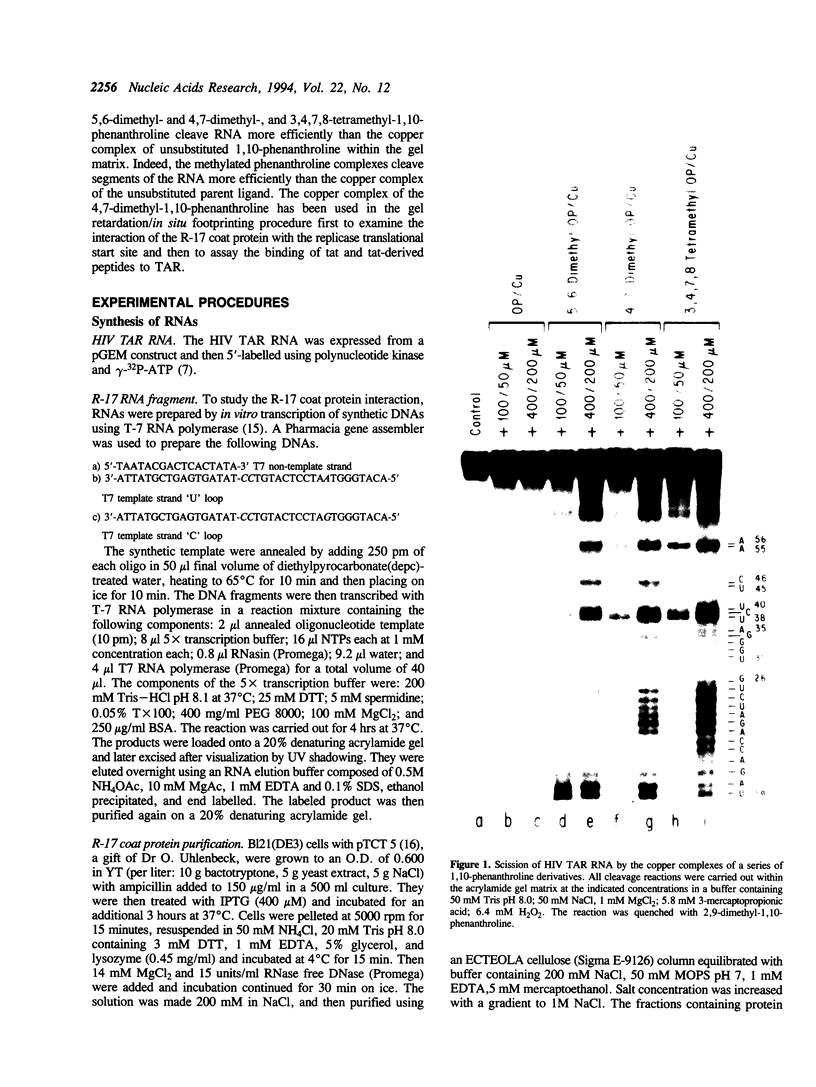
Abstract
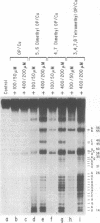
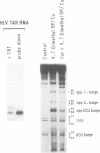
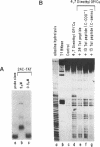
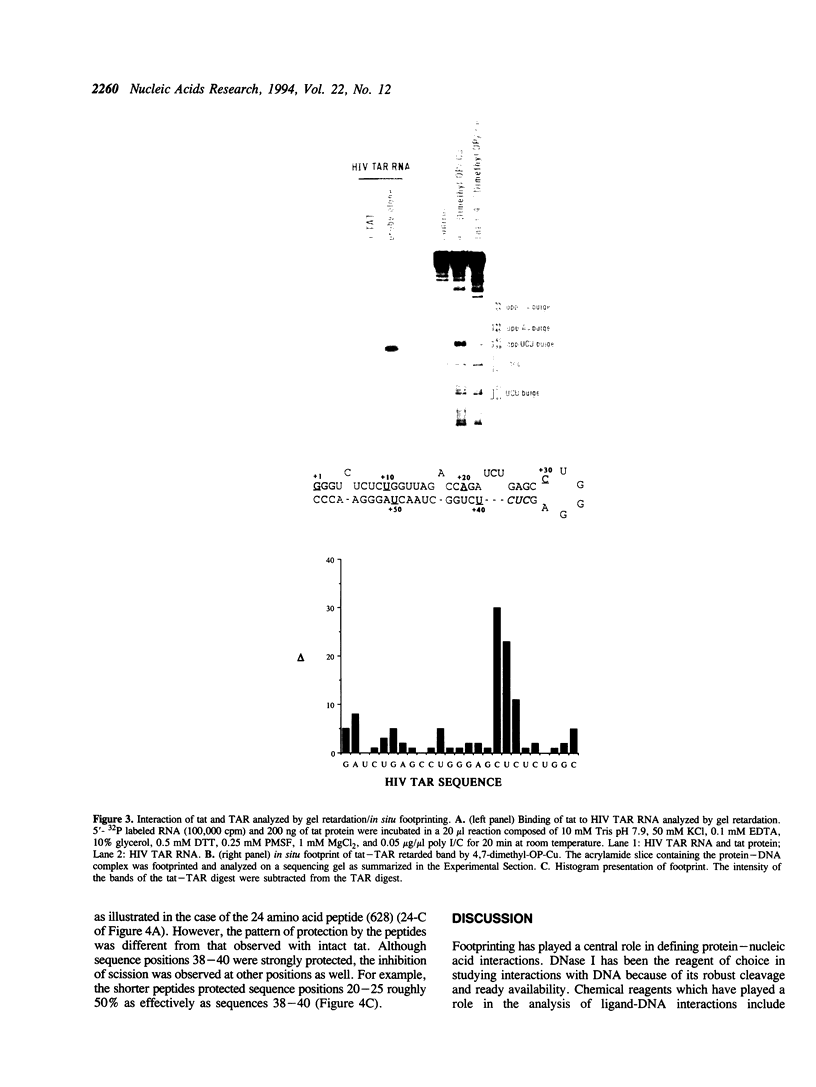
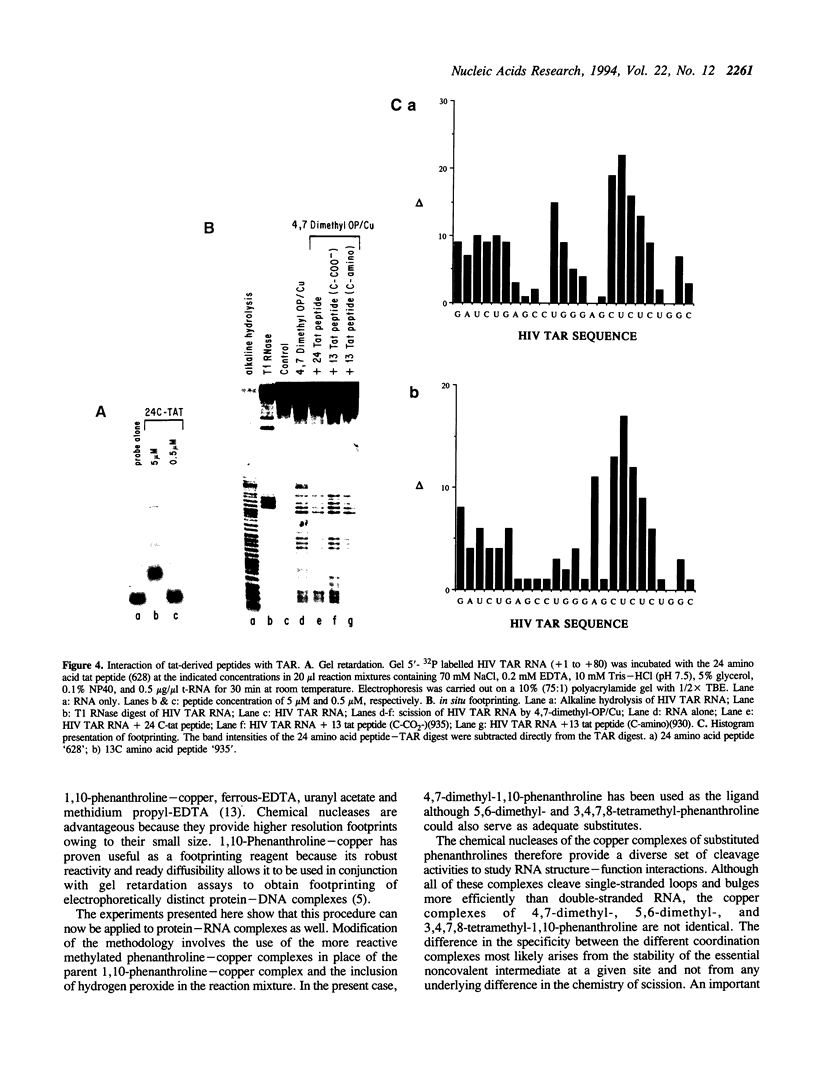
RNA-protein complexes isolated following a gel retardation assay can be footprinted within the gel matrix using the chemical nuclease activities of 4,7-dimethyl-, 5,6-dimethyl-, and 3,4,7,8-tetramethyl-1,10-phenanthroline-copper. These complexes are more reactive than 1,10-phenanthroline-copper but share its reaction preference for bulges and loops. The interaction of the coat protein of R-17 with its viral RNA target and tat- and tat-derived peptides with HIV TAR RNA have been studied. In both cases, the RNA sequence opposite a 2-3 nucleotide bulge are protected. Tat-derived peptides inhibit cleavage at sites which intact tat does not protect. These results are consistent with transcription studies which have suggested that truncation of tat increases nonspecific binding.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhattacharyya A., Murchie A. I., Lilley D. M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990 Feb 1;343(6257):484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- Carey J., Lowary P. T., Uhlenbeck O. C. Interaction of R17 coat protein with synthetic variants of its ribonucleic acid binding site. Biochemistry. 1983 Sep 27;22(20):4723–4730. doi: 10.1021/bi00289a017. [DOI] [PubMed] [Google Scholar]
- Churcher M. J., Lamont C., Hamy F., Dingwall C., Green S. M., Lowe A. D., Butler J. G., Gait M. J., Karn J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J Mol Biol. 1993 Mar 5;230(1):90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- Darsillo P., Huber P. W. The use of chemical nucleases to analyze RNA-protein interactions. The TFIIIA-5 S rRNA complex. J Biol Chem. 1991 Nov 5;266(31):21075–21082. [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990 Dec;9(12):4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Gaynor R., Soultanakis E., Kuwabara M., Garcia J., Sigman D. S. Specific binding of a HeLa cell nuclear protein to RNA sequences in the human immunodeficiency virus transactivating region. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4858–4862. doi: 10.1073/pnas.86.13.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott J. M., Willis M. C., Koch T. H., Uhlenbeck O. C. A specific, UV-induced RNA-protein cross-link using 5-bromouridine-substituted RNA. Biochemistry. 1991 Jun 25;30(25):6290–6295. doi: 10.1021/bi00239a030. [DOI] [PubMed] [Google Scholar]
- Graeble M. A., Churcher M. J., Lowe A. D., Gait M. J., Karn J. Human immunodeficiency virus type 1 transactivator protein, tat, stimulates transcriptional read-through of distal terminator sequences in vitro. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6184–6188. doi: 10.1073/pnas.90.13.6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamy F., Asseline U., Grasby J., Iwai S., Pritchard C., Slim G., Butler P. J., Karn J., Gait M. J. Hydrogen-bonding contacts in the major groove are required for human immunodeficiency virus type-1 tat protein recognition of TAR RNA. J Mol Biol. 1993 Mar 5;230(1):111–123. doi: 10.1006/jmbi.1993.1129. [DOI] [PubMed] [Google Scholar]
- Jayasena S. D., Johnston B. H. Site-specific cleavage of the transactivation response site of human immunodeficiency virus RNA with a tat-based chemical nuclease. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3526–3530. doi: 10.1073/pnas.89.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Graeble M. A. New insights into the mechanism of HIV-1 trans-activation. Trends Genet. 1992 Nov;8(11):365–368. doi: 10.1016/0168-9525(92)90284-b. [DOI] [PubMed] [Google Scholar]
- Kuwabara M. D., Sigman D. S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987 Nov 17;26(23):7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- Leibold E. A., Munro H. N. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5' untranslated region of ferritin heavy- and light-subunit mRNAs. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary P. T., Uhlenbeck O. C. An RNA mutation that increases the affinity of an RNA-protein interaction. Nucleic Acids Res. 1987 Dec 23;15(24):10483–10493. doi: 10.1093/nar/15.24.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder A., Chen C. B., Gaynor R., Sigman D. S. 1,10-Phenanthroline-copper, a footprinting reagent for single-stranded regions of RNAs. Biochem Biophys Res Commun. 1992 Sep 30;187(3):1503–1509. doi: 10.1016/0006-291x(92)90472-w. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa G. J., Chen C. H., Kuwabara M. D., Nierlich D. P., Sigman D. S. Scission of RNA by the chemical nuclease of 1,10-phenanthroline-copper ion: preference for single-stranded loops. Nucleic Acids Res. 1989 Jul 11;17(13):5361–5375. doi: 10.1093/nar/17.13.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa G. J., Nierlich D. P. Mapping the lacZ ribosome binding site by RNA footprinting. Biochemistry. 1989 Oct 3;28(20):8067–8072. doi: 10.1021/bi00446a015. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tan R., Calnan B. J., Frankel A. D., Williamson J. R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992 Jul 3;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Sigman D. S. Chemical nucleases. Biochemistry. 1990 Oct 2;29(39):9097–9105. doi: 10.1021/bi00491a001. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Chen C. H. Chemical nucleases: new reagents in molecular biology. Annu Rev Biochem. 1990;59:207–236. doi: 10.1146/annurev.bi.59.070190.001231. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Kuwabara M. D., Chen C. H., Bruice T. W. Nuclease activity of 1,10-phenanthroline-copper in study of protein-DNA interactions. Methods Enzymol. 1991;208:414–433. doi: 10.1016/0076-6879(91)08022-a. [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Sczekan S. R., Theil E. C. Structure of the 5' untranslated regulatory region of ferritin mRNA studied in solution. Nucleic Acids Res. 1990 Aug 11;18(15):4463–4468. doi: 10.1093/nar/18.15.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA binding assays for Tat-derived peptides: implications for specificity. Biochemistry. 1992 Oct 27;31(42):10281–10287. doi: 10.1021/bi00157a015. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- Witherell G. W., Gott J. M., Uhlenbeck O. C. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acid Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]