Abstract
Background
Staphylococcus epidermidis is an opportunistic pathogen involved in hospital-acquired infections, particularly in those related to medical devices. This study characterized 50 genetically unrelated S. epidermidis isolates from bloodstream infections (BSIs, n = 31) and nares (n = 19) of neonates in relation to staphylococcal chromosomal cassette mec (SCCmec) type, biofilm production and associated genes, and the arginine catabolic mobile elements (ACME), in order to detect virulence factors that could discriminate a potential invasiveness isolate or predict an increasing pathogenicity.
Results
Isolates from both groups showed no difference for biofilm production and ACME genes detection. However, BSI isolates harbored more frequently the sdrF and sesI genes (p < 0.05), whereas biofilm producer isolates were associated with presence of the aap gene. The sdrF gene was also significantly more in the biofilm producer isolates from BSI. The SCCmec type IV and the ccr2 complex were related to BSI isolates (p < 0.05), while 83% of the nasal isolates were non-typeable for the SCCmec elements, with the mec complex and ccr undetectable as the most frequent profile.
Conclusions
Despite the great clonal diversity displayed by S. epidermidis isolates from neonates, BSI isolates harbored more frequently the sdrF and sesI adhesin genes, while nasal isolates were very variable in SCCmec composition. These aspects could be advantageous to improve colonization in the host increasing its pathogenicity.
Keywords: S. epidermidis, Neonates, Bloodstream infection, Nasal, SCCmec, Virulence
Background
Staphylococcus epidermidis, a common human commensal microorganism that colonizes skin and mucosal surfaces, has become an opportunistic pathogen, due to its ability to colonize invasive medical devices causing bloodstream infections (BSI) [1]. Some of the interventions used to treat neonates, particularly those admitted to neonatal intensive care units (NICUs), including prolonged antibiotic use and invasive procedures that disrupt the skin integrity, may expose neonates to the risk of developing S. epidermidis infections [2].
A wide range of surface proteins with adhesive properties improves the ability of S. epidermidis to adhere to different surfaces [1]. The Bhp protein (Bap homologue protein) and the autolisin/adhesin AtlE (autolysin of S. epidermidis) mediate the initial adhesion through hydrophobic interactions [3]. Almost at the same time, human extracellular matrix components bound and cover the polymeric surface, and a group of microbial proteins called Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs), like SdrF, SdrG (also known as Fbe) and Embp (extracellular matrix-binding protein) can specifically bind to collagen, fibrinogen and fibronectin, respectively [4–6]. Additionally, AtlE and Aae (autolysin/adhesin of S. epidermidis) proteins bind nonspecifically to fibrinogen, fibronectin and vitronectin [7, 8]. Other S. epidermidis proteins have also been described as putative adhesins, like the GehD lipase that binds to collagen [9] and the S. epidermidis surface (Ses) proteins, among them SesI, has gained attention due to their immunogenic properties [10] and its association with invasive isolates [11]. Many S. epidermidis isolates carry the icaADBC operon that encodes proteins involved in the synthesis of the exopolysaccharide PIA (polysaccharide intercellular adhesin), which connects the bacteria cells in the biofilm [12]. PIA together with Embp, Bhp and Aap (accumulation-associated proteins) are responsible for the intercellular adhesion and accumulation, enabling the biofilm formation [6, 13, 14].
S. epidermidis isolates have presented methicillin resistance, which is determined by the acquisition of the mecA gene, carried by a genetic mobile element known as staphylococcal chromosomal cassette mec (SCCmec). The mecA gene encodes a modified penicillin-binding protein (PBP2a) that presents low affinity for beta-lactam antibiotics [15]. Eleven types (I to XI) of SCCmec have been assigned for Staphylococcus aureus based on the classes of the mec gene complex and the types of the ccr gene complex [16]. In S. epidermidis these elements are very diverse and most of the isolates are defined as non-typeable [17–19].
The presence of the arginine catabolic mobile element (ACME) among S. epidermidis isolates has been receiving more attention since it may provide advantages in host colonization by staphylococcal cells [20]. This genetic element is composed of two gene clusters, the arc-operon, encoding a secondary arginine deiminase system and the opp3-operon that encodes a putative oligopeptide permease system [21]. It has been proposed that the ccr recombinase of the SCCmec element could be responsible by the ACME mobilization, suggesting that the horizontal transfer of these two elements may be linked [20].
Despite the increased number of studies involving the S. epidermidis species, there are still few studies that have detected characteristics that could distinguish infection and colonization isolates, especially among isolates from neonates. In this study we evaluated 50 genetically unrelated S. epidermidis isolates from bloodstream infections and nasal colonization of neonates in relation to SCCmec types, biofilm formation and associated genes, and the presence of ACME in order to detect virulence factors that could distinguish a potential invasiveness isolate or predict an increasing pathogenicity.
Methods
Clinical isolates
One hundred twenty-six S. epidermidis isolates from 126 neonates admitted in NICUs of four hospitals at Rio de Janeiro, Brazil, between May 2007 and March 2012 and belonging to the laboratory collection were characterized. Among them, 54 were recovered from blood cultures and related to bloodstream infections (BSIs) following the Center of Disease Control (CDC) criteria (2008). The other 72 isolates were obtained from nasal swabs by the infection control commission professionals. One isolate per patient was included in this study. After identification of all isolates as S. epidermidis by the simplified phenotypic [22] and PCR methods [23], they were characterized by pulsed field gel electrophoresis (PFGE) to exclude the clonality of isolates. Bacterial DNA was extracted and digested with the SmaI enzyme [24] and the restriction fragments were separated using a BioRad CHEF DR III apparatus, the PFGE profiles obtained were analyzed with Bio-Numerics software. Similarity percentage was identified on a dendrogram derived from the unweighted pair group method using arithmetic averages and based on Dice coefficients. Isolates showing a similarity coefficient < 80% or differences of five or more bands were considered genetically unrelated. For the 126 S. epidermidis isolates evaluated 50 different PFGE genetic backgrounds were identified, corresponding to 31 BSI isolates and 19 isolates from nares (Fig. 1). One representative isolate of each PFGE genotype was randomly selected for this study.
Fig. 1.
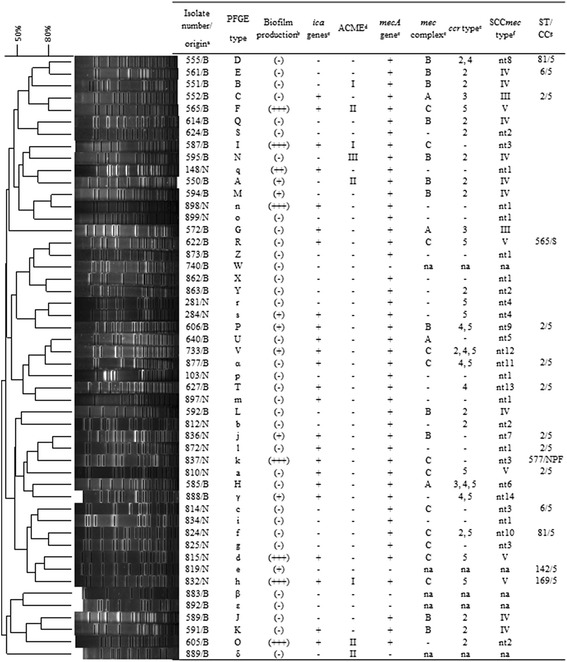
Dendrogram of the pulsed-field gel electrophoresis (PFGE) profiles of SmaI-digested genomic DNA of 50 genetically unrelated Staphylococcus epidermidis isolates and associated characteristics. Similarities percentage is identified on a dendrogram derived from the unweighted pair group method using arithmetic averages and based on Dice coefficients. a B: Bloodstream infection isolates, N: Nasal isolates; b (+++): strong; (++): moderate; (+): weak: (−): non-biofilm producer; c +: presence; −: absence; d I: arcA+/opp3AB+, II: arcA+/opp3AB-, III: arcA-/opp3AB+; −: negative; e −: not-detectable; na: not-applicable (methicillin-sensitive isolate); f na: not-applicable (methicillin-sensitive isolate); nt: non-typeable; g ST: sequence type; CC: clonal complex; S: singleton; NPF: none predicted founder
mecA gene detection and SCCmec typing
Bacterial DNA was extracted as previously described by Pitcher [25] and then detection of the mecA gene and SCCmec typing were performed according with Del Vecchio et al. and Kondo et al., respectively [26, 27]. The latter method consists of two multiplex PCR to detect the ccr complex (encoding for recombinases) and the mec complex (encoding for beta-lactam resistance). The combination of the types of ccr and mec class allowed the identification of the type of SCCmec (I to IX), and verify non-typeable isolates. The following S. aureus reference strains and clinical isolates were used as positive controls for SCCmec typing: EMRSA-3/Cordobés (SCCmec I) [28], Mu50 (SCCmec II) [29], HU25 (SCCmec III) [24], 527a (SCCmec IV) [30] and 557a (SCCmec V) [31].
Phenotypic detection of biofilm formation
Biofilm formation was determined according to Iorio et al. [18]. The S. epidermidis strains ATCC 35984 (formerly RP62A) and ATCC 12228 were used as positive and negative controls, respectively. All isolates were classified into the following categories: strong, moderate, weak and non-biofilm producer.
PCR assays for biofilm associated genes and ACME elements
The detection of the virulence genes aae, atlE, aap, bhp, embp, fbe, gehD, sdrF, sesI, the icaADB operon and ACME allotypes was performed by PCR. ACME allotypes were classified as: ACME-I contains both the arc and the opp-3 gene clusters; ACME-II contains arc but not opp-3; and ACME-III contains opp-3 without arc [20]. Bacterial DNA was extracted as previously described [25]. The primers and PCR conditions are summarized in the Table 1. Staphylococcus spp. reference strains used as positive controls were S. epidermidis ATCC 35984 (aae, aap, bhp, embP, gehD, icaABD and sesI genes), S. epidermidis ATCC 12228 (sdrF gene), S. epidermidis ATCC 14490 (atlE and fbe genes) and a clinical isolate of S. aureus number 526a/USA300 (arcA and opp3AB genes) [30].
Table 1.
Genes, oligonucleotide primers and PCR conditions used in this study
| Protein or genetic element | Gene | Primers 5’ → 3’a | Amplicon size (bpb) |
PCR conditions |
References |
|---|---|---|---|---|---|
| Aae | aae | F: GAGGAGGATTTTAAAGTGC R: AACATGACCATAGTAACC |
858 | 94 °C, 3 min; 40 cycles of: 94 °C, 90s; 55 °C, 1 min; 72 °C, 90s; final extension 72 °C, 5 min. | [8] |
| Aap | aap | F: ATACAACTGGTGCAGATGGTTG R: GTAGCCGTCCAAGTTTTACCAG |
400 | 94 °C, 3 min; 30 cycles of: 94 °C, 1 min; 50 °C, 1 min; 72 °C, 1 min; final extension 72 °C, 5 min. | [52] |
| AtlE | atlE | F: CAACTGCTCAACCGAGAACA R: TTTGTAGATGTTGTGCCCCA |
682 | 94 °C, 3 min; 30 cycles of: 94 °C, 1 min; 62 °C, 1 min; 72 °C, 1 min; final extension 72 °C, 5 min. | [33] |
| Bhp | bhp | F: ACGGACAATATCGTCTCTCAA R:: AACTTCAGCCGTTCCCTT |
1917 | 94 °C, 2 min; 40 cycles of: 94 °C, 30s; 55 °C, 30s; 72 °C, 75 s; final extension 72 °C, 5 min. | [10] |
| Embp | embp | F: AGCGGTACAAATGTCAATATC R: AGAAGTGCTCTAGCATCATCC |
455 | 94 °C, 3 min; 30 cycles of: 94 °C, 1 min; 62 °C, 1 min; 72 °C, 1 min; final extension 72 °C, 5 min. | [6] |
| GehD | gehD | F: TTTGAATTCTGCGCAAGCTCAATATAA R: TTTGCGGCCGCTATCGCTACTTACGTGTAA |
1179 | 94 °C, 2 min; 30 cycles of: 94 °C, 30s; 55 °C, 30s; 72 °C, 75 s; final extension 72 °C, 5 min. | [9] |
| SdrF | sdrF | F: GCTGAAGACAATCAATTAG R: TTAATATCCCCCTGTGCTG |
1875 | 94 °C, 4 min; 30 cycles of: 94 °C, 2 min; 60 °C, 1 min; 72 °C, 2 min; final extension 72 °C, 5 min. | [10] |
| SdrG | fbe | F: TAAACACCGACGATAATAACCAAA R: GGTCTAGCCTTATTTTCATATTCA |
496 | 94 °C, 3 min; 30 cycles of: 94 °C, 1 min; 62 °C, 1 min; 72 °C, 1 min; final extension 72 °C, 5 min. | [5] |
| SesI | sesI | F: GCTGATTATGTAAATGACTCAAAT R: AGCTTTTGTTGTTTGAGCTTC |
408 | 94 °C, 3 min; 30 cycles of: 94 °C, 1 min; 50 °C, 1 min; 72 °C, 1 min; final extension 72 °C, 5 min. | [11] |
| IcaADB | icaADB | F: TTATCAATG CCGCAGTTGTC R: GTTTAACGCGAGTGCGCTAT |
546 | 94 °C, 3 min; 30 cycles of: 94 °C, 1 min; 58 °C, 1 min; 72 °C, 1 min; final extension 72 °C, 5 min. | [33] |
| ACME | arcA | F: CTAACACTGAACCCCAATG R: GAGCCAGAAGTACGCGAG |
1946 | 95 °C, 10 min; 30 cycles of: 94 °C, 1 min; 52 °C, 1 min; 72 °C, 2 min; final extension 72 °C, 5 min. | [43] |
| opp3AB | F: GCAAATCTGTAAATGGTCTGTTC R: GAAGATTGGCAGCACAAAGTG |
1183 |
a F: Forward; R: Reverse
b bp: base pairs
Multilocus sequence typing
Among the 126 S. epidermidis isolates previously analyzed by PFGE, 15 isolates (seven BSI and eight nasal) that represented genotypes clustering five or more isolates were selected for characterization by MLST in the present study [32]. The PCR products were purified using the commercial system “GTX PCR and band purification” (GE 50 Healthcare, Buckinghamshire, England) according to the manufacturer’s specifications. The purified products were sequenced using the automated DNA sequencer ABI3100 (Applied Biosystems, Foster, CA, USA). Sequence types (ST) were determinate using the MLST database (http://www.mlst.net/) and characterized as singletons or members of a clonal complex (CC) by the eBURST algorithm (accessible at http://eburst.mlst.net/). Numbers for new ST reported here were assigned by the S. epidermidis MLST database curator.
Statistical methods
All comparisons were performed using the χ 2 test or the Fischer’s exact test. Differences were considered statistically significant when values of p < 0.05 were obtained.
Results
Biofilm production and genes associated
The ability to produce biofilm was analyzed for 50 S. epidermidis neonatal isolates from BSI (31 isolates) and nasal colonization (19 isolates). Biofilm formation was positive for 16 isolates, 8 (25.8%) from BSI and 8 (42.1%) from nares (Table 2); no significant difference between the groups was observed. Among the BSI isolates, three were classified as strong and five as weak biofilm producers. Among nasal isolates, four were strong, one moderate and three were weak biofilm producers. All the S. epidermidis isolates, irrespective to the biofilm production, carried at least three of the ten biofilm-associated genes investigated in this study, and 86% of them harbored five or more of these genes. The icaADB genes were present in 81.2% of the biofilm producer isolates and were detected in all isolates classified as moderate and strong producers and in the majority (5/8; 62.5%) of the weak biofilm producers. These genes were detected in 14 (45.2%) of the BSI and 10 (52.6%) of the nasal isolates. The aae, atlE, embp and fbe genes were frequently found in both groups of isolates, ranging from 74 to 100%. However, the sdrF and sesI genes were more commonly found among BSI isolates (p = 0.001 and p = 0.02, respectively). The sdrF gene was also significantly more associated with the biofilm producer isolates from BSI (p = 0.007). Furthermore, the presence of the aap gene was more frequent among biofilm producer isolates (15/16; 93.7%) than among non-biofilm producers (19/34; 55.9%) (p = 0.009).
Table 2.
Virulence genes and biofilm production in 50 Staphylococcus epidermidis isolates from bloodstream infection and nasal colonization
| Biofilm-associated virulence genes |
Number (%) of isolates | Number (%) of biofilm producer isolates | ||||
|---|---|---|---|---|---|---|
| BSIa
(n = 31) |
Nasalb
(n = 19) |
p value | BSIa
(n = 8) |
Nasalb
(n = 8) |
p value | |
| aae | 31 (100) | 19 (100) | 1 | 8 (100) | 8 (100) | 1 |
| aap | 18 (58) | 16 (84) | 0.068 | 7 (88) | 8 (100) | 1 |
| atlE | 30 (97) | 19 (100) | 1 | 8 (100) | 8 (100) | 1 |
| bhp | 1 (3) | 1 (5) | 1 | 0 (0) | 0 (0) | 1 |
| embp | 28 (90) | 19 (100) | 0.279 | 8 (100) | 8 (100) | 1 |
| fbe | 23 (74) | 16 (84) | 0.498 | 7 (88) | 6 (75) | 1 |
| gehD | 23 (74) | 11 (58) | 0.349 | 6 (75) | 6 (75) | 1 |
| sdrF | 20 (65) | 2 (11) | 0.001c | 6 (75) | 0 (0) | 0.007c |
| sesI | 12 (39) | 0 (0) | 0.02c | 3 (38) | 0 (0) | 0.2 |
| icaADB | 14 (45) | 10 (53) | 0.772 | 6 (75) | 7 (88) | 0.6 |
| arcA | 6 (19) | 1 (5) | 0.229 | 4 (50) | 1(13) | 0.282 |
| opp3AB | 3 (10) | 1 (5) | 1 | 1 (13) | 1 (13) | 1 |
a BSI: Bloodstream infection isolates
b Nasal: Nasal isolates
c: results with statistical significance
Taken together, the detection of all biofilm associated genes showed 28 different genetic profiles (Table 3). Seven of them are shared by isolates of both groups and included 22 (44%) isolates. Seventeen profiles were exclusive for the BSI isolates and four for the nasal colonizers. The majority of the profiles (78.6%) included only 1 or 2 S. epidermidis isolates, demonstrating a wide diversity of virulence genes profiles in this staphylococci species.
Table 3.
Virulence genes profiles and SCCmec types identified among 50 Staphylococcus epidermidis isolates from bloodstream infection and nasal colonization
| S. epidermidis isolates source (n)a | Isolates | Biofilm-associated virulence genes c | SCCmec types (n) b, d |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (n)b | icaADB | aae | aap | atlE | bhp | embp | fbe | gehD | sdrF | sesI | ||
| BSI and Nasal (22) |
6 (1B + 5 N) | + | + | + | + | - | + | + | + | - | - | V (1B + 2 N), nt1, nt3, nt7 |
| 4 (1B + 3 N) | - | + | + | + | - | + | + | + | - | - | IV (1B), nt1, nt10, na | |
| 3 (2B + 1 N) | - | + | + | + | - | + | + | - | - | - | IV (1B), nt3, na | |
| 3 (1B + 2 N) | + | + | + | + | - | + | + | - | - | - | nt3 (1B), nt1 (2 N) | |
| 2 (1B + 1 N) | - | + | - | + | - | + | + | + | - | - | IV (1B), nt3 | |
| 2 (1B + 1 N) | + | + | + | + | - | + | + | + | + | - | nt2 (1B), V | |
| 2 (1B + 1 N) | - | + | + | + | - | + | - | - | - | - | nt2 (1B), nt4 | |
| BSI (23) |
4 | + | + | + | + | - | + | + | + | + | + | III, nt5, nt9, nt12 |
| 3 | - | + | + | + | - | + | + | + | + | + | IV (2), nt8 | |
| 2 | - | + | + | + | - | + | + | + | + | - | IV, na | |
| 1 | + | + | - | + | - | + | + | + | + | + | III | |
| 1 | - | + | + | + | - | + | + | + | - | + | IV | |
| 1 | + | + | - | + | - | + | + | + | + | - | IV | |
| 1 | - | + | - | + | - | + | + | + | + | + | IV | |
| 1 | + | + | - | + | - | + | - | + | - | - | V | |
| 1 | - | + | - | + | - | + | - | - | + | + | nt2 | |
| 1 | + | + | - | + | - | - | + | + | + | + | nt6 | |
| 1 | + | + | - | + | - | - | + | - | - | - | nt13 | |
| 1 | - | + | - | + | + | + | - | + | + | - | na | |
| 1 | - | + | - | + | - | + | - | + | - | - | nt1 | |
| 1 | - | + | - | + | - | + | - | + | + | - | nt1 | |
| 1 | + | + | + | + | - | + | + | - | + | - | nt11 | |
| 1 | - | + | - | - | - | - | - | + | + | - | na | |
| 1 | + | + | - | + | - | + | - | - | + | - | nt14 | |
| Nasal (5) |
2 | - | + | - | + | - | + | + | - | - | - | nt1 |
| 1 | + | + | + | + | - | + | - | - | - | - | nt4 | |
| 1 | + | + | + | + | - | + | - | + | - | - | nt1 | |
| 1 | - | + | + | + | + | + | + | - | + | - | nt2 | |
a BSI: Bloodstream infection isolates
b B: Bloodstream infection isolates; N: Nasal isolates
c +: presence; -: absence
d III: mec complex A/ccr 3; IV: mec complex B/ccr 2; V: mec complex C/ccr 5; nt: non-typeable; na: not-applicable (methicillin-sensitive isolate); nt1: mec complex -/ccr -; nt2: mec complex -/ccr 2; nt3: mec complex C/ccr -; nt4: mec complex -/ccr 5; nt5: mec complex A/ccr -; nt6: mec complex A/ccr 3,4 and 5; nt7: mec complex B/ccr -; nt8: mec complex B/ccr 2 and 4; nt9: mec complex B/ccr 4 and 5; nt10: mec complex C/ccr 2 and 5; nt11: mec complex C/ccr 4 and 5; nt12: mec complex C/ccr 2,4 and 5; nt13: mec complex -/ccr 4; nt14: mec complex -/ccr 4 and 5
ACME detection
Among the BSI isolates two of them harbored the arcA and opp3AB genes (ACME I), four had only the arcA gene (ACME II) and one had only the opp3AB (ACME III) (Table 2). ACME elements were detected only in one nasal isolate, corresponding to ACME I. Despite the frequent presence of the ACME elements among the BSI isolates (seven isolates), no statistical significance was verified. Four of seven strong biofilm producer isolates harbored the ACME, however no association between the presence of this genetic island and biofilm production (p = 0.092) or the strong biofilm production (p = 0.106) was detected. Of eight ACME positive isolates, five were included in SCCmec types IV (three isolates) or V (2).
Detection of the mecA gene and SCCmec typing
The mecA gene was detected in 27/31 (87.1%) and 18/19 (94.7%) of the BSI and nasal isolates, respectively. For the 27 methicillin-resistant S. epidermidis (MRSE) isolates from BSI that were analyzed for composition of SCCmec elements, 13 (48.1%) were typeable: 9 (69.2%) harbored the SCCmec type IV, 2 (15.4%) the type III and 2 (15.4%) the type V (Fig. 1, Table 3). Many of the isolates from BSI (14/51.9%) were classified as non-typeable (nt), which possessed more than one ccr allotype; no ccr allotype or mec complex detectable; or no ccr allotype and mec complex detectable. Among the 18 MRSE nasal isolates only 3 (16.7%) were classified into a SCCmec type and harbored the type V, whereas the other 15 (83.3%) isolates were nt.
While the BSI isolates harbored more frequently the SCCmec type IV (33.3%) (p = 0.007) or the ccr complex 2 (51.9%) (p = 0.013), the nasal isolates showed an undetectable ccr complex (11 isolates; 61.1%) (p = 0.003) that included seven isolates of the prevalent nt1 profile (no mec complex and no ccr detectable) (p = 0.019).
Diversity and MLST characterization
The dendrogram obtained for the 50 S. epidermidis evaluated showed a distribution of isolates in seven larger clusters and three of them were composed exclusively by BSI or nasal isolates (Fig. 1). It was also possible identify eight pair of isolates that group together with about 80% of similarity. Among them the isolates of each of the pairs 555-561 and 552-565 belonged to BSI isolates from the same hospital and were recovered with about four months of difference. Although of this fact the isolates presented different genetic and phenotypic characteristics, including composition of the SCCmec elements, ST and virulence genes. The isolates of each of the pairs 836-872, 281-284 and 824-825 belonged to the same clinical origin (nasal) and NICU and had similar characteristics between each of them, but they showed differences in the composition of their SCCmec types or virulence. The remaining three pairs of isolates (622-873, 640-733 and 594-898) were from different NICUs and periods of isolation, and presented remarkable differences between the isolates within the pairs.
For seven BSI isolates evaluated by MLST, 4 STs were identified: ST2 (four isolates), ST6, ST81 and ST565 (Fig. 1). Among the eight nasal isolates evaluated, ST2 was also the most frequent (three isolates), whereas the STs 6, 81, 142, 169 and a new ST577 were also identified. The majority (13/15) of the isolates analyzed were included into CC5, the major CC of S. epidermidis. Two S. epidermidis isolates were not classified into CC5, the ST577 with no predicted founder and a singleton ST565.
Discussion
Several extrinsic factors associated with the nosocomial environment may disturb the delicate host-microbe balance of the neonates, resulting in a lifestyle conversion of S. epidermidis from mutualism to pathogenicity [2]. This species has become the focus of studies that attempt to understand which bacterial features can help the establishment of such infections. In this study, we evaluated different molecular characteristics associated with virulence and resistance in 50 genetically unrelated S. epidermidis isolates from neonates, 31 from BSIs and 19 from nasal colonization. We found some characteristics that differentiate isolates of these two groups, showing that some aspects could provide advantages to the pathogen to increase its colonization and pathogenicity.
Some authors have proposed that the ability to produce biofilm in combination with the presence of the ica operon could be used as pathogenesis markers to distinguish invasive from commensal isolates [33, 34]. Our results and other studies [35, 36] demonstrated no significant differences on biofilm production or presence of the ica genes between isolates of both groups. On the other hand, it should be noted that only 16 isolates of this study showed phenotypically biofilm production, while 45% of the BSI and 53% of the nasal isolates harbored the ica genes. It is possible that this fact was changed by addition of supplementary factors to the culture medium, such as glucose or NaCl, which could lead to or increase biofilm production [37, 38].
This is the first report to show a differentiated distribution of the biofilm-associated sdrF gene, found almost exclusively in BSI isolates. In order to confirm this result other 40 nasal isolates were also tested and only 25% were positive for the gene, maintaining a significant difference in relation to invasive isolates. Some studies have demonstrated that the SdrF protein is able to mediate alone the adherence of S. epidermidis to a wide variety of plastic materials, such as catheters and other prosthetic devices, participating in the initial adhesion through ionic interactions and then to the collagen, through specific receptor-ligand interactions [4, 39]. Taking this into consideration, S. epidermidis isolates carrying the sdrF gene would have a greater potential to attach to medical devices, aspect considerably advantageous for the establishment of invasive infections, including BSI.
Similarly to our results, Söderquist and coworkers [11] found a significant association between the presence of the sesI gene in invasive isolates and its absence among commensal ones, suggesting that this gene may be a possible virulence marker. To confirm these results, we tested other 40 isolates from neonate nares and found only two S. epidermidis isolates carrying sesI gene among them (data not shown), reinforcing the near absence of this gene among commensal isolates. However, Bowden and coworkers [10] already had detected the sesI gene in 45% of the S. epidermidis isolates from neonates BSI and in 29% of the contaminants isolates, as well as in 34% of the skin isolates from these patients. According to Söderquist et al. [11] since healthy people did not regularly carry sesI positive isolates the patients could acquire these isolates from the hospital environment after admission and become colonized. In addition, invasive S. epidermidis isolates can be selected during hospitalization, and this period responsible for changing a commensal bacterial population by isolates with greater virulence. However, the correlation of this gene with pathogenicity is not clear yet and further studies are necessary to understand the contribution of the SesI protein to the pathogenic potential of S. epidermidis.
In this study the presence of the aap gene was associated with biofilm producer isolates, and this association may be explained considering the important functions of the Aap protein in adhesion and intercellular aggregation, allowing the biofilm formation. In this line, two studies involving S. epidermidis ica operon deficient isolates identified the Aap protein as the major adhesin responsible for biofilm composed mainly of proteinaceous factors [13, 40]. This fact could explain how three ica operon-deficient isolates but aap gene-positive from the present study were able to produce biofilm, supporting the important role of this gene on ica-independent biofilm formation. Concerning the presence almost ubiquitous of the atlE, aae, fbe and embp genes, as well as the low frequency of the bhp gene observed in this study, other authors had also found similar results in S. epidermidis isolates [10, 33–35, 41].
The acquisition of the genetic island ACME by the staphylococci species seems to provide advantages in terms of host colonization, rather than an enhanced pathogenicity [20]. In the present study, only 8 (16%) of the 50 isolates harbored this element, which do not support other studies that found a high prevalence of ACME elements among S. epidermidis isolates [20, 42]. Svensson and coworkers [43] evaluated S. epidermidis isolates from neonates and detected this genetic element in 43% of blood isolates, while Granslo et al. [44] detected it in 23% of blood isolates, similarly to our results in relation to the BSI isolates (7/31; 22.6%). However, these authors also detected ACME in many isolates considered as contaminants and they concluded that this genetic element do not seem to be associated with increased pathogenicity of S. epidermidis. Geographic location could interfere in its occurrence, but additional studies are needed to clarify these findings. It has also been proposed that the ccrAB complex of the SCCmec element could be implicated in ACME mobilization [20]. However, in our study no association was found between any SCCmec type or ccr complexes and the ACME elements among the S. epidermidis isolates analyzed.
The high proportion of BSI S. epidermidis isolates harboring the mecA gene has become an expected fact. In our study, this gene was detected in 94.7% of the nasal isolates, which contrasts with the low frequencies described by Cherifi and coworkers [45] for their set of commensal isolates. However, it is important recognize the difference between commensal isolates from healthy individuals, without contact with a hospital and those obtained from hospitalized patients, as described by Rohde and coworkers [35]. These authors found great similarities in relation to the detection of the mecA gene and virulence factors between invasive and commensal isolates from patients with bone marrow transplantation, but with a striking difference from those obtained from healthy volunteers. This can be explained by the selective pressure that the hospital environment exerts, thus could led to the acquisition of genetic elements, including resistance and/or virulence genes, which can ensure the survival of the microorganism.
Concerning the SCCmec types, it was observed that the type IV was prevalent in BSI isolates (p = 0.007), while the type III was detected only in two of them, in contrast with the study of Pinheiro and coworkers [46] that detected 53.2% of their MRSE isolates from blood cultures carrying the SCCmec type III. However, the high frequency of the SCCmec IV in clinical isolates of S. epidermidis was already observed previously [47–49], demonstrating that this SCCmec type has become a common occurrence among hospital isolates. In addition, it was observed in our study that the ccr2 complex that comprises the SCCmec type IV was also associated with BSI isolates. Similarly, Svensson and coworkers [43] also reported a major frequency of ccr2 complex in their set of S. epidermidis isolates from blood cultures of neonates, while, Barbier et al. [42] found this association type with isolates recovered from nares. These data suggest that S. epidermidis appear to be efficient to acquire the ccr2 complex and could be the major reservoir for this type of genetic element, regardless of the origin of isolates.
A high proportion of isolates was SCCmec non-typeable (64.4% in this study) and this finding was not surprising, since this fact has been frequently reported among CNS isolates, independent of the clinical origin [36, 43, 48, 50]. However, in our study non-typeable profiles were more frequently associated with nasal isolates (83.3%), including the nt1 prevalent profile (ccr-mec complex undetectable). These data show that S. epidermidis isolates from the nasal origin can present a high genetic diversity of the SCCmec elements. This diversity was shown by Conlan and coworkers [51] who observed that S. epidermidis commensal isolates have an open pan-genome with considerable diversity between isolates, even when derived from a single individual or body site.
In this study, the MLST results for some selected isolates showed that the ST2 was the most frequent lineage, independent of the clinical origin. Moreover, this ST is included into the CC5 that was designated for 13 of 15 S. epidermidis isolates analyzed. ST2 has been reported to be the most widely disseminated hospital-associated ST type among S. epidermidis isolates [17, 18, 47, 51]. According to Li et al. [17], the successful spread of this lineage may be associated with the fact that, by recombination, ST2 generates novel phenotypic and genotypic variants, such as ica genes-positive isolates, which makes ST2 isolates easily able to spread in the hospital environment.
The greatest limitation of this study is the small number of isolates tested. In order to have as much diversity as possible and the results obtained be not biased since some isolates belonged to the same clonal group, the final selection based on the results of PFGE greatly reduced this number. However, for the virulence genes that had demonstrated significant differences, an extra number of isolates was tested and the results found were confirmed. On the other hand, the inclusion of nasal S. epidermidis isolates from healthy volunteers without any relation to the hospital environment could give us an overview of the presence of the virulence genes, such as sdrF, to confirm the importance of this gene as a significant marker.
Conclusions
This study showed that despite the great clonal diversity displayed by the S. epidermidis isolates from neonates, those from BSI harbored more frequently the sdrF and sesI biofilm-associated genes. Moreover, mostly of BSI isolates carried the SCCmec type IV or the ccr2 complex, while 83.3% of nasal isolates were non-typeable, showing more diversity for the SCCmec elements composition. It is interesting to note that even being our set of S. epidermidis isolates from patients in contact with the hospital environment we found significant differences in two genes that eventually could be used as markers of invasiveness. However, S. epidermidis possesses a great genetic plasticity that allow acquire, lose or regulate genetic elements that provide advantages to improve its colonization in the host increasing its pathogenicity.
Acknowledgements
We are indebted to Dr Dennis de Carvalho Ferreira (Universidade Estácio de Sá) for providing some of the staphylococcal isolates.
Funding
This work was supported by grants from Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordenação de Aperfeiçoamento Pessoal de Nível Superior, Fundação Universitária José Bonifácio, and Programa de Núcleos de Excelência.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Authors’ contributions
VCS carry out experiments, analyzed the data and wrote the article, NLPI contributed to the experimental design, data interpretation and manuscript writing, MCF and RCCh carry out the experiments, KRNS designed the study, supervised field work and revised the manuscript. All authors read and approved the final version for publication.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of Secretaria Municipal de Saúde da Prefeitura da Cidade do Rio de Janeiro under numbers 239A/2007 e 372A/2010.
Abbreviations
- Aae
Autolysin/adhesin of S. epidermidis
- Aap
Accumulation-associated protein
- ACME
Arginine catabolic mobile element
- ATCC
American Type Culture Collection
- AtlE
Autolysin of S. epidermidis
- B or BSI
Bloodstream infection
- Bhp
Bap homologue protein
- bp
Base pairs
- CC
Clonal complex
- CDC
Centers for Disease Control and Prevention
- CNS
Coagulase negative staphylococci
- Embp
Extracellular matrix-binding protein
- F
Forward
- GehD
Extracellular lipase of S. epidermidis
- MLST
Multilocus sequence typing
- MRSE
Methicillin-resistant Staphylococcus epidermidis
- MSCRAMMS
Microbial surface components recognizing adhesive matrix molecules
- N
Nasal
- na
Not-applicable
- NICUs
Neonatal intensive care units
- NPF
None predicted founder
- nt
Non-typeable
- PBP
Penicillin-binding protein
- PCR
Polymerase chain reaction
- PFGE
Pulsed field gel electrophoresis
- PIA
Polysaccharide intercellular adhesin
- R
Reverse
- SCCmec
Staphylococcal chromosomal cassette mec
- SdrF
Serine/aspartate repeat protein F
- SdrG/Fbe
Serine/aspartate repeat protein G/fibrinogen-binding protein
- SesI
S. epidermidis surface protein I
- ST
Sequence type
References
- 1.Otto M. Staphylococcus epidermidis--the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7(8):555–67. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong Y, Speer CP. The role of Staphylococcus epidermidis in neonatal sepsis: guarding angel or pathogenic devil? Int J Med Microbiol. 2014;304(5-6):513–20. doi: 10.1016/j.ijmm.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Vacheethasanee K, Temenoff JS, Higashi JM, Gary A, Anderson JM, Bayston R, Marchant RE. Bacterial surface properties of clinically isolated Staphylococcus epidermidis strains determine adhesion on polyethylene. J Biomed Mater Res. 1998;42(3):425–32. doi: 10.1002/(SICI)1097-4636(19981205)42:3<425::AID-JBM12>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Arrecubieta C, Lee MH, Macey A, Foster TJ, Lowy FD. SdrF, a Staphylococcus epidermidis surface protein, binds type I collagen. J Biol Chem. 2007;282(26):18767–76. doi: 10.1074/jbc.M610940200. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson M, Frykberg L, Flock JI, Pei L, Lindberg M, Guss B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect Immun. 1998;66(6):2666–73. doi: 10.1128/iai.66.6.2666-2673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams RJ, Henderson B, Sharp LJ, Nair SP. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect Immun. 2002;70(12):6805–10. doi: 10.1128/IAI.70.12.6805-6810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heilmann C, Hussain M, Peters G, Götz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24(5):1013–24. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 8.Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekötter A, Peters G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology. 2003;149(Pt 10):2769–78. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- 9.Bowden MG, Visai L, Longshaw CM, Holland KT, Speziale P, Hook M. Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J Biol Chem. 2002;277(45):43017–23. doi: 10.1074/jbc.M207921200. [DOI] [PubMed] [Google Scholar]
- 10.Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, Speziale P, Höök M. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151(Pt 5):1453–64. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- 11.Söderquist B, Andersson M, Nilsson M, Nilsdotter-Augustinsson A, Persson L, Friberg O, Jacobsson S. Staphylococcus epidermidis surface protein I (SesI): a marker of the invasive capacity of S. epidermidis? J Med Microbiol. 2009;58(Pt 10):1395–7. doi: 10.1099/jmm.0.008771-0. [DOI] [PubMed] [Google Scholar]
- 12.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20(5):1083–91. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 13.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JK, Heilmann C, Herrmann M, Mack D. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55(6):1883–95. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 14.Tormo MA, Knecht E, Götz F, Lasa I, Penadés JR. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology. 2005;151(Pt 7):2465–75. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 15.Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001;9(10):486–93. doi: 10.1016/S0966-842X(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 16.Intenational Working Group on the Sthaphylococcal Cassette Chromossome Elements (IWG-SCC). http://www.sccmec.org/Pages/SCC_TypesEN.html. Accessed 6 Dec 2016.
- 17.Li M, Wang X, Gao Q, Lu Y. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J Med Microbiol. 2009;58(Pt 4):456–61. doi: 10.1099/jmm.0.007567-0. [DOI] [PubMed] [Google Scholar]
- 18.Iorio NL, Caboclo RF, Azevedo MB, Barcellos AG, Neves FP, Domingues RM, dos Santos KR. Characteristics related to antimicrobial resistance and biofilm formation of widespread methicillin-resistant Staphylococcus epidermidis ST2 and ST23 lineages in Rio de Janeiro hospitals, Brazil. Diagn Microbiol Infect Dis. 2012;72(1):32–40. doi: 10.1016/j.diagmicrobio.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Salgueiro VC, Azevedo MB, Iorio NL, Amorim Ede L, dos Santos KR. Staphylococcal cassette chromosome mec elements in methicillin-resistant coagulase-negative staphylococci from a Brazilian neonatal care unit. Pediatr Infect Dis J. 2014;33(10):1089–90. doi: 10.1097/INF.0000000000000440. [DOI] [PubMed] [Google Scholar]
- 20.Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, Higashi J, Sullam PM, Lin J, Wong KI, King KA, Otto M, Sensabaugh GF, Diep BA. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One. 2009;4(11):e7722. doi: 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367(9512):731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 22.Iorio NL, Ferreira RB, Schuenck RP, Malvar KL, Brilhante AP, Nunes AP, Bastos CC, Dos Santos KR. Simplified and reliable scheme for species-level identification of Staphylococcus clinical isolates. J Clin Microbiol. 2007;45(8):2564–9. doi: 10.1128/JCM.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iorio NL, Azevedo MB, Frazão VH, Barcellos AG, Barros EM, Pereira EM, de Mattos CS, dos Santos KR. Methicillin-resistant Staphylococcus epidermidis carrying biofilm formation genes: detection of clinical isolates by multiplex PCR. Int Microbiol. 2011;14(1):13–7. doi: 10.2436/20.1501.01.130. [DOI] [PubMed] [Google Scholar]
- 24.Vivoni AM, Diep BA, de Gouveia Magalhães AC, Santos KR, Riley LW, Sensabaugh GF, Moreira BM. Clonal composition of Staphylococcus aureus isolates at a Brazilian university hospital: identification of international circulating lineages. J Clin Microbiol. 2006;44(5):1686–91. doi: 10.1128/JCM.44.5.1686-1691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitcher DG, Sauders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8(9):1551–6. [Google Scholar]
- 26.Del Vecchio VG, Petroziello JM, Gress MJ, McCleskey FK, Melcher GP, Crouch HK, Lupski JR. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J Clin Microbiol. 1995;33(8):2141–4. doi: 10.1128/jcm.33.8.2141-2144.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51(1):264–74. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teixeira MM, Araújo MC, Silva-Carvalho MC, Beltrame CO, Oliveira CC, Figueiredo AM, Oliveira AG. Emergence of clonal complex 5 (CC5) methicillin-resistant Staphylococcus aureus (MRSA) isolates susceptible to trimethoprim-sulfamethoxazole in a Brazilian hospital. Braz J Med Biol Res. 2012;45(7):637–43. doi: 10.1590/S0100-879X2012007500065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40(1):135–6. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 30.Schuenck RP, Nouér SA, Winter Cde O, Cavalcante FS, Scotti TD, Ferreira AL, Giambiagi-de Marval M, dos Santos KR. Polyclonal presence of non-multiresistant methicillin-resistant Staphylococcus aureus isolates carrying SCCmec IV in health care-associated infections in a hospital in Rio de Janeiro, Brazil. Diagn Microbiol Infect Dis. 2009;64(4):434–41. doi: 10.1016/j.diagmicrobio.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Cavalcante FS, Schuenck RP, Ferreira DC, da Costa CR, Nouér SA, dos Santos KR. Meticillin-resistant Staphylococcus aureus: spread of specific lineages among patients in different wards at a Brazilian teaching hospital. J Hosp Infect. 2014;86(2):151–4. doi: 10.1016/j.jhin.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, Enright MC. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J Clin Microbiol. 2007;45(2):616–9. doi: 10.1128/JCM.01934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frebourg NB, Lefebvre S, Baert S, Lemeland JF. PCR-Based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J Clin Microbiol. 2000;38(2):877–80. doi: 10.1128/jcm.38.2.877-880.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mekni MA, Bouchami O, Achour W, Ben HA. Strong biofilm production but not adhesion virulence factors can discriminate between invasive and commensal Staphylococcus epidermidis strains. APMIS. 2012;120(8):605–11. doi: 10.1111/j.1600-0463.2012.02877.x. [DOI] [PubMed] [Google Scholar]
- 35.Rohde H, Kalitzky M, Kröger N, Scherpe S, Horstkotte MA, Knobloch JK, Zander AR, Mack D. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J Clin Microbiol. 2004;42(12):5614–9. doi: 10.1128/JCM.42.12.5614-5619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hellmark B, Berglund C, Nilsdotter-Augustinsson A, Unemo M, Söderquist B. Staphylococcal cassette chromosome mec (SCCmec) and arginine catabolic mobile element (ACME) in Staphylococcus epidermidis isolated from prosthetic joint infections. Eur J Clin Microbiol Infect Dis. 2013;32(5):691–7. doi: 10.1007/s10096-012-1796-2. [DOI] [PubMed] [Google Scholar]
- 37.Potter A, Ceotto H, Giambiagi-Demarval M, dos Santos KR, Nes IF, Bastos MC. The gene bap, involved in biofilm production, is present in Staphylococcus spp. strains from nosocomial infections. J Microbiol. 2009;47(3):319–26. doi: 10.1007/s12275-009-0008-y. [DOI] [PubMed] [Google Scholar]
- 38.Calà C, Amodio E, Di Carlo E, Virruso R, Fasciana T, Giammanco A. Biofilm production in Staphylococcus epidermidis strains, isolated from the skin of hospitalized patients: genetic and phenotypic characteristics. New Microbiol. 2015;38(4):521–9. [PubMed] [Google Scholar]
- 39.Toba FA, Visai L, Trivedi S, Lowy FD. The role of ionic interactions in the adherence of the Staphylococcus epidermidis adhesin SdrF to prosthetic material. FEMS Microbiol Lett. 2013;338(1):24–30. doi: 10.1111/1574-6968.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hennig S, Nyunt Wai S, Ziebuhr W. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int J Med Microbiol. 2007;297(2):117–22. doi: 10.1016/j.ijmm.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Barbieri R, Pesce M, Franchelli S, Baldelli I, De Maria A, Marchese A. Phenotypic and genotypic characterization of Staphylococci causing breast peri-implant infections in oncologic patients. BMC Microbiol. 2015;10(15):26. doi: 10.1186/s12866-015-0368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbier F, Lebeaux D, Hernandez D, Delannoy AS, Caro V, François P, Schrenzel J, Ruppé E, Gaillard K, Wolff M, Brisse S, Andremont A, Ruimy R. High prevalence of the arginine catabolic mobile element in carriage isolates of methicillin-resistant Staphylococcus epidermidis. J Antimicrob Chemother. 2011;66(1):29–36. doi: 10.1093/jac/dkq410. [DOI] [PubMed] [Google Scholar]
- 43.Svensson K, Hellmark B, Söderquist B. Characterization of SCCmec elements in methicillin-resistant Staphylococcus epidermidis isolated from blood cultures from neonates during three decades. APMIS. 2011;119(12):885–93. doi: 10.1111/j.1600-0463.2011.02801.x. [DOI] [PubMed] [Google Scholar]
- 44.Granslo HN, Klingenberg C, Fredheim EG, Rønnestad A, Mollnes TE, Flaegstad T. Arginine catabolic mobile element is associated with low antibiotic resistance and low pathogenicity in Staphylococcus epidermidis from neonates. Pediatr Res. 2010;68(3):237–41. doi: 10.1203/PDR.0b013e3181eb01e0. [DOI] [PubMed] [Google Scholar]
- 45.Cherifi S, Byl B, Deplano A, Nonhoff C, Denis O, Hallin M. Comparative epidemiology of Staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J Clin Microbiol. 2013;51(5):1541–7. doi: 10.1128/JCM.03378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinheiro L, Brito CI, Pereira VC, Oliveira A, Bartolomeu AR, Camargo CH, Cunha ML. Susceptibility Profile of Staphylococcus epidermidis and Staphylococcus haemolyticus Isolated from Blood Cultures to Vancomycin and Novel Antimicrobial Drugs over a Period of 12 Years. Microb Drug Resist. 2016;22(4):283–93. doi: 10.1089/mdr.2015.0064. [DOI] [PubMed] [Google Scholar]
- 47.Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189(6):2540–52. doi: 10.1128/JB.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ibrahem S, Salmenlinna S, Lyytikäinen O, Vaara M, Vuopio-Varkila J. Molecular characterization of methicillin-resistant Staphylococcus epidermidis strains from bacteraemic patients. Clin Microbiol Infect. 2008;14(11):1020–7. doi: 10.1111/j.1469-0691.2008.02080.x. [DOI] [PubMed] [Google Scholar]
- 49.Martínez-Meléndez A, Morfín-Otero R, Villarreal-Treviño L, Camacho-Ortíz A, González-González G, Llaca-Díaz J, Rodríguez-Noriega E, Garza-González E. Molecular epidemiology of coagulase-negative bloodstream isolates: detection of Staphylococcus epidermidis ST2, ST7 and linezolid-resistant ST23. Braz J Infect Dis. 2016;20(5):419–28. doi: 10.1016/j.bjid.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soroush S, Jabalameli F, Taherikalani M, Amirmozafari N, Fooladi AA, Asadollahi K, Beigverdi R, Emaneini M. Investigation of biofilm formation ability, antimicrobial resistance and the staphylococcal cassette chromosome mec patterns of methicillin resistant Staphylococcus epidermidis with different sequence types isolated from children. Microb Pathog. 2016;93:126–30. doi: 10.1016/j.micpath.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 51.Conlan S, Mijares LA, Comparative Sequencing Program NISC, Becker J, Blakesley RW, Bouffard GG, Brooks S, Coleman H, Gupta J, Gurson N, Park M, Schmidt B, Thomas PJ, Otto M, Kong HH, Murray PR, Segre JA. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 2012;13(7):R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandecasteele SJ, Peetermans WE, Merckx R, Rijnders BJ, Van Eldere J. Reliability of the ica, aap and atlE genes in the discrimination between invasive, colonizing and contaminant Staphylococcus epidermidis isolates in the diagnosis of catheter-related infections. Clin Microbiol Infect. 2003;9(2):114–9. doi: 10.1046/j.1469-0691.2003.00544.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.


