Abstract
Objectives
Attaining high quality RNA from the tissues or organs of deceased donors used for research can be challenging due to physiological and logistical considerations. In this investigation,
Methods
RNA Integrity Number (RIN) was determined in pancreatic samples from 236 organ donors and utilized to define high (≥6.5) and low (≤4.5) quality RNA. Logistic regression was used to evaluate the potential effects of novel or established organ and donor factors on RIN.
Results
Univariate analysis revealed donor cause of death (Odds Ratio [OR]=0.35, 95% Confidence Interval [CI]=0.15–0.77, p=0.01), prolonged tissue storage prior to RNA extraction (OR=0.65, 95%CI 0.52–0.79, p<0.01), pancreas region sampled (multiple comparisons, p<0.01), and sample type (OR=0.32, 95%CI 0.15–0.67, p<0.01) negatively influenced outcome. Conversely, duration of final hospitalization (OR=3.95, 95%CI 1.59–10.37, p<0.01) and sample collection protocol (OR=8.48, 95%CI 3.96–19.30, p<0.01) positively impacted outcome. Islet RNA obtained via laser capture microdissection improved RIN when compared to total pancreatic RNA from the same donor (∆RIN=1.3, 95%CI 0.6–2.0, p<0.01).
Conclusions
A multivariable model demonstrates that autopsy- and biopsy-free human pancreata received, processed, and preserved at a single center, using optimized procedures, from organ donors dying of anoxia with normal lipase levels increase the odds of obtaining high-quality RNA.
Keywords: pancreas, human, RNA quality, organ donors, diabetes, RIN
INTRODUCTION
Studies employing the use of RNA are routine and widespread. A measure has been broadly accepted that standardizes RNA quality evaluation,1 and isolation protocols have been commercially available and in use for several years. However, even in the presence of uniform sample treatment, handling, and RNA storage methods, the quality of RNA can vary extensively in human tissue specimens.2 This is particularly true of the pancreas, where elevated levels of ribonuclease1 (RNAse1), a double-stranded RNA-degrading enzyme, are present.3–5 For example, when compared to a panel of tissues taken from different donors6 or the same individual,2 RNA quality of most pancreas samples were found to be lower than ½ to ¾ of all tissue types tested, respectively.
A limited number of studies using the human pancreas have sought to identify the factors that may contribute to RNA quality outcomes.2,6–8 Tauriainen et al. found that the storage of pancreas tissue up to 6 years in RNAlater, a medium that inhibits RNAse activity, preserved the quality of RNA once extracted.7 Likewise, Rudloff et al. did not find any quality differences in RNA extracted from samples stored for different time periods in Optimal Cutting Temperature (OCT) compound, a medium that is used for tissue embedding prior to sample sectioning.8 Zeugner et al. showed that vacuum-sealing of tissue for short-term storage prior to RNA extraction can minimize the drop in RNA quality.6 The Genotype-Tissue Expression (GTEx) Consortium reported that RNA quality dropped in samples with longer ischemia times,2 a finding contrary to Taurianinen7 and Rudloff8; however, sample types and preservation methods differed between studies.
Conversely, using non-pancreas human tissue, several factors have been examined. Variables involving time have consistently been shown to negatively affect RNA quality outcomes, including longer post-mortem interval,9–11 biopsy sample12 or tissue recovery13 periods, and sample processing times.13,14 The manner of death can also affect RNA quality; high agonal factor scores, a quantification of certain conditions and their duration during death, are associated with degraded RNA.15 A collection of other factors have been reported to be important in maximizing RNA quality outcomes, including neutral to basic tissue pH,16 tissue with normal pathology,11 sample preservation in RNAlater,17 and the use of alternative nucleic acid extraction methods.18
Although these studies are informative, the applicability of the findings to a biobank,19 primarily composed of human pancreas samples from deceased organ donors, is unknown. Most of the available research is based on samples obtained from surgical biopsy,6,12,18 autopsy donations,9–11,13,15,16 or the placenta14,17; very few studies include the use of pancreata from human organ donations.2,7 This is important because the logistics and timing of organ or tissue recovery are very different depending on the source of the samples. Moreover, many studies used only tumor6,8 or diseased11,12,16 tissue, did not access RNA quality in a comparable manner,7,10,15 nor were measures of outcome statistically quantified.2,6,7,10 The importance of these previously established, and newly proposed, variables in maximizing human pancreas RNA quality have not been evaluated alone or in combination with one another. Therefore, this study was performed to identify predictors that increase the odds of obtaining high quality human pancreatic RNA in samples made available through the Network for Pancreatic Organ Donors with Diabetes (nPOD) program.20,21 This initiative has been in operation for over 9 years and includes a biobank composed mainly of pancreatic tissue obtained from deceased whole-organ donors in the United States recovered under stringent standard operating procedures.19
MATERIALS AND METHODS
Human Pancreatic Donors
Following acquisition of informed research consent from next of kin, from October 16, 2006 to October 24, 2014, 297 human pancreata were obtained from deceased donors in the United States and centrally shipped to the nPOD program21 at the University of Florida for processing, as previously described.19,22,23 Excluded from consideration were 22 autopsy cases and 12 organ donors with insufficient samples. Of the remaining 263 organ donors, pancreatic samples were available and stored for RNA extraction as fresh frozen blocks embedded in OCT media (Sakura Finetek, Torrance, Calif.) or minced tissue preserved in liquid nitrogen with RNAse inhibitor (RNAlater; Ambion, Foster City, Calif.). RNA extraction and quality quantification was successfully performed from 236 donors (n=242 samples). All experiments were conducted under the approval of the University of Florida Institutional Review Board.
Total RNA Extraction
Approximately 25 mg of tissue from each sample was disrupted and homogenized using VWR® disposable pellet mixer. Total pancreatic RNA extraction was performed, as per manufacturer’s instructions, using the RNeasy Plus Mini Kit (Qiagen, Germantown, Md.) and included residual genomic DNA removal using gDNA eliminator columns. Following extraction, RNA was eluted in RNAse/DNAse-free water. Purity and concentration of extracted RNA were assessed using Epoch Microplate Spectrophotometer (BioTek, Winooski, Vt.). Tissue RNA extraction was performed at the University of Florida, and done in the same manner for both vials and blocks.
Laser-capture was conducted on OCT slides from 37 of the same 45 blocks used for total pancreatic RNA extraction. Slides were fixed in cold 100% methanol for 1 min, then dehydrated with a graded ethanol series (75%, 95%, and 100%) for 1 minute each, followed by Xylene for 5 minutes, then left to air-dry for 10 minutes. Laser-capture microscopy was conducted on an Arcturus Pixcell II Laser-Capture Microdissection (LCM) system (Arcturus Bioscience, Mountain View, Calif.). Methods described by Marselli et al. were used to take advantage of the natural auto-fluorescence of human islets to identify and capture only the islet tissue from these pancreatic sections.24 RNA was extracted from each LCM sample using the Arcturus PicoPure RNA Isolation Kit, per manufacturer’s protocol (Applied Biosystems, Grand Island, N.Y.). Islet RNA extraction was performed at the University of Tennessee.
Determination of RNA Integrity Number (RIN)
RNA integrity, expressed as RIN, was uniformly determined using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, Calif.). Briefly, 1µl of each RNA sample was electrophoretically separated on a microfluidic RNA 6000 Nano chip according to the manufacturer’s protocol. RIN was calculated by the 2100 Bioanalyzer Expert software (version: B.02.08.S1 648 [SR2]) using an algorithm based on a quantification estimate, that includes the ratio of present ribosomal RNA fragments (28S and18S), height of the 28S peak, fast area ratio (the fraction of the area in the fast region compared to the total area under the curve), and marker height, as previously described.1
Study Outcome
The primary outcome variable is RIN, originally designed to assess RNA quality on a scale of 1–10, where 1 represents complete degradation and 10 perfectly intact.1 To help define ‘low’ and ‘high’ RIN values, samples were grouped into tertiles as follows: first tertile (1.9–4.5 RIN; n=78 samples), second tertile (4.6–6.3 RIN; n=88 samples), and third tertile (6.4–8.7 RIN; n=76 samples). Samples in the second tertile were excluded from further analysis, consistent with studies that show RIN values within this range represent partially degraded RNA.25,26 As such, the analysis focused on factors that associated with high RIN values. Based on this approach, RNA was considered low-quality if RIN ≤4.5 (n=75 samples) and high-quality if RIN >6.5 (n=68 samples). A reduction in number from 78 to 75 and 69 to 68, in the low and high-quality groups, respectively, was due to the exclusion of donors sampled more than once; All other samples represented 1 donor each. A total of 143 pancreatic samples were analyzed to determine factors that may increase the odds of obtaining high-quality RNA.
Datasets
This study used data provided by the nPOD program and the Organ Procurement and Transplantation Network (OPTN). The OPTN data system includes data on all donor, wait-listed candidates, and transplant recipients in the US, submitted by the members of the OPTN. The Health Resources and Services Administration (HRSA), U.S. Department of Health and Human Services provides oversight to the activities of the OPTN contractor. All analyzed data can be accessed online, at https://npoddatashare.coh.org, to those with verified login credentials.
Statistical Analysis
Percentages are reported for all categorical variables. For continuous variables, the measure of central tendency was described using either the mean ± 1 standard deviation (SD) or median plus range (min, max), based on the distribution of values and evaluation of skewness and kurtosis.
Univariate logistic regression (ULR) model testing was used for all variables considered for analysis. Chi-square p-values, the corresponding odds ratio (OR) and the 95% confidence interval (95% CI) are reported for all categorical variables. In the case of continuous variables, the OR and 95% CI are defined based on units indicated in the parenthesis for each factor examined, unless otherwise noted. The profile likelihood method was used to generate the reported statistical measures.
Multivariable logistic regression (MLR) was used to test for the simultaneous effects of multiple predictors. Candidate variables included all those from the ULR analysis with p<0.20. To identify collinear terms in candidate models, a matrix of pairwise correlations was generated for all continuous variables, defined as any two factors with p<0.05 and Pearson's correlation coefficient (r) ≥0.80. Multicollinearity for continuous variables was defined when the variance inflation factor (VIF) was abnormally larger than 1, as previously described.27 Multiclass categorical variables were independently screened for relationships using either a chi-square, fisher’s exact, or the freeman-halton extension of the fisher’s exact test. Association between categorical variables was defined as any two factors with p<0.05 and a Cramer’s V value of >0.30 (for nominal versus nominal or multi-class nominal versus ordinal data) or a Kendall’s Tau –b (for a × a tables) or -c (for a × b tables) value of <−0.30 or >0.30 (for ordinal versus ordinal data or dichotomous nominal versus ordinal). Interaction terms were generated using a priori hypotheses for greater-than-additive effects on the dependent variable. The MLR model was constructed using a 10-fold cross-validation approach. Model performance was evaluated via 1) the area under the receiver operator characteristic curve (AUROC),28 and 2) the Hosmer-Lemeshow goodness of fit test29,30 using the penalized likelihood method of Firth31 to reduce bias in maximum likelihood parameter estimates where sample size is limited. An AUROC=0.5 is considered as having no discriminatory power, 0.70≤AUROC≤0.8 as acceptable, 0.8≤AUROC≤0.9 as excellent, and AUROC≥0.9 as outstanding, as previously defined.30 A Hosmer-Lemeshow p-value>0.05 indicates a good model fit. A previously developed SAS macro was adopted for implementation of the 10-fold cross-validation approach.32 All reported p-values are 2-sided, and considered significant if <0.05. All statistical analysis was performed using SAS software version 9.4 TS Level 1M1 (SAS Institute, Cary, N.C.).
RESULTS
Samples from 143 organ donors (Supplemental Digital Content, Table S1) were used in this study to help determine predictors that increase the odds of obtaining high quality total RNA from the pancreas.
Univariate Analysis
Only one donor variable had a significant correlation with high quality RNA (Table 1). Overall, cause of death was determined to be an important factor (p=0.03); relative to those succumbing to anoxia, samples from individuals with head trauma were less likely to result in high-quality RNA (OR=0.35, p=0.01).
Table 1.
Donor Variables Examined for Univariate Association with RNA Quality Outcome
| Name | ≤4.5 RIN | ≥6.5 RIN | Odds Ratio |
95% CI | p-value | ||
|---|---|---|---|---|---|---|---|
| n | % or ValueA | n | % or ValueA | ||||
| Cause of Death | 0.03 | ||||||
| Anoxia | 20 | 37% | 34 | 63% | - | - | - |
| Cerebrovascular/StrokeB | 19 | 56% | 15 | 44% | 0.46 | (0.19–1.11) | 0.09 |
| Head Trauma | 29 | 63% | 17 | 37% | 0.35 | (0.15–0.77) | 0.01 |
| OtherC | 6 | 75% | 2 | 25% | 0.20 | (0.03–0.94) | 0.06 |
| C-peptideD,E (ng/mL) | 69 | 2.01 (0.00,26.42) | 64 | 0.44 (0.00,23.18) | 0.95 | (0.88–1.01) | 0.10 |
| Pancreas LipaseE,F (u/L) | |||||||
| Normal | 41 | 51% | 39 | 49% | - | - | - |
| Elevated or Critical | 16 | 67% | 8 | 33% | 0.526 | (0.19–1.34) | 0.18 |
| Pancreas AmylaseE,F (u/L) | 0.59 | ||||||
| Normal | 41 | 58% | 30 | 42% | - | - | - |
| Low | 6 | 43% | 8 | 57% | 1.82 | (0.58–6.06) | 0.31 |
| Elevated or Critical | 10 | 56% | 8 | 44% | 1.09 | (0.38–3.10) | 0.87 |
| Kidney CreatinineE,F (mg/dL) | 0.71 | ||||||
| Normal | 34 | 56% | 27 | 44% | - | - | - |
| Low | 3 | 43% | 4 | 57% | 1.68 | (0.34–9.13) | 0.52 |
| Elevated or Critical | 29 | 59% | 20 | 41% | 0.87 | (0.40–1.86) | 0.72 |
| Blood pHE | 72 | 7.4 (±0.2) | 64 | 7.4 (±0.1) | 2.09 | (0.17–67.36) | 0.57 |
| HematocritE,F (%) | |||||||
| Normal | 9 | 69% | 4 | 31% | - | - | - |
| Low | 63 | 51% | 60 | 49% | 2.14 | (0.66–8.25) | 0.21 |
Number and % are reported for all categorical variables. Use of mean (± 1 SD) or median (min, max) for continuous variables.
Donors with cause of death originally noted as cerebral edema (1) and other:cardiovascular (1) were grouped into this category.
Includes perinatal condition/birth defect (2), meningitis (2), spinal cord injury (1), cystic fibrosis (1), respiratory distress/failure (1), and diabetic ketoacidosis (1).
A fill value of 0.001 was used when assay results <0.05 (lower limit of detection)
Taken within 24 hrs prior to or at death
Time also played a key role in obtaining RNA of high quality (Table 2). The length of the final hospitalization stay was an important factor (p=0.01); relative to those dying < 3 days after being admitted, samples from donors who stayed in the hospital > 6 days were more likely to yield high-quality RNA (OR=3.95, p<0.01). Tissue storage time was also found to influence RNA quality outcome. For every year spent in storage prior to RNA extraction, there was a decrease in the odds of obtaining high-quality RNA (OR=0.65, p<0.01).
Table 2.
Univariate Influence of Time on RNA Quality Outcome
| Name | ≤4.5 RIN | ≥6.5 RIN | Odds Ratio |
95% CI | p-value | ||
|---|---|---|---|---|---|---|---|
| n | % or ValueA | n | % or ValueA | ||||
| DowntimeB (minutes) | 66 | 0(0,75) | 63 | 2(0,90) | 1.02 | (1.00–1.04) | 0.06 |
| Length of Final Hospitalization StayC (days) |
0.01 | ||||||
| <3 | 30 | 61% | 19 | 39% | - | - | - |
| ≥3 to <6 | 25 | 52% | 23 | 48% | 1.46 | (0.65–3.28) | 0.36 |
| ≥6 | 10 | 29% | 25 | 71% | 3.95 | (1.59–10.37) | <0.01 |
| Warm Ischemia TimeD (min) | 11 | 24.2 (±12.1) | 13 | 18.8 (±5.8) | 0.94 | (0.83–10.30) | 0.16 |
| Pancreas Transport TimeE (hrs) | 70 | 14.7 (±5.4) | 68 | 15.1 (±7.3) | 1.01 | (0.96–1.07) | 0.6825 |
| Tissue Storage Time Prior to RNA Extraction (years) |
75 | 2.3 (0.0,6.8) | 68 | 0.5 (0.0,6.2) | 0.65 | (0.52–0.79) | <0.01 |
Number and % are reported for all categorical variables. Use of mean (± 1 SD) or median (min, max) for continuous variables.
Downtimes were noted in13 charts as greater than X; in those cases, the value of X was taken as the downtime.
Defined as the time from admission to aortic cross-clamp. Grouping based upon a previously published report55
Defined as the time from cardiac death to aortic cross-clamp, applicable only to non-heart beating donors. The remaining samples were from brain-dead donors with no elapsed time prior to organ retrieval.
Indicates time from aortic cross-clamp to laboratory receipt.
Once samples were processed for biobank storage in the laboratory, factors related to the type of sample and preservation procedures used were found to influence RNA quality outcome (Table 3). Overall, pancreas region was shown to be an important variable (p<0.01); in particular, relative to the head, samples taken from either the body (OR=0.11, p<0.01) or the tail (OR=0.20, p<0.01) had deceased odds of obtaining high-quality RNA. Samples from tissue blocks resulted in deceased odds of obtaining high-quality RNA (OR=0.32, p<0.01), relative to minced tissue stored in vials. The order of sample preservation was shown to be critical; the preparation of vials (containing RNALater) followed by tissue blocks (preserved in OCT or formalin-fixed paraffin embedded media) was more likely to result in high-quality RNA (OR=8.48, p<0.01) versus when performed in the opposite order.
Table 3.
Univariate Effects of Sample Processing on RNA Quality Outcome
| Name | ≤4.5 RIN | ≥6.5 RIN | Odds Ratio |
95% CI | p-value | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| Region of Pancreas SampledA | <0.01 | ||||||
| Head | 23 | 34% | 45 | 66% | - | - | - |
| Body | 14 | 82% | 3 | 18% | 0.11 | (0.02–0.38) | <0.01 |
| Tail | 35 | 71% | 14 | 29% | 0.20 | (0.09–0.45) | <0.01 |
| Sample TypeB | |||||||
| Vials | 43 | 44% | 55 | 56% | - | - | - |
| Blocks | 32 | 71% | 13 | 29% | 0.32 | (0.15–0.67) | <0.01 |
| Staff Performing RNA Extraction | 0.53 | ||||||
| #1 | 50 | 49% | 51 | 51% | - | - | - |
| #2 | 23 | 59% | 16 | 41% | 0.68 | (0.32–1.43) | 0.32 |
| #3 | 2 | 67% | 1 | 33% | 0.49 | (0.02–5.27) | 0.57 |
| Sample Collection ProceduresC | |||||||
| Original Protocol | 63 | 71% | 26 | 29% | - | - | - |
| New Protocol | 12 | 22% | 42 | 78% | 8.48 | (3.96–19.30) | <0.01 |
Excluded 3 cases where region sampled was unknown
Vials contain minced tissue preserved in RNAlater; Blocks are composed of fresh frozen tissue embedded in OCT media. Vials and blocks represent total pancreatic RNA extracts. Laser capture islet-RNA was not included here.
Major difference is that, initially, after cleaning the pancreas, vials were routinely prepared following tissue block preservation but occasionally performed simultaneously, i.e. original protocol. Later, beginning with nPOD caseID 6170, vials were prepared first, before tissue block preservation, i.e. new protocol.
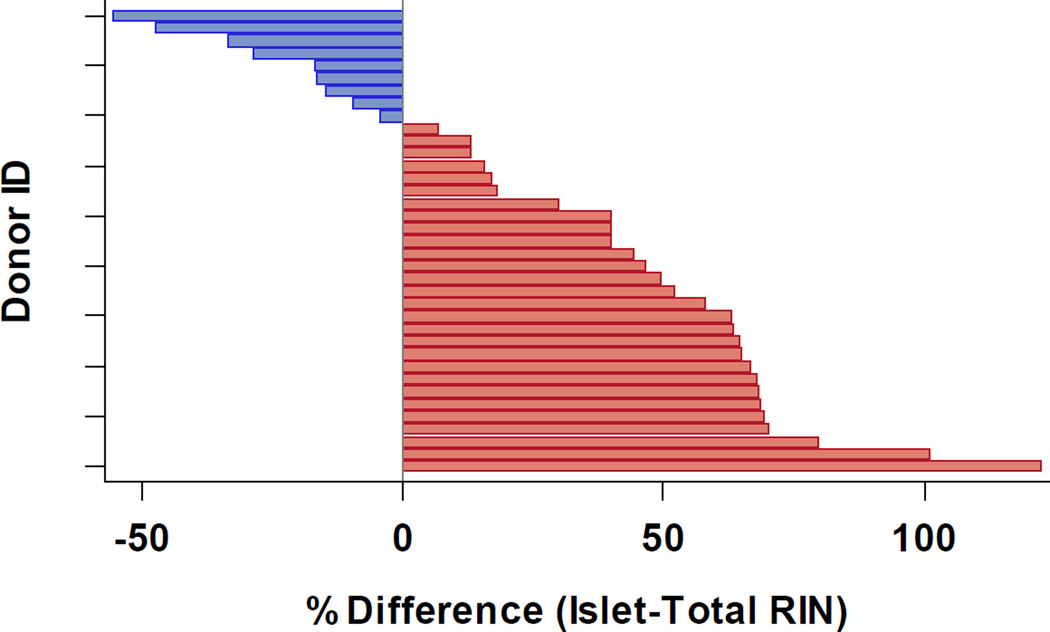
Overall RNA quality was also found to be higher in LCM islet samples versus total pancreatic tissue taken from the same donor (Figure 1). There was a statistically significant increase in mean RIN values when using islet (5.7±1.2) versus total (4.4 ±2.1) RNA obtained from the same donor (1.3, 95%CL 0.6 to 2.0, p<0.01; two-tailed paired t-test).
Figure 1. Comparison of RIN Values from Islet and Total RNA using the Same Donor Pancreas.
RIN values from total RNA (both low and high quality) were compared to those from LCM islets obtained from the same pancreas (n=37 donors). In 9 cases (blue), RIN from total RNA was found to be a median of 16.7 absolute percentage points higher (4.3 min, 55.6 max) than that of islet RNA. In 28 cases (red), RIN from islet RNA was found to be 55.3 percentage points higher (6.9, 122.4) than that of total RNA.
Multivariable Analysis
To determine if a multifactorial influence on RNA quality was present, all variables from tables 1–3 with p<0.20 were considered as candidate factors in an MLR model. Two exceptions were made: 1) warm ischemia time was excluded due to small sample size, and 2) pancreas transport time was considered further because organ transit time was found to be significant in a previous study.9
A total of 10 variables were identified as candidate factors and screened for collinearity, multicollinearity, and interactions. Relationships existed between region of pancreas sampled and sample type (p<0.01, Cramer’s V=0.72), region of pancreas sampled and lipase (p<0.01, Cramer’s V=0.32), region of pancreas sampled and order of sample collection (p<0.01, Cramer’s V=0.44), lipase and sample type (p<0.01, tau-b=−0.38), and sample type and order of sample collection (p<0.01, tau-b=0.47). Region of pancreas sampled and sample type were thus, dropped from consideration as candidate variables in the MLR model. No other collinear or multicollinear terms were identified. Interactions were identified and judiciously chosen, based, when available, on evidence from the literature; terms included tissue storage time by sample collection procedures and downtime by length of hospitalization stay. A total of 8 variables and 2 interaction terms were considered for MLR model building.
An MLR model was created using a 10-fold cross validation approach (Table 4). There were 85 samples with complete data for all candidate variables used in the MLR model, a reduction from 143 used in the ULR testing. Interactions terms were initially included, found not to be significant (p>0.10), and removed before running the final model. The AUROC for the model was found to be 0.78 (±0.15) and the Hosmer-Lemeshow p-value 0.91, indicating acceptable discrimination and good calibration (i.e. fit), respectively. A total of 3 factors were found to affect the odds of obtaining high-quality RNA (Table 4). Similar to the univariate analysis, cause of death altered the odds of obtaining RNA with high RIN values; donors succumbing to head trauma (OR=0.18, p=0.02) decreased the odds of obtaining high-quality RNA, when compared to the anoxic group. Samples from donors with elevated or critical levels of lipase decreased the odds of obtaining high-quality RNA (OR=0.22, p=0.05). Finally, the order and method of tissue preservation affected the odds of obtaining high-quality RNA; compared to tissue block (1st) vial (2nd) preparation, when vials were prepared prior to blocks, the odds of obtaining high-quality RNA in both sample types increased (OR=4.70, p=0.02).
Table 4.
Multivariable Logistic Regression Model of Factors Affecting RNA Quality Outcome
| NameB,D | ≤4.5 RIN | ≥6.5 RIN | Odds RatioE |
95% CI | p-value | ||
|---|---|---|---|---|---|---|---|
| n | % or ValueA | n | % or ValueA | ||||
| Cause of Death | |||||||
| Anoxia | 11 | 35% | 20 | 65% | - | - | - |
| Cerebrovascular/Stroke | 10 | 50% | 10 | 50% | 0.28 | (0.05–1.30) | 0.13 |
| Head Trauma | 22 | 71% | 9 | 29% | 0.18 | (0.04–0.71) | 0.02 |
| Other | 2 | 67% | 1 | 33% | 0.37 | (0.02–4.47) | 0.49 |
| C-peptide (ng/mL) | 45 | 1.17 (0.00,16.76) |
40 | 0.59 (0.00,20.55) |
0.97 | (0.87–1.08) | 0.59 |
| Pancreas Lipase (u/L) | |||||||
| Normal | 31 | 47% | 35 | 53% | - | - | - |
| Elevated or Critical | 14 | 74% | 5 | 26% | 0.22 | (0.04–0.89) | 0.05C |
| Downtime (minutes) | 45 | 0 (0,75) | 40 | 5 (0,90) | 1.02 | (0.98–1.05) | 0.37 |
| Length of Final Hospitalization Stay (days) |
|||||||
| <3 | 20 | 67% | 10 | 33% | - | - | - |
| ≥3 to <6 | 20 | 59% | 14 | 41% | 1.21 | (0.35–4.23) | 0.77 |
| ≥6 | 5 | 24% | 16 | 76% | 3.21 | (0.75–15.24) | 0.14 |
| Pancreas Transport Time (hrs) | 45 | 15.0 (±5.3) | 40 | 15.2 (±7.6) | 1.01 | (0.92–1.10) | 0.88 |
| Tissue Storage Time Prior to RNA Extraction (years) |
45 | 1.5 (0.0,6.8) | 40 | 0.5 (0.0,6.1) | 0.82 | (0.53–1.20) | 0.34 |
| Sample Collection Procedures | |||||||
| Original Protocol | 36 | 69% | 16 | 31% | - | - | - |
| New Protocol | 9 | 27% | 24 | 73% | 4.70 | (1.32–17.96) | 0.02 |
Number and % are reported for all categorical variables. Use of mean (± 1 SD) or median (min, max) for continuous variables.
Actual value 0.0496
Interaction terms storage time by protocol and downtime by hospital stay were not found to be significant (p>0.10) and excluded from the final model.
Represents MLR model-based OR values
DISCUSSION
The objective of this study was to investigate the influence of established and novel factors in obtaining high-quality RNA, dichotomized as either high or low RIN values, from donated human pancreata of deceased organ donors in the United States. We developed a multivariable model, based on univariate screening of predictors, that exhibits acceptable discrimination of RNA quality between samples and was shown to be a good fit. The data demonstrate that there are conditions and procedures that contribute to the degradation of RNA prior to extraction, but before or during tissue processing.
This is the first study to show that samples from pancreas donors succumbing to head trauma were less likely to lead to high quality RNA versus those dying from anoxia. Although, to our knowledge, this relationship has never been evaluated, a limited number of studies have examined the influence of organ donor cause of death on other outcomes.33,34 Kaddis et al. found that pancreata from donors who died of head trauma were less likely to lead to successful human islet isolation when compared to the cerebrovascular/stroke group, and although those in the anoxic group did better when compared to the same reference group, the relationship was not found to be statistically significant.34 Beyond the pancreas, Singhal et al. showed that lung and heart transplant recipients had lower incidence of rejection when the donor died of anoxia versus head trauma, and that anoxia was a significant donor predictor of lung recipient outcomes in a multivariable model.33 Sustained stability of the organ donor over time may be required to preserve organ function; marked pathophysiological changes, that affect multiple organs, can take place both before and after donor brain death.35 We found that donors dying from anoxia stayed in the hospital longer than those suffering from head trauma (data not shown), and that the odds of obtaining high quality RNA increased, univariately, when using donors who remained in the hospital for longer periods of time. There was a statistically significant association between cause of death and length of hospitalization stay; however, it did not meet our pre-defined criteria for collinearity and was thus not reported. It may also be that donors dying from head trauma suffer terminal brain injuries that compromise the vasomotor system, thus leading to potentially poor blood circulation and, in turn, an inability to provide oxygen and nutrients to organs and tissues. For example, Belzberg et al. evaluated the hemodynamic patterns of head trauma patients who eventually were declared brain dead and found that measures of heart performance and tissue perfusion values to be low during the pre-brain death period36; others have reported similar findings.37 It is unknown if and how the hemodynamic status of cerebrovascular/stroke donors differs from those dying from head trauma; however, this cause of death was not found to be a statistically significant factor in our study.
The multivariable analysis also identified the importance of pancreatic lipase levels. Critical levels of lipase, as how was defined in this study, are indicative of pancreatic inflammation in the acute form, but can also be representative of other conditions.38 Repeated episodes of inflammation can lead to scar tissue formation, atrophy of the pancreas, and loss of organ function, as can be seen in chronic pancreatitis.39 Thus, the detrimental effects of elevated lipase levels on RNA quality seen here may be suggestive of disease or damage to the pancreas before the time of death.
The only modifiable factor found in this study, under the complete control of a biobank, and statistically significant in both forms of the analysis, is the order in which samples are preserved. Using a previously described method for processing of human pancreata,23 i.e. our “new” protocol, tissue vials were prepared prior to blocks. The process of pancreatic tissue block preparation is time consuming, requiring slicing of the pancreas, from head to tail, into blocks that must be frozen, embedded (alternating between OCT and formalin-fixed paraffin-embedded), and then stored. In contrast, vial preparation occurs rapidly, whereby minced tissue pieces are simply placed into cryovials with or without RNALater, allowed to sit at room temperature for a brief period of time (in vials with RNALater), and rapidly frozen. RNA quality from tissue sitting at room temperature, following a prolonged period of cold storage, has not been examined in detail; however, degradation generally occurs as a function of time to tissue preservation,2,13 although these studies did not account for tissue storage time in the analysis. Nonetheless, by collecting vials first, the time to vial preservation was reduced significantly, with only a negligible increase in the time to block preservation. Unfortunately, the timing of vial and block preparation was not routinely recorded, and thus not analyzable in this study.
Although found to be relevant elsewhere, this study was not able to establish a relationship to outcome of warm ischemia time, pancreas transport time, or C-peptide. The warm ischemia time reported in this study was minimal, and similar to that described by Markmann and colleagues, who found that successful human islet transplantation was possible from such pancreas donors.40 However, prolonged warm ischemia time, not seen here, can result in functionally damaged organs due to the accumulation of metabolites, inadequate oxygenation, and depletion of energy stores.41–43 Next, the GTEx Consortium reported a drop in RNA quality with longer ischemia times, a finding that contradicts previous reports7,8 and this study. The major difference between that study and ours is the use of a cold preservation solution to slow down the organ degradation process and hence provide adequate time for transport without compromising the quality of the pancreas. Moreover, GTEx collects the mid-portion of the pancreas; however, samples for this study were obtained from a biobank that recovers the entire pancreas, often times with the spleen, duodenum, and adipose tissue attached. This, too, may play a role in preserving tissue quality. Finally, although C-peptide is a clinical marker of beta cell function, it was not found to be predictive of pancreatic RNA quality.
There were challenges and limitations in this study. All of the data in Table 1, excluding cause of death and C-peptide, were aggregated from individual laboratories across the United States, each with potentially different normal cut-off values. To address this, previously published general reference value ranges were used for pediatric and adult populations.44,45 Second, due to the rare and limited number of samples in the biobank, several variables and interactions were not forced into the multivariable model despite their clinical relevance; the factors and terms included amylase,46 creatinine,47 hematocrit,48,49 lipase by amylase,50,51 lipase or amylase by creatinine, and lipase or amylase by hematocrit. Additionally, due to collinearity issues, sample type and region of pancreas sampled were excluded from the multivariable modeling, despite statistical significance, but cannot be overlooked. For example, studies from Taurianinen7 and Rudloff8, that examined sample type, found no differences in RNA quality of samples stored for long periods of time in a single tissue storage media; however, when directly comparing RNAlater to OCT, this investigation found that the odds of obtaining high-quality RNA was greater in vials versus blocks, respectively. Likewise, data from Wang52 and Rahier53 suggest regional differences in islet and beta cell volume density distributions across the human pancreas, respectively; if and how this might contribute to RNA quality outcomes, as seen in this study, is an intriguing, yet unexplored area of research. Third, besides the order of sample preservation, another major difference between the collection procedures is that the new protocol routinely involved the use of 3 staff members, versus 1–2 individuals in the old. However, the names and roles of staff members that participated in processing each case, as well as time spent, was not consistently recorded and could not be analyzed, but may have also played a role in preserving RNA quality. Finally, additional studies will need to be performed to determine if the improvements seen in RIN values with the use of LCM-obtained islet vs. total RNA extracts are attributable to sample preparation methods, cell-type specific differences, or other factors.
In conclusion, this study demonstrates that using pancreata from cadaveric organ donors dying of anoxia with normal lipase levels increase the odds of obtaining high quality RNA. Using an optimized tissue collection and processing protocol is not only a key factor in determining RNA quality outcomes, but also important in facilitating the comparison of data across multiple laboratories.
Supplementary Material
Acknowledgments
The authors thank Maria Peterson for abstracting downtime data from donor medical charts and Dr. Amanda Posgai for editing and formatting the manuscript. Organ Procurement Organizations (OPO) partnering with nPOD to provide research resources are listed at http://www.jdrfnpod.org/for-partners/npod-partners. Some of the data reported here have been supplied by the UNOS as the contractor for the OPTN. The interpretation and reporting of these data are the responsibility of the author(s) and in no way should be seen as an official policy of or interpretation by the OPTN or the U.S. Government.
Source of Funding: This research was performed with the support of the nPOD, a collaborative type 1 diabetes research project, and sponsored by the Juvenile Diabetes Research Foundation (grant numbers 25-2013-268, 25-2012-380, and 25-2007-874 to MAA, including a subcontract to JSK; 47-2013-520 to ICG and Clayton Mathews) and the National Institute of Diabetes Digestive and Kidney Diseases (grant number DK104155-01 to ICG and MCT).
Abbreviations used throughout
- AUROC
Area under the receiver operator characteristic curve
- GTEx
Genotype-Tissue Expression
- LCM
Laser-Capture Microdissection
- MLR
Multivariable logistic regression
- nPOD
Network for Pancreatic Organ Donors with Diabetes
- OCT
Optimal Cutting Temperature
- OPTN
Organ Procurement and Transplantation Network
- RIN
RNA Integrity Number
- ULR
Univariate logistic regression
- VIF
Variance Inflation factor
Footnotes
Conflicts of Interest: The authors of this manuscript have conflicts of interest to disclose as described by Pancreas. MAA and AP serve as executive directors of the nPOD program, and JSK directs its data management core. The authors declare that there are no other conflicts of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
Table S1. General Organ Donor Characteristics of Assessed nPOD Case Samples.
REFERENCES
- 1.Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weickmann JL, Glitz DG. Human ribonucleases. Quantitation of pancreatic-like enzymes in serum, urine, and organ preparations. J Biol Chem. 1982;257:8705–8710. [PubMed] [Google Scholar]
- 4.Futami J, Tsushima Y, Murato Y, et al. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 1997;16:413–419. doi: 10.1089/dna.1997.16.413. [DOI] [PubMed] [Google Scholar]
- 5.Sorrentino S. The eight human “canonical” ribonucleases: Molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010;584:2194–2200. doi: 10.1016/j.febslet.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 6.Zeugner S, Mayr T, Zietz C, et al. RNA quality in fresh-frozen gastrointestinal tumor specimens-experiences from the tumor and healthy tissue bank TU Dresden. Recent Results Cancer Res. 2015;199:85–93. doi: 10.1007/978-3-319-13957-9_9. [DOI] [PubMed] [Google Scholar]
- 7.Tauriainen S, Salmela K, Rantala I, et al. Collecting high-quality pancreatic tissue for experimental study from organ donors with signs of β-cell autoimmunity. Diabetes Metab Res Rev. 2010;26:585–592. doi: 10.1002/dmrr.1129. [DOI] [PubMed] [Google Scholar]
- 8.Rudloff U, Bhanot U, Gerald W, et al. Biobanking of human pancreas cancer tissue: impact of ex-vivo procurement times on RNA quality. Ann Surg Oncol. 2010;17:2229–2236. doi: 10.1245/s10434-010-0959-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birdsill AC, Walker DG, Lue L, et al. Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank. 2011;12:311–318. doi: 10.1007/s10561-010-9210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen J, Lesnikova I, Funder AM, et al. DNA and RNA analysis of blood and muscle from bodies with variable postmortem intervals. Forensic Sci Med Pathol. 2014;10:322–328. doi: 10.1007/s12024-014-9567-2. [DOI] [PubMed] [Google Scholar]
- 11.Sheedy D, Say M, Stevens J, et al. Influence of liver pathology on markers of postmortem brain tissue quality. Alcohol Clin Exp Res. 2012;36:55–60. doi: 10.1111/j.1530-0277.2011.01580.x. [DOI] [PubMed] [Google Scholar]
- 12.Dev H, Rickman D, Sooriakumaran P, et al. Biobanking after robotic-assisted radical prostatectomy: a quality assessment of providing prostate tissue for RNA studies. J Transl Med. 2011;9:121. doi: 10.1186/1479-5876-9-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim BJ, Sprehe N, Morganti A, et al. The effect of postmortem time on the RNA quality of human ocular tissues. Mol Vis. 2013;19:1290–1295. [PMC free article] [PubMed] [Google Scholar]
- 14.Jobarteh ML, Moore SE, Kennedy C, et al. The effect of delay in collection and processing on RNA integrity in human placenta: Experiences from rural Africa. Placenta. 2014;35:72–74. doi: 10.1016/j.placenta.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomita H, Vawter MP, Walsh DM, et al. Effect of Agonal and Postmortem Factors on Gene Expression Profile: Quality Control in Microarray Analyses of Postmortem Human Brain. Biol Psychiatry. 2004;55:346–352. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stan AD, Ghose S, Gao XM, et al. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123:1–11. doi: 10.1016/j.brainres.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fajardy I, Moitrot E, Vambergue A, et al. Time course analysis of RNA stability in human placenta. BMC Mol. Biol. 2009;10:21. doi: 10.1186/1471-2199-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali SA, Alman B. RNA extraction from human articular cartilage by chondrocyte isolation. Anal. Biochem. 2012;429:39–41. doi: 10.1016/j.ab.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Campbell-Thompson M, Wasserfall C, Kaddis J, et al. Network for Pancreatic Organ Donors with Diabetes (nPOD): developing a tissue biobank for type 1 diabetes. Diabetes Metab Res Rev. 2012;28:608–617. doi: 10.1002/dmrr.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaddis JS, Pugliese A, Atkinson MA. A run on the biobank: what have we learned about type 1 diabetes from the nPOD tissue repository? Curr Opin Endocrinol Diabetes Obes. 2015;22:290–295. doi: 10.1097/MED.0000000000000171. [DOI] [PubMed] [Google Scholar]
- 21.Pugliese A, Yang M, Kusmarteva I, et al. The Juvenile Diabetes Research Foundation Network for Pancreatic Organ Donors with Diabetes (nPOD) Program: goals, operational model and emerging findings. Pediatr Diabetes. 2014;15:1–9. doi: 10.1111/pedi.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell-Thompson ML, Heiple T, Montgomery E, et al. Staining protocols for human pancreatic islets. J Vis Exp. 2012:e4068. doi: 10.3791/4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell-Thompson ML, Montgomery EL, Foss RM, et al. Collection protocol for human pancreas. J Vis Exp. 2012:e4039. doi: 10.3791/4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marselli L, Thorne J, Ahn YB, et al. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab. 2008;93:1046–1053. doi: 10.1210/jc.2007-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleige S, Walf V, Huch S, et al. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett. 2006;28:1601–1613. doi: 10.1007/s10529-006-9127-2. [DOI] [PubMed] [Google Scholar]
- 26.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Mansfield ER, Helms BP. Detecting Multicollinearity. Am Stat. 1982;36:158–160. [Google Scholar]
- 28.Metz CE. Basic principles of ROC analysis. Semin Nucl Med. 1978;8:283–298. doi: 10.1016/s0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]
- 29.Hosmer DW, Hosmer T, Le Cessie S, et al. A comparison of goodness-of-fit tests for the logistic regression model. Stat Med. 1997;16:965–980. doi: 10.1002/(sici)1097-0258(19970515)16:9<965::aid-sim509>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 30.Hosmer DW, Lemeshow S. Applied Logistic Regression. 2nd. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2000. [Google Scholar]
- 31.Firth D. Bias Reduction of Maximum Likelihood Estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 32.Gu P. Welcome to My SAS Showcase: ROC comparison using stratified k-fold cross-validation in logistic regression adjusting for oversampling. [Accessed May 27, 2015];2013 Dec 30; Available at: https://sasshowcase.wordpress.com/category/sas-macro. [Google Scholar]
- 33.Singhal AK, Sheng X, Drakos SG, et al. Impact of donor cause of death on transplant outcomes: UNOS registry analysis. Transplant Proc. 2009;41:3539–3544. doi: 10.1016/j.transproceed.2009.06.192. [DOI] [PubMed] [Google Scholar]
- 34.Kaddis JS, Danobeitia JS, Niland JC, et al. Multicenter analysis of novel and established variables associated with successful human islet isolation outcomes. Am J Transplant. 2010;10:646–656. doi: 10.1111/j.1600-6143.2009.02962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKeown DW, Bonser RS, Kellum JA. Management of the heartbeating brain-dead organ donor. Br J Anaesth. 2012;108(suppl 1):i96–i107. doi: 10.1093/bja/aer351. [DOI] [PubMed] [Google Scholar]
- 36.Belzberg H, Shoemaker WC, Wo CC, et al. Hemodynamic and oxygen transport patterns after head trauma and brain death: implications for management of the organ donor. J Trauma. 2007;63:1032–1042. doi: 10.1097/01.ta.0000235995.86162.d2. [DOI] [PubMed] [Google Scholar]
- 37.Colombo J, Shoemaker WC, Belzberg H, et al. Noninvasive monitoring of the autonomic nervous system and hemodynamics of patients with blunt and penetrating trauma. J Trauma. 2008;65:1364–1373. doi: 10.1097/TA.0b013e31818cc307. [DOI] [PubMed] [Google Scholar]
- 38.Yadav D, Agarwal N, Pitchumoni CS. A critical evaluation of laboratory tests in acute pancreatitis. Am J Gastroenterol. 2002;97:1309–1318. doi: 10.1111/j.1572-0241.2002.05766.x. [DOI] [PubMed] [Google Scholar]
- 39.Witt H, Apte MV, Keim V, et al. Chronic pancreatitis: challenges and advances in pathogenesis, genetics, diagnosis, and therapy. Gastroenterology. 2007;132:1557–1573. doi: 10.1053/j.gastro.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Markmann JF, Deng S, Desai NM, et al. The use of non-heart-beating donors for isolated pancreatic islet transplantation. Transplantation. 2003;75:1423–1429. doi: 10.1097/01.TP.0000061119.32575.F4. [DOI] [PubMed] [Google Scholar]
- 41.Steinbrook R. Organ Donation after Cardiac Death. New Engl J Med. 2007;357:209–213. doi: 10.1056/NEJMp078066. [DOI] [PubMed] [Google Scholar]
- 42.Dunne K, Doherty P. Donation after circulatory death. Contin Educ Anaesth Crit Care. 2011;11:82–86. [Google Scholar]
- 43.Watson CJE, Dark JH. Organ transplantation: historical perspective and current practice. Br J Anaesth. 2012;108(suppl 1):i29–i42. doi: 10.1093/bja/aer384. [DOI] [PubMed] [Google Scholar]
- 44.Colantonio DA, Kyriakopoulou L, Chan MK, et al. Closing the gaps in pediatric laboratory reference intervals: a CALIPER database of 40 biochemical markers in a healthy and multiethnic population of children. Clin Chem. 2012;58:854–868. doi: 10.1373/clinchem.2011.177741. [DOI] [PubMed] [Google Scholar]
- 45.Pagana KD, Pagana TJ, Pagana TN. Mosby's Diagnostic and Laboratory Test Reference. 12th. St. Louis, Mo.: Elsevier: Mosby; 2014. [Google Scholar]
- 46.Keller J, Layer P. Human pancreatic exocrine response to nutrients in health and disease. Gut. 2005;54(Suppl 6):vi1–vi28. doi: 10.1136/gut.2005.065946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Royse VL, Jensen DM, Corwin HL. Pancreatic enzymes in chronic renal failure. Arch Intern Med. 1987;147:537–539. [PubMed] [Google Scholar]
- 48.Lankisch PG, Mahlke R, Blum T, et al. Hemoconcentration: an early marker of severe and/or necrotizing pancreatitis? A critical appraisal. Am J Gastroenterol. 2001;96:2081–2085. doi: 10.1111/j.1572-0241.2001.03966.x. [DOI] [PubMed] [Google Scholar]
- 49.Banks PA, Freeman ML. Practice Guidelines in Acute Pancreatitis. Am J Gastroenterol. 2006;101:2379–2400. doi: 10.1111/j.1572-0241.2006.00856.x. [DOI] [PubMed] [Google Scholar]
- 50.Lankisch PG, Burchard-Reckert S, Lehnick D. Underestimation of acute pancreatitis: patients with only a small increase in amylase/lipase levels can also have or develop severe acute pancreatitis. Gut. 1999;44:542–544. doi: 10.1136/gut.44.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keim V, Teich N, Fiedler F, et al. A comparison of lipase and amylase in the diagnosis of acute pancreatitis in patients with abdominal pain. Pancreas. 1998;16:45–49. doi: 10.1097/00006676-199801000-00008. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Misawa R, Zielinski MC, et al. Regional Differences in Islet Distribution in the Human Pancreas - Preferential Beta-Cell Loss in the Head Region in Patients with Type 2 Diabetes. PLoS ONE. 2013;8:e67454. doi: 10.1371/journal.pone.0067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahier J, Guiot Y, Goebbels RM, et al. Pancreatic β-cell mass in European subjects with type 2 diabetes. Diabetes Obes Metab. 2008;10:32–42. doi: 10.1111/j.1463-1326.2008.00969.x. [DOI] [PubMed] [Google Scholar]
- 54.Sutton PA, Humes DJ, Purcell G, et al. The Role of Routine Assays of Serum Amylase and Lipase for the Diagnosis of Acute Abdominal Pain. Ann R Coll Surg Engl. 2009;91:381–384. doi: 10.1308/003588409X392135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.In't Veld P, De Munck N, Van Belle K, et al. β-Cell Replication Is Increased in Donor Organs From Young Patients After Prolonged Life Support. Diabetes. 2010;59:1702–1708. doi: 10.2337/db09-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.