Abstract
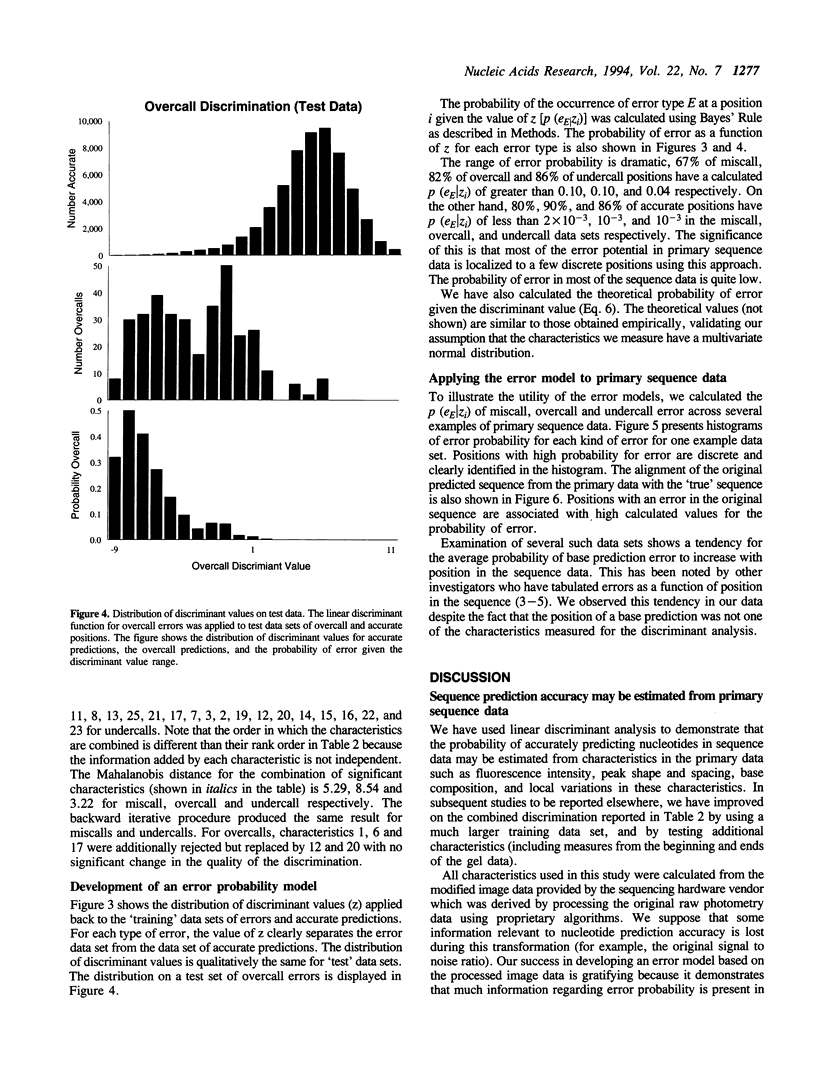
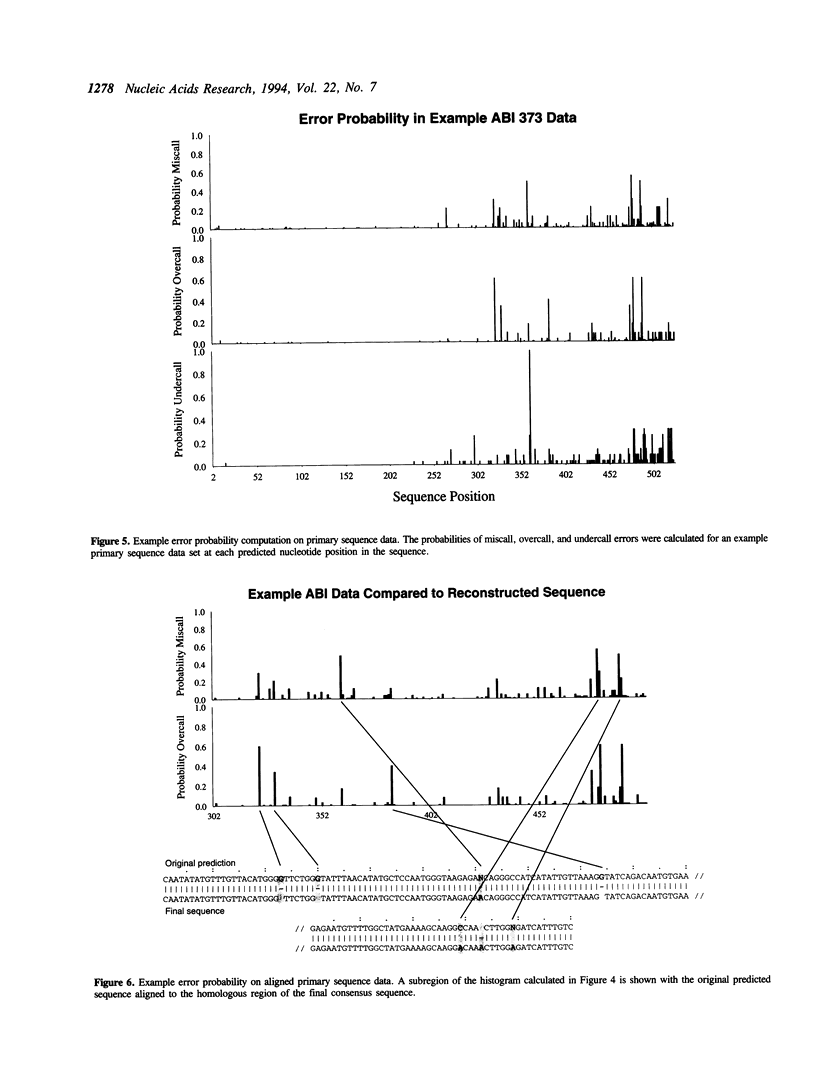
DNA sequence predicted from polyacrylamide gel-based technologies is inaccurate because of variations in the quality of the primary data due to limitations of the technology, and to sequence-specific variations due to nucleotide interactions within the DNA molecule and with the gel. The ability to recognize the probability of error in the primary data will be useful in reconstructing the target sequence of a DNA sequencing project, and in estimating the accuracy of the final sequence. This paper describes the use of linear discriminant analysis to assign position-specific probabilities of incorrect, over- and under-prediction of nucleotides for each predicted nucleotide position in primary sequence data generated by a gel-based DNA sequencing technology. Using this method, most of the error potential in primary sequence data can be assigned to a limited number of discrete positions. The use of probability values in the sequence reconstruction process, and in estimating the accuracy of consensus sequence determination is described.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowling J. M., Bruner K. L., Cmarik J. L., Tibbetts C. Neighboring nucleotide interactions during DNA sequencing gel electrophoresis. Nucleic Acids Res. 1991 Jun 11;19(11):3089–3097. doi: 10.1093/nar/19.11.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. Q., Hunkapiller T. Sequence accuracy of large DNA sequencing projects. DNA Seq. 1992;2(6):335–342. doi: 10.3109/10425179209020814. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Waterman M. S. The accuracy of DNA sequences: estimating sequence quality. Genomics. 1992 Sep;14(1):89–98. doi: 10.1016/s0888-7543(05)80288-5. [DOI] [PubMed] [Google Scholar]
- Huang X. A contig assembly program based on sensitive detection of fragment overlaps. Genomics. 1992 Sep;14(1):18–25. doi: 10.1016/s0888-7543(05)80277-0. [DOI] [PubMed] [Google Scholar]
- Krawetz S. A. Sequence errors described in GenBank: a means to determine the accuracy of DNA sequence interpretation. Nucleic Acids Res. 1989 May 25;17(10):3951–3957. doi: 10.1093/nar/17.10.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen T., Lopez R., Prydz H. An estimate of the sequencing error frequency in the DNA sequence databases. DNA Seq. 1992;2(6):343–346. doi: 10.3109/10425179209020815. [DOI] [PubMed] [Google Scholar]
- Posfai J., Roberts R. J. Finding errors in DNA sequences. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4698–4702. doi: 10.1073/pnas.89.10.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- States D. J., Botstein D. Molecular sequence accuracy and the analysis of protein coding regions. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5518–5522. doi: 10.1073/pnas.88.13.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Du Z., Thomas K., Wilson R., Hillier L., Staden R., Halloran N., Green P., Thierry-Mieg J., Qiu L. The C. elegans genome sequencing project: a beginning. Nature. 1992 Mar 5;356(6364):37–41. doi: 10.1038/356037a0. [DOI] [PubMed] [Google Scholar]