Summary
During eukaryotic DNA interstrand cross-link (ICL) repair, cross-links are resolved (“unhooked”) by nucleolytic incisions surrounding the lesion. In vertebrates, ICL repair is triggered when replication forks collide with the lesion, leading to FANCI-FANCD2-dependent unhooking and formation of a double-strand break (DSB) intermediate. Using Xenopus egg extracts, we describe here a replication-coupled ICL repair pathway that does not require incisions or FANCI-FANCD2. Instead, the ICL is unhooked when one of the two N-glycosyl bonds forming the cross-link is cleaved by the DNA glycosylase NEIL3. Cleavage by NEIL3 is the primary unhooking mechanism for psoralen- and abasic site-ICLs. When N-glycosyl bond cleavage is prevented, unhooking occurs via FANCI-FANCD2-dependent incisions. In summary, we identify an incision-independent unhooking mechanism that avoids DSB formation and represents the preferred pathway of ICL repair in a vertebrate cell-free system.
Introduction
DNA replication and transcription require DNA unwinding. Therefore, chemical agents that create DNA interstrand cross-links (ICLs) are extremely cytotoxic. ICLs are induced by diverse chemical agents (Deans and West, 2009), including nitrogen mustards, platinum compounds, mitomycin C, and psoralen, which are used in chemotherapy. ICLs can also be generated by endogenous agents, most notably formaldehyde, acetaldehyde, and malondialdehyde (Duxin and Walter, 2015). In addition, the deoxyribose sugar of an abasic (AP) site (generated by spontaneous hydrolysis or the action of a DNA glycosylase) can isomerize to a ring-open aldehyde that is attacked by a base on the opposite strand, generating an ‘AP-ICL’ (Price et al., 2014). Given that mammalian cells contain ~100,000 AP sites at steady state (Nakamura and Swenberg, 1999), AP-ICLs may form in vivo. ICLs are chemically and structurally diverse. Cisplatin links guanines on opposing strands through their N7 nitrogens. The resulting ICL is so distorting that the cytosines opposite the crosslinked guanines are flipped out of the DNA helix. Upon exposure to UV light, psoralen links thymines on opposite strands, but the resulting ICL is not highly distorting. Reactive aldehydes generally induce cross-links between the exocyclic amines of bases, but little is known about their structure. An important question is whether these chemically diverse ICLs are repaired via similar or distinct strategies.
The vertebrate ICL repair pathway that contributes most to cell survival is coupled to DNA replication (Clauson et al., 2013; Kottemann and Smogorzewska, 2013; Zhang and Walter, 2014). This pathway involves structure-specific endonucleases, DNA recombinases, and translesion synthesis (TLS) polymerases. In addition, it requires 19 ‘FANC’ proteins, mutations in which cause the bone marrow failure and cancer predisposition syndrome Fanconi anemia. A key factor in this pathway is the FANCI-FANCD2 heterodimer (FANCI-D2), whose mono-ubiquitylation protects cells from ICL-inducing agents. Importantly, the Fanconi pathway protects cells from the genotoxic effects of endogenous aldehydes, though the nature of the relevant DNA lesions is presently unclear (Duxin and Walter, 2015; Garaycoechea et al., 2012; Langevin et al., 2011; Pontel et al., 2015).
By replicating a plasmid containing a site-specific cisplatin-ICL in Xenopus egg extracts, we previously described a mechanism of replication-coupled ICL repair (Figure S1A; Raschle et al., 2008). The leading strands of converging replication forks initially stall ~20 nucleotides from the lesion due to steric hindrance by the replicative CMG helicase, which encircles and travels along the leading strand template ahead of DNA polymerase (Fu et al., 2011). The first known event in repair is removal of the stalled CMGs, which requires fork convergence, the tumor suppressor BRCA1-BARD1, and ubiquitin signaling (Long et al., 2014; Zhang et al., 2015). Subsequently, the leading strand is extended to within one nucleotide of the ICL (“−1” position). Ubiquitylated FANCI-D2 then binds near the ICL and helps recruit the XPF-ERCC1-SLX4 complex, which promotes incisions surrounding the ICL (Klein Douwel et al., 2014; Knipscheer et al., 2009). This ‘unhooking’ reaction creates a double-stranded DNA break in one sister chromatid and a mono-adduct in the other strand, which is bypassed with ~98% accuracy using the TLS polymerase REV1-Pol ζ (Budzowska et al., 2015). The DSB is repaired by RAD51-dependent homologous recombination (Long et al., 2011). The remaining mono-adduct is not removed in egg extracts, although this may occur in cells. A similar mechanism of repair was observed for a nitrogen mustard-like ICL (Raschle et al., 2008). Because this cell-free system requires all the major proteins implicated in ICL repair in cells, it likely recapitulates physiological repair. Importantly, all eukaryotic mechanisms of ICL repair described so far, including those operating outside of S phase (Williams et al., 2013), involve nucleolytic incisions to unhook the cross-link.
We wanted to address whether chemically diverse ICLs are repaired by similar or different mechanisms in Xenopus egg extracts. We therefore prepared plasmids containing a psoralen- or AP-ICL and incubated them in egg extract alongside a cisplatin-ICL plasmid. Strikingly, although repair of psoralen- and AP-ICLs requires replication fork convergence, as seen for the cisplatin-ICL, the downstream repair events are completely different. Specifically, one of the two N-glycosyl bonds that form the psoralen- or AP-ICL is cleaved by the NEIL3 DNA glycosylase, unhooking the cross-link in the absence of incisions. This reaction does not require FANCI-D2 or CMG unloading. After unhooking, gaps are filled in via TLS polymerases. When N-glycosyl bond cleavage is inhibited, repair switches to FANCI-D2-dependent incisions. Thus, N-glycosyl cleavage by NEIL3 represents the primary unhooking pathway for psoralen- and AP-ICLs, whereas incisions represent a back-up mechanism. We conclude that ICLs can be unhooked via two independent mechanisms in S phase, one of which avoids DSB formation and thus minimizes the potential for gross chromosomal rearrangements.
Results
pICLPso repair in the absence of DSB formation
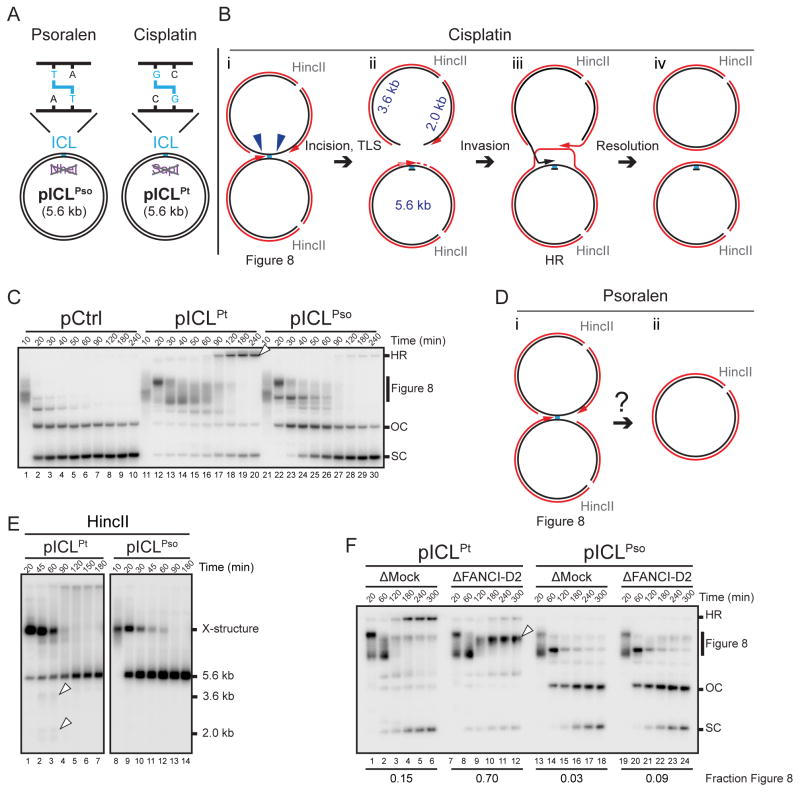
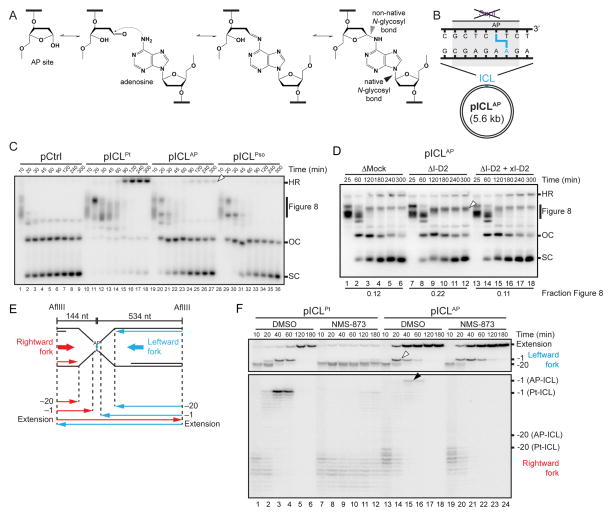
To examine the mechanism of psoralen-ICL repair, we prepared a 5.6 kb plasmid containing a site-specific psoralen-ICL (Figures 1A and S1B; pICLPso) and replicated it in Xenopus egg extracts containing [α-32P]dATP alongside undamaged (pCtrl) and cisplatin-ICL containing plasmids (Figure 1A; pICLPt). Replication intermediates were examined by native gel electrophoresis and autoradiography. pCtrl was rapidly converted to open circular and supercoiled replication products (Figure 1C, lanes 1–10). In contrast, on pICLPt and pICLPso, replication forks converged on the ICL, leading to the accumulation of “Figure 8” structures (Figure 1C, Figures 1Bi, 1Di; Raschle et al., 2008). During pICLPt repair, dual incisions of the Figure 8 intermediate generated a DSB that is repaired via HR with the other sister (Figure 1Biii; Long et al., 2011). The resulting HR intermediates were detected near the top of the gel (Figure 1C, arrowhead; Long et al., 2011). Concurrently, supercoiled (SC) molecules accumulated (Figure 1C, lanes 15–20). Surprisingly, during pICLPso repair, HR intermediates were scarce, and Figure 8 structures appeared to be directly converted into open circular molecules (nicked or gapped; OC) and then supercoiled species (Figure 1C, lanes 21–30). As seen for pICLPt (Raschle et al., 2008), unhooking and error-free repair of pICLPso was replication-dependent (Figure S1C–F), although the efficiency of repair was low (Figure S1G; see Discussion). Furthermore, when only a single fork encountered the psoralen-ICL, it failed to trigger ICL processing (Zhang et al., 2015). The data above indicate that psoralen-ICL repair, although dependent on replication fork convergence, might involve a novel mechanism.
Figure 1. A psoralen-ICL is unhooked by an incision-independent pathway that does not require FANCI-D2.
(A) Cartoons of pICLPso and pICLPt. NheI and SapI restriction sites coincide with the psoralen-and cisplatin-ICLs, respectively.
(B) Cartoon of pICLPt replication intermediates digested with HincII.
(C) pCtrl, pICLPt, or pICLPso was replicated in egg extracts with [α-32P]dATP. Replication intermediates were separated on a native agarose gel and visualized by autoradiography. SC, supercoiled. OC, open circular. HR, homologous recombination intermediate, also denoted by white arrowhead.
(D) Cartoon of pICLPso replication intermediates digested with HincII.
(E) pICLPt or pICLPso was replicated in egg extracts with [α-32P]dATP. Replication intermediates were digested with HincII, separated on a native agarose gel, and visualized by autoradiography. White arrowheads, 3.6 kb and 2.0 kb incision products depicted in (B).
(F) pICLPt or pICLPso was replicated in mock or FANCI-D2 depleted egg extracts in the presence of [α-32P]dATP, and the repair intermediates were analyzed as in (C). White arrowhead, Figure 8 DNA structure. The fraction Figure 8 indicates the proportion of Figure 8 structures relative to total species at 300 min.
Given the apparent absence of HR products during pICLPso repair, we examined whether this process involves a DSB intermediate. During pICLPt repair, FANCI-D2-promotes incisions around the ICL (Figure 1Bi–ii; Klein Douwel et al., 2014; Knipscheer et al., 2009; Raschle et al., 2008) that are detected as 3.6 kb and 2.0 kb fragments after digestion with HincII (Figure 1Bii and Figure 1E, arrowheads). These fragments appear transiently, likely because they undergo resection and/or strand invasion into the sister chromatid (Figure 1Biii). In contrast, no 3.6 kb and 2.0 kb incision products were detected during pICLPso repair (Figure 1E, lanes 8–14). As shown previously (Knipscheer et al., 2009), when FANCI-D2 was immunodepleted from egg extracts (Figure S1H), pICLPt accumulated as Figure 8 DNA structures, and supercoiled product was less abundant, consistent with an incision defect (Figure 1F, lanes 7–12). In contrast, FANCI-D2 depletion had almost no effect on the accumulation of open circular and supercoiled pICLPso products (Figure 1F, lanes 19–24), even though the psoralen-ICL induced FANCD2 ubiquitylation (Figure S1I). In summary, the data indicate that a psoralen-ICL is unhooked by an incision-independent pathway that does not involve a DSB intermediate or require FANCI-D2.
Translesion DNA synthesis is required for gap filling of both daughter molecules
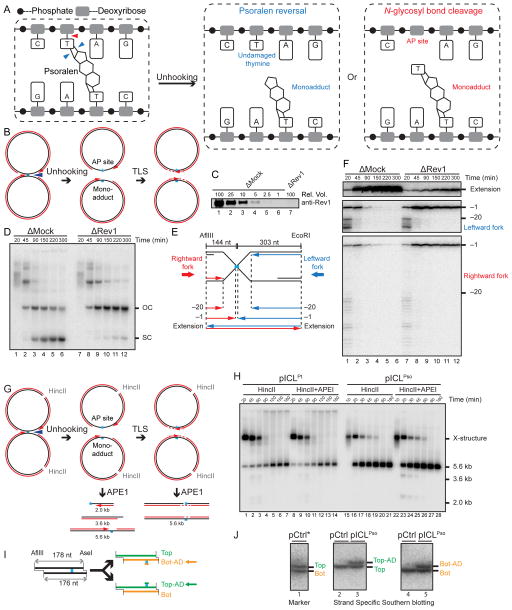
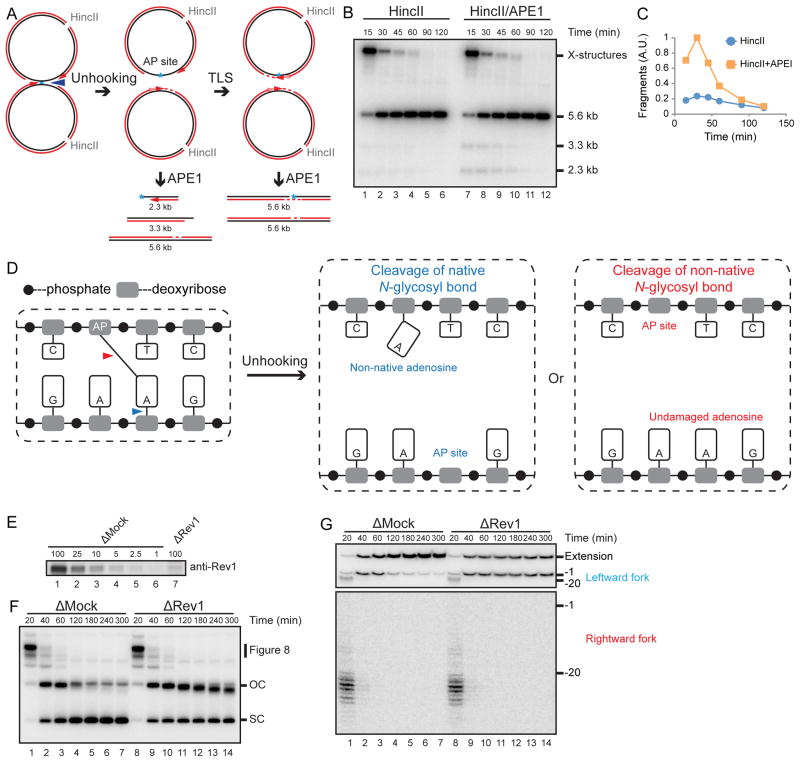
We envisioned two mechanisms of psoralen-ICL unhooking without incisions. First, in a process analogous to photoreversal (Cimino et al., 1986), the cyclobutane ring between the thymine and psoralen is broken (Figure 2A, left panel, blue arrowheads). This reaction would yield a psoralen mono-adduct on one strand and an undamaged thymine on the other strand (Figure 2A, middle panel). Alternatively, one of the two N-glycosyl bonds between a damaged thymine and a sugar is cleaved (Figure 2A, left panel, red arrowhead), generating an AP site on one parental strand and a thymine-psoralen mono-adduct on the other strand (Figure 2A, right panel). The first model predicts that after unhooking, gap filling on only one daughter molecule should require translesion DNA synthesis (not shown), whereas the second model predicts gap filling on both daughters requires TLS (Figure 2B). To distinguish between the models, we immunodepleted the TLS DNA polymerase Rev1, which is required for ICL repair (Budzowska et al., 2015). In Rev1-depleted egg extract (Figure 2C), the vast majority of pICLPso accumulated as gapped plasmids (Figure 2D), consistent with the second model. To examine where the leading strands stalled after Rev1 depletion, we digested repair intermediates with AflIII and EcoRI (Figure 2E), which allowed us to monitor rightward and leftward leading strands on the same denaturing gel (Figure 2F). After Rev1 depletion, rightward and leftward leading strands both stalled at or near the −1 position (Figure 2F, middle and bottom panels), and there was a strong reduction in extension products (Figure 2F, top panel), consistent with a lesion bypass defect on both strands. We conclude that gap filling of both daughter molecules is dependent on TLS, consistent with the N-glycosyl bond cleavage model (Figure 2A, right panel).
Figure 2. Psoralen-ICL is unhooked via the cleavage of a DNA N-glycosyl bond.
(A) Two possible incision-independent unhooking pathways for a psoralen-ICL. Blue arrowheads (left panel) show unhooking of a psoralen-ICL via breakage of the cyclobutane ring (psoralen reversal), resulting in an undamaged thymine and a monoadduct (middle panel). The red arrowhead (left panel) shows psoralen-ICL unhooking via cleavage of a DNA N-glycosyl bond, resulting in an AP site and a monoadduct (right panel).
(B) Model for pICLPso unhooking via cleavage of a DNA N-glycosyl bond, which necessitates gap filling of both daughter molecules by TLS.
(C) Mock-depleted and Rev1-depleted NPE were analyzed by Rev1 Western blotting. A relative volume of 100 corresponds to 0.25 μl NPE.
(D) pICLPso was replicated in mock or Rev1 depleted egg extracts in the presence of [α-32P]dATP. Repair intermediates were analyzed as in Figure 1C.
(E) Schematic illustration of nascent leading strands liberated after digestion of pICLPso with AflIII and EcoRI.
(F) pICLPso was replicated in mock- or Rev1-depleted egg extracts in the presence of [α-32P]dATP. Samples were purified and digested with AflIII and EcoRI before separation on a denaturing polyacrylamide gel. Three portions of the autoradiograph are shown with different contrasts for optimal display.
(G) Expected species after digestion of pICLPso replication intermediates with HincII and APE1, including 3.6 kb and 2.0 kb fragments.
(H) pICLPt or pICLPso was replicated in egg extracts with [α-32P]dATP. Replication intermediates were digested with HincII and APE1, separated on a native agarose gel, and visualized by autoradiography. Quantification of 3.6 kb and 2.0 kb fragments is presented in Figure S1J.
(I) Depiction of final repair products after AflIII and AseI digestion. AflIII and AseI generate different sized overhangs, allowing separation of top (178 nt) and bottom (176 nt) strands. Depending on which N-glycosyl bond is cleaved, the mono-adduct is present on the bottom (Bot-AD) or top (Top-AD) strands.
(J) Detection of pICLPso mono-adducts. pCtrl or pICLPso was replicated for 3 hours in egg extract. DNA was isolated, digested with AflIII and AseI, separated on a denaturing sequencing gel, and analyzed by strand-specific Southern blotting to visualize the top strand (middle panel) or bottom strand (right panel). The absence of bottom (lane 2) or top strands (lane 4) in the Southern blot of pCtrl indicates the strand specificity of the blotting protocol. To generate size markers for the top (178 nt) and bottom (176 nt) strands (left panel, lane 1), pCtrl was replicated in the presence of [α-32P]dATP (pCtrl*) and analyzed on the same sequencing gel after AflIII and AseI digestion.
The psoralen-ICL is unhooked via N-glycosyl bond cleavage
The N-glycosyl bond cleavage model predicts that an AP site is generated during psoralen-ICL unhooking (Figure 2A, right panel). To test this prediction, we recovered pICLPso repair intermediates and digested them with recombinant APE1, which cleaves the phosphodiester bond 5′ to an AP site. If an AP site is present near the ICL, simultaneous digestion with APE1 and HincII should yield 3.6 and 2.0 kb DNA fragments (Figure 2G). As shown in Figure 2H, APE1 and HincII digestion led to a substantial increase in 3.6 and 2.0 kb fragments relative to HincII digestion alone (lanes 15–28, quantified in Figure S1J). Disappearance of these fragments at later times (Figure 2H, lanes 27–28) resulted from translesion DNA synthesis past the AP site, after which APE1 cleavage of one strand does not create a DSB (Figure 2G). In contrast, during cisplatin-ICL repair, APE1 digestion did not increase the basal level of 3.6 and 2.0 kb DNA fragments that arise from FANCI-D2-dependent incisions (Figure 2H, lanes 1–14, and Figure S1J). We conclude that AP sites are specifically generated near the lesion during psoralen-ICL repair.
We also examined whether there is a mono-adduct attached to the unhooked parental strand, as predicted for N-glycosyl bond cleavage (Figure 2A, right panel). If the two N-glycosyl bonds are cleaved with equal probability, the mono-adduct should be detected on both parental strands. To test this idea, we digested final pICLPso repair products with AflIII and AseI, so that the unmodified top and bottom strands differed by 2 nt (Figure 2I), allowing us to differentiate them on a denaturing polyacrylamide gel (Figure 2J, lane 1). After replication of pICLPso and digestion with AflIII and AseI, we detected the top and bottom parental strands, respectively, by strand-specific Southern blotting. As shown in lanes 3 and 5 of Figure 2J, both parental strands contained a mono-adduct. Collectively, our data strongly argue that the psoralen-ICL is unhooked via cleavage of one of the two N-glycosyl bonds forming the cross-link.
Switching between repair pathways
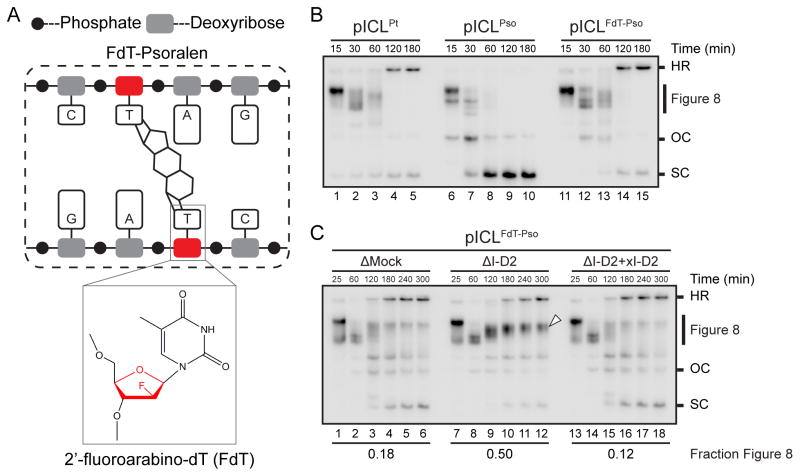
Although we detected APE1 cleavage products in every pICLPso repair reaction examined, the amount varied between experiments (data not shown). Therefore, to further examine whether the N-glycosyl bond is cleaved (Figure 2A), we made a new plasmid in which the psoralen cross-link is formed between two 2′-fluoroarabino-dT nucleotides (pICLFdT-Pso; Figure 3A). The 2′ fluorine renders the thymidine N-glycosyl bond resistant to enzymatic cleavage (Maiti et al., 2009), and should therefore inhibit the N-glycosyl bond cleavage pathway. Strikingly, when pICLFdT-Pso was replicated in egg extracts, Figure 8 structures were still resolved, but the repair intermediates included HR structures and thus much more closely resembled those of pICLPt than pICLPso (Figure 3B). Accordingly, in FANCI-D2-depleted extract (Figure S2A), pICLFdT-Pso accumulated as Figure 8 structures (Figure 3C, arrowhead), as seen for pICLPt (Figure S2B; Knipscheer et al., 2009), and leading strands stalled at the −1 position (Figure S2C). These data confirm that a psoralen-ICL formed between normal thymidines is unhooked via cleavage of the N-glycosyl bond. Moreover, when the N-glycosyl bond cannot be cleaved, the psoralen-ICL is unhooked via FANCI-D2 dependent incisions, which appear to function as a back-up repair pathway (Figure S3).
Figure 3. Psoralen-ICL undergoes FANCI-D2 dependent processing when the N-glycosyl bond cannot be cleaved.
(A) Schematic of a psoralen-ICL formed between two 2′-fluoroarabino-dT (FdT) nucleosides (inset).
(B) pICLPt, pICLPso, or pICLFdT-Pso was replicated in egg extracts in the presence of [α-32P]dATP. Repair intermediates were analyzed as in Figure 1C.
(C) pICLFdT-Pso was replicated in mock-depleted egg extract, FANCI-D2 depleted extract (ΔI-D2), or ΔI-D2 extract supplemented with xFANCI-D2 (ΔI-D2 + xI-D2) in the presence of [α-32P]dATP, and the repair intermediates were analyzed as in Figure 1C. White arrowhead, Figure 8 DNA structure that persists in the absence of FANCI-D2. The fraction Figure 8 indicates the proportion of Figure 8 structures relative to total species at 300 min.
CMG unloading is not required for psoralen-ICL unhooking
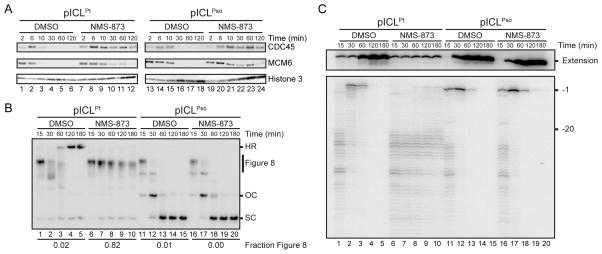
We previously showed that during pICLPt replication, CMG unloading is required to allow leading strand approach to the −1 position and for ICL repair (Long et al., 2014). To address whether CMG unloading is also required for psoralen-ICL repair, we first sought a convenient and specific means to inhibit CMG unloading. Because the p97 ATPase is required for CMG unloading during DNA replication termination (Maric et al., 2014; Moreno et al., 2014), we reasoned it might also extract CMG from forks stalled at ICLs. To test this idea, we added NMS-873, an allosteric inhibitor of p97 (Magnaghi et al., 2013), to egg extracts. As shown by chromatin pull-down, NMS-873 blocked the unloading of CMG from an undamaged plasmid, as expected from a defect in termination (Figure S4A), and from pICLPt (Figure 4A, left panel). Under these conditions, pICLPt repair intermediates accumulated as Figure 8 structures (Figure 4B, lanes 6–10), and leading strands stalled at the −20 position (Figure 4C, lanes 6–10). Strikingly, although NMS-873 also prevented CMG unloading from pICLPso (Figure 4A, right panel), unhooking was unaffected (Figure 4B, lanes 11–20). In the presence of NMS-873, leading strands also did not arrest at the −20 position (Figure 4C, lanes 16–20), presumably because psoralen-ICL unhooking allowed CMG to travel beyond the ICL, permitting extension of the leading strand to the ICL (Figure S3i–ii). On the other hand, NMS-873 efficiently blocked unhooking and leading strand extension on pICLFdT-Pso (Figure S4B–C), demonstrating that repair of this template requires CMG unloading, as expected given its dependence on FANCI-D2-dependent incisions. Thus, CMG unloading is essential for ICL repair via the incision pathway but not the N-glycosyl bond cleavage pathway.
Figure 4. CMG unloading is not required for psoralen-ICL unhooking.
(A) pICLPt or pICLPso (containing 48 lacO repeats) was replicated in the presence or absence of NMS-873 and, at the indicated times, pulled down using LacI-coated beads. Western blotting was performed on chromatin samples with the indicated antibodies.
(B) pICLPt and pICLPso were replicated in the presence or absence of NMS-873 with [α-32P]dATP, and repair intermediates were analyzed as in Figure 1C. The fraction Figure 8 indicates the proportion of Figure 8 structures relative to total species at 180 min.
(C) DNA samples from (B) were digested with AflIII and EcoRI before separation on a denaturing polyacrylamide gel. Extension products and rightward fork products (see Figure 2E) are shown with different contrast for optimal display.
In conclusion, a psoralen-ICL is repaired in Xenopus egg extract via cleavage of the N-glycosyl bond, CMG bypass, and TLS-dependent gap filling (Figure S3i–iii). The AP site is probably then removed via endogenous AP endonuclease and lyase activities (Figure S3iv) while the monoadduct may be removed via base excision repair (Couve et al., 2009). When the N-glycosyl bond cannot be cleaved, psoralen-ICL repair proceeds via the classic, FANCI-D2-dependent incision pathway that requires CMG unloading (Figure S3v–ix).
Repair of an AP-ICL by incision-dependent and incision-independent pathways
We next wished to examine an ICL that might form in cells. AP sites in DNA comprise an equilibrating mixture of structures, including a ring-opened aldehyde that can react with the exocyclic amine of a nucleobase on the opposite strand, generating an “AP-ICL” through a nonnative N-glycosyl bond (Figure 5A; Price et al., 2015). AP sites react most efficiently with adenines on the other strand that are displaced by one residue to the 3′ side (Figure 5B). Given the high abundance of AP sites in cells and considerable stability of AP-ICLs (t1/2 ~ 3 days at 37 °C; Price et al., 2015), AP-ICLs may represent physiologically relevant impediments to DNA replication.
Figure 5. An AP-ICL is repaired by an incision-independent pathway that does not require FANCI-D2 or CMG unloading.
(A) Mechanism of AP-ICL formation.
(B) Cartoon of an AP-ICL containing plasmid (pICLAP), in which the SapI restriction site coincides with the ICL.
(C) pCtrl, pICLPt, pICLAP, or pICLPso was replicated in egg extracts and analyzed as in Figure 1C. White arrowhead, homologous recombination intermediate that accumulates for pICLAP.
(D) pICLAP was replicated in mock-depleted egg extract, FANCI-D2-depleted extract (ΔI-D2), or ΔI-D2 extract supplemented with xFANCI-D2 (ΔI-D2+xI-D2), and analyzed as in Figure 1C. White arrowhead, Figure 8 structures that persist in the absence of FANCI-D2. The fraction Figure 8 indicates the proportion of Figure 8 structures relative to total species at 300 min.
(E) Schematic illustration of nascent leading strands liberated by digestion of pICLAP with AflIII.
(F) pICLPt and pICLAP were replicated with [α-32P]dATP in the presence or absence of NMS-873. The nascent strands were purified and digested with AflIII before separation on a denaturing polyacrylamide gel. Two portions of the autoradiograph are shown with different contrasts for optimal display. White arrowhead, leftward -1 products. Black arrowhead, rightward -1 products.
We generated a plasmid containing a site-specific AP-ICL (pICLAP; Figure 5B and Figure S1B) and incubated it in egg extract. Both unhooking and error-free repair of the AP-ICL was replication-dependent (Figure S1C and D and Figure S5A and B). The amount of pICLAP repair products (43%) was much greater than the amount (5%) observed for pICLPso (compare Figures S5B and S1G; see Discussion). As seen for pICLPso, the majority of pICLAP Figure 8 molecules was converted directly to gapped and supercoiled species (Figure 5C). However, a minor fraction accumulated in the well as HR products (arrowhead). These data suggest that a minority of pICLAP molecules is unhooked via incisions (as for pICLPt), and a majority might be unhooked via the N-glycosyl bond cleavage pathway (as for pICLPso). Consistent with this interpretation, we detected a modest accumulation of Figure 8 molecules in FANCI-D2-depleted egg extract (Figure 5D, arrowhead and Figure S5C). However, FANCI-D2 depletion had almost no effect on the formation of nicked and supercoiled molecules (Figure 5D), indicating that the bulk of pICLAP unhooking is FANCI-D2- and incision-independent. We conclude that a small fraction of pICLAP is unhooked via FANCI-D2-dependent incisions, whereas the majority is unhooked by an alternative mechanism.
To gain additional insight into pICLAP repair, we examined nascent strand products (Figure 5E). As shown in Figure 5F, both the leftward and rightward leading strands stalled transiently near the −20 position before being converted to longer products (lanes 13–18). For the leftward leading strand, which encounters the AP-side of the ICL (Figure 5E), stalling near −20 was followed by a strong but transient arrest at the −1 position (Figure 5F, white arrowhead), suggesting that unhooking of pICLAP generates a lesion whose bypass requires TLS. For the rightward leading strand, which encounters the adenine-side of the ICL (Figure 5E), stalling near −20 was also followed by a −1 arrest (black arrowhead). However, these −1 products accumulated more slowly than those of the leftward fork, and to a lesser extent. This suggests that the rightward −1 products might derive from the small fraction of pICLAP molecules that undergo CMG unloading, incisions, and TLS bypass of the resulting mono-adduct (compare accumulation of rightward −1 products for pICLPt [Figure 5F, lanes 1 to 4] and pICLAP [Figure 5F, lanes 13 to 16]). To test this idea, we added the p97 inhibitor NMS-873 to suppress the incision pathway. As seen for pICLPso (Figure 4B), NMS-873 strongly inhibited CMG unloading from pICLAP (Figure S5D) but had no effect on unhooking (Figure S5E, lanes 19–24). Importantly, while NMS-873 had no significant effect on the leftward leading strand, it completely abolished −1 arrest of the rightward leading strand (Figure 5F, lanes 19–24), consistent with formation of this product requiring CMG unloading and incisions. In support of the −1 product entering the incision pathway, FANCI-D2 depletion led to its persistence (Figure S5F). Our results show that a small fraction of pICLAP unhooking involves CMG unloading, leading strand advance to −1, and FANCI-D2-dependent incisions, as seen for pICLPt and pICLPso-FdT. However, pICLAP is unhooked predominantly by an incision-independent pathway that does not require CMG unloading.
We next addressed whether incision-independent AP-ICL unhooking involves enzymatic cleavage of an N-glycosyl bond. Consistent with this idea, pICLAP repair intermediates were sensitive to APE1 digestion (Figure 6A–C), indicating that unhooking of pICLAP generates an AP site. The AP-ICL contains two distinct N-glycosyl bonds, either of which could be cleaved (Figures 5A and 6D). To distinguish which N-glycosyl bond is cleaved, we depleted Rev1 from egg extract (Figure 6E), and examined pICLAP replication in the presence of NMS-873 to suppress the incision pathway. As shown in Figure 6F, in the absence of Rev1, roughly half of the unhooked plasmids accumulated as open circular molecules, indicating that synthesis of only one nascent strand requires translesion synthesis, as predicted for cleavage of the nonnative N-glycosyl bond (Figure 6D, red pathway). Strikingly, in the absence of Rev1, the leftward leading strand, which encounters the AP-side of the ICL, stalled at the −1 position (Figure 6G, upper panel), whereas the rightward leading stand, which encounters the adenine-side of the ICL, exhibited no −1 arrest (Figure 6G, lower panel). These data further argue that the AP-ICL is unhooked via enzymatic cleavage of the non-native N-glycosyl bond to regenerate an AP site and an undamaged adenosine (Figure 6D, red pathway). As seen for cisplatin- and psoralen-ICLs (Zhang et al., 2015), AP-ICL unhooking requires replication fork convergence (Figure S6).
Figure 6. An AP-ICL is unhooked via cleavage of the non-native N-glycosyl bond.
(A) Expected species after digestion of pICLAP with HincII and APE1, including 3.3 kb and 2.3 kb fragments.
(B) pICLAP was replicated in egg extracts and then digested with HincII and APE1 as in Figure 2H.
(C) The 3.3 kb and 2.3 kb fragments in (B) were quantified and plotted.
(D) Two possible incision-independent unhooking pathways for an AP-ICL. Cleavage of the native N-glycosyl bond (blue arrowhead) yields a non-native adenosine and an AP site (middle panel), whereas cleavage of the non-native N-glycosyl bond (red arrowhead) yields an AP site and a normal adenosine (right panel).
(E) Mock-depleted and Rev1-depleted NPE were analyzed by Rev1 Western blotting. A relative volume of 100 corresponds to 0.66 μl NPE.
(F) pICLAP was replicated in mock- or Rev1-depleted egg extract in the presence of [α-32P]dATP and NMS-873 and analyzed as in Figure 1C.
(G) pICLAP was replicated in mock or Rev1 depleted egg extract in the presence of [α-32P]dATP and NMS-873. Samples were purified and digested with AflIII before separation on a denaturing polyacrylamide gel. Two portions of the autoradiograph are displayed with different contrast for optimal display.
NEIL3 unhooks psoralen- and AP-ICLs
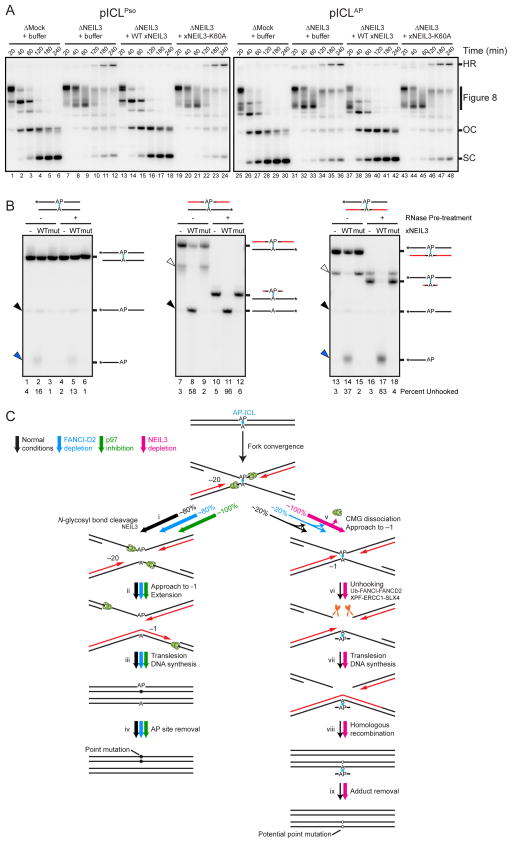
We next sought to identify the DNA glycosylase that unhooks psoralen- and AP-ICLs. In an unrelated mass spectrometry analysis of undamaged plasmid replicating in egg extracts, we identified NEIL3 as a possible replisome component (unpublished results). NEIL3 is one of eleven annotated glycosylases in the Xenopus genome and a member of the Fpg/Nei glycosylase family that has been implicated in the removal of oxidized bases from ssDNA (Liu et al., 2013a). We raised an antibody to Xenopus NEIL3 and depleted it from egg extracts (Figure S7A). Replication of undamaged or cisplatin-ICL containing plasmids was unaffected by NEIL3-depletion (Figure S7C). In contrast, depletion of NEIL3 led to a reduction in pICLPso and pICLAP open circular and supercoiled DNA products, and caused accumulation of these plasmids as Figure 8 structures and slowly migrating species resembling the HR reaction intermediates of pICLPt (Figure 7A, lanes 7 to 12 and 31 to 36). The effect of NEIL3-depletion was reversed by recombinant wild-type NEIL3 but not catalytically deficient NEIL3-K60A (Figure 7A, lanes 13 to 24 and 37 to 48, and Figure S7B; Krokeide et al., 2013). Thus, despite being structurally distinct, both psoralen- and AP-ICLs are unhooked by NEIL3. When NEIL3-depleted extracts were supplemented with NMS-873 to inhibit CMG unloading and the incision pathway, the low mobility intermediates were abolished and Figure 8 structures persisted for an extended period (Figure S7D, arrowheads). This result indicates that incisions and N-glycosyl bond cleavage represent the two major pICLPso and pICLAP unhooking pathways in egg extracts.
Figure 7. NEIL3 unhooks psoralen- and AP-ICLs.
(A) pICLPso or pICLAP was replicated in mock-depleted-, NEIL3-depleted-, or NEIL3-depleted egg extract supplemented with 300 nM recombinant NEIL3 in the presence of [α-32P]dATP. Repair intermediates were analyzed as in Figure 1C. As shown in Figure S7E, 15 nM recombinant NEIL3 also rescued pICLAP unhooking in NEIL3-depleted egg extract.
(B) Oligonucleotide substrates containing an AP-ICL were incubated with recombinant wild-type (WT) or K60A mutated (mut) NEIL3, separated on a denaturing polyacrylamide gel, and visualized by autoradiography. Black lines, DNA. Red lines, RNA. Cyan, AP-ICL. Asterisks indicate the 32P radiolabel. Black arrowheads, unhooked ssDNA. Blue arrowheads, β-elimination cleavage product. Bands marked by white arrowheads likely represent substrate that was not denatured during electrophoresis.
(C) Model for AP-ICL repair by incision-independent (left branch) or incision-dependent (right branch) pathways. Black lines, parental DNA. Red lines, nascent leading strands. Cyan, AP-ICL. Green ovals, CMG helicase. Pathway utilization is illustrated for unmodified extract (black arrows), FANCI-D2 depleted extract (blue arrows), extract supplemented with NMS-873 (green arrows), and NEIL3-depleted extract (pink arrows). Closed circles indicate point mutations that arise due to bypass of an AP site. Open circles indicate potential point mutations that may be introduced due to bypass of an AP-ICL-derived adenine monoadduct.
To determine whether NEIL3 is sufficient to unhook an ICL, we incubated recombinant NEIL3 with different AP-ICL containing oligonucleotide substrates and looked for the appearance of ssDNA (Figure 7B, black arrowheads). When the AP-ICL was located in duplex DNA, NEIL3-dependent unhooking was inefficient (Figure 7B, lane 2). To address whether the presence of ssDNA might enhance NEIL3 activity, we engineered ribonucleotides on one strand surrounding the ICL (Figure 7B, red lines), and optionally digested the substrates with RNase before addition of NEIL3. As shown in Figure 7B, the presence of RNA increased NEIL3-dependent unhooking (lane 8), and RNase digestion stimulated this activity further (lane 11). Catalytically inactive NEIL3-K60A had no activity on these substrates (Figure 7B, mut). Therefore, NEIL3 possesses intrinsic ICL unhooking activity that is stimulated by unusual double helical structure and/or ssDNA character. Notably, when the strand containing the AP site was labeled, a rapidly migrating species was detected (Figure 7B, blue arrowheads). This species arises from the AP lyase activity of NEIL3 (Liu et al., 2010), which cleaves the phosphodiester backbone, yielding a shortened ssDNA product. However, NEIL3’s AP lyase activity appears to be largely irrelevant for ICL repair because we find no evidence of DSB formation during AP-ICL unhooking in egg extract (see Discussion). Importantly, no shortened ssDNA product was detected when the bottom strand was labeled (Figure 7B, lanes 8 and 11), indicating that AP lyase activity is targeted exclusively to the top strand. Therefore, recombinant NEIL3 cleaves the non-native N-glycosyl bond (Figure 6D, red arrowhead), as observed in egg extracts.
Our results show that AP-ICLs are unhooked via two pathways. The minor pathway involves CMG unloading and FANCI-D2-dependent incisions (Figure 7C, thin black arrows). The major pathway, which is analogous to psoralen-ICL unhooking (Figure S3), involves cleavage of the non-native N-glycosyl bond by NEIL3 (Figure 7C, thick black arrows). Unhooking likely allows converging CMGs to pass over the lesion, after which the remaining gaps are filled (Figure 7Cii–iii). For the strand containing the AP site, gap filling requires Rev1-dependent TLS (Figure 7Ciii). Finally, the AP site is probably removed by an AP endonuclease (Figure 7Civ).
Discussion
We previously showed that Xenopus egg extracts repair cisplatin-ICLs via FANCI-D2-dependent incisions at stalled replication forks. Using the same cell-free system, we now report a mechanism of S phase ICL repair that operates on AP-ICLs (Figure 7Ci–iv), which may form in vivo, as well as psoralen-ICLs (Figure S3i–iv). In this pathway, after forks converge on the ICL, the lesion is unhooked via N-glycosyl bond cleavage by NEIL3. N-glycosyl cleavage is faster than incision and avoids DSB formation. When N-glycosyl cleavage is prevented, unhooking occurs via incisions, establishing highly flexible pathway choice. Our results identify a new mechanism of S phase ICL repair that avoids the risk of gross chromosomal rearrangements.
NEIL3 as an ICL glycosylase
NEIL3 is a vertebrate-specific Fpg/Nei family DNA glycosylase. Although NEIL3 acts preferentially on further oxidation products of 8-oxo-dG in vitro, its biological function has remained elusive (Liu et al., 2013a). Our work now identifies a role for NEIL3 in replisome bypass of ICLs in S phase. Consistent with this idea, NEIL3 is specifically expressed in the S and G2 phases of proliferating tissues including bone marrow, spleen, neuronal progenitor cells, and tumours (Hildrestrand et al., 2009; Neurauter et al., 2012; Torisu et al., 2005).
Most DNA glycosylases cleave the N-glycosyl bond only after the damaged base is rotated by ~180° into the enzyme’s catalytic pocket (“eversion”; Brooks et al., 2013). The question arises how NEIL3 unhooks an ICL, given that the bases of an ICL cannot be readily everted. Notably, the structure of NEIL3’s glycosylase domain is virtually superimposable on that of NEIL1 (Liu et al., 2013a), a related glycosylase that removes oxidized bases via classic base eversion. However, NEIL3 contains two features that suggest a unique mode of action. First, NEIL3 lacks two of the three NEIL1 residues that stabilize the nucleobase opposite the everted base, and it lacks the loop that stabilizes the everted base (Liu et al., 2013b). Second, the DNA binding cleft of NEIL3 contains two negatively charged patches that disfavor interactions with the DNA strand opposite the damaged base (Liu et al., 2013b), which explains NEIL3’s strong preference for single-stranded DNA damage (Liu et al., 2010). Based on these considerations, we speculate that replication fork convergence places ICLs in a single-stranded DNA context, allowing partial eversion of the cross-linked base into the active site of NEIL3, which is unusually open. As such, NEIL3’s mechanism is probably distinct from that of AlkD, a DNA glycosylase recently shown to cleave bulky base lesions in the complete absence of base eversion (Mullins et al., 2015). A recent report that Streptomyces orf1 DNA glycosylase can act on azinomycin B ICLs (Wang et al., 2016) suggests that ICL unhooking by DNA glycosylases is widely conserved in evolution.
NEIL3 is bifunctional, possessing both DNA glycosylase and AP lyase activities (Figure 7B; Krokeide et al., 2013; Liu et al., 2010). However, in the context of replication-coupled ICL repair, AP lyase activity after unhooking would create a DSB. We observed little or no evidence of DSB formation during psoralen- or AP-ICL repair, especially when NEIL3 was the sole unhooking activity (Figure 4B and S5E), implying that NEIL3’s AP lyase activity is suppressed during ICL repair. We speculate that once NEIL3 has unhooked the ICL, CMG or some other factor displaces NEIL3 from DNA, preventing the lyase reaction.
Pathway choice during ICL repair
We showed that upon chemical stabilization of N-glycosyl bonds or depletion of NEIL3, psoralen- and AP-ICLs switched pathways and were now processed via FANCI-D2-dependent incisions (Figure S3 and Figure 7C, pink arrows). Conversely, when incisions were blocked during AP-ICL repair (with NMS-873), all repair occurred via N-glycosyl cleavage (Figure 7C, green arrows). Therefore, for these two lesions, there is considerable flexibility in which pathway is utilized for repair. NEIL3-mediated N-glycosyl cleavage is the primary mechanism, probably because it does not require CMG unloading or FANCI-D2 activation, and therefore occurs more rapidly than incisions. We previously reported that, like a cisplatin ICL, a nitrogen mustard-like ICL is unhooked via incisions (Raschle et al., 2008). However, this ICL was generated between 7-deaza-2′-deoxyguanosines to counteract the inherent instability of the N-glycosyl bond in N7-alkylated guanines (Guainazzi et al., 2010). This stabilized nitrogen mustard-ICL might resist the action of an ICL glycosylase, channeling the lesion into the incision-dependent pathway. Therefore, of the four ICLs we have examined so far, cisplatin-ICLs are the only ones that are clearly refractory to N-glycosyl bond cleavage. This feature of cisplatin-ICLs might result from the massive distortion of the DNA induced by the lesion. It should be noted that Neil3-nullizygous MEFs exhibit modest sensitivity to cisplatin (Rolseth et al., 2013), suggesting that a portion of cisplatin-ICLs may be processed by NEIL3 in cells. More ICLs will have to be examined to determine whether the degree of distortion is a primary factor in determining pathway choice.
Given our observation that N-glycosyl bond cleavage is an efficient means of repairing psoralen-ICLs, why does FANCD2 deficiency cause hyper-sensitivity to psoralen in C. elegans, mouse, and human cells (Houghtaling et al., 2005; Lee et al., 2007; Rothfuss and Grompe, 2004)? There are several possibilities. First, the number of ICLs generated in clonogenic survival experiments may overwhelm NEIL3, leading to a requirement for FANCI-D2-dependent unhooking. Second, psoralen-ICLs may usually be processed via N-glycosyl unhooking, but a fraction may not be compatible with this pathway due to local DNA or chromatin structure, requiring incisions. This concept is illustrated by AP-ICL processing in egg extracts, which occurs by both pathways (Figure 7C). Even in the case of pICLPso, FANCI-D2 depletion stabilized a small population of Figure 8 molecules (Figure 1F, lanes 19–24), indicating the incision pathway also operates on a minor fraction of these lesions. This observation is consistent with the recruitment of FANC proteins to chromatin in a recent proteomic analysis of psoralen-ICL repair (Raschle et al., 2015). Finally, FANCD2 may play at least one other role in ICL repair that is independent of unhooking. Importantly, the severe phenotype observed in humans lacking FANC genes (Joenje and Patel, 2001) shows that glycosylase-mediated unhooking does not fully substitute for incisions in the processing of endogenous lesions. It is important to note that like FANC-deificient mice, Neil3-nullizygous mice exhibit no severe phenotypes (Sejersted et al., 2011; Torisu et al., 2005), raising the possibility that these two repair systems have overlapping functions. Future experiments will address the relative roles of incision versus N-glycosyl bond cleavage in the response to exogenous and endogenous ICLs.
Mutagenic potential of different ICL repair strategies
We showed previously that cisplatin-ICL repair is ~98% error-free because after incisions, bypass of the liberated mono-adduct is quite accurate (Budzowska et al., 2015). This can be explained if bypass utilizes DNA pol eta, which prefers to insert cytosines across from guanine cisplatin adducts (Alt et al., 2007). Although cisplatin-ICL repair appears to generate relatively little point mutagenesis, it involves a DSB intermediate, which can lead to gross chromosomal rearrangements, a hallmark of cancer. The mutagenic potential of the NEIL3 pathway is very different. After N-glycosyl bond cleavage of a psoralen-ICL, one parental strand retains an AP site whereas the other contains a thymine-psoralen mono-adduct (Figure S3, black arrows). Although we have not determined the fidelity of psoralen-ICL repair, it is likely to be low since the AP site is non-instructive (Liao et al., 2007). Consistent with this prediction, the efficiency of error-free repair, as measured by restriction-site regeneration, is only ~5%: the mono-adduct (which is not removed in egg extracts) blocks the restriction site in half the unhooked plasmids, and random nucleotides are inserted across from the AP site in the other half (Figure S3iii). Repair should be further reduced by the fact that not all AP sites are removed after lesion bypass (Figure S3iv). In contrast, after N-glycosyl unhooking of the AP-ICL, one sister carries an AP site and the other a normal adenine (Figure 7Ci). Bypass of the AP site should be highly mutagenic, as for the psoralen-ICL, but the adenine will be copied accurately without TLS (Figure 7Cii). Accordingly, error-free AP-ICL repair is much more efficient than error-free psoralen-ICL repair, approaching 50%. In summary, cisplatin-ICL repair involves low point mutagenesis but a high potential for gross chromosomal rearrangements, whereas N-glycosyl bond unhooking likely involves high point mutagenesis while avoiding translocations.
N-glycosyl bond cleavage and CMG dynamics
Cisplatin-ICL unhooking requires replication fork convergence to allow CMG unloading and FANCI-D2-dependent incision by XPF-ERCC1 (Klein Douwel et al., 2014; Knipscheer et al., 2009; Zhang et al., 2015). Neil3-dependent unhooking of psoralen- (Zhang et al., 2015) and AP-ICLs (Figure S6) also requires fork convergence. However, in this case, CMG unloading is not essential for repair. Instead, CMG may actually be required for N-glycosyl bond cleavage. When CMG unloading was blocked during AP-ICL repair with p97 inhibitor, the incision pathway was abolished, and all the ICLs were processed via N-glycosyl bond cleavage (Figure 7C, green arrows). In contrast, in FANCI-D2-depleted extract (where CMG unloading is unaffected; Figure S5F), unincised AP-ICLs remained trapped as Figure 8 structures and were not processed by NEIL3 (Figure 7C, thin blue arrow). We speculate that once CMG dissociation commits a lesion to the incision pathway, NEIL3 cannot cleave the N-glycosyl bond. In this scenario, CMG might recruit NEIL3 to the ICL and/or maintain an appropriate DNA structure for NEIL3 action. The dependence of unhooking on converged CMGs would insure that NEIL3 does not accidentally attack bases at single, active replication forks. After N-glycosyl bond cleavage, CMGs probably translocate past the unhooked ICL until they reach the downstream Okazaki fragment of the converged fork, whereupon they dissociate via the mechanism that operates during termination (Figure S3i–iii and Figure 7Ci–iii; Dewar et al., 2015).
Conclusion
In contrast to the current view that eukaryotic ICL unhooking necessarily involves endonucleolytic incisions, we show that ICLs can also be unhooked via N-glycosyl cleavage by the NEIL3 glycosylase. This reaction does not require CMG unloading, mobilization of the Fanconi pathway, or a DSB break intermediate, and is therefore fast while also avoiding the risk of gross chromosomal rearrangements. Therapeutically, inactivation of NEIL3 in cancer patients might enhance the efficacy of chemotherapy whereas upregulation of NEIL3 may ameliorate the symptoms of Fanconi anemia.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Johannes Walter, johannes_walter@hms.harvard.edu
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Egg extracts were prepared using Xenopus laevis (Nasco Cat #LM0053MX). All experiments involving animals were approved by the Harvard Medical Area Institutional Animal Care and Used Committee and conform to relevant regulatory standards.
METHOD DETAILS
All experiments were performed at least twice, and a representative result is shown.
Preparation of pICL
The preparation of pICL was performed as previously described (note that the psoralen-ICL plasmids used in this paper contain an array of 48 lacO repeats in the vector backbone, as do AP-ICL plasmids, except those used in Figures 5, S5, and 6; Enoiu et al., 2012; Zhang et al., 2015). Briefly, the parental plasmid was digested with BbsI. The purified, crosslinked duplex oligonucleotide was then ligated into the tandem BbsI sites of the corresponding backbone plasmid. To make psoralen-ICL plasmids, complementary primers containing only one thymidine (or 2′-fluoroarabino-dT (FdT) for pICLFdT-Pso) were annealed in annealing buffer (100 mM potassium acetate, 30 mM HEPES-KOH (pH 7.4), and 2 mM magnesium acetate) at a concentration of 50 μM each. DNA-trioxsalen (referred as psoralen in the main text) crosslinking was carried out using 2.6 μM annealed DNA in crosslinking buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 50 mM NaCl) and 87.6 μM trioxsalen (Sigma). The reaction was exposed to 365 nm UVA light for six periods of 15 minutes each, at a power of 4 mW/cm2. After every cycle, fresh trioxsalen was added to 87.6 μM. Crosslinked DNA was purified from a 20% polyacrylamide, 8 M urea gel. 2′-fluoroarabino-dT (FdT) was synthesized by LINK technology (Maiti et al., 2009). DNA oligonucleotides containing FdT were synthesized and purified at the Keck Foundation Biotechnology Resource Laboratory at Yale University.
To make AP-ICL plasmids, an oligonucleotide containing a single deoxyuracil (IDT) was crosslinked to its complement essentially as described (Price et al., 2014). Briefly, the oligonucleotides were annealed in 30 mM HEPES-KOH (pH 7.0), 100 mM NaCl by heating to 95 °C for 5 min and cooling at 1 °C/min to 18 °C. The annealed duplex was then treated with uracil gylcosylase (NEB) in UDG buffer (20 mM Tris-HCl, 10 mM DTT, 10 mM EDTA [pH 8.0]) for 2 h at 37 °C followed by extraction with phenol:chloroform:isoamyl alcohol (25:24:1; pH 8) and ethanol precipitation. The duplex was then dissolved in 50 mM HEPES-KOH (pH 7.0), 100 mM NaCl and incubated at 37 °C for 120 h to allow crosslink formation. Crosslinked DNA was purified from a 20% polyacrylamide, 8 M urea gel.
To confirm the presence of the cross-links within the ligated plasmids, 150 ng pICL was digested with 5 U NotI (NEB) in 1× NEB buffer 3.1 for 120 min at 37 °C. The resulting digestion fragments were then treated with 5 U calf intestinal phosphatase (NEB) for 60 min at 37 °C followed by extraction with phenol:chloroform:isoamyl alcohol (25:24:1; pH 8) and ethanol precipitation. The fragments were then 5′ end radiolabeled with [γ-32P]dATP and T4 PNK (NEB), separated on a 15% polyacrylamide (19:1), 8 M urea, 1× TBE gel, and visualized by autoradiography.
Psoralen crosslinked duplexes (with the cross-link between the two highlighted Ts: thymidine for a normal psoralen-ICL, 2′-fluoroarabino-dT for a FdT-Psoralen-ICL):
5′-CCCCGGGGCTAGCC-3′
5′-GCACGGCTAGCCCC-3′
Abasic site crosslinked oligo duplexes (the cross-link occurs between the positions in bold; the ICL was generated in two orientations [AP site on top strand vs AP site on bottom strand] by swapping the underlined overhangs that are complementary to the pICL backbone):
5′-CCCTCTTCCGCTCdUTCTTTC-3′
5′-GCACGAAAGAAGAGCGGAAG-3′
Xenopus egg extracts and DNA replication
Xenopus egg extracts were prepared as described (Lebofsky et al., 2009). For DNA replication, plasmids were first incubated in a high-speed supernatant (HSS) of egg cytoplasm (final concentration of 7.5 ng DNA/μL extract) for 20–30 minutes at room temperature to license the DNA, followed by the addition of two volumes of nucleoplasmic egg extract (NPE) to initiate replication. Where indicated, NPE was supplemented with ~310 nM recombinant Xenopus FANCI-D2 complex or the indicated amount of recombinant Xenopus NEIL3. In all figures, the 0 minute time point refers to the time of NPE addition. For DNA labeling, reactions were supplemented with [α-32P]dATP, which is incorporated into nascent strands during replication. Where indicated, NMS-873 p97 inhibitor (Sigma) was used at a concentration of 200 μM in NPE. For replication assays without digestion, replication was stopped by adding 0.5 μl of each reaction to 10 μl of replication stop solution A (5% SDS, 80 mM Tris pH 8.0, 0.13% phosphoric acid, 10% Ficoll) supplemented with 1 μl Proteinase K (20 mg/ml) (Roche). Samples were incubated for 1 hour at 37 °C prior to separation by 0.8% native agarose gel electrophoresis. DNA samples were then detected using a phosphorimager (Lebofsky et al., 2009). For all other applications, replication reactions were stopped in 10 volumes of 50 mM Tris (pH 7.5), 0.5% SDS, 25 mM EDTA, and replication intermediates were purified as previously described (Raschle et al., 2008). Where indicated, replication intermediates were digested with 0.08 U/μL HincII (NEB) and/or 0.08 U/μL APE1 (NEB).
Antibodies and immunodepletions
Immunodepletions using antibodies against FANCD2 (Rabbit 20019; Knipscheer et al., 2009), FANCI (Rabbit 26864; Duxin et al., 2014), and Rev1 (Rabbits 714 and 1010; Budzowska et al., 2015) were performed as described previously (Budzowska et al., 2015). Briefly, protein A Sepharose Fast Flow beads (GE Healthcare) were incubated with antibodies overnight. Two rounds of FANCI-D2 co-depletion were performed at room temperature for 20 minutes each and three rounds of Rev1 depletion were performed at 4 degrees for 1 hour each. For NEIL3 immunodepletion, 2.5 volumes of affinity purified antibodies against NEIL3 (1 mg/mL; New England Peptide project #3470) were incubated with 1 volume Sepharose Fast Flow beads overnight at 4 °C. Three rounds of NEIL3 depletion were then performed by incubating five volumes of egg extract with 1 volume antibody-bound beads for 1 hour at 4 °C. Western blots were probed using antibodies against Rev1 (Rabbit 1010), FANCD2 (Rabbit 20019), FANCI (Rabbit 26651), MCM6 (New England Peptide project #2926), CDC45 (Rabbit 534; Walter and Newport, 2000), Histone H3 (Cell Signaling Technology #9715), and NEIL3 (New England Peptide project #3470).
Nascent strand analysis
Nascent strand analysis was performed as described (Raschle et al., 2008). Briefly, pICL plasmids were replicated in the presence of [α-32P]dATP as described above, and purified replication products were digested with AflIII or AflIII and EcoRI, as indicated, followed by addition of 0.5 volumes Gel loading Buffer II (Life Technologies). Radiolabeled nascent strands were then separated on a 7% denaturing polyacrylamide gel, transferred to filter paper, dried, and visualized by autoradiography. Sequencing gel markers (not shown) were generated using the Thermo Sequenase Cycle Sequencing kit (USB Corporation) with a primer (5′-CATGTTTTACTAGCCAGATTTTTCCTCCTCTCCTG-3′) that anneals to pICL 149 nucleotides upstream of the cross-link.
Error free repair assay
The error free repair assay was performed as described (Raschle et al., 2008). Briefly, pICLPso or pICLAP was replicated in the presence or absence of geminin (400 nM in HSS). A 1.2 kb ApaLI or 0.9 kb HindIII pCtrl digestion fragment was added before extraction as the loading control. The replication products were harvested and digested with HincII/NheI (for pICLPso) or HincII/SapI (for pICLAP), and separated on a 1% agarose gel. Southern blotting was subsequently performed. DNA was fragmented in 0.25 N HCl for 10 min and then transferred (1.5 M NaCl, 0.4 M NaOH transfer buffer) onto a Hybond-N+ membrane (Amersham) by capillary action overnight. After transfer, the membrane was washed in 4× SSC for 5 minutes, and UV irradiated to crosslink the DNA to the membrane. Pre-hybridization was performed with 25 ml of hybridization buffer (4× SSC, 2% SDS, 1× Blocking reagent (Roche), 0.1 mg/ml Salmon sperm DNA (Life Technologies) for 30 minutes at 45 °C. Hybridization was carried out overnight with 25 μl of probe prepared with Roche random labeling kit (Roche) using pCtrl as a template. After overnight hybridization, the membrane was washed 4 times with 0.5× SSC, 0.25% SDS for 15 minutes at 45 °C. The dried membrane was exposed to a phosphoimager screen, and the repair products were quantified.
Strand-specific Southern blot
Strand-specific Southern blot was performed as previously described (Raschle et al., 2008; Zhang et al., 2015). Briefly, AflIII and AseI digested replication intermediates were separated on a 7% polyacrylamide gel and transferred to a Hybond-N+ membrane (Amersham). After transfer, the membrane was rinsed in 4× SSC for 5 minutes, and UV irradiated to crosslink the DNA to the membrane. The membrane was then pre-hybridized with 25 ml Ultrahyb buffer (Ambion) for at least 3 hours at 42 °C. Strand-specific probes generated by a PCR based primer extension reaction (Raschle et al., 2008) were added to the hybridization buffer and incubated with the membrane at 42 °C overnight. The membrane was washed 2 times with 2× SSC, 0.1% SDS for 5 minutes at 42 °C, dried, and exposed to a phosphorimager screen. In Figure 2J, the original images were converted into a log scale for display by applying the function f(p)=log (p)*255/log (255) to each pixel (p) in the images.
Replication-dependent unhooking assay
pICLPso or pICLAP was replicated in the presence or absence of geminin (400 nM in HSS). The replication products were isolated, digested with HincII, and separated on an alkaline 0.9% agarose gel (50 mM NaOH, 1 mM EDTA [pH 8.0]). Southern blotting was subsequently performed. DNA was fragmented in 0.25 N HCl for 10 min and transferred (1.5 M NaCl, 0.4 M NaOH transfer buffer) to a Hybond-N+ membrane (Amersham) by capillary action overnight. After transfer, the membrane was washed in 4× SSC for 5 minutes, and UV irradiated at 120,000 μJ/cm2 to crosslink the DNA to the membrane. Pre-hybridization was performed in 20 mL of Ultrahyb buffer (Ambion) for 18 h at 42 °C. Hybridization was carried out overnight at 42 °C with 25 μl of probe prepared with Roche Random Primed DNA Labeling Kit (Roche) using pCtrl as a template. The membrane was then washed twice with 1× SSC, 0.6% SDS for 15 minutes at 45 °C and twice with 0.1× SSC, 0.6% SDS for 15 min at 65 °C and then visualized by autoradiography.
Plasmid pull down
The plasmid pull down assay was performed as described (Budzowska et al., 2015). Briefly, streptavidin-coupled magnetic beads (Invitrogen; 6 μl per pull-down) were washed three times with 50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA pH 8, 0.02% Tween-20. Biotinylated LacI was added to the beads (12 pmol per 6 μl beads) and incubated at room temperature for 40 min. The beads were then washed four times with 10 mM Hepes (pH 7.7), 50 mM KCl, 2.5 mM MgCl2, 250 mM sucrose, 0.25 mg/ml BSA, 0.02% Tween-20 and resuspended in 40 μl of the same buffer. The bead suspension was stored on ice until needed. At the indicated times, 8 μl samples of the replication reaction were withdrawn and gently mixed with LacI-coated streptavidin Dynabeads. The suspension was immediately placed on a rotating wheel and incubated for 30 min at 4°C. The beads were washed three times with 10 mM Hepes (pH 7.7), 50 mM KCl, 2.5 mM MgCl2, 0.25 mg/ml BSA, 0.03% Tween-20. All residual buffer was removed, and the beads were resuspended in 40 μl of 2× Laemmli sample buffer. Equal volumes of the protein samples were blotted with the indicated antibodies.
Purification of recombinant Xenopus FANCI-FANCD2 complex and NEIL3
Xenopus FANCI-FANCD2 complex was purified from baculovirus infected Sf9 cells as described previously (Knipsheer et al., 2009). NEIL3 was PCR amplified from a cDNA library prepared from Xenopus laevis eggs (a gift from T. G. W. Graham) using primers A and B below. The NEIL3 cDNA was then re-amplified using primers C and D to introduce a C-terminal FLAG epitope tag, digested with EcoRI and XhoI, and ligated into a similarly digested pFastBac1 vector (ThermoFisher). The NEIL3-K60A mutation was introduced by Quikchange mutagenesis using primers E and F and confirmed by Sanger sequencing. Baculoviruses expressing NEIL3 were then prepared using the Bac-to-Bac system (ThermoFisher) according to the manufacturer’s protocols. NEIL3 protein was expressed in 250 mL suspension cultures of Sf9 insect cells (ThermoFisher) by infection with baculovirus expressing NEIL3-FLAG for 72 h. Sf9 cells were collected and suspended in 10 mL lysis buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10% glycerol, 1× Roche EDTA-free Complete protease inhibitor cocktail, 0.5 mM PMSF, 0.2% Triton X-100). Cells were lysed by sonication and the soluble fraction was collected by spinning the lysate at 25,000 rpm in a Beckman SW41 rotor for 1 hour. The soluble lysate was incubated with 200 μL anti-FLAG M2 affinity resin (Sigma) for 90 min at 4 °C. The resin was washed once with 10 mL lysis buffer, twice with wash buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10% glycerol, 0.2% Triton X-100), and three times with buffer A (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10% glycerol). NEIL3-FLAG protein was eluted from the resin with buffer A containing 100 μg/mL 3× FLAG peptide (Sigma). Elution fractions containing NEIL3-FLAG protein were pooled and dialyzed against 50 mM HEPES-KOH (pH 7.0), 300 mM NaCl, 1 mM DTT, 20% glycerol at 4 °C for 12 hours and then dialyzed against 50 mM HEPES-KOH (pH 7.0), 150 mM NaCl, 1 mM DTT, 15% glycerol at 4 °C for 3 hours. Aliquots of protein (~0.15 mg/mL) were stored at −80 °C.
Primer A: CAATTATGGTGGAGGGTCCGGG
Primer B: CTGTTTCACGTCTACTCTGTTTTTGCCC
Primer C: GCGCGCGGAATTCACCATGGTGGAGGGTCCGGGCTG
Primer D: CCAGCCCTCGAGCGTCTACTTGTCGTCATCGTCTTTGTAGTCACATGCCCAGTGCTCTTC
Primer E: GCTACGCTGGAGTGGAAACGCTGGGAGCGGAGCTTTTTATAT
Primer F: ATATAAAAAGCTCCGCTCCCAGCGTTTCCACTCCAGCGTAGC
NEIL3 glycosylase assay
AP-ICLs between complementary DNA or DNA/RNA chimeric oligonucleotides (IDT) were prepared and gel purified as described above. AP-ICLs were digested with 0.16 mg/mL RNase A (Sigma) and 0.2 U/μL RNase H (NEB) in 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT at 37 °C for 120 min and then purified to remove RNA. To monitor unhooking of AP-ICLs, 2.5 nM 5′ radiolabeled cross-linked substrate was incubated with 20 nM recombinant NEIL3 in 20 mM HEPES-KOH (pH 7.0), 50 mM NaCl, 1 mM DTT, 0.1 mg/mL BSA for 60 min at 37 °C (Liu et al., 2010). Reactions were quenched with 1 volume of 2× formamide buffer (86% formamide, 2× TBE, 20 mM EDTA [pH 8.0]), separated on a denaturing polyacrylamide and visualized by autoradiography.
Sequences of oligonucleotides for NEIL3 glycosylase assay (positions of cross-links are indicated in bold):
AP-ICL top (DNA): GCCATAGTAAGAAGAGCCGAATGC
AP-ICL top (DNA/RNA): rGrCrCrArUrArGrUrArAGAArGrArGrCrCrGrArArUrGrC
AP-ICL bottom (DNA): GCATTCGGCTCdUTCTTACTATGGC
AP-ICL bottom (DNA/RNA): rGrCrArUrUrCrGrGrCrUCdUTrCrUrUrArCrUrArUrGrGrC
QUANTIFICATION AND STATISTICAL ANALYSIS
Autoradiographs were quantified using ImageJ.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-FANCD2 | Knipscheer et al., 2009 | Rabbit 20019 |
| Rabbit polyclonal anti-FANCI | Duxin et al., 2014 | Rabbits 26651 and 26864 |
| Rabbit polyclonal anti-CDC45 | Walter and Newport, 2000 | Rabbit 534 |
| Rabbit polyclonal anti-MCM6 | New England Peptide | Project #2926 |
| Rabbit polyclonal anti-H3 | Cell Signaling Technology | Cat #9715 |
| Rabbit polyclonal anti-Rev1 | Budzowska et al., 2015 | Rabbits 714 and 1010 |
| Rabbit polyclonal anti-NEIL3 | New England Peptide; this study | Project #3470 |
| Rabbit polyclonal anti-NEIL3 | Abgent; this study | Rabbit 57141 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Xenopus laevis NEIL3-FLAG | this study | N/A |
| Recombinant Xenopus laevis NEIL3-FLAG K60A | this study | N/A |
| Recombinant Xenopus laevis FANCI-FANCD2 | Knipscheer et al., 2009 | N/A |
| 2'-fluoroarabino-dT (FdT) | LINK Technologies | N/A |
| NMS-873 | Sigma | Cat #SML1128 |
| Trioxsalen | Sigma | Cat #T6137 |
| Critical Commercial Assays | ||
| Thermo Sequenase Cycle Sequencing Kit | USB | Cat#785001KT |
| Random Primed DNA Labeling Kit | Roche | Cat#11004760001 |
| Bac to Bac Expression System | ThermoFisher | Cat#10359016 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Sf9 insect cells | ThermoFisher | Cat#B82501 |
| Experimental Models: Organisms/Strains | ||
| Xenopus laevis | Nasco | Cat #LM0053MX |
| Recombinant DNA | ||
| pICL-lacOPt | Zhang et al., 2015 | N/A |
| pICL-lacOPso | Zhang et al., 2015 | N/A |
| pICL-lacOAP | this study | N/A |
| pICL-lacOAP-reverse | this study | N/A |
| pCtrl-lacO | Dewar et al., 2015 | N/A |
| pICLPt | Raschle et al., 2008 | N/A |
| pICLAP | this study | N/A |
| pCtrl | Raschle et al., 2008 | N/A |
| pFastBac1-NEIL3-FLAG | this study | N/A |
| pFastBac1-NEIL3K60A-FLAG | this study | N/A |
| Sequence-Based Reagents | ||
| See Table S1 for sequences of primers and oligonucleotides used in the preparation of pICLs and AP-ICL unhooking substrates | this study | N/A |
| Software and Algorithms | ||
| ImageJ | Abramoff et al., 2004 | N/A |
| Other | ||
Supplementary Material
Acknowledgments
We thank James Dewar and Markus Raschle for sharing unpublished mass spectrometry results, Martin Cohn and Anna Motnenko for the protocol of psoralen-ICL preparation, Markus Raschle for feedback on the manuscript, and members of the Walter lab for discussions. This work was supported by a Jane Coffin Childs postdoctoral fellowship to D.R.S., a Human Frontiers Science Program long-term fellowship (LT000773/2010-L) and a European Molecular Biology Organization long-term fellowship (ALTF 742-2009) to M.B., NIH grants HL98316 and GM62267 to J.C.W, and NIH grant GM072711 to A.C.D. J.C.W is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
J.Z. first discovered incision-independent unhooking and characterized psoralen-ICL repair. D.R.S. characterized unhooking of the AP-ICL and identified NEIL3 as the ICL glycosylase. M.B. performed the experiments in Figure 2, C–F. A.C.D. supplied 2′-fluoroarabino-dT (FdT) and consulted on data interpretation. J.C.W., J.Z., and D.R.S. designed experiments, analyzed the data, and wrote the paper.
References
- Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzowska M, Graham TG, Sobeck A, Waga S, Walter JC. Regulation of the Rev1-pol zeta complex during bypass of a DNA interstrand cross-link. EMBO J. 2015;34:1971–1985. doi: 10.15252/embj.201490878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino GD, Shi YB, Hearst JE. Wavelength dependence for the photoreversal of a psoralen-DNA cross-link. Biochemistry. 1986;25:3013–3020. doi: 10.1021/bi00358a042. [DOI] [PubMed] [Google Scholar]
- Clauson C, Scharer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol. 2013;5:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couve S, Mace-Aime G, Rosselli F, Saparbaev MK. The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure. J Biol Chem. 2009;284:11963–11970. doi: 10.1074/jbc.M900746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Dewar JM, Budzowska M, Walter JC. The mechanism of DNA replication termination in vertebrates. Nature. 2015;525:345–350. doi: 10.1038/nature14887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Dewar JM, Yardimci H, Walter JC. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014;159:346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia? Curr Opin Cell Biol. 2015;37:49–60. doi: 10.1016/j.ceb.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoiu M, Ho TV, Long DT, Walter JC, Scharer OD. Construction of plasmids containing site-specific DNA interstrand cross-links for biochemical and cell biological studies. Methods Mol Biol. 2012;920:203–219. doi: 10.1007/978-1-61779-998-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. Selective Bypass of a Lagging Strand Roadblock by the Eukaryotic Replicative DNA Helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- Guainazzi A, Campbell AJ, Angelov T, Simmerling C, Scharer OD. Synthesis and molecular modeling of a nitrogen mustard DNA interstrand crosslink. Chemistry. 2010;16:12100–12103. doi: 10.1002/chem.201002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildrestrand GA, Neurauter CG, Diep DB, Castellanos CG, Krauss S, Bjoras M, Luna L. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 2009;10:45. doi: 10.1186/1471-2202-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling S, Newell A, Akkari Y, Taniguchi T, Olson S, Grompe M. Fancd2 functions in a double strand break repair pathway that is distinct from non-homologous end joining. Hum Mol Gen. 2005;14:3027–3033. doi: 10.1093/hmg/ddi334. [DOI] [PubMed] [Google Scholar]
- Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokeide SZ, Laerdahl JK, Salah M, Luna L, Cederkvist FH, Fleming AM, Burrows CJ, Dalhus B, Bjoras M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst) 2013;12:1159–1164. doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, Walter JC. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- Lee KY, Yang I, Park JE, Baek OR, Chung KY, Koo HS. Developmental stage- and DNA damage-specific functions of C. elegans FANCD2. Biochem Biophys Res Comm. 2007;352:479–485. doi: 10.1016/j.bbrc.2006.11.039. [DOI] [PubMed] [Google Scholar]
- Liao S, Matsumoto Y, Yan H. Biochemical reconstitution of abasic DNA lesion replication in Xenopus extracts. Nucleic Acids Res. 2007;35:5422–5429. doi: 10.1093/nar/gkm552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci USA. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Holmes A, Averill AM, Cannan W, Wallace SS. Expression and purification of active mouse and human NEIL3 proteins. Protein Expr Purif. 2012;84:130–139. doi: 10.1016/j.pep.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Doublie S, Wallace SS. Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat Res. 2013a;743–744:4–11. doi: 10.1016/j.mrfmmm.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Imamura K, Averill AM, Wallace SS, Doublie S. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure. 2013b;21:247–256. doi: 10.1016/j.str.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol Cell. 2014;56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi P, D’Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- Maiti A, Morgan MT, Drohat AC. Role of two strictly conserved residues in nucleotide flipping and N-glycosylic bond cleavage by human thymine DNA glycosylase. The J Biol Chem. 2009;284:36680–36688. doi: 10.1074/jbc.M109.062356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric M, Maculins T, De Piccoli G, Labib K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 2014;346:1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SP, Bailey R, Campion N, Herron S, Gambus A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science. 2014;346:477–481. doi: 10.1126/science.1253585. [DOI] [PubMed] [Google Scholar]
- Mullins EA, Shi R, Parsons ZD, Yuen PK, David SS, Igarashi Y, Eichman BF. The DNA glycosylase AlkD uses a non-base-flipping mechanism to excise bulky lesions. Nature. 2015;527:254–258. doi: 10.1038/nature15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- Neurauter CG, Luna L, Bjoras M. Release from quiescence stimulates the expression of human NEIL3 under the control of the Ras dependent ERK-MAP kinase pathway. DNA Repair (Amst) 2012;11:401–409. doi: 10.1016/j.dnarep.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Pontel LB, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, Arends MJ, Chandrasekaran G, Broecker V, Wei W, Liu L, et al. Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Mol Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NE, Catalano MJ, Liu S, Wang Y, Gates KS. Chemical and structural characterization of interstrand cross-links formed between abasic sites and adenine residues in duplex DNA. Nucleic Acids Res. 2015;43:3434–3441. doi: 10.1093/nar/gkv174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NE, Johnson KM, Wang J, Fekry MI, Wang Y, Gates KS. Interstrand DNA-DNA cross-link formation between adenine residues and abasic sites in duplex DNA. J Am Chem Soc. 2014;136:3483–3490. doi: 10.1021/ja410969x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Smeenk G, Hansen RK, Temu T, Oka Y, Hein MY, Nagaraj N, Long DT, Walter JC, Hofmann K, et al. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science. 2015;348:1253671. doi: 10.1126/science.1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolseth V, Krokeide SZ, Kunke D, Neurauter CG, Suganthan R, Sejersted Y, Hildrestrand GA, Bjoras M, Luna L. Loss of Neil3, the major DNA glycosylase activity for removal of hydantoins in single stranded DNA, reduces cellular proliferation and sensitizes cells to genotoxic stress. Biochim Biophys Acta. 2013;1833:1157–1164. doi: 10.1016/j.bbamcr.2012.12.024. [DOI] [PubMed] [Google Scholar]
- Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejersted Y, Hildrestrand GA, Kunke D, Rolseth V, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Asegg M, Fleming AM, Saugstad OD, et al. Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proc Natl Acad Sci USA. 2011;108:18802–18807. doi: 10.1073/pnas.1106880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D’Andrea AD. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J Biochem. 2005;138:763–772. doi: 10.1093/jb/mvi168. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Wang S, Liu K, Xiao L, Yang L, Li H, Zhang F, Lei L, Li S, Feng X, Li A, et al. Characterization of a novel DNA glycosylase from S. sahachiroi involved in the reduction and repair of azinomycin B induced DNA damage. Nucleic Acids Res. 2016;44:187–197. doi: 10.1093/nar/gkv949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HL, Gottesman ME, Gautier J. The differences between ICL repair during and outside of S phase. Trends Biochem Sci. 2013;38:386–393. doi: 10.1016/j.tibs.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC. DNA interstrand cross-link repair requires replication-fork convergence. Nat Struct Mol Biol. 2015;22:242–247. doi: 10.1038/nsmb.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.