensates for BBSome defects as ectocytic removal of GPR161, a negative regulator of Hedgehog signaling, permits the appropriate transduction of Hedgehog signals in Bbs mutants. Finally, ciliary receptors that lack retrieval determinants such as the anorexigenic GPCR NPY2R undergo signal-dependent ectocytosis in wild-type cells. Our data show that signal-dependent ectocytosis regulates ciliary signaling in physiological and pathological contexts.
Graphical Abstract
INTRODUCTION
The cilium is a compartment 5,000 times smaller than the rest of the cell that concentrates receptors for Hedgehog signaling, vision, olfaction and body weight homeostasis (Schou et al., 2015). By maintaining its own concentrations of the second messengers cAMP and Ca2+, the cilium provides unique reaction conditions to signaling molecules that dynamically enter and exit cilia upon pathway activation (Nachury, 2014). For example, activation of the Hedgehog pathway triggers the ciliary exit of the G Protein Coupled Receptor (GPCR) GPR161 and the resulting decrease in ciliary cAMP levels facilitates subsequent signaling steps (Mukhopadhyay et al., 2013). Signal-dependent ciliary exit extends to several phototransduction molecules (Calvert et al., 2006) and to the GPCRs Somatostatin Receptor 3 (SSTR3) and Neuropeptide Y Receptor 2 (NPY2R) (McIntyre et al., 2016). Yet, the functional importance of exit to the appropriate transduction of signals remains controversial (Kim et al., 2015; Yue et al., 2014).
The conventional pathway for retrieval of activated GPCRs from cilia back into the cell relies on the conformational sensor β-arrestin 2 and on the BBSome, a complex of Bardet-Biedl Syndrome (BBS) proteins that forms a membranous coat with the Arf-like GTPase Arl6/Bbs3 (Jin et al., 2010; Lechtreck et al., 2013; McIntyre et al., 2016; Pal et al., 2016). Meanwhile, ectocytosis has recently emerged as an alternative ciliary exit route in the unicellular green alga Chlamydomonas (Cao et al., 2015; Wood et al., 2013). During ectocytosis, the localized outward curving of ciliary or plasma membranes buds an extracellular vesicle (EV) termed ectosome or microvesicle (Colombo et al., 2014; Wood and Rosenbaum, 2015). While ciliary ectocytosis might represent a specialization of Chlamydomonas where the cilium bears the only membrane exposed to the extracellular environment, analysis of mammalian and nematode EVs has detected several proteins found in part in cilia, in particular the polycystic kidney disease (PKD) proteins fibrocystin/PKHD1 and Polycystin-2/PKD2 and the protrusion-specific protein prominin (Wood and Rosenbaum, 2015). However, EVs can also originate from multivesicular body secretion and this type of EVs (termed exosomes) may deliver proteins to cilia upon fusion with the ciliary membrane. Membranous buds associated with cilia could thus represent either exosomes fusing with cilia or ectosomes budding from cilia (Wang and Barr, 2016). Given the multiple hypotheses for the origin of cilia-related EVs in worms and mammals, the evolutionary conservation of ciliary ectocytosis remains to be determined. Similarly, the regulatory triggers and molecular mechanisms of ciliary ectocytosis as well as the interplay between retrieval and ectocytosis are largely unknown. Finally, the range of functions carried out by EVs remains an open question. While EVs were initially thought to represent a form of cellular disposal (Pan et al., 1985), this hypothesis has been largely supplanted by the idea that EVs carry information across cells (Desrochers et al., 2016), as echoed by reports of bioactivity in ciliary ectosomes and cilia-related EVs (Cao et al., 2015; Wang et al., 2014; Wood et al., 2013).
While imaging the signal-dependent removal of GPCRs from mammalian cilia in real time, we found that activated GPCRs become concentrated at the ciliary tip and ectocytosed when retrieval fails. Signal-dependent ciliary ectocytosis is a remarkably efficient and specific means to remove activated GPCRs from cilia. Surprisingly, a defined actin-regulated machinery triggers ectosome release from the tip of cilia. Functionally, ciliary ectocytosis is required for the appropriate regulation of Hedgehog signaling in retrieval mutants. Our results show that signal-dependent ectocytosis mirrors receptor-mediated endocytosis as an evolutionarily conserved process that removes activated receptors from the cell surface to alter signaling processes.
RESULTS
Failure to retrieve GPCRs from cilia results in ectocytosis from the tip
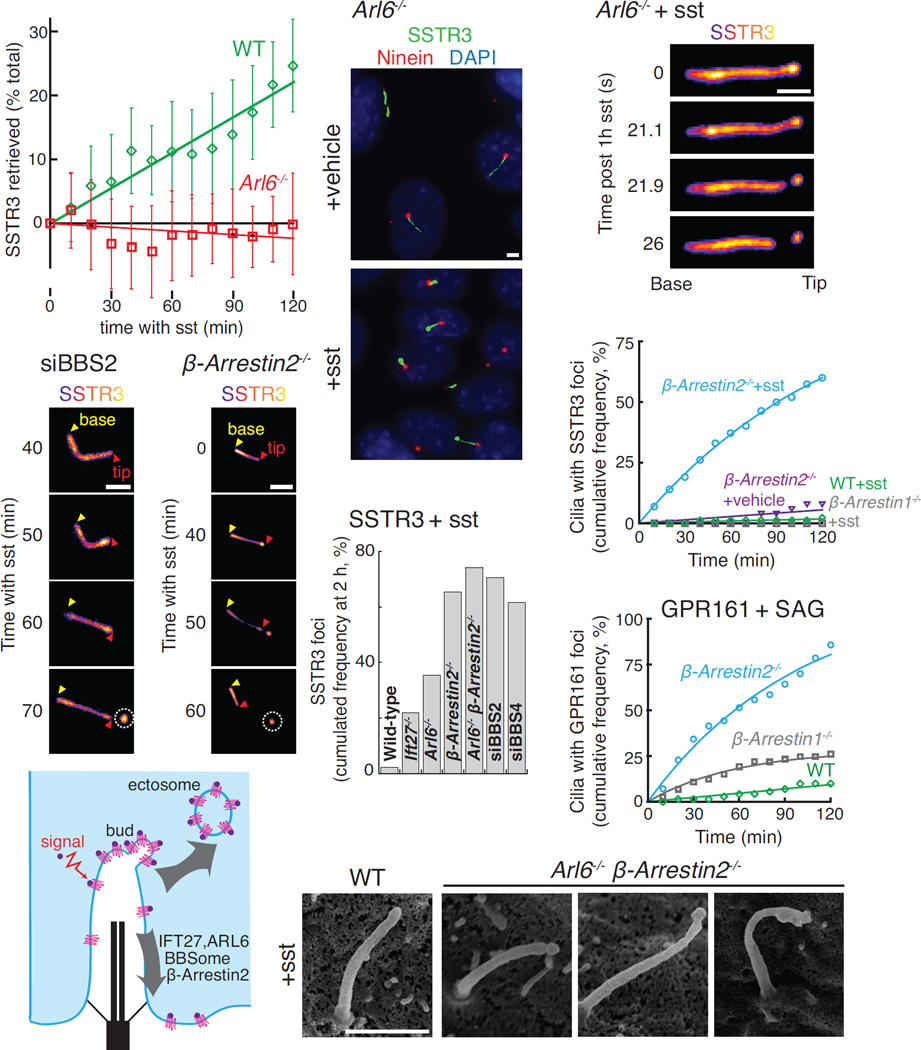
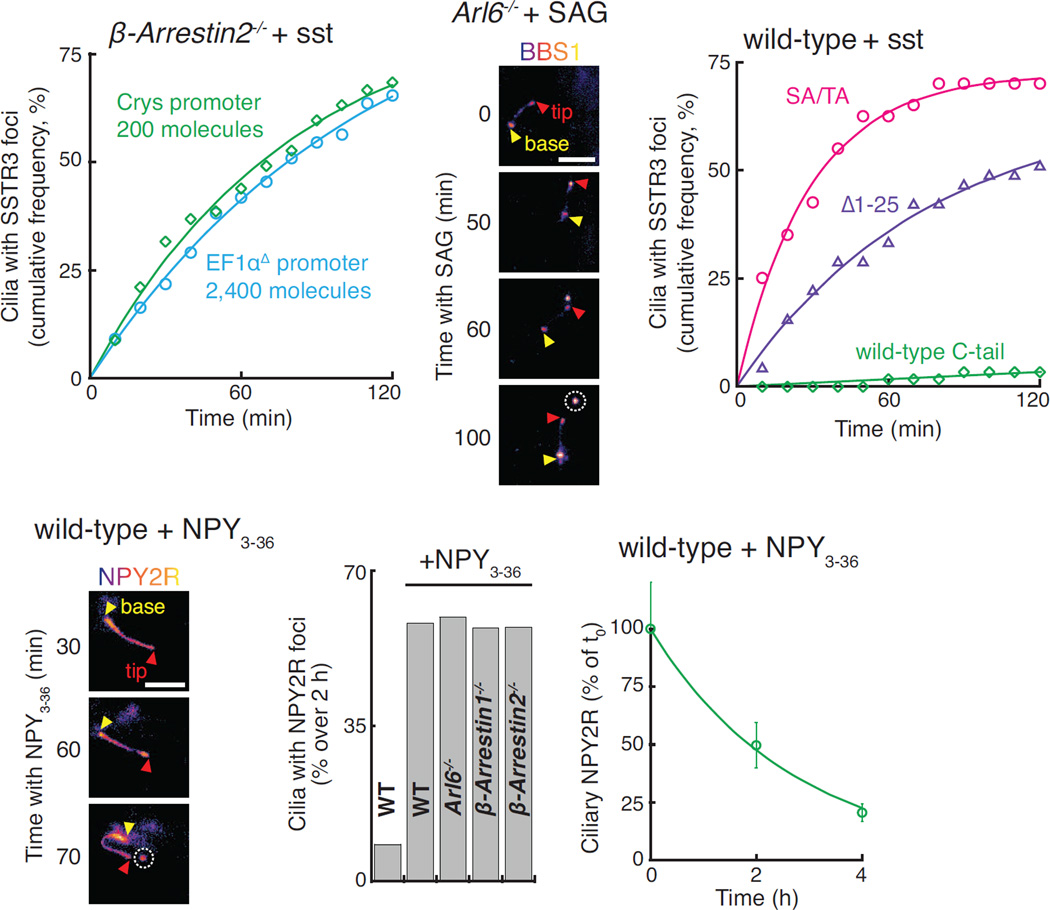
SSTR3 is a well-characterized Gαi-coupled receptor that localizes to cilia of hippocampal neurons and undergoes agonist-dependent exit (McIntyre et al., 2016). By leveraging endogenous-strength promoters, we reconstituted the agonist-dependent exit of SSTR3 in Inner Medullar Collecting Duct (IMCD3) kidney cells (Movie S1). SSTR3 was imaged either with an intracellular NeonGreen tag or through an extracellular fusion to a biotinylated acceptor peptide (AP) revealed with fluorescently labelled monovalent streptavidin (SA647). This system allowed us to demonstrate that the BBSome/Arl6 coat is required for agonist-dependent retrieval of SSTR3 from cilia (Fig. 1A), consistent with the BBSome requirement for Hedgehog-dependent exit of GPR161 from cilia (Eguether et al., 2014; Liew et al., 2014). Surprisingly, in the course of imaging Arl6−/− cells treated with somatostatin (sst), we noticed that the failure to retrieve SSTR3 resulted in a striking accumulation of SSTR3 at the cilia tips (Fig. 1B). Live imaging revealed that the tip foci of SSTR3 rapidly underwent scission, resulting in the release of EVs p ackaged with SSTR3 (Fig. 1C, S1A and Movie S2). These SSTR3 EVs clearly resided outside of the cell as they moved freely and diffused away from the cilium into the medium (Movie S2). Signal-dependent accumulation of activated SSTR3 at the tips and subsequent ectocytosis were observed in all instances of failed retrieval including cells depleted of the BBSome subunits BBS2 and BBS4 (Fig. 1D and F) or cells lacking β-arrestin2 (Fig. 1E) or the BBSome regulators Arl6 and Ift27 (Fig. 1C and 1F, S1A–B, and Movie S3). Under the same conditions, very little tip concentration or ectocytosis of SSTR3 was observed in wild-type cells (Fig. 1F, and Movie S1).
Figure 1. Un-retrieved GPCRs are ectocytosed.
(A) ARL6 is necessary for retrieval of activated SSTR3. APSSTR3NG was stably expressed in IMCD3 cells under the control of an EF1α promoter lacking the TATA box (pEF1αΔ). APSSTR3NG was pulse-labeled with SA647 before addition of sst to WT and Arl6−/− cells and fluorescence intensity was tracked in individual cilia. Ectocytic-based removal of SSTR3 was not counted in order to measure only retrieval (see STAR Methods for details). Data were linearly fitted. Error bars: SD. (n=14–35 cilia)
(B) Activated SSTR3 accumulates at the ciliary tip in Arl6−/− cells. IMCD3-[APSSTR3NG];Arl6−/− cells were treated with vehicle or sst for 6 h, fixed, and immunostained for ninein to mark the base of the cilium.
(C) Release of a SSTR3-rich ectosome from the cilium tip. SA647-labeled IMCD3-[APSSTR3];Arl6−/− cells were treated with sst for 1 h and then imaged at 1.21 Hz.
(D–E) SSTR3 ectocytosis from BBS2-depleted (D) and β-arrestin2−/− knockout (E) cells. IMCD3-[APSSTR3NG] cells were imaged in the NeonGreen channel every 10 min following addition of sst. In all subsequent time series, yellow and red triangles point to the base and tip of cilia respectively and the ectosome is circled.
(G) SSTR3 foci formation in various retrieval mutants and knockdowns. Data was acquired as in panels D and E and the cumulated foci frequency over 2 h was plotted. (n=50–87 cilia)
(H) Formation of SSTR3-enriched tip foci requires active signaling and defective retrieval. IMCD3-[APSSTR3NG] cells were imaged every 10 min following addition of sst or vehicle and tip foci were scored in the NeonGreen channel. Data were fitted to a simple exponential. (n=50–76 cilia).
(I) Activation of the Hedgehog pathway in β-arrestin2−/− cells leads to the formation of GPR161 tip foci (see also Fig. 3A). IMCD3-[pCrys-APGPR161NG3] cells, either wild-type or knockout for β-Arrestin 1 or 2, were imaged following addition of SAG and scored and fitted as in panel (G). (n=71–73 cilia).
(J) Model illustrating the competing removal modalities of activated GPCRs.
(K) Scanning EM reveals buds at the ciliary tip in retrieval-defective cells. WT or Arl6−/− β-arrestin2−/− IMCD3-[APSSTR3NG] cells were treated with sst for 1 h before fixation. Additional examples and counting statistics are in Fig. S1C–D. All scale bars are 2 µm.
The pre-ectocytic concentration of SSTR3 at the ciliary tip was dependent on receptor activation. Within the span of 2 h, SSTR3 accumulations at the tips of cilia were seen in more than two-thirds of β-arrestin2−/− cilia treated with sst but in less than 5% of vehicle-treated β-arrestin2−/− cilia (Fig. 1G). Similarly, GPR161 becomes concentrated at the tips of cilia and undergoes ectocytosis when retrieval mutants are treated with the Hedgehog pathway agonist SAG. Activation of the Hedgehog pathway in β-arrestin2−/− cells led to the widespread production of ectosomes highly enriched for GPR161 with over 85% of cilia producing a tip focus after 2 h (Fig. 1H and Movie S4). Thus, activation of ciliary signaling receptors in retrieval-incompetent cells drives ectosome formation and we propose that ciliary retrieval and ectocytosis represent competing processes to remove activated signaling receptors from cilia (Fig. 1I).
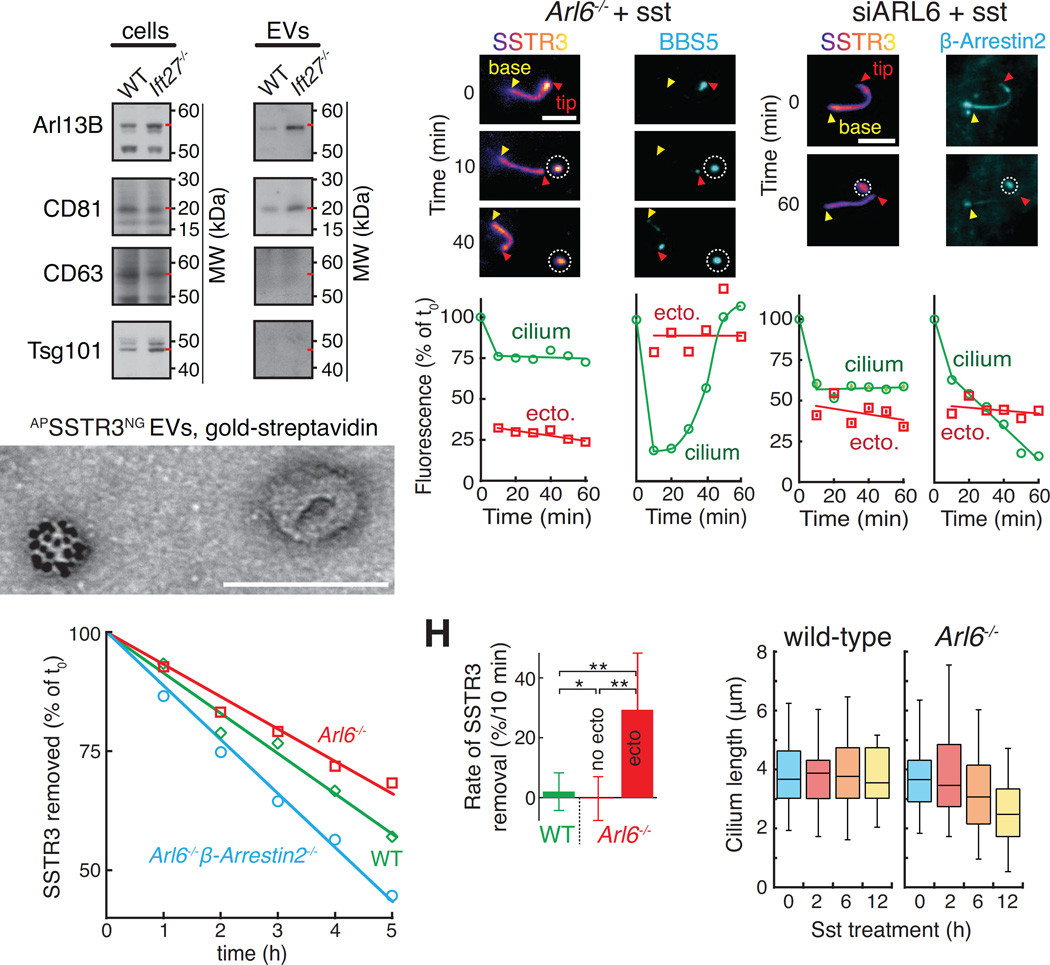
Scanning electron microscopy (EM) of retrieval mutant cells treated with sst revealed distinct tip structures with small and large buds often displaying a constriction at their base suggestive of scission in progress (Fig. 1J and S1C). Importantly, no buds were observed in wild-type cells (Fig. 1J and S1D). To follow the fate of ectocytosed GPCRs, we generated EV preparations from the supernatant of serum-starved IMCD3 cells (Fig. S2A). These EVs were positive for the cilia-localized proteins Arl13B and CD81 but negative for the exosome marker CD63 (Kowal et al., 2016) and the ESCRT-1 protein Tsg101 (Nabhan et al., 2012) (Fig. 2A). Upon stimulation of ciliary ectocytosis, streptavidin-gold detection of GPCRs tagged with an extracellular AP tag reveled a small number of EVs packaged with SSTR3 or GPR161 (Fig. 2B and S2B). No staining was detected without expression of AP-tagged GPCRs. (Fig. S2C and E). The diameter of ciliary EVs was just over 100 nm, a typical value for small EVs (Fig. S2D and (Kowal et al., 2016)). We conclude that retrieval mutant cells release ciliary EVs packaged with GPCRs.
Figure 2. Efficiency of ciliary ectocytosis.
(A–B) Characterization of EV preparations from ciliated cells. (A) Immunoblots of EVs purified from the culture supernatant of IMCD3 cells treated with SAG for 16h. Red dashes point to the known molecular weights of each antigen. Lysate and EV lanes contained 1 and 100 cell equivalents, respectively. The CD63 and Tsg101 blots of EV fractions were exposed longer to emphasize the absence of bands. (B) Imaging of EVs reveals a population of vesicles packaged with ciliary GPCRs. EVs from sst-stimulated IMCD3-[APSSTR3NG]; β-arrestin2−/− cells were labeled with streptavidin-conjugated gold particles and imaged by negative stain EM. See Fig. S2A for diagram and S2B–C for controls and additional examples. Scale bar: 100 nm.
(C–D) SSTR3 and the BBSome are lost by ectocytosis. (C) A representative cilium from a sst-treated and SA647-labeled IMCD3-[APSSTR3,NG3BBS5];Arl6−/− cell was tracked for 1 h following ectosome release. The ectosome is circled. Fluorescence levels for the cilium and ectosome are quantified in (D). No photobleaching corrections were applied. Scale bar: 4 µm.
(E–F) Significant amounts of ciliary β-Arrestin 2 are lost by ectocytosis. (E) SA647-labeled Arl6-depleted IMCD3-[APSSTR3, β-Arrestin2GFP] cells were imaged every 10 min for the 2 h following addition of sst. Fluorescence levels for the cilium and ectosome are quantified in (F). No photobleaching corrections were applied. Scale bar: 4 µm.
(G) APSSTR3NG was pulse-labeled with SA647 before addition of sst to WT, Arl6−/−, and Arl6−/−/β-arrestin2−/− IMCD3-[APSSTR3NG] cells and tracking of fluorescence in individual cilia to measure removal. Data were linearly fitted. (n=27–33 cilia).
(H) Substantial amounts of SSTR3 are lost in each round of ectocytosis. For all cilia tracked in Fig. 1A, we systematically measured the decrease in ciliary SSTR3 fluorescence between two successive frames (10 min frame rate) to calculate the instantaneous removal rate. For Arl6−/− cells, we separately analyzed transitions were no ectosome release event occurred (no ecto) from those where an ectocytosis event was detected (ecto). See STAR Methods for details. Error bars: SD. n= 167 for WT, 369 for Arl6−/− no ecto and 23 for Arl6−/− ecto. * = p<2×10−4, and ** = p<2×10−8 (Student’s t test).
(I) Pervasive ectocytosis results in cilia shortening. The length of cilia in IMCD3-[APSSTR3NG] WT or Arl6−/− cells was tracked for 12 h following addition of sst by imaging the NeonGreen channel. The complete time course is shown in Fig. S3A. (n=45–62 cilia).
Ectocytosis results in significant losses of ciliary material
To gain insights into the possible roles of signal-dependent ectocytosis, we estimated the quantities of ciliary material lost by ectocytosis. In a representative event from an Arl6−/− cell, more than 30% of ciliary SSTR3 was lost through the release of a single tip focus (Fig. 2C and D, and Movie S3). Coincidental with SSTR3 ectocytosis, substantial amounts of the BBSome (Fig. 2C and D) and β-arrestin 2 were lost (Fig. 2E and F and Movie S3). Thus, the retrieval machinery is efficiently co-ectocytosed with the cargoes it fails to retrieve.
Remarkably, ectocytosis removed SSTR3 from Arl6−/− cilia with a similar efficiency as retrieval in wild-type cells (Fig. 2G). However, ectocytosis and retrieval differ markedly in their instantaneous behavior. Retrieval is a steady and fairly inefficient process, removing 2‰ of ciliary SSTR3 per min (Fig. 2H). Meanwhile, ectocytosis is an extremely efficient but rather infrequent process. On average, sst-treated Arl6−/− cells experienced 0.6‰ ectocytosis event per min but each ectocytosis event removed 31% of ciliary SSTR3 (Fig. 2H), yielding an aggregate removal rate of 1.9‰ per min.
It is notable that ectocytosis removes ciliary material with sufficient magnitude to result in a progressive decrease of cilia length in Arl6−/− cells treated with sst (Fig. 2I and S3A) and a drastic increase in cilium length variability (Fig. S3B). Thus, while signal-dependent ciliary ectocytosis is an effective disposal modality, it consumes more ciliary material than retrieval.
Ectocytosis results in selective losses of ciliary material
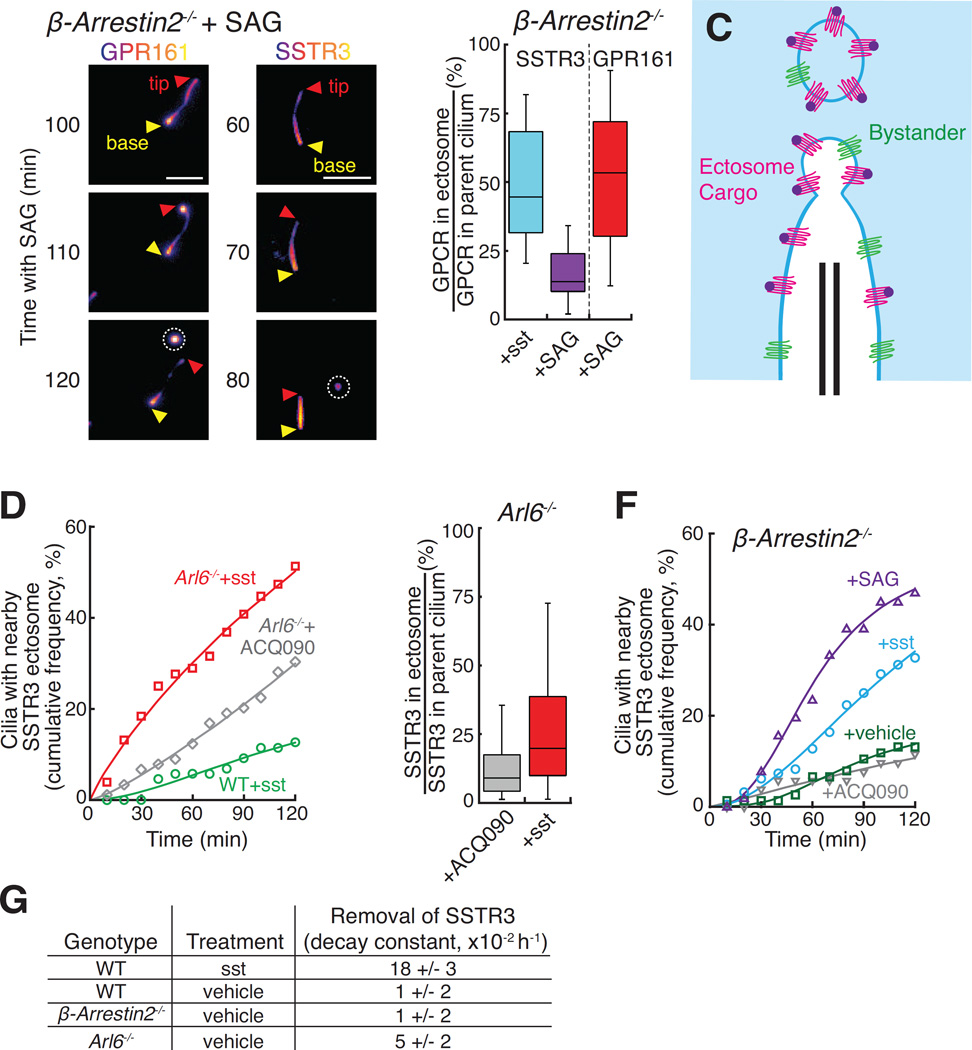
The shortening of Arl6−/− cilia under signaling conditions suggested that ectocytosis may indiscriminately remove ciliary proteins. To directly assess the specificity of ectocytosis, we measured efficiencies for off-pathway and on-pathway packaging of GPR161 and SSTR3 into ectosomes. On-pathway packaging was extremely efficient: Hedgehog pathway activation in β-arrestin2−/− cells generated ectosomes containing 51% of ciliary GPR161 (Fig. 3A and B, S3C, and Movie S4), and sst addition to β-arrestin2−/− cells produced ectosomes containing 49% of ciliary SSTR3 (Fig. 1D and 3B, S3D, and Movie S3). Consistent with a high packaging efficiency for specific cargoes, we calculate that the membrane density of SSTR3 is at least 5-fold higher in ectosomes than in the cilium (see Methods). By contrast, only 16% of ciliary SSTR3 found its way into SAG-induced ectosomes (Fig. 3A and B, S3D, and Movie S4). Thus, activated receptors are actively concentrated into ectosomes and ciliary membrane proteins that are not excluded from the forming ectosome undergo a low level of ectocytosis (Fig. 3C).
Figure 3. Specificity of ciliary ectocytosis.
(A–B) Comparison of on- and off-pathway ectocytosis. (A) β-arrestin2−/− IMCD3-[APGPR161NG3] and β-arrestin2−/− IMCD3-[APSSTR3NG] cells were tracked in the NeonGreen channel following Hh pathway activation. GPR161 but not SSTR3 is highly enriched in ectosomes compared to cilia. Ectosomes are circled. Scale bar: 4 µm. (B) The NeonGreen intensity of ectosomes observed in experiments presented in Fig. 3A and 1E was quantified and normalized to the total NeonGreen intensity of the parent cilium. (n=24 ectosomes).
(C) Diagram illustrating how activated GPCRs (red) are enriched in ectosomes while bystander proteins (green) undergo bulk flow ectocytosis.
(D) Arl6−/− cells undergo both signal-dependent and constitutive ectocytosis. SA647-labeled WT or Arl6−/− IMCD3-[APSSTR3NG] cells were imaged following addition of sst or antagonist (ACQ090). The number of cilia with a nearby SSTR3 focus was scored. Data was fit to a Hill equation of no theoretical significance. (n=76–89 cilia).
(E) Packaging efficiencies of constitutive and signal-dependent ectocytosis. Ectosomes observed in experiments presented in Fig. 3D were analyzed following the procedure used in Fig. 3B. (n=38–43 ectosomes).
(F) Signal-dependent ectocytosis from β-arrestin2−/− cells. Ectosomes from SA647-labeled IMCD3-[APSSTR3NG]; β-arrestin2−/− cells were counted following addition of sst, SAG, ACQ090, or vehicle. Analysis and fitting was as in panel (D). (n=51–141 cilia).
(G) Constitutive ectocytosis removes measurable levels of ciliary proteins. APSSTR3NG was pulse-labeled by SA647 and analyzed by fixed imaging after 6 or 9 h treatment with sst, vehicle, or SAG (see Fig. S3F). Data were fit to an exponential decay and the rate constants for SSTR3 removal are shown in the table. (n=222–327 cilia measured per rate constant).
The selectivity of ectocytosis was also seen when comparing constitutive and signal-dependent ectocytosis. In Arl6−/− cells, we observed some ectocytosis even when SSTR3 was kept inactive by addition of the SSTR3 antagonist ACQ090 (Fig. 3D). This signal-independent ectocytosis is likely to reflect the existence of ciliary proteins that are constitutively cleared from cilia by the BBSome – the viral receptor CAR (Mick et al., 2015), Syntaxin 3 and Syntaxin BP1 (Datta et al., 2015) or phospholipase D (Lechtreck et al., 2013) – and become ectocytosed in Bbs mutants. Nevertheless, constitutive ectocytosis is neither as pervasive nor as efficient as signal-dependent ectocytosis as SSTR3 activation enhanced ectosome production in Arl6−/− cells beyond the constitutive level (Fig. 3D) and ectosomes released from sst-stimulated cells contained twice as much SSTR3 as ectosomes released from antagonist-treated cells (Fig. 3E). Meanwhile, SSTR3-positive ectosomes were only rarely released from β-arrestin2−/− cilia in the absence of sst or SAG stimulation (Fig. 3F), likely reflecting a specific function of β-arrestin 2 in the retrieval of activated GPCRs but not of other BBSome cargoes. Similar to the SSTR3-poor ectosomes released from SAG-treated β-arrestin2−/− cells (Fig. 3B), the few ectosomes released from wild-type cells (Fig. 3D) contained very low levels of SSTR3 (Fig. S3E). In conclusion, inactive SSTR3 can become incorporated into constitutive ectosomes as a bystander rather than a packaged cargo.
In this context, it is worth noting that the ciliary levels of SSTR3 decreased even in the absence of sst addition in Arl6−/− cells but remained constant in wild-type and β-arrestin2−/− cells (Fig. 3G and S3F). Constitutive ectocytosis thus removes measurable amounts of bystander proteins from cilia, leading to the counter-intuitive conclusion that ciliary retrieval defects can result in the loss rather than the accumulation of ciliary proteins.
Ectocytosis is not limited to pathological or artificial settings
The concern that ciliary membrane protein overexpression might be responsible for ectocytosis was ruled out because SSTR3 tip foci were observed in sst-treated β-arrestin2−/− cells expressing from 2,400 (near-endogenous) down to 200 molecules of SSTR3 per cilium (Fig. 4A). Moreover, even in the absence of any transgene encoding a ciliary membrane protein, SAG-dependent ectosomes were clearly detected in Arl6−/− cells by their packaging of BBSome (Fig. 4B).
Figure 4. NPY2R exits wild-type cilia by ectocytosis.
(A) β-arrestin2−/− cells expressing APSSTR3NG3 under the control of the δ-crystallin (Crys) or pEF1αΔ-APSSTR3NG were imaged in the NeonGreen channel every 10 min for the 2 h following addition of sst. The number of molecules in cilia was determined using a fluorescently-tagged virus calibrator (see STAR methods). SSTR3 tip foci were scored and analyzed as in Fig. 1G. (n=55–57 cilia).
(B) IMCD3-[NG3BBS1];Arl6−/− cells were imaged every 10 min for the 2 h following addition of SAG. Ectocytosis is evidenced by release of a NG3BBS1 focus as in Fig. 2C.
(C) SSTR3 variants lacking retrieval determinants are ectocytosed from WT cells. IMCD3 cells expressing variants of APSSTR3 (described in Fig. S4A) were pulse-labeled with SA647 and imaged every 10 min for the 2 h following addition of sst. SSTR3 tip foci were scored and analyzed as in Fig. 1G. (n=40–60 cilia).
(D–F) NPY2R is efficiently ectocytosed from WT cells. (D) IMCD3-[pEF1αΔ-NPY2RNG] cells were imaged every 10 min following treatment with the agonist NPY3–36. All scale bars are 4 µm. (E) The cumulated frequency of tip foci was scored by imaging IMCD3-[NPY2RNG] cell lines every 10 min for 2 h following addition of NPY3–36. (n=28–79 cilia). (F) IMCD3-[NPY2RNG] cells were pre-treated with the translation inhibitor emetine to prevent the appearance of newly synthesized NPY2RNG in cilia. Cells were then treated with NPY3–36 for 0, 2, or 4 h before fixation and quantitation of ciliary NeonGreen. Error bars: SEM. (n=30–54 cilia).
Secondly, since we did not observe substantial ectocytosis of GPR161 or SSTR3 in wild-type cells, the possibility remained that ciliary ectocytosis represented a safety valve whose opening was limited to pathological cases of failed retrieval. To determine whether ectocytosis can take place in wild-type cilia, we took advantage of SSTR3Δ1–25 and SSTR3SA/TA, two SSTR3 variants unable to interact with β-arrestin (Roth et al., 1997) and that fail signal-dependent ciliary retrieval (Fig. S4A). Remarkably, SSTR3SA/TA and SSTR3Δ1–25 were efficiently ectocytosed upon sst treatment of wild-type cells (Fig. 4C). Thus, GPCR variants that lack retrieval determinants undergo signal-dependent ectocytosis from wild-type cilia. This finding predicts that endogenous ciliary GPCRs that lack recognition features for the BBSome and β-arrestin will be ectocytosed in wild-type cells upon activation. The Neuropeptide Y Receptor 2 (NPY2R), a ciliary GPCR that participates in the hypothalamic regulation of feeding behavior, binds poorly to β-arrestin and lacks a canonical BBSome-binding motif in its third intracellular loop (Loktev and Jackson, 2013; Marion et al., 2006). As predicted, addition of the NPY2R agonist Neuropeptide Y 3–36 (NPY3–36) to wild-type IMCD3 cells led to ectocytosis of NPY2R (Fig. 4D, S4B and Movie S5). Interfering with BBSome or β-arrestin function did not increase NPY2R ectocytosis (Fig. 4E and S4C), strongly suggesting that the BBSome and β-arrestin do not participate in NPY2R removal from cilia and that ectocytosis represents the sole means of removing NPY2R from cilia. Importantly, the kinetics of ectocytosis-mediated NPY2R exit from cilia of IMCD3 cells (Fig. 4F) were similar to those of endogenous NPY2R exiting hypothalamic neuron cilia (Loktev and Jackson, 2013) and faster than the retrieval-mediated exit of SSTR3 from wild-type cilia (Fig. 1A). Signal-dependent ectocytosis is thus not limited to the pathological context of retrieval mutants and efficiently removes activated signaling receptors from cilia in physiological settings such as NPY2R and the ciliary adhesion receptor SAG1 in mating Chlamydomonas gametes (Cao et al., 2015).
F-actin is required for ectocytosis but dispensable for retrieval
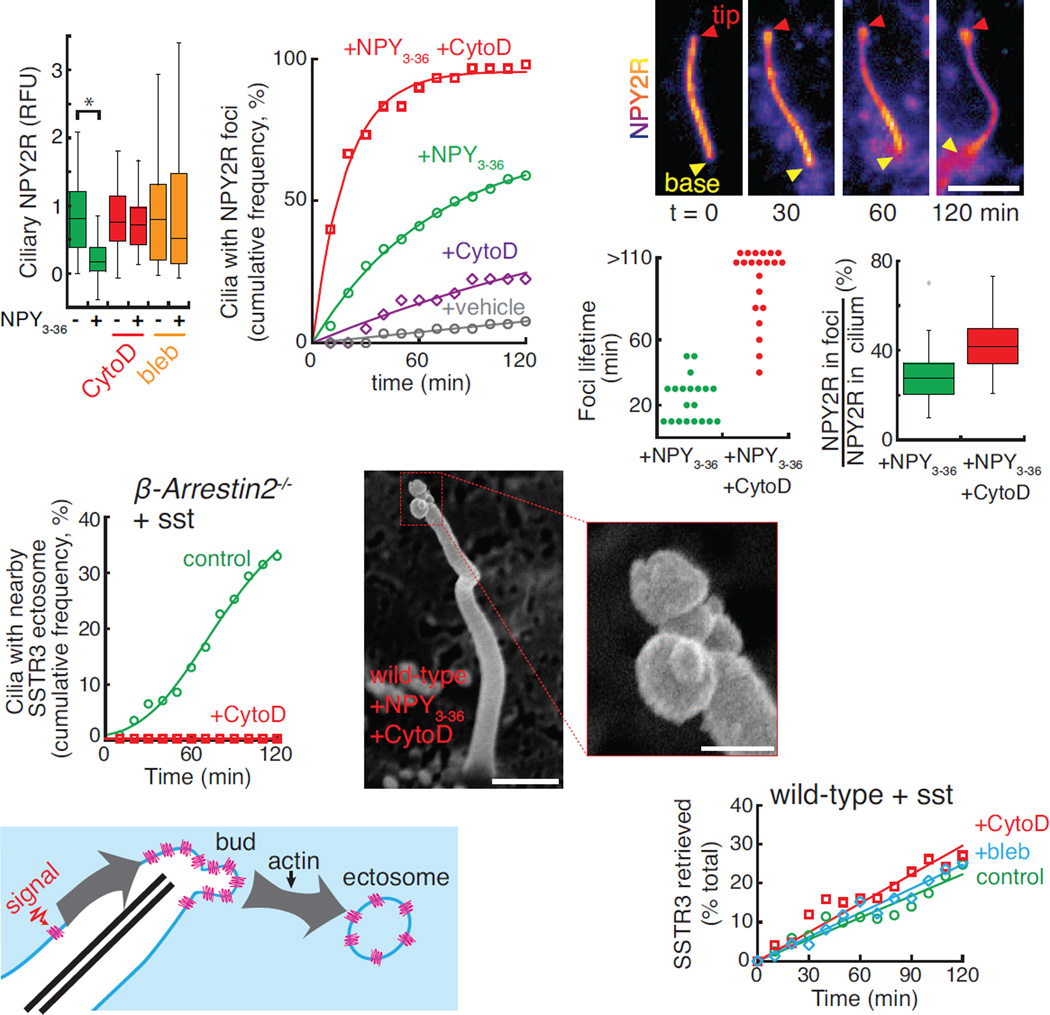
The existence of physiological cargoes for signal-dependent ectocytosis and the coordinated nature of the process suggested that specific molecular machines execute ectocytosis. Our recent ciliary proteomics profiling of the retrieval mutant Ift27−/− (Mick et al., 2015) revealed several actin regulators enriched in the proteome of retrieval-incompetent cilia, thus hinting at a possible role for actin in ciliary ectocytosis. Remarkably, removal of activated NPY2R from cilia was abolished by the myosin inhibitor blebbistatin or by low doses of the actin poison cytochalasin D (Fig. 5A). The step blocked by actin poisons was downstream of tip accumulation as nearly every cilium experienced a tip focus of NPY2R after 1 h in the presence of cytochalasin D and NPY3–36 (Fig. 5B). Congruently, instead of rapidly detaching from cilia (Fig. 4D), NPY2R tip foci remained at the ciliary tips for extended times in the presence of cytochalasin D (Fig. 5C). Whereas NPY2R tip foci normally lasted for 10 to 30 min before ectocytic release, addition of cytochalasin D led to the persistence of NPY2R tip foci for over 100 min (Fig. 5D and Movie S6). Consistent with a blockage at the step of ectosome release, the amount of NPY2R accumulating in buds was significantly increased by actin poisoning (Fig. 5E). Similarly, sst-induced ectocytosis of SSTR3 from β-arrestin2−/− cilia was abolished by cytochalasin D (Fig. 5F) and Arl6−/− cells treated with cytochalasin D and sst accumulated SSTR3-rich foci with lifetimes of several hours (Fig. S4D). Most strikingly, scanning EM demonstrated the accumulation of partially pinched buds emanating from the tips of cilia in cells treated with cytochalasin D (Fig. 5G). Collectively, these results indicate that actin polymerization is required for a late step in the release of ectosomes from the ciliary tip, most likely vesicle scission (Fig. 5H).
Figure 5. Ectosome release requires F-actin.
(A) NPY2R exit requires actomyosin activity. Emetine-pretreated IMCD3-[NPY2RNG] cells were treated with NPY3–36 or vehicle and cytochalasin D (CytoD) or blebbistatin (bleb) for 4 h before fixation and quantitation of ciliary NeonGreen as in Fig. 4F. * indicates p<0.01 (Mann-Whitney U test). (n=30–67 cilia).
(B) Actin poisoning causes an increased incidence of NPY2RNG tip foci. NPY2RNG tip foci were scored and analyzed as in Fig. 1G. (n=30–47 cilia).
(C–E) Cytochalasin D stabilizes NPY2RNG tip foci. (C) Cytochalasin D-treated IMCD3-[NPY2RNG] cells were imaged every 10 min for 2 h following addition of NPY3–36. A tip focus appears at 30 min and lasts for the remainder of the observation period. (D) Dot plot showing the time between appearance and ectocytosis of NPY2R foci for either NPY3–36 or NPY3–36 plus cytochalasin D-treated cells. Most foci persist beyond the 2 h observation period in the presence of cytochalasin D. (E) The NeonGreen fluorescence of tip foci and cilia were measured and ratioed to plot the fraction of ciliary NeonGreen signal inside the foci. Failure to release ectosomes in cytochalasin D-treated cells results in hyperaccumulation of NPY2R at the tip. (n=20 foci).
(F) SSTR3 ectocytosis is blocked by cytochalasin D. Ectosomes from untreated or cytochalasin D-treated IMCD3-[APSSTR3NG];β-arrestin2−/− cells were counted following addition of sst. Data were fit to a Hill equation of no theoretical significance. (n=55–141 cilia).
(G) Cytochalasin D treatment results in the accumulation of multiple tip buds. IMCD3-[NPY2RNG] cells were treated with cytochalasin D and NPY3–36 for 1 h, fixed, and analyzed by scanning EM. Inset shows a magnification of the tip. Scale bars: 0.5 µm (main panel), 125 nm (inset).
(H) Diagram illustrating the ectocytosis step mediated by actin.
(I) SSTR3 retrieval remains unaffected by blebbistatin or cytochalasin D. SSTR3 removal from wild-type cilia is 95% reliant on retrieval (see STAR Methods). APSSTR3NG was pulse-labeled with SA647 before addition of sst to cells and tracking of fluorescence in individual cilia. (n=14–15 cilia).
One potential caveat of the above experiments is that actin participates in many cellular processes and actin poisons may indirectly impinge on ectocytosis. First, ectocytosis was blocked but cells moved normally when treated with low doses of actomyosin poisons (Fig. S4E). Combined with the observation of ectocytosis on vertical cilia that did not contact other cells (Fig. S4F), these results rule out the hypothesis that ciliary fragments are sheared as cells move and cilia remain stuck to cells or glass. More importantly, SSTR3 retrieval and SSTR3 endocytosis from the plasma membrane were unaffected by actomyosin poisons (Fig. 5I and S4G and H). Thus, it is likely that the observed effects of actin poisons on ectosome scission were direct.
Drebrin and Myosin 6 are specific mediators of ectocytosis
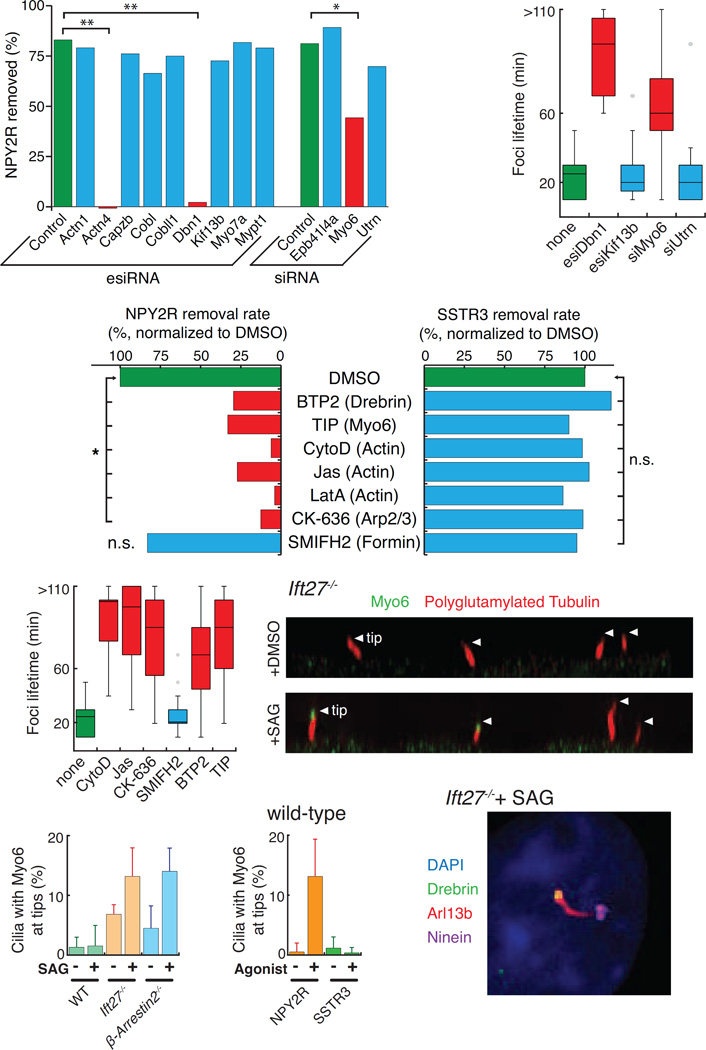
To further determine whether actin plays a direct role in ectosome scission and to gain insights into the mechanisms of ectocytosis, the actin-related hits from the proteomics profiling of Ift27−/− cilia were tested in the NPY2R exit assay (Fig. 4F). Those hits were the actin nucleator cordon-bleu (Cobl) and its paralogue cordon-bleu-like 1 (Cobll1), the microtubule-actin crosslinker drebrin (Dbn1), the unconventional motor myosin 6 (Myo6), the myosin phosphatase targeting subunit Mypt1, the dystrophin-related protein utrophin (Utrn), the band 4.1 like 4A (Epb41l4a), the capping protein Capzb and α-actinin 1 and 4 (Actn1, Actn4). In addition, we tested myosin 7a (Myo7a) which localizes and functions at the photoreceptor connecting cilia (Wolfrum et al., 1998) and the kinesin-3 Kif13b, a binding partner of utrophin and the mammalian orthologue of C.elegans KLP-6 which is required for the release of PKD-positive EVs in nematodes (Wang et al., 2014). While siRNA-mediated depletion of most hits did not affect ciliary removal of NPY2R, depletion of drebrin, myosin 6, or α-actinin 4 significantly diminished NPY2R exit (Fig. 6A and S5A). Congruently, the tip foci lifetime was increased in drebrin and myosin 6-depleted cells but not in utrophin, Kif13b, or control-depleted cells (Fig. 6B). Depletion of drebrin and Kif13b was nearly complete at the protein level and a small amount of myosin 6 protein remained after siRNA treatment (Fig. S5B–D). These data strongly suggest that Kif13b is not required for ciliary exit of NPY2R, leading us to conclude that signal-dependent ciliary ectocytosis in mammalian cells is likely different from the PKD-EV secretion pathway reported in nematodes (Wang et al., 2014).
Figure 6. An actin network mediates EV release at the ciliary tip.
(A) Myosin 6, Drebrin, and α-Actinin 4 are required for NPY2R removal from cilia. IMCD3-[NPY2RNG] cells were depleted of the indicated proteins before measuring the decrease in median ciliary fluorescence of NPY2R upon 4 h treatment with NPY3–36 (see Fig. S5A for detailed distribution). To compare knockdowns, the relative amount of NPY2R lost from cilia was plotted (1 − CiliaryFluorescenceNPY/CiliaryFluorescencevehicle). Multiple regression analysis was used to identify significant decreases in amount of NPY2R removed: * indicates p < 0.02, and ** p < 0.002. (n=72–255 cilia). See STAR Methods for details.
(B) Drebrin and Myosin 6 are necessary for ectosome release. NPY2R foci lifetimes were measured as in Fig. 5D (n=11–31 foci).
(C–D) An actin-regulated machinery is required for ciliary ectocytosis but dispensable for retrieval. (C) GPCR removal was imaged in IMCD3-[NPY2RNG] or IMCD3-[APSSTR3NG] cells treated with 0.5 µM cytochalasin D (CytoD), 15 µM jasplakinolide (Jas), 1 µM latrunculin A (LatA), 1 µM BTP2, 25 µM TIP, 50 µM of the Arp2/3 inhibitor CK-636 or 25 µM of Formin 2 inhibitor SMIFH2 (Fig. S6A–E). Total removal rates were calculated as in Fig. 6A (NPY2R) and Fig. 2G (SSTR3) and normalized to the value for DMSO-treated cells. Statistical tests were as in Fig. 6A (n=123–298 cilia for NPY2R fixed imaging, n=11–14 cilia for SSTR3 live-cell imaging). See STAR Methods for details. (D) NPY2R foci lifetimes were measured as in Fig. 5D. Most foci persist beyond the 2 h observation period in the presence of cytochalasin D, jasplakinolide, CK-636, BTP2, or TIP. (n=20–39 foci).
(E–G) Myosin 6 localizes to the tip of cilia in cells undergoing ectocytosis. WT, Ift27−/−, and β-arrestin2−/− cells were treated with DMSO or SAG for 1 h, fixed, and stained for endogenous myosin 6. (D) x-z projection showing myosin 6 at the tip of upward pointing Ift27−/− cilia in the presence of SAG but not with DMSO. White triangles point to the tips of cilia. (E) shows the frequency of myosin 6 at the tip of cilia for each genotype with or without SAG. (G) compares the frequency of cilia with myosin 6 at the tip in cells expressing NPY2RNG or APSSTR3NG and treated with the respective agonists. Error bars: SD between microscope fields. (n=78–322 cilia).
(H) Drebrin localizes to cilia. Ift27−/− cells were treated with SAG for 1 h, fixed, and stained for endogenous drebrin, Arl13B and ninein. Drebrin localized near the tip, distal to the basal body marker ninein. All scale bars are 4 µm.
The availability of pharmacological agents that target myosin 6 and drebrin made it possible to assess whether the effects of myosin 6 and drebrin depletion on NPY2R exit could be recapitulated by acute chemical interference. Remarkably, acute treatment with the drebrin inhibitor BTP2 (Law et al., 2015) strongly reduced the rates of ectocytosis-mediated NPY2R removal from cilia but did not affect the retrieval-mediated ciliary removal of SSTR3 (Fig. 6C and S6A). Since BTPs have been reported to interfere with store-operated calcium entry, we tested known blockers of store-operated calcium channel and found that they had no effect on NPY2R exit (Fig. S6B). Similarly, the myosin 6 inhibitor 2,4,6-triiodophenol (TIP) (Heissler et al., 2012) greatly reduced ectocytosis-mediated exit without affecting retrieval-mediated exit (Fig. 6C and S6C). The comparison of NPY2R and SSTR3 exit further established the specific role of actin in ectocytosis as three actin poisons acting through different mechanisms all interfered with ectocytosis without affecting retrieval. Finally, consistent with the enrichment of Arp3 in the ciliary proteome of Ift27 mutants (Mick et al., 2015), our chemical screening strategy showed that the Arp2/3 complex is required for ectocytosis but not for retrieval and that formin activity is dispensable for both ectocytosis and retrieval (Fig. 6C and S6D and E). In conclusion, we find that six different drugs targeting specific elements of the actin cytoskeleton all interfere with NPY2R ectocytosis without affecting SSTR3 retrieval. In further support of this conclusion, the lifetime of NPY2R tip foci was significantly increased when cells were treated with small molecules targeting actin, myosin 6, drebrin or the Arp2/3 complex but not when formins were inhibited (Fig. 6D).
The accumulation of uncleaved buds at the tips of cilia treated with cytochalasin D suggested that actin specifically acts at the site of ectosome scission. While the considerable levels of cytoplasmic actin precluded the imaging of actin in cilia, staining for endogenous myosin 6 revealed a clear and specific tip localization in the retrieval mutants Ift27−/− and β-arrestin2−/− but not in wild-type cells (Fig. 6E and S6F). Moreover, activation of the Hh pathway in retrieval mutants increased the number of myosin 6-positive cilia (Fig. 6F). Further linking myosin 6 tip accumulation with ectocytosis, myosin 6 was observed at the tip of wild-type cilia upon activation of NPY2R but not SSTR3 (Fig. 6G). Similarly, endogenous drebrin was found in cilia with a tip-biased distribution (Fig. 6H and S6G). Together, these results establish the existence of a specific actin network that acts at the tips of cilia to mediate ectosome release.
Ectocytosis compensates for retrieval defects to ensure appropriate Gli3 processing
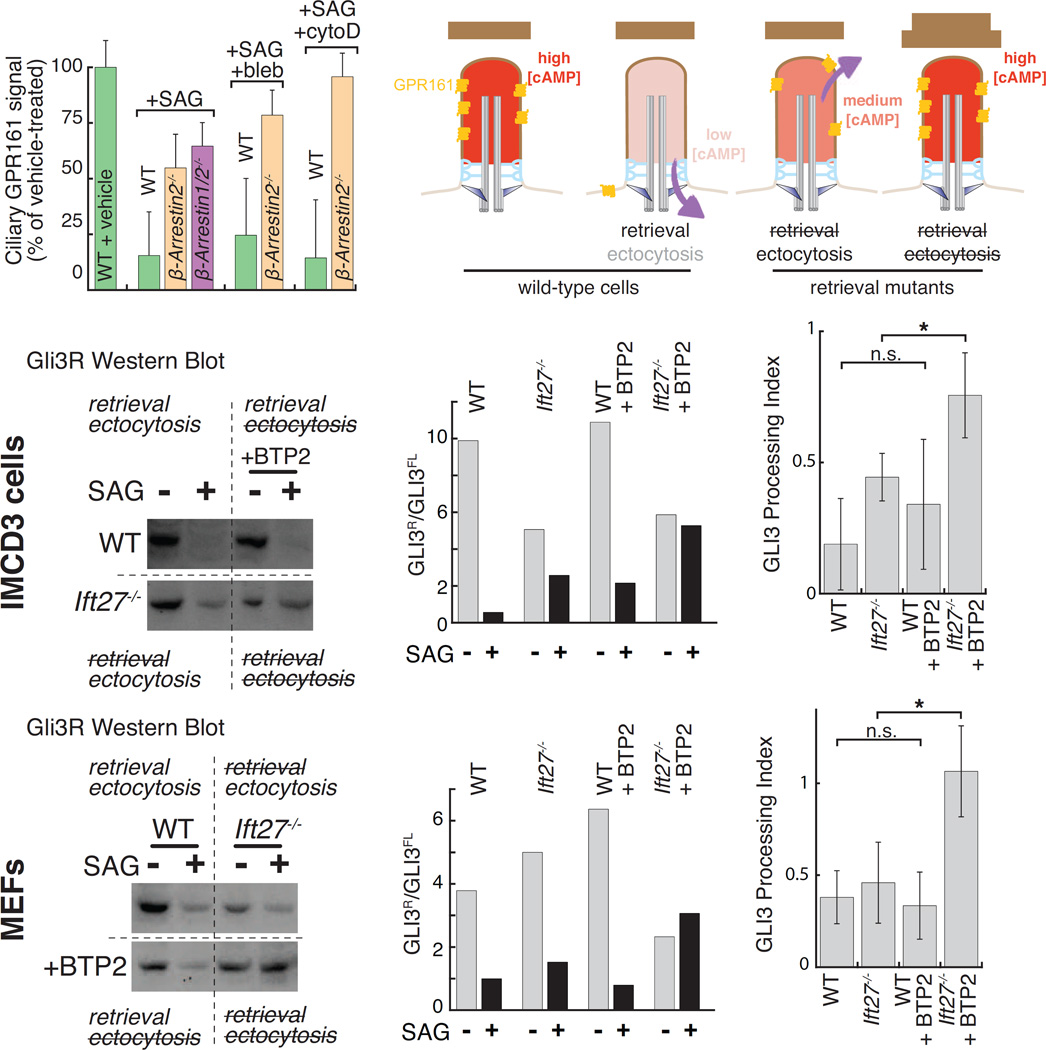
Given that ectocytosis removes large quantities of signaling receptors from cilia, ectocytosis is likely to affect ciliary signaling processes. In particular, our finding that AP- and NeonGreen-tagged GPR161 is ectocytosed in retrieval mutants (Fig. 3A and S3C) suggested that ectocytosis may affect the transduction of Hedgehog signals. We first sought to test the importance of ectocytosis in the removal of endogenous GPR161 by leveraging our newly discovered pharmacological inhibitors of ectocytosis. After a 2h activation of the Hh pathway, the average ciliary levels of endogenous GPR161 decreased by 80% in wild-type cells (Fig. 7A) (Mukhopadhyay et al., 2013)). Consistent with the recent finding that β-arrestins are required for Hh-induced retrieval of GPR161 from cilia (Pal et al., 2016), nearly all β-arrestin1/2−/− and β-arrestin2−/− cilia remained positive for GPR161 after 2h SAG treatment (Fig. S7A). Nonetheless, measurement of fluorescence signals showed that the ciliary levels of GPR161 decreased in β-arrestin1/2−/− and β-arrestin2−/− cells in response to Hh pathway activation, albeit with a reduced efficiency compared to wild-type cilia (Fig. 7A, “+SAG”). To test whether ectocytosis was responsible for the signal-induced removal of GPR161 from retrieval mutant cilia, we treated cells with actin poisons. The removal of GPR161 from β-arrestin2−/− cilia was significantly impaired by blebbistatin and completely abolished by cytochalasin D (Fig. 7A). Consistent with actin poisons leaving retrieval intact, the SAG-dependent exit of GPR161 from WT cilia was unaffected by cytochalasin D or blebbistatin (Fig. 7A). We conclude that endogenous GPR161 is removed from β-arrestin2−/− cilia by ectocytosis.
Figure 7. Ciliary exit pathways are required for the appropriate regulation of Hedgehog signaling.
(A) Endogenous GPR161 is lost by ectocytosis in retrieval-defective cells. WT, β-arrestin2−/−, or Arrb1/2−/− cells were treated with vehicle or SAG, fixed, immunostained for GPR161 and ciliary fluorescence levels measured. As indicated, cells were treated with blebbistatin (bleb) or cytochalasin D (CytoD). Mean values were normalized to the vehicle condition. Error bars: SEM. (n=61–163 cilia).
(B) Hedgehog-dependent removal of GPR161 is predicted to lower ciliary cAMP levels. In retrieval mutants, GPR161 is removed by ectocytosis still permitting a partial reduction in ciliary cAMP. Simultaneous blockage of retrieval and ectocytosis release is predicted to keep ciliary cAMP levels constant irrespective of Hedgehog pathway activation.
(C–H) Retrieval and ectocytosis ensure appropriate Gli3 processing. Wild-type or Ift27−/− (C–E) IMCD3 or (F–H) MEF cells were starved for 24 h, and then treated with vehicle or SAG and either DMSO or the Drebrin inhibitor BTP2 for 8 h. (C and F) show Western blots for Gli3R (see Fig. S7B–C for full blot). (D and G) plot ratios of Gli3R to Gli3FL from representative experiments. (E and H) report the Gli3 processing index from 3 biological replicates. The Gli3 processing index is the fractional change of the Gli3R:Gli3FL ratio upon SAG treatment (Gli3R:Gli3FLSAG/Gli3R:Gli3FLvehicle). Error bars: SD.
The current model for GPR161 function posits that GPR161 maintains a tonic level of cAMP in cilia through coupling to Gαs and ciliary adenylate cyclases (Mukhopadhyay et al., 2013). This high level of ciliary cAMP activates PKA inside cilia (Mick et al., 2015) and results in the efficient processing of the Hh effector and transcription factor Gli3 into repressor (Gli3R). Upon Hh pathway activation, ciliary exit of GPR161 results in a decrease of ciliary cAMP concentration and a consequent reduction in the amount of Gli3R produced (Fig. 7B). To assess whether GPR161 ectocytosis affects ciliary cAMP levels and Hh signaling, we measured the levels of Gli3R produced under conditions that block either retrieval, ectocytosis or both (Fig. 7B). While the ratio of Gli3R to Gli3FL was normally reduced 5-fold upon Hh pathway stimulation in wild-type IMCD3 cells, adding SAG to Ift27−/− cells only resulted in a 2-fold decrease of Gli3R levels (Fig. 7C–E and S7B).Thus, as previously reported, blockage of GPR161 retrieval mildly interferes with Hh-dependent Gli3 processing (Eguether et al., 2014), most likely because the magnitude of Hh-induced decrease in ciliary cAMP is reduced (Fig. 7B). Ectocytosis inhibition in retrieval mutant cells led to considerably more pronounced effects on Hh-dependent Gli3 processing. Addition of the drebrin inhibitor BTP2 to Ift27−/− cells completely abolished the Hh-dependent decrease of Gli3R levels (Fig. 7C–E). Similarly, the myosin 6 inhibitor TIP further dampened the magnitude of the Hh-dependent reduction in Gli3R levels in Ift27−/− cells (Fig. S7B). To further validate these results, we turned to immortalized mouse embryonic fibroblasts (MEFs), a well-accepted model of Hh signaling (Fig. 7F and S7C). While the blockage of BBSome-mediated retrieval did not have a pronounced effect on Hh-dependent Gli3 processing in MEFs, inhibition of ectocytosis in Ift27−/− cells completely abolished the Hh-dependent decrease of Gli3R levels (Fig. 7F–H). Consistent with GPR161 retrieval remaining unaffected by BTP2, this inhibitor did not significantly affect the SAG-dependent reduction in Gli3R levels in wild-type cells (Fig. 7F–H). Ectocytosis thus disposes of excess GPR161 from cilia in retrieval mutants to enable a partial reduction of ciliary cAMP levels. In this context, ectocytosis plays the role of a functional safety valve for the cilium, providing a means to downregulate ciliary PKA signaling when retrieval fails.
DISCUSSION
Receptor packaging into ectosomes at the tips of cilia
We report here that, like the Chlamydomonas adhesion receptor SAG1, the anorexigenic GPCR NPY2R becomes dynamically concentrated at the tip of the cilium before getting packaged into outward buds for ectocytosis. In addition, GPR161 and SSTR3 exit cilia through the ectocytic pathway when the retrieval route is blocked. The packaging of signaling receptors into ciliary ectosomes is a efficient and selective as SSTR3 concentration is increased at least 5-fold within ectosome membranes compared to donor membranes (Fig. S2E and Methods). Similarly, SAG1 is significantly concentrated in Chlamydomonas ectosomes as compared to cilia (Cao et al., 2015). The driving force that packages a signaling receptor into ectosomes is unknown but clustering of signaling receptors at the tips may be sufficient for cargo selection.
A major question relates to the sculpting of the outward membrane buds. While abundant evidence exists for the ESCRT complexes budding intraluminal vesicles into the lumen of the multivesicular body, tetraspanins have been proposed to impart outward membrane curvature to forming exosomes (Colombo et al., 2014) and the pentaspanin prominin may act similarly in ciliary ectocytosis given its concentration in buds near the tips of cilia (Dubreuil et al., 2007). More generally, lipid-based microdomains may initiate ectosome budding from the plasma membrane (Desrochers et al., 2016).
Actin and cilia
We find that actin polymerization is required for the release of ectosomes from the ciliary tip. As cytochalasin D treatment leads to cilium lengthening (Kim et al., 2010), ectocytosis may participate in the constitutive turnover of ciliary membrane. Mechanistically, actin polymerization directly triggers the scission of membrane tubules by reorganizing lipid domains during clathrin-independent endocytosis (Römer et al., 2010). Actin polymerization may thus promote ectosome scission (see Fig. 5G). Alternatively, localized actin polymerization at the ciliary tip may extrude membranous tubes that fragment into EVs (see Fig. S1B, cell 6), a phenomenon known as pearling (McConnell and Tyska, 2007).
The requirement for myosin 6, drebrin, the Arp2/3 complex and α-actinin 4 in the ectocytic process indicates that a coordinated actin machinery triggers ectosome release. Myosin 6 participates in several trafficking processes, recognizing cargoes through ubiquitin and various adaptors. Interestingly, drebrin couples dynamic microtubules to F-actin to trigger the elongation of filipodia at the axonal growth cone (Geraldo et al., 2008). Dynamic microtubules, such as those induced by mating signaling in Chlamydomonas (Mesland et al., 1980), may thus recruit drebrin to the tips of cilia. α-actinin 4 is a Ca2+-sensitive F-actin bundling protein that anchors actin to membrane proteins and may regulate trafficking through direct interactions with the ESCRT-0 protein Hrs and the ciliopathy protein ALMS1 (Collin et al., 2012). Finally, since actin filaments, α-actinin and myosin VIIa have all been found at the site of disc formation in photoreceptors (Wolfrum et al., 1998), actin may perform other membrane-shaping functions inside cilia besides ectocytosis.
Pervasive ectocytosis accounts for the loss of signaling receptors from Bbs cilia
While specific proteins are actively packaged into nascent ectosomes, ectocytosis also removes bystander proteins because they are not excluded from the forming ectosome. This enrichment of specific cargoes and incorporation of non-retained bystanders into vesicular bulk flow is seen in other examples of vesicle budding (Barlowe and Helenius, 2016) with the notable difference that in intracellular transport, non-specific losses can be recovered by retrieval pathways. Our finding that constitutive ectocytosis removes GPCRs from cilia of Bbs mutants (Fig. 3G and S3F) may explain why the levels of SSTR3, NPY2R, PC1 and SMO appear to decrease in Bbs mutant cilia (Berbari et al., 2008; Loktev and Jackson, 2013; Su et al., 2014). Similarly, the considerable losses of BBSome by ectocytosis in Arl6−/− cell (Fig. 2C and D, and Movie S3) are likely to account for the previously reported decrease in ciliary BBSome abundance in Arl6-depeted cells (Jin et al., 2010).
Generality of signal-dependent ectocytosis
Several groups have found the PKD proteins PKD2 and PKHD1 in EVs (Wang and Barr, 2016). Although the origin of PKD-EVs remains ambiguous, the published literature does not reject the hypothesis that PKD-EVs are the product of signal-dependent ciliary ectocytosis (Wood and Rosenbaum, 2015). Meanwhile in Chlamydomonas the packaging of the adhesion receptor SAG1 and of the sporangium wall lytic protease into ectosomes originate at the tips of cilia and may be regulated by signaling or cell cycle status (Cao et al., 2015; Wood et al., 2013). Thus, while PKD-EVs, SAG1-EVs, lytic protease-EVs and GPCR-EVs may originate from distinct processes, it is appealing to consider that signal-dependent ciliary ectocytosis is a widely conserved mechanism that packages activated signaling molecules into EVs.
Ciliary ectosomes: gold or garbage?
The first report of EVs found that exosome secretion rids the maturing reticulocyte of transferrin receptor (Pan et al., 1985). Consequently, EV release was initially regarded as a way of eliminating membrane material from cells, or cell compartments such as synapses, that lack lysosomes (Bartheld and Altick, 2011). Ciliary ectocytosis provides another example of EVs as disposal vessels and can compensate for the loss of retrieval activity. While the BBSome normally removes the repressors of Hh signaling GPR161 and Patched-1 from cilia, ectocytosis permits a partial transduction of Hh signals by disposing of ciliary GPR161 (and possibly Patched-1) when retrieval is compromised (Fig. 7). These results demonstrate the functional importance of exit from cilia and suggest that the mild phenotypes of Bbs animals compared to mutants that lack cilia will be exacerbated by treatment with pharmacological inhibitors of ectocytosis.
Recently, attention has shifted to the idea that ectosomes and other types of EVs transmit signals from one cell to another. Plasma membrane ectocytosis packages Ras and activated EGFR into EVs and may spread neoplastic behavior through tissues (Desrochers et al., 2016). Trypanosomes produce ciliary EVs that fuse with other parasites to transfer resistance to killing by the host (Szempruch et al., 2016). Chlamydomonas and nematode PKD-EVs are also bioactive and mammalian PKD-EVs specifically adhere and fuse with cilia (Wang and Barr, 2016). The packaging of NPY2R in ciliary ectosomes may similarly enable the transmission of signals across cells. Alternatively, signal-dependent ectocytosis may modulate NPY2R signaling in a cell-autonomous manner. In the future, the mechanistic dissection of ciliary ectocytosis will help determine whether signal-dependent ectocytosis only serves as a disposal strategy or if it also acts to spread signals across cells in mammalian organisms.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagents generated in this study please contact Maxence Nachury (nachury@gmail.com ).
Experimental Model and Subject Details
All IMCD3 cell lines were generated from a parental mouse IMCD3-FlpIn line. IMCD3 Flp-In cells were cultured in DMEM/F12, HEPES media (11330-057, Gibco) supplemented with 5% FBS (Gemini Bio-Products), 100 U/ml penicillin-streptomycin (15140-122, Gibco), and 2 mM L-glutamine (400-106, Gemini Bio-Products). To reconstitute agonist-dependent exit, SSTR3 and GPR161 fusion proteins were expressed at near-endogenous levels by using a single-copy low-expression cassette. The FlpIn system (ThermoFischer Scientific) was used for reliable, single-site integration at a defined genomic locus.
Immortalized mouse embryonic fibroblasts from wild-type and Ift27−/− mice (gift from Gregory Pazour; (Eguether et al., 2014)). Briefly, fibroblasts from embryos (embryonic day 14) were immortalized by SV40 Large T Antigen and cultured in DMEM (11995-073, Gibco) supplemented with 10% FBS, 100 U/ml penicillin-streptomycin, and 2 mM L-glutamine.
Cells were passaged using 0.05% Trypsin-EDTA (25300-120, ThermoFisher Scientific).
Methods
Plasmid construction and Generation of stable cell lines
Cell lines expressing BBS1/5, CSAP, and GPCRs were generated using the Flp-In System (ThermoFisher Scientific). Incorporation of low-expression promoters and additional expression cassettes were done by modification of the pEF5B/FRT plasmid. Briefly, site-directed mutagenesis was used to introduce an NsiI site in the pEF5B/FRT plasmid between the Ampicillin cassette and EF1α promoter. By traditional cloning in the NsiI and SpeI site, the EF1α promoter was replaced with either a TATA-box mutant (EF1αΔ) or a minimal chicken lens δ-crystallin (Crys) promoter to reduce protein expression. For EF1αΔ, the sequence TATAA was changed to TCCCC. Subsequent cloning into the NsiI site was used to incorporate NsiI-pPGK-BirA-ER, or NsiI-pEF1αΔ-APSSTR3–IRES–BirA-ER followed by a HSV polyadenylation signal. Open reading frames for Gateway cloning were cloned into pENTR vectors from cDNAs for human BBS1 and BBS5 (gifts from Val Sheffield), BirA-ER (gift from Alice Ting), human CSAP (gift from Iain Cheeseman), mouse GPR161 (BC028163, MGC, Dharmacon), human NPY2R (BC075052, MGC, Dharmacon), and mouse SSTR3 (gift from Kirk Mykytyn). BBS1, BBS5, and CSAP were expressed by the EF1α promoter, NPY2R and SSTR3 the EF1αΔ, and GPR161 by the Crys promoter. As indicated, NeonGreen (NG) and TandemTomato were used in fusion proteins. Assembled pEF5B/FRT plasmids were then transfected into IMCD3 Flp-In cells along with a plasmid encoding Flp-Recombinase (pOG44), and stable transformants were selected by Blasticidin resistance. β-Arrestin2GFP (gift from Mark Scott) was stably expressed from a pCMV-based plasmid (pEGFP-N) that was transfected into IMCD3 cells and selected using Neomycin resistance. For CRISPR-based genome editing, Cas9 and guide RNAs were transiently expressed from pX330 (Addgene, Feng Zhang). Guide RNAs targeted the first open reading of either mouse Arrb1 (ACTCACCCACGGGGTCCACG) or Arrb2 (TCTAGGCAAACTTACCCACA). The efficacy of each guide RNA was confirmed by amplifying the target locus and assessing editing frequency by the Surveyor Mutation Detection Kit (706020, Transgenomic). Clones were then isolated by limited dilution, and analyzed by western blotting. Knockouts were then confirmed by DNA sequencing of five independently cloned isolates of the genomic locus using CloneJET PCR cloning kit (K1231, ThermoFisher Scientific).
Drug treatments
The following reagents were used at the indicated concentrations: 20 µM ACQ090, 100 mM 2-APB, 50 µM (−)-Blebbistatin, 1 µM BTP2, 50 µM CK-636, 0.5 µM Cytochalasin D, 50 µM Emetine, 15 µM Jasplakinolide, 1 µM LaCl3, 1 µM Latrunculin A, 1 µM Neuropeptide Y 3–36, 200 µM SAG, 25 µM SMIFH2, 10 µM Somatostatin 14, 0.5 µM Taxol, and 25 µM TIP. Peptides were dissolved in DMEM/F12 media, HEPES, no phenol red (termed vehicle, 11039-021, Gibco). Small molecules were dissolved in DMSO (276855, Sigma-Aldrich). GPCR ligands were used 100 to 1,000 fold above KD.
As indicated in figure legends, (−)-Blebbistatin was added concurrently with GPCR agonist (t = 0). 2-APB, LaCl3 or TIP was added 10 min prior to agonist or vehicle addition. Jasplakinolide was added 30 min prior. BTP2, Cytochalasin D or SMIFH2 was added 1 h prior. Latrunculin A was added 10 min prior to fixed imaging experiments or 1 h prior to live-cell imaging. CK-636 or Emetine was added 2 h prior to agonist or vehicle addition. For fixed imaging pulse chases involving emetine, all cells were treated with emetine for the same duration. For example, in Fig. 4F, cells were treated with emetine for 2 h, then vehicle and/or NPY3–36 for 4 h.
Knockdowns and transfections
Plasmid transfections were performed using Lipofectamine 2000 (11668027, ThermoFisher Scientific), and siRNAs and esiRNAs were by Lipofectamine RNAiMAX (13778-075, ThermoFisher Scientific). First, the transfection reagent was diluted in Optimem (31985070, ThermoFisher Scientific) and incubated at room temperature for 5 min. Nucleic acid reagents were then added to the diluted transfection reagent for 20 min before addition to suspended cells. The transfection-cell mixture was then plated in a cell culture dish. For plasmid transfections, the transfection reagent was replaced by fresh DMEM/F12 medium after 4 h. For all transfected cells, cells were switched to starvation media after 48 h, and analyzed 16 h later.
Cell imaging
For fixed imaging, 70,000 cells were seeded on acid-washed 12 mm #1.5 cover glass (Fisherbrand) in a 24-well plate. After 24 h of growth, cells were starved for 16 h and fixed in Phosphate Buffer Saline (PBS) containing 4% sucrose and 4% paraformaldehyde for 15 min at room temperature. Cells were then permeabilized in PBS containing 0.1% Triton-X100 (BP151-500, ThermoFisher Scientific), 5% normal donkey serum (017-000-121, Jackson Immunoresearch Labs), and 3% bovine serum albumin (BP1605-100, ThermoFisher Scientific) for 30 min. Permeabilized cells were incubated with the specified primary antibodies for 1 h, washed three times with PBS, and incubated with dye-coupled secondary antibodies (Jackson Immunoresearch Labs) for 30 min. Cells were then washed two times with PBS, stained with Hoechst DNA dye, and washed twice more with PBS before mounting slides with Fluoromount G (Electron Microscopy Sciences).
For live-cell imaging, 400,000 cells were seeded on acid-washed 25 mm cover glass (Electron Microscopy Sciences) in a 6 cm dish. After 24 h of growth, cells were starved for 16 h and transferred to the DeltaVision stage for imaging at 37 °C inside an environmental chamber. Cells were imaged in DMEM/F12 media, HEPES, no phenol red (11039-021, Gibco). For all >1 h imaging experiments, the imaging chamber was overlaid with a petri dish containing a moist towel to maintain the imaging volume and pH. To accurately measure removal of APSSTR3NG, the AP tag of SSTR3 was pulse-labeled with Alexa647-labeled monovalent streptavidin (SA647) for 5 min (Howarth and Ting, 2008).
Cells were imaged on either an AxioImager.M1 microscope (Carl Zeiss) equipped with a PlanApo 63x/1.4NA objective lens, or by a DeltaVision system (Applied Precision) equipped with a PlanApo 60x/1.40 objective lens (Olympus), a CoolSNAP HQ2 camera (Photometrics), an EMCCD camera, and a solid state illumination module (InsightSSI), a TIRF module and a quantifiable laser module. Image analysis was done with Fiji. In most experiments, Z-stacks with 0.5-µm separation were acquired but only the most in-focus plane was used for image analysis. For imaging cilia pointing upwards (‘dorsal cilia’), a 0.3-µm separation Z-stack was acquired and Z-stacks were deconvolved using SoftWoRx. The base of the cilium was identified by marker localization (Ninein, CSAP, BBS1/5), the accumulation of intracellular GPCR-positive vesicles around the basal body, the change in focal plane for GPCR fluorescence as the base of the cilium bends towards the cell, and orientation away from the cell body for top/dorsal cilia.
Extracellular vesicle preparation
IMCD3 cells were cultured to near confluency in a 15 cm dish. The medium was then replaced with DMEM/F12 medium containing 0.2% FBS plus SAG, somatostatin, or DMSO. Media was harvested after 16 h and extracellular vesicles were purified as diagramed in Fig. S2A (Kowal et al., 2016). First, the cell culture medium was transferred to a 50 mL conical tube. The cells were then washed with 10 mL PBS, and the washed media was added to the conical. Second, the conical was centrifuged at 300 RCF for 10 min at 4C to remove cell debris. The supernatant was then transferred to a new 50 mL conical tube and centrifuged at 2000 RCF for 20 min at 4C. The resulting supernatant was then transferred to a Beckman JA-20 Fixed Angle Rotor, and centrifuged at 10,000 RCF for 40 min at 4C to produce the P10 pellet. The supernatant was then ultracentrifuged at 100,000 RCF for 90 min at 4C in a Beckman SW-28 Swinging Bucket Rotor. The pellet was washed in PBS and ultracentrifuged at 100,000 RCF for 90 min at 4C in a Beckman TLS-55 to produce the P100 pellet.
Electron microscopy
Cells cultured on cover glass and treated with blebbistatin for 1 h before fixation. Blebbistatin reduced the rate of SSTR3 ectocytosis. Cells were fixed with 3.5% glutaraldehyde (16120, Electron Microscopy Sciences) in 0.1 M Phosphate Buffer (PB) for 1 h at room temperature, washed with PB, post-fixed with 1% osmium tetroxide (Electron Microscopy Sciences) in PB for 45 min at room temperature, rinsed with deionized water, and dehydrated first in ethanol, then with CO2 by the critical point drying method. The samples were coated with gold/palladium alloy by sputter coating. Primary cilia on the surface of cells were studied under a Hitachi S-4800 scanning electron microscope using Quantax 400 software (Bruker Corporation).
EVs (p10 and P100) were resuspended in room temperature PBS and incubated for 1h on UV-irradiated carbon/formvar copper grids. Grids were then washed with 50 µl PBS for 5 min, and biotinylated EVs were labeled with streptavidin-conjugated 3-nm gold nanoparticles for 30 min. Streptavidin-conjugated gold nanoparticles were diluted 100 times in PBS. Grids were washed 5 times in PBS, negatively stained with 1.5% uranyl acetate for 5 min, and dried in a dust-free chamber. The EVs were visualized using a JEOL 1400 electron microscope (120 kV) at 10,000 × or 25,000 × magnification.
Quantification and Statistical Analysis
Image analysis and curve fitting
Data analysis, curve fitting, and presentation were done with ImageJ, Excel, Matlab, and Kaleidagraph. Integrated ciliary intensities were measured on ImageJ by a segmented line (width = 3), and were corrected for background by subtracting the fluorescence immediately adjacent. Background can include nonspecific antibody or label staining, detector noise, out-of-focus fluorescence, and plasma membrane-localized receptors. For live-cell imaging of APSSTR3 and APSSTR3NG, a mathematical background correction was applied rather than subtracting a unique background measurement for each time point. Briefly, the background was determined by multiplying the average nonciliary fluorescence by the measured area of the cilium. Live-cell imaging data was also corrected for photobleaching. The rate of fluorescence decay from photobleaching was determined by imaging cilia at 2 Hz in the buffer used for the relevant experiment. Experiments were designed to have <15% photobleaching. Measurements were corrected using the equation:
where λ is the exponential decay constant for photobleaching, n is the number of image acquisitions take, Fn is the integrated fluorescence measured for image acquisition ‘n’, F1 is the integrated fluorescence at the first measurement, and Fcorrected is the reported integrated fluorescence.
Retrieval of SA647-labeled APSSTR3 or APSSTR3NG was defined as reduction of ciliary SA647 or NeonGreen fluorescence without observed ectocytosis. To deconvolve removal of SSTR3 by retrieval versus ectocytosis, the instantaneous change in SA647 between each time point was analyzed and sorted based on whether an ectosome was released or not (see Fig. 2G, Arl6−/−). For reporting retrieval rates (see Fig. 1A), all instantaneous velocities describing ectocytosis events were discarded. The remaining instantaneous velocities were ordered by time point, averaged, and used to describe a theoretical, ectocytosis-free experiment (see Fig. 1A, Arl6−/− + sst).
All box plots are Tukey boxplots. The box displays the second, third, and fourth quartiles. The whiskers represent values within the 1.5× the interquartile range. Outliers exceeding the whiskers are plotted as points or omitted. Box plots and medians are frequently used to describe the GPCR abundance as, for both endogenous and reconstituted GPCRs, infrequent cilia with very high GPCR content distort the population average. Removal of NPY2R in Fig. 6A and C is expressed by the change in median ciliary fluorescence (F) such that:
For Fig. 6C, removal was normalized to DMSO-treated cilia to compare NPY2R and SSTR3 removal.
Statistically significant differences in the rate of GPCR removal (Fig. 6A and C) were identified by a z-statistic comparing regressions. Briefly, data as in Fig. S6A–E were linearly fit (ΔFluorescence = m * time + c) in Kaleidagraph. A z-statistic was used to compare a condition to the control:
where sedifference is the propagated standard error of the slopes:
The resulting z-statistics were then converted to a p-values for interpretation and identifying conditions for further investigation.
Electron microscopy images of EVs were analyzed by ImageJ. Gold-positive EVs were defined as vesicle-shaped structures containing multiple 3-nm diameter electron-dense nanoparticles. On ImageJ, a polyline was drawn on the edge of the vesicles to measure the the area and Feret diameter. The number of gold particles per vesicles was counted manually and divided by the vesicle area to calculate the gold bead density per µm2.
Quantitative western blots were imaged with a ChemiDoc Touch (Bio-Rad). The resulting images were quantified using Image Lab (Bio-Rad).
Calculation of SSTR3 density in extracellular vesicles
To estimate the packaging of SSTR3 into EVs, we compared the density of SSTR3 in ciliary versus EV membranes. While the GFP fluorescence of ciliary APSSTR3GFP can be used to quantify the number of SSTR3 molecules per cilium and estimate the physical proportions of cilia (see below), this approach cannot estimate the SSTR3 density in EV membranes as the diameter of an EV is below the resolution of conventional microscopy. Therefore, we used the density of streptavidin-conjugate gold molecules in electron microscopy images to gauge the density of biotinylated APSSTR3NG (see below and Fig. 2B). These estimates find a 5.3-fold increase in SSTR3 membrane density in EVs versus cilia with 614 SSTR3 per µm2 in ciliary membranes and 3274 SSTR3 per µm2 in EV membranes. However, estimating the SSTR3 density in EVs by streptavidin-conjugate gold molecules is inherently an underestimate as (1) a single streptavidin-conjugate gold molecule can bind many SSTR3 molecules and (2) not all APSSTR3NG molecules are biotinylated or accessible to streptavidin.
To measure the number of SSTR3 molecules in cilia of IMCD3-[pEF1αΔ-APSSTR3GFP] cells, we compared the fluorescence of IMCD3-[pEF1αΔ-APSSTR3GFP] cilia to that of IMCD3-[pEF1α-APSSTR3GFP] cilia. The number of SSTR3 molecules in IMCD3-[pEF1α-APSSTR3GFP] cilia had been previously measured using a rotavirus-like particle with 120 GFP molecules as a calibrator (Breslow et al., 2013). GFP-tagged viral particles and IMCD3-[pEF1α-APSSTR3GFP] cells were imaged on a spinning-disc confocal (CSU10, Yokogawa Corporation of America on a Ecplise TE2000, Nikon) with a 63x, 1.4 NA Plan Apochromat objective, a 488 nm laser (Innova 70C-Spectrum, Coherent), and a charge-coupled device camera (Cascade 512B, Photometrics). Data was collected using MetaMorph software (Molecular Devices). Rotavirus-like particle intensity was measured in a summed z-stack projection. Adjacent background was subtracted, and the resulting mean particle intensity of the virus was divided by 120 to estimate the fluorescence intensity of a single GFP molecule under the imaging conditions used. Similarly, the summed z-projection intensities for IMCD3-[pEF1α-APSSTR3GFP] cilia were measured, and the SSTR3-GFP intensity per unit length was converted to the number of SSTR3-GFP molecules per micrometer length of the ciliary membrane. As the average IMCD3-[pEF1α-APSSTR3GFP] cilium is 7.7 µm by fluorescence measurement, there are 50,360 SSTR3 molecules per cilium, and 6,540 SSTR3 molecules per micrometer length.
The total GFP fluorescence in IMCD3-[pEF1α-SSTR3GFP] cilia was measured to be 20.55-fold greater than that in IMCD3-[pEF1αΔ-APSSTR3GFP] cilia. Both cell lines were imaged through a PlanApo 60x/1.40 objective (Olympus) on a DeltaVision system (Applied Precision). A solid state module (InsightSSI) provided illumination and images were acquired with a sCMOS camera (Applied Precision) using SoftWoRx software (Applied Precision). Z-stacks with 0.5-µm separation were acquired and the most in-focus plane was used for image analysis. The integrated fluorescence density of GFP for over 30 cilia was measured with Fiji, and the fluorescence of the adjacent area was subtracted as background. Thus, IMCD3-[pEF1αΔ-APSSTR3GFP] cilia have 616 SSTR3 molecules per micrometer length. As the average IMCD3-[pEF1αΔ-APSSTR3GFP] cilium is 4 µm by fluorescence measurement, there are 2,450 SSTR3 molecules per cilium.
The density of SSTR3 molecules in the ciliary membrane was estimated by modeling the membrane area of the cilium as a one-capped cylinder:
where radius equals the diameter of the ciliary membrane (300 nm) divided by two, and height is the median length measured in Fig. 2H, 4 µm. For a cilium with 3.8 µm2 membrane area, the density of SSTR3 is 614 SSTR3 per µm2 of ciliary membrane.
For EVs, gold particles were counted and divided by the EV membrane area:
where radius equals the median diameter of an SSTR3-containing P10 EV (83 nm) divided by two. As the average gold-positive EV had 20.31 gold particles, an EV with 0.022 µm2 membrane area would have 939 gold particles per µm2. We estimate that 29% of APSSTR3NG molecules can be labeled by monovalent streptavidin, by comparing absolute counting of SA647 to NeonGreen in cilia of IMCD3-[pEF1αΔ-APSSTR3NG] cells. Thus, by correcting for APSSTR3NG that are not detectable by streptavidin, we estimate 3274 SSTR3 per µm2 in EV membranes.
Supplementary Material
(A) Ectosomes are secreted from the tip of cilia. IMCD3-[pEF1αΔ-APSSTR3, pEF1α-NG3CSAP] cells were pulse-labeled with SA647 and imaged every 10 min following addition of sst. The base-proximal marker NG3CSAP illustrates that SA647-labeled APSSTR3 ectosomes originate from the ciliary tip.
(B) Additional examples of SSTR3 ectocytosis from Arl6−/−/β-arrestin2−/− cells. IMCD3-[pEF1αΔ-APSSTR3NG] cells were imaged every 10 min in the NeonGreen channel following addition of sst. Identical results were obtained by imaging in the SA647 channel. Yellow and red triangles point to the base and tip of cilia respectively and ectosomes are circled. Cells 1 to 4 depict typical ectocytosis events. Cell 5 depicts the occasional release of large ciliary ectosome that appeared attached to the coverslip. Cell 6 corresponds to the occasional cilia observed to form multiple SSTR3 tip foci that were ectocytosed as a group, suggestive of pearling. A model is shown to aid visualization. Scale bars: 4 µm. Yellow and red triangles point to the base and tip of cilia and ectosomes are circled.
(C) Additional examples of scanning electron microscopy of ectocytic buds at the ciliary tip in retrieval-defective cells. IMCD3-[APSSTR3NG];Arl6−/−/β-arrestin2−/− cells were treated with sst for 1 h before processing for scanning electron microscopy. Scale bar: 1 µm.
(D) Counting of tip buds observed by scanning electron microscopy in Fig. 1J and S1C. The rightmost example in Fig. 1J was categorized as a large bud. (n=26–48 cilia).
(A) Diagram of EV purification protocol. Cell culture supernatants were from 10 million cells starved for 16h in the presence of SAG or sst. The pellets from the 10,000 and 100,000 × g spins (termed P10 and P100) were analyzed by EM or immunoblotting.
(B–C) Imaging of EVs reveals a population of vesicles packaged with ciliary GPCRs. EVs from the P10 (B) and P100 (C) fractions purified from the supernatant of SAG-stimulated β-arrestin2−/− IMCD3 and IMCD3-[pCrys-APGPR161NG3] or sst-stimulated IMCD3-[pEF1αΔ-APSSTR3NG] cells were fixed, labeled with streptavidin-conjugated 3-nm gold nanoparticles, and imaged by negative stain EM. The top panel of (B) shows two EVs from APGPR161NG3-expressing cells, one densely labeled with gold particles (white insets) and the other unlabeled, as well as a gold particle non-specifically adhering to the grid (black inset). Densely decorated EVs were produced by cells expressing AP-tagged GPCRs but not by control IMCD3 cells. All scale bars are 50 nm.
(D) The diameter of EVs with 3 or more gold particles was measured in ImageJ (Methods). Error bars: SD. (n=37–47 EVs per GPCR).
(E) Measurement of GPCR surface density in EVs. EVs with 1 or more gold particles were selected from P10 micrographs and the number of gold particles per EV was manually counted and divided by the surface area of the EV. Although occasional gold particles are detected on control EVs, the very low density suggests that that the gold particles were non-specifically bound (n=37–47 EVs per GPCR).
(A) Signal-dependent ectocytosis results in cilium shortening. WT and Arl6−/− IMCD3-[pEF1αΔ-APSSTR3NG] cells were monitored for 12 h following addition of sst. At each hour, the length of cilia was measured by SSTR3 fluorescence. (n=23–62 measurements per box).
(B) Summary statistics for changes in the mean (top) and standard deviation (bottom) of cilia length for data shown in panel (A). Error bars represent the propagated standard error. (n=211–257 cilia per comparison).
(C–D) GPR161 is packaged into ectosomes produced upon Hedgehog pathway activation while SSTR3 remains a passenger of those ectosomes. IMCD3-[pCrys-APGPR161NG3];β-arrestin2−/− cells (C) or IMCD3-[pEF1αΔ-APSSTR3NG];β-arrestin2−/− cells (D) were imaged in the NeonGreen channel every 10 min following addition of SAG or sst. Yellow and red triangles point to the base and tip of cilia and the ectosome is circled.
(E) The NeonGreen intensity of ectosomes observed in experiments presented in Fig. 3A–E was quantified and normalized to the total NeonGreen intensity of the parent cilium. (n=11–43 ectosomes).
(F) Prolonged signaling and pervasive ectocytosis result in depletion of ciliary SSTR3. APSSTR3NG was pulse-labeled by SA647 and analyzed by fixed imaging after vehicle, sst, or SAG treatment for 6 or 9 h. Error Bars: 95% Confidence Interval. * indicates p<0.03 and ** for p<0.001 decrease after 9 h (Mann-Whitney U test). (n=50–140 cilia per point).
(A) Diagram of the carboxyterminal tail (C-tail) of SSTR3 highlighting truncations and mutations used in Fig. 4C. The third intracellular loop is labeled ‘i3’ and the C-tail is boxed with the amino acid sequence displayed to the right. C-tail residues were numbered 1–99 starting immediately after the NPxxY(x)5F motif which terminates the last transmembrane segment. SSTR3Δ1–25 indicates that the first 25 residues of the C-tail were deleted. SSTR3SA/TA is a variant where all 13 C-tail serine and threonine residues (asterisks) are mutated to alanine.
(B) Additional example of NPY2R ectocytosis from a wild-type cell. IMCD3-[pEF1αΔ-NPY2RNG] cells were imaged by NeonGreen fluorescence every 10 min for the 2 h following addition of NPY3–36. Yellow and red triangles point to the base and tip of cilia and the ectosome is circled.
(C) Activated NPY2R accumulates into tip foci irrespective of genotype. To score the frequency of tip foci, IMCD3-[NPY2RNG] cell lines were imaged by NeonGreen fluorescence every 10 min for 2 h following addition of NPY3–36 or vehicle and tip foci scored. Data were fit to a simple exponential. (n=28–79 cilia).
(D) SSTR3 tip foci are stabilized by cytochalasin D. Cytochalasin D-treated IMCD3-[pEF1αΔ-APSSTR3NG];Arl6−/− cells were imaged every hour by NeonGreen following addition of sst. Top: representative image of a long-live tip focus. Bottom: quantitation of 4 similar foci plotted as the fraction of ciliary SSTR3 in the focus.
(E) Genotypes and drug treatments used in this study do not significantly affect cilia movement in IMCD3 cells. IMCD3-[APSSTR3NG] or IMCD3-[NPY2RNG] cells with the indicated genotype or drug treatment were imaged by NeonGreen fluorescence every 10 min for 2 h following addition of agonist (sst or NPY3–36). The bar graph reports the average distance traversed by the ciliary base during 2 h in the presence of agonist. Error bars represent SD. No condition was statistically different by ANOVA. (n=30–41 cilia).
(F) GPR161 ectocytosis as seen on vertically oriented cilia. The dorsal surface of IMCD3-[pCrys-APGPR161NG3];β-arrestin2−/− cells was imaged every 10 min for the 2 h following addition of SAG. Tip foci and ectosomes were frequently observed, and ectosomes were rapidly lost into the media. The cilia appear misshaped as they span several focal planes at an angle. Yellow and red triangles point to the base and tip of cilia and the ectosome is circled.
(G) Cytochalasin D does not block SSTR3 endocytosis from the plasma membrane. Wild-type, cytochalasin D-treated, and β-arrestin1/2−/− IMCD3-[APSSTR3NG] cells were imaged by NeonGreen fluorescence immediately after addition of sst (t0) and 10 min later (t10). The brightest signal in each image is from cilia-localized SSTR3 but the contrast is adjusted to reveal SSTR3 on the plasma membrane and internal structures. After 10 min of sst, endocytosis results in more SSTR3 at internal structures in wild-type and cytochalasin D-treated while all SSTR3 signal remains at the surface of β-arrestin1/2−/− cells. Examples of internalized SSTR3 structures are circled. All scale bars are 4 µm.
(H) Counting of endocytic vesicles observed in experiments presented in Fig. S4G. Internal structures were counted by NeonGreen fluorescence immediately after addition of sst (t0) and 10 min later (t10). (n=25 cells). These results are in agreement with previous observations that actin poisons have only mild effects on clathrin-mediated endocytosis in mammalian cells unless membrane tension is high or very high doses of inhibitors are used (Mooren et al., 2012)
(A) Screening for proteins necessary for removing NPY2R from cilia. IMCD3-[pEF1αΔ-NPY2RNG] cells were depleted of the indicated proteins before treatment with Emetine, and NPY3–36 or vehicle for 4 h followed by fixation and measurement of ciliary NeonGreen levels. (n=34–121 cilia).
(B–D) Immunoblots of IMCD3 cells treated with siRNAs targeting Drebrin (B), Kif13b (C), Myosin 6 (D), or non-targeting controls for 48 h prior to 16 h starvation. Annexin V and HSP60 were used as loading controls.
(A–E) F-actin and several actin-regulated proteins are required for removal of NPY2R but not SSTR3. Emetine-treated IMCD3-[pEF1αΔ-NPY2RNG] or SA647 pulse-labeled IMCD3-[pEF1αΔ-APSSTR3NG] cells were assayed for GPCR removal. For NPY2R, cells were treated with vehicle or NPY3–36 for 4 h before fixation and quantification of ciliary NeonGreen (box plots). For SSTR3, cilia were imaged by SA647 every 10 min for 2 h following addition of sst. Data were linearly fitted (scatter plots). Cells were treated with (A) DMSO or the Drebrin inhibitor BTP2, (B) the Ca2+ channel inhibitors LaCl3 or 2-APB, (C) the myosin 6 inhibitor TIP, (D) the actin poisons jasplakinolide (Jas) or latrunculin A (LatA), the Arp2/3 inhibitor CK-636, or (E) the Formin 2 inhibitor SMIFH2. For (E), IMCD3-[pEF1αΔ-NPY2RtandemTomato] cells were used as SMIFH2 resulted in greater background fluorescence in the NeonGreen channel. For axis scaling, outliers were omitted. (n=76–187 cilia for NPY2R fixed imaging, n=11–13 cilia for SSTR3 live-cell imaging).
(F) Myosin 6 antibody staining of the ciliary tip is specific. Ift27−/− cells were treated with siRNA targeting myosin 6 or a control siRNA for 48 h. Cells were then starved for 16 h and treated with DMSO or SAG for 1 h, fixed, and immunostained for endogenous myosin 6. Myosin 6 tip localization on dorsal cilia was scored. Error bars: SD between microscope fields. (n=185–186 cilia).
(G) Drebrin localizes to the tips of cilia. β-arrestin2−/− cells were starved for 16 h then treated with SAG for 1 h, fixed, and immunostained for endogenous drebrin. Drebrin localized at the cilia tip, distal to the basal body marker ninein. Scale bar: 4 µm.
(A) β-Arrestin2 is necessary for efficient removal of GPR161 from cilia. WT, β-arrestin2−/− or β-arrestin1/2−/− IMCD3 cells were starved for 24 h, treated with either vehicle or SAG for 2 h, fixed, and immunostained for endogenous GPR161. GPR161-positive cilia were manually counted. The quantitation of GPR161 ciliary levels within the same data set (Fig. 7A) demonstrates that, while GPR161 remains detectable in nearly all cilia of SAG-treated β-arrestin1/2−/− and β-arrestin2−/− IMCD3 cells, the ciliary levels of GPR161 are reduced upon SAG addition to β-arrestin1/2−/− and β-arrestin2−/− IMCD3 cells. (n=93–151 cilia).
(B and C) Retrieval and ectocytosis ensure appropriate Gli3 processing. Wild-type or Ift27−/− cells were starved for 24 h, and then treated with vehicle or SAG and either DMSO, the drebrin inhibitor BTP2 or the myosin 6 inhibitor TIP for 8 h. Lysates from (B) IMCD3 cells or (C) MEF were immunoblotted for Gli3 and Annexin 5 used as a loading control.
Highlights.
Signal-dependent exit of GPCRs from cilia proceeds through retrieval or ectocytosis
GPCRs that cannot be retrieved back into the cell are instead ectocytosed
Actin, myosin 6 and drebrin mediate the scission of ectosomes from cilia tips
Hedgehog signaling fails when both ectocytosis and retrieval are blocked
In Brief.
The tips of cilia are transit points where proteins can be removed by actin-mediated vesicle release rather than undergoing retrograde transport to the cell body.
Acknowledgments
We thank Drs. Bornens, Buss, Janke, Mukhopadhyay, Scales and Chishti for the gifts of antibodies; Drs. Cheeseman, Mykytyn, Scott, Sheffield and Ting for the gifts of cDNAs; Novartis for the gift of ACQ090; the Nachury lab, Rich Lewis, Bill Snell, David Williams and Tim Stearns for helpful discussion. This work was supported by grants to M.V.N. (GM089933), J.M.G-V. (PROMETEOII/2014/075), Stanford Microscopy Facility (1S10RR02678001) and a Fayez Sarofim/Damon Runyon Fellowship to A.R.N. (DRG 2160-13).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, Formal Analysis, Writing – Original Draft, A.R.N. and M.V.N.; Investigation, A.R.N., J.S.G., V.H-P., D.P., and F.Y.; Methodology, Writing – Review & Editing, All Authors; Funding Acquisition, M.V.N., A.R.N. and J.M.G-V.; Resources, Supervision, M.V.N. and J.M.G-V.
REFERENCES
- Barlowe C, Helenius A. Cargo Capture and Bulk Flow in the Early Secretory Pathway. Annu Rev Cell Dev Biol. 2016;32:197–222. doi: 10.1146/annurev-cellbio-111315-125016. [DOI] [PubMed] [Google Scholar]
- Bartheld Von CS, Altick AL. Multivesicular bodies in neurons: distribution, protein content, and trafficking functions. Prog. Neurobiol. 2011;93:313–340. doi: 10.1016/j.pneurobio.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci USA. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol. 2013;203:129–147. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Kendrick-Jones J, Lionne C, Knight AE, Côté GP, Paul Luzio J. The localization of myosin VI at the golgi complex and leading edge of fibroblasts and its phosphorylation and recruitment into membrane ruffles of A431 cells after growth factor stimulation. J Cell Biol. 1998;143:1535–1545. doi: 10.1083/jcb.143.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Arshavsky VY. Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 2006;16:560–568. doi: 10.1016/j.tcb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Cao M, Ning J, Hernandez-Lara CI, Belzile O, Wang Q, Dutcher SK, Liu Y, Snell WJ. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. Elife. 2015;4:183. doi: 10.7554/eLife.05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin GB, Marshall JD, King BL, Milan G, Maffei P, Jagger DJ, Naggert JK. The Alström syndrome protein, ALMS1, interacts with α-actinin and components of the endosome recycling pathway. PLoS ONE. 2012;7:e37925. doi: 10.1371/journal.pone.0037925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- Datta P, Allamargot C, Hudson JS, Andersen EK, Bhattarai S, Drack AV, Sheffield VC, Seo S. Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc Natl Acad Sci U S A. 2015;112:E4400–E4409. doi: 10.1073/pnas.1510111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrochers LM, Antonyak MA, Cerione RA. Extracellular Vesicles: Satellites of Information Transfer in Cancer and Stem Cell Biology. Dev Cell. 2016;37:301–309. doi: 10.1016/j.devcel.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V, Marzesco A-M, Corbeil D, Huttner WB, Wilsch-Bräuninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T, San Agustin JT, Keady BT, Jonassen JA, Liang Y, Francis R, Tobita K, Johnson CA, Abdelhamed ZA, Lo CW, et al. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell. 2014;31:279–290. doi: 10.1016/j.devcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S, Khanzada UK, Parsons M, Chilton JK, Gordon-Weeks PR. Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat Cell Biol. 2008;10:1181–1189. doi: 10.1038/ncb1778. [DOI] [PubMed] [Google Scholar]
- Heissler SM, Selvadurai J, Bond LM, Fedorov R, Kendrick-Jones J, Buss F, Manstein DJ. Kinetic properties and small-molecule inhibition of human myosin-6. FEBS Lett. 2012;586:3208–3214. doi: 10.1016/j.febslet.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M, Ting AY. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat Protoc. 2008;3:534–545. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blanc P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, C Hsia EY, Brigui A, Plessis A, Beachy PA, Zheng X. The role of ciliary trafficking in Hedgehog receptor signaling. Science Signaling. 2015;8:ra55–ra55. doi: 10.1126/scisignal.aaa5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M, Lee Y, Morales JL, Ning G, Huang W, Pabon J, Kannan AK, Jeong A-R, Wood A, Carter C, et al. Cutting Edge: Drebrin-Regulated Actin Dynamics Regulate IgE-Dependent Mast Cell Activation and Allergic Responses. J Immunol. 2015;195:426–430. doi: 10.4049/jimmunol.1401442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Brown JM, Sampaio JL, Craft JM, Shevchenko A, Evans JE, Witman GB. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol. 2013;201:249–261. doi: 10.1083/jcb.201207139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew GM, Ye F, Nager AR, Murphy JP, Lee JS, Aguiar M, Breslow D, Gygi SP, Nachury MV. The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev Cell. 2014;31:265–278. doi: 10.1016/j.devcel.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loktev AV, Jackson PK. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep. 2013;5:1316–1329. doi: 10.1016/j.celrep.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Marion S, Oakley RH, Kim K-M, Caron MG, Barak LS. A beta-arrestin binding determinant common to the second intracellular loops of rhodopsin family G protein-coupled receptors. J Biol Chem. 2006;281:2932–2938. doi: 10.1074/jbc.M508074200. [DOI] [PubMed] [Google Scholar]
- McConnell RE, Tyska MJ. Myosin-1a powers the sliding of apical membrane along microvillar actin bundles. J Cell Biol. 2007;177:671–681. doi: 10.1083/jcb.200701144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JC, McIntyre JC, Hege MM, Berbari NF. Trafficking of ciliary G protein-coupled receptors. Methods Cell Biol. 2016;132:35–54. doi: 10.1016/bs.mcb.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Mesland DA, Hoffman JL, Caligor E, Goodenough UW. Flagellar tip activation stimulated by membrane adhesions in Chlamydomonas gametes. J Cell Biol. 1980;84:599–617. doi: 10.1083/jcb.84.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, Nachury MV. Proteomics of Primary Cilia by Proximity Labeling. Dev Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, Jackson PK. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV. How do cilia organize signalling cascades? Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2014;369:20130465–20130465. doi: 10.1098/rstb.2013.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal K, Hwang S-H, Somatilaka B, Badgandi H, Jackson PK, DeFea K, Mukhopadhyay S. Smoothened determines β-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J Cell Biol. 2016;212:861–875. doi: 10.1083/jcb.201506132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Kreienkamp HJ, Meyerhof W, Richter D. Phosphorylation of four amino acid residues in the carboxyl terminus of the rat somatostatin receptor subtype 3 is crucial for its desensitization and internalization. J Biol Chem. 1997;272:23769–23774. doi: 10.1074/jbc.272.38.23769. [DOI] [PubMed] [Google Scholar]
- Römer W, Pontani L-L, Sorre B, Rentero C, Berland L, Chambon V, Lamaze C, Bassereau P, Sykes C, Gaus K, et al. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell. 2010;140:540–553. doi: 10.1016/j.cell.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Schou KB, Pedersen LB, Christensen ST. Ins and outs of GPCR signaling in primary cilia. EMBO Rep. 2015;16:1099–1113. doi: 10.15252/embr.201540530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Driscoll K, Yao G, Raed A, Wu M, Beales PL, Zhou J, Zhou J. Bardet-Biedl syndrome proteins 1 and 3 regulate the ciliary trafficking of polycystic kidney disease 1 protein. Hum Mol Genet. 2014;23:5441–5451. doi: 10.1093/hmg/ddu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szempruch AJ, Sykes SE, Kieft R, Dennison L, Becker AC, Gartrell A, Martin WJ, Nakayasu ES, Almeida IC, Hajduk SL, et al. Extracellular Vesicles from Trypanosoma brucei Mediate Virulence Factor Transfer and Cause Host Anemia. Cell. 2016;164:246–257. doi: 10.1016/j.cell.2015.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Barr MM. Ciliary Extracellular Vesicles: Txt Msg Organelles. Cell. Mol. Neurobiol. 2016:1–9. doi: 10.1007/s10571-016-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Silva M, Haas LA, Morsci NS, Nguyen KCQ, Hall DH, Barr MM. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum U, Liu X, Schmitt A, Udovichenko IP, Williams DS. Myosin VIIa as a common component of cilia and microvilli. Cell Motil Cytoskeleton. 1998;40:261–271. doi: 10.1002/(SICI)1097-0169(1998)40:3<261::AID-CM5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Wood CR, Rosenbaum JL. Ciliary ectosomes: transmissions from the cell's antenna. Trends Cell Biol. 2015;25:276–285. doi: 10.1016/j.tcb.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue S, Tang L-Y, Tang Y, Tang Y, Shen Q-H, Ding J, Chen Y, Zhang Z, Yu T-T, Zhang YE, et al. Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. Elife. 2014;3:128. doi: 10.7554/eLife.02555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Ectosomes are secreted from the tip of cilia. IMCD3-[pEF1αΔ-APSSTR3, pEF1α-NG3CSAP] cells were pulse-labeled with SA647 and imaged every 10 min following addition of sst. The base-proximal marker NG3CSAP illustrates that SA647-labeled APSSTR3 ectosomes originate from the ciliary tip.
(B) Additional examples of SSTR3 ectocytosis from Arl6−/−/β-arrestin2−/− cells. IMCD3-[pEF1αΔ-APSSTR3NG] cells were imaged every 10 min in the NeonGreen channel following addition of sst. Identical results were obtained by imaging in the SA647 channel. Yellow and red triangles point to the base and tip of cilia respectively and ectosomes are circled. Cells 1 to 4 depict typical ectocytosis events. Cell 5 depicts the occasional release of large ciliary ectosome that appeared attached to the coverslip. Cell 6 corresponds to the occasional cilia observed to form multiple SSTR3 tip foci that were ectocytosed as a group, suggestive of pearling. A model is shown to aid visualization. Scale bars: 4 µm. Yellow and red triangles point to the base and tip of cilia and ectosomes are circled.
(C) Additional examples of scanning electron microscopy of ectocytic buds at the ciliary tip in retrieval-defective cells. IMCD3-[APSSTR3NG];Arl6−/−/β-arrestin2−/− cells were treated with sst for 1 h before processing for scanning electron microscopy. Scale bar: 1 µm.
(D) Counting of tip buds observed by scanning electron microscopy in Fig. 1J and S1C. The rightmost example in Fig. 1J was categorized as a large bud. (n=26–48 cilia).
(A) Diagram of EV purification protocol. Cell culture supernatants were from 10 million cells starved for 16h in the presence of SAG or sst. The pellets from the 10,000 and 100,000 × g spins (termed P10 and P100) were analyzed by EM or immunoblotting.
(B–C) Imaging of EVs reveals a population of vesicles packaged with ciliary GPCRs. EVs from the P10 (B) and P100 (C) fractions purified from the supernatant of SAG-stimulated β-arrestin2−/− IMCD3 and IMCD3-[pCrys-APGPR161NG3] or sst-stimulated IMCD3-[pEF1αΔ-APSSTR3NG] cells were fixed, labeled with streptavidin-conjugated 3-nm gold nanoparticles, and imaged by negative stain EM. The top panel of (B) shows two EVs from APGPR161NG3-expressing cells, one densely labeled with gold particles (white insets) and the other unlabeled, as well as a gold particle non-specifically adhering to the grid (black inset). Densely decorated EVs were produced by cells expressing AP-tagged GPCRs but not by control IMCD3 cells. All scale bars are 50 nm.
(D) The diameter of EVs with 3 or more gold particles was measured in ImageJ (Methods). Error bars: SD. (n=37–47 EVs per GPCR).
(E) Measurement of GPCR surface density in EVs. EVs with 1 or more gold particles were selected from P10 micrographs and the number of gold particles per EV was manually counted and divided by the surface area of the EV. Although occasional gold particles are detected on control EVs, the very low density suggests that that the gold particles were non-specifically bound (n=37–47 EVs per GPCR).
(A) Signal-dependent ectocytosis results in cilium shortening. WT and Arl6−/− IMCD3-[pEF1αΔ-APSSTR3NG] cells were monitored for 12 h following addition of sst. At each hour, the length of cilia was measured by SSTR3 fluorescence. (n=23–62 measurements per box).
(B) Summary statistics for changes in the mean (top) and standard deviation (bottom) of cilia length for data shown in panel (A). Error bars represent the propagated standard error. (n=211–257 cilia per comparison).
(C–D) GPR161 is packaged into ectosomes produced upon Hedgehog pathway activation while SSTR3 remains a passenger of those ectosomes. IMCD3-[pCrys-APGPR161NG3];β-arrestin2−/− cells (C) or IMCD3-[pEF1αΔ-APSSTR3NG];β-arrestin2−/− cells (D) were imaged in the NeonGreen channel every 10 min following addition of SAG or sst. Yellow and red triangles point to the base and tip of cilia and the ectosome is circled.
(E) The NeonGreen intensity of ectosomes observed in experiments presented in Fig. 3A–E was quantified and normalized to the total NeonGreen intensity of the parent cilium. (n=11–43 ectosomes).
(F) Prolonged signaling and pervasive ectocytosis result in depletion of ciliary SSTR3. APSSTR3NG was pulse-labeled by SA647 and analyzed by fixed imaging after vehicle, sst, or SAG treatment for 6 or 9 h. Error Bars: 95% Confidence Interval. * indicates p<0.03 and ** for p<0.001 decrease after 9 h (Mann-Whitney U test). (n=50–140 cilia per point).
(A) Diagram of the carboxyterminal tail (C-tail) of SSTR3 highlighting truncations and mutations used in Fig. 4C. The third intracellular loop is labeled ‘i3’ and the C-tail is boxed with the amino acid sequence displayed to the right. C-tail residues were numbered 1–99 starting immediately after the NPxxY(x)5F motif which terminates the last transmembrane segment. SSTR3Δ1–25 indicates that the first 25 residues of the C-tail were deleted. SSTR3SA/TA is a variant where all 13 C-tail serine and threonine residues (asterisks) are mutated to alanine.
(B) Additional example of NPY2R ectocytosis from a wild-type cell. IMCD3-[pEF1αΔ-NPY2RNG] cells were imaged by NeonGreen fluorescence every 10 min for the 2 h following addition of NPY3–36. Yellow and red triangles point to the base and tip of cilia and the ectosome is circled.
(C) Activated NPY2R accumulates into tip foci irrespective of genotype. To score the frequency of tip foci, IMCD3-[NPY2RNG] cell lines were imaged by NeonGreen fluorescence every 10 min for 2 h following addition of NPY3–36 or vehicle and tip foci scored. Data were fit to a simple exponential. (n=28–79 cilia).
(D) SSTR3 tip foci are stabilized by cytochalasin D. Cytochalasin D-treated IMCD3-[pEF1αΔ-APSSTR3NG];Arl6−/− cells were imaged every hour by NeonGreen following addition of sst. Top: representative image of a long-live tip focus. Bottom: quantitation of 4 similar foci plotted as the fraction of ciliary SSTR3 in the focus.
(E) Genotypes and drug treatments used in this study do not significantly affect cilia movement in IMCD3 cells. IMCD3-[APSSTR3NG] or IMCD3-[NPY2RNG] cells with the indicated genotype or drug treatment were imaged by NeonGreen fluorescence every 10 min for 2 h following addition of agonist (sst or NPY3–36). The bar graph reports the average distance traversed by the ciliary base during 2 h in the presence of agonist. Error bars represent SD. No condition was statistically different by ANOVA. (n=30–41 cilia).
(F) GPR161 ectocytosis as seen on vertically oriented cilia. The dorsal surface of IMCD3-[pCrys-APGPR161NG3];β-arrestin2−/− cells was imaged every 10 min for the 2 h following addition of SAG. Tip foci and ectosomes were frequently observed, and ectosomes were rapidly lost into the media. The cilia appear misshaped as they span several focal planes at an angle. Yellow and red triangles point to the base and tip of cilia and the ectosome is circled.
(G) Cytochalasin D does not block SSTR3 endocytosis from the plasma membrane. Wild-type, cytochalasin D-treated, and β-arrestin1/2−/− IMCD3-[APSSTR3NG] cells were imaged by NeonGreen fluorescence immediately after addition of sst (t0) and 10 min later (t10). The brightest signal in each image is from cilia-localized SSTR3 but the contrast is adjusted to reveal SSTR3 on the plasma membrane and internal structures. After 10 min of sst, endocytosis results in more SSTR3 at internal structures in wild-type and cytochalasin D-treated while all SSTR3 signal remains at the surface of β-arrestin1/2−/− cells. Examples of internalized SSTR3 structures are circled. All scale bars are 4 µm.
(H) Counting of endocytic vesicles observed in experiments presented in Fig. S4G. Internal structures were counted by NeonGreen fluorescence immediately after addition of sst (t0) and 10 min later (t10). (n=25 cells). These results are in agreement with previous observations that actin poisons have only mild effects on clathrin-mediated endocytosis in mammalian cells unless membrane tension is high or very high doses of inhibitors are used (Mooren et al., 2012)
(A) Screening for proteins necessary for removing NPY2R from cilia. IMCD3-[pEF1αΔ-NPY2RNG] cells were depleted of the indicated proteins before treatment with Emetine, and NPY3–36 or vehicle for 4 h followed by fixation and measurement of ciliary NeonGreen levels. (n=34–121 cilia).
(B–D) Immunoblots of IMCD3 cells treated with siRNAs targeting Drebrin (B), Kif13b (C), Myosin 6 (D), or non-targeting controls for 48 h prior to 16 h starvation. Annexin V and HSP60 were used as loading controls.
(A–E) F-actin and several actin-regulated proteins are required for removal of NPY2R but not SSTR3. Emetine-treated IMCD3-[pEF1αΔ-NPY2RNG] or SA647 pulse-labeled IMCD3-[pEF1αΔ-APSSTR3NG] cells were assayed for GPCR removal. For NPY2R, cells were treated with vehicle or NPY3–36 for 4 h before fixation and quantification of ciliary NeonGreen (box plots). For SSTR3, cilia were imaged by SA647 every 10 min for 2 h following addition of sst. Data were linearly fitted (scatter plots). Cells were treated with (A) DMSO or the Drebrin inhibitor BTP2, (B) the Ca2+ channel inhibitors LaCl3 or 2-APB, (C) the myosin 6 inhibitor TIP, (D) the actin poisons jasplakinolide (Jas) or latrunculin A (LatA), the Arp2/3 inhibitor CK-636, or (E) the Formin 2 inhibitor SMIFH2. For (E), IMCD3-[pEF1αΔ-NPY2RtandemTomato] cells were used as SMIFH2 resulted in greater background fluorescence in the NeonGreen channel. For axis scaling, outliers were omitted. (n=76–187 cilia for NPY2R fixed imaging, n=11–13 cilia for SSTR3 live-cell imaging).
(F) Myosin 6 antibody staining of the ciliary tip is specific. Ift27−/− cells were treated with siRNA targeting myosin 6 or a control siRNA for 48 h. Cells were then starved for 16 h and treated with DMSO or SAG for 1 h, fixed, and immunostained for endogenous myosin 6. Myosin 6 tip localization on dorsal cilia was scored. Error bars: SD between microscope fields. (n=185–186 cilia).
(G) Drebrin localizes to the tips of cilia. β-arrestin2−/− cells were starved for 16 h then treated with SAG for 1 h, fixed, and immunostained for endogenous drebrin. Drebrin localized at the cilia tip, distal to the basal body marker ninein. Scale bar: 4 µm.
(A) β-Arrestin2 is necessary for efficient removal of GPR161 from cilia. WT, β-arrestin2−/− or β-arrestin1/2−/− IMCD3 cells were starved for 24 h, treated with either vehicle or SAG for 2 h, fixed, and immunostained for endogenous GPR161. GPR161-positive cilia were manually counted. The quantitation of GPR161 ciliary levels within the same data set (Fig. 7A) demonstrates that, while GPR161 remains detectable in nearly all cilia of SAG-treated β-arrestin1/2−/− and β-arrestin2−/− IMCD3 cells, the ciliary levels of GPR161 are reduced upon SAG addition to β-arrestin1/2−/− and β-arrestin2−/− IMCD3 cells. (n=93–151 cilia).
(B and C) Retrieval and ectocytosis ensure appropriate Gli3 processing. Wild-type or Ift27−/− cells were starved for 24 h, and then treated with vehicle or SAG and either DMSO, the drebrin inhibitor BTP2 or the myosin 6 inhibitor TIP for 8 h. Lysates from (B) IMCD3 cells or (C) MEF were immunoblotted for Gli3 and Annexin 5 used as a loading control.