Abstract
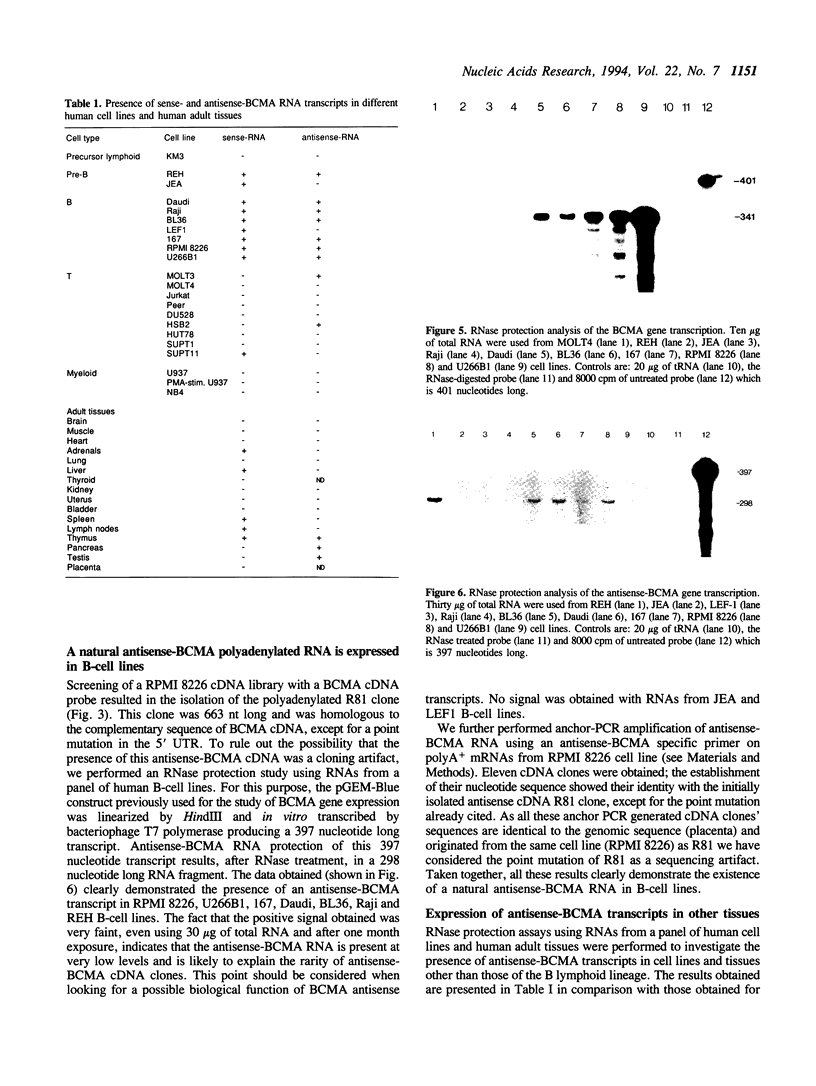
In a previous study of a t(4;16)(q26;p13) translocation, found in a human malignant T-cell lymphoma the BCMA gene, located on chromosome band 16p13.1, has been characterized. In this study we show that the BCMA gene is organized into three exons and its major initiation transcription site is located 69 nucleotides downstream of a TATA box. RNase protection assays demonstrated that the BCMA gene is preferentially expressed in mature B cells, suggesting a role for this gene in the B-cell developmental process. A cDNA complementary to the BCMA cDNA was cloned and sequenced and its presence was assessed by RNase protection assay and anchor-PCR amplification. This antisense-BCMA RNA is transcribed from the same locus as BCMA, and exhibits mRNA characteristic features, e.g. polyadenylation and splicing. It also contains an ORF encoding a putative 115 aa polypeptide, presenting no homology with already known sequences. RNase protection assays demonstrated the simultaneous expression of natural sense and antisense-BCMA transcripts in the majority of human B-cell lines tested.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelman J. P., Bond C. T., Douglass J., Herbert E. Two mammalian genes transcribed from opposite strands of the same DNA locus. Science. 1987 Mar 20;235(4795):1514–1517. doi: 10.1126/science.3547652. [DOI] [PubMed] [Google Scholar]
- Alting-Mees M. A., Short J. M. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989 Nov 25;17(22):9494–9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanidis C., de Jong P. J. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 1990 Oct 25;18(20):6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988 Dec 23;55(6):1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Bonner T. I., Oppermann H., Seeburg P., Kerby S. B., Gunnell M. A., Young A. C., Rapp U. R. The complete coding sequence of the human raf oncogene and the corresponding structure of the c-raf-1 gene. Nucleic Acids Res. 1986 Jan 24;14(2):1009–1015. doi: 10.1093/nar/14.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Burrows P. D., Cooper M. D. Regulated expression of cell surface antigens during B cell development. Semin Immunol. 1990 May;2(3):189–195. [PubMed] [Google Scholar]
- Chen C. N., Malone T., Beckendorf S. K., Davis R. L. At least two genes reside within a large intron of the dunce gene of Drosophila. Nature. 1987 Oct 22;329(6141):721–724. doi: 10.1038/329721a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Dolnick B. J. Cloning and characterization of a naturally occurring antisense RNA to human thymidylate synthase mRNA. Nucleic Acids Res. 1993 Apr 25;21(8):1747–1752. doi: 10.1093/nar/21.8.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Edwards J. B., Delort J., Mallet J. Oligodeoxyribonucleotide ligation to single-stranded cDNAs: a new tool for cloning 5' ends of mRNAs and for constructing cDNA libraries by in vitro amplification. Nucleic Acids Res. 1991 Oct 11;19(19):5227–5232. doi: 10.1093/nar/19.19.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fend F., Nachbaur D., Oberwasserlechner F., Kreczy A., Huber H., Müller-Hermelink H. K. Phenotype and topography of human thymic B cells. An immunohistologic study. Virchows Arch B Cell Pathol Incl Mol Pathol. 1991;60(6):381–388. doi: 10.1007/BF02899570. [DOI] [PubMed] [Google Scholar]
- Fu X. D., Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992 Apr 24;256(5056):535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- Fuchs R. MacPattern: protein pattern searching on the Apple Macintosh. Comput Appl Biosci. 1991 Jan;7(1):105–106. doi: 10.1093/bioinformatics/7.1.105. [DOI] [PubMed] [Google Scholar]
- Grausz D., Lanotte M., Valensi F., Hillion J., Chen S. J., Chen Z., Morinet F., Berger R. A new Ph1+, bcr cell line derived from a patient with ALL-L1 gained autonomy in culture concomitant to CD23 expression. Leukemia. 1990 May;4(5):359–364. [PubMed] [Google Scholar]
- Guglielmi P., Davi F. Expression of a novel type of immunoglobulin C lambda transcripts in human mature B lymphocytes producing kappa light chains. Eur J Immunol. 1991 Feb;21(2):501–508. doi: 10.1002/eji.1830210237. [DOI] [PubMed] [Google Scholar]
- Hahn S., Pinkham J., Wei R., Miller R., Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol Cell Biol. 1988 Feb;8(2):655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun R. S., Serventi I. M., Moss J. Rapid, reliable ligation-independent cloning of PCR products using modified plasmid vectors. Biotechniques. 1992 Oct;13(4):515–518. [PubMed] [Google Scholar]
- Henikoff S., Keene M. A., Fechtel K., Fristrom J. W. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell. 1986 Jan 17;44(1):33–42. doi: 10.1016/0092-8674(86)90482-4. [DOI] [PubMed] [Google Scholar]
- Hofmann W. J., Momburg F., Möller P. Thymic medullary cells expressing B lymphocyte antigens. Hum Pathol. 1988 Nov;19(11):1280–1287. doi: 10.1016/s0046-8177(88)80282-x. [DOI] [PubMed] [Google Scholar]
- Isaacson P. G., Norton A. J., Addis B. J. The human thymus contains a novel population of B lymphocytes. Lancet. 1987 Dec 26;2(8574):1488–1491. doi: 10.1016/s0140-6736(87)92622-5. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Kimelman D., Kirschner M. W. An antisense mRNA directs the covalent modification of the transcript encoding fibroblast growth factor in Xenopus oocytes. Cell. 1989 Nov 17;59(4):687–696. doi: 10.1016/0092-8674(89)90015-9. [DOI] [PubMed] [Google Scholar]
- Kishimoto A., Nishiyama K., Nakanishi H., Uratsuji Y., Nomura H., Takeyama Y., Nishizuka Y. Studies on the phosphorylation of myelin basic protein by protein kinase C and adenosine 3':5'-monophosphate-dependent protein kinase. J Biol Chem. 1985 Oct 15;260(23):12492–12499. [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Korsmeyer S. J., Arnold A., Bakhshi A., Ravetch J. V., Siebenlist U., Hieter P. A., Sharrow S. O., LeBien T. W., Kersey J. H., Poplack D. G. Immunoglobulin gene rearrangement and cell surface antigen expression in acute lymphocytic leukemias of T cell and B cell precursor origins. J Clin Invest. 1983 Feb;71(2):301–313. doi: 10.1172/JCI110770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lanotte M., Martin-Thouvenin V., Najman S., Balerini P., Valensi F., Berger R. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood. 1991 Mar 1;77(5):1080–1086. [PubMed] [Google Scholar]
- Lazar M. A., Hodin R. A., Cardona G., Chin W. W. Gene expression from the c-erbA alpha/Rev-ErbA alpha genomic locus. Potential regulation of alternative splicing by opposite strand transcription. J Biol Chem. 1990 Aug 5;265(22):12859–12863. [PubMed] [Google Scholar]
- Lazar M. A., Hodin R. A., Darling D. S., Chin W. W. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol Cell Biol. 1989 Mar;9(3):1128–1136. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laâbi Y., Gras M. P., Carbonnel F., Brouet J. C., Berger R., Larsen C. J., Tsapis A. A new gene, BCM, on chromosome 16 is fused to the interleukin 2 gene by a t(4;16)(q26;p13) translocation in a malignant T cell lymphoma. EMBO J. 1992 Nov;11(11):3897–3904. doi: 10.1002/j.1460-2075.1992.tb05482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993 Dec 3;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lerner A., D'Adamio L., Diener A. C., Clayton L. K., Reinherz E. L. CD3 zeta/eta/theta locus is colinear with and transcribed antisense to the gene encoding the transcription factor Oct-1. J Immunol. 1993 Sep 15;151(6):3152–3162. [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Marshall R. D. Glycoproteins. Annu Rev Biochem. 1972;41:673–702. doi: 10.1146/annurev.bi.41.070172.003325. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe S. H., Dong X. F. Heterogeneous nuclear ribonucleoprotein A1 catalyzes RNA.RNA annealing. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):895–899. doi: 10.1073/pnas.89.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe S. H., Lazar M. A. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J Biol Chem. 1991 Nov 25;266(33):22083–22086. [PubMed] [Google Scholar]
- Portman D. S., Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994 Jan 1;13(1):213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Rebagliati M. R., Melton D. A. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell. 1987 Feb 27;48(4):599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. D., McFall P., Morgan R., Link M., Hecht F., Cleary M., Sklar J. Long-term growth of malignant thymocytes in vitro. Blood. 1989 Jun;73(8):2182–2187. [PubMed] [Google Scholar]
- Smith S. D., Morgan R., Link M. P., McFall P., Hecht F. Cytogenetic and immunophenotypic analysis of cell lines established from patients with T cell leukemia/lymphoma. Blood. 1986 Mar;67(3):650–656. [PubMed] [Google Scholar]
- Sureau A., Soret J., Vellard M., Crochet J., Perbal B. The PR264/c-myb connection: expression of a splicing factor modulated by a nuclear protooncogene. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11683–11687. doi: 10.1073/pnas.89.24.11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K. M., Inouye M. Antisense RNA. Crit Rev Biochem Mol Biol. 1990;25(3):155–184. doi: 10.3109/10409239009090608. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Vellard M., Sureau A., Soret J., Martinerie C., Perbal B. A potential splicing factor is encoded by the opposite strand of the trans-spliced c-myb exon. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2511–2515. doi: 10.1073/pnas.89.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk R., Köster M., Pöting A., Hartmann L., Knöchel W. An antisense transcript from the Xenopus laevis bFGF gene coding for an evolutionarily conserved 24 kd protein. EMBO J. 1989 Oct;8(10):2983–2988. doi: 10.1002/j.1460-2075.1989.tb08448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. W., Nishikura K. Cell cycle expression of RNA duplex unwindase activity in mammalian cells. Mol Cell Biol. 1988 Feb;8(2):770–777. doi: 10.1128/mcb.8.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990 Jul;15(7):277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- Wilusz J., Pettine S. M., Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1989 May 25;17(10):3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgett J. R., Gould K. L., Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986 Nov 17;161(1):177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Duin M., van Den Tol J., Hoeijmakers J. H., Bootsma D., Rupp I. P., Reynolds P., Prakash L., Prakash S. Conserved pattern of antisense overlapping transcription in the homologous human ERCC-1 and yeast RAD10 DNA repair gene regions. Mol Cell Biol. 1989 Apr;9(4):1794–1798. doi: 10.1128/mcb.9.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]