Abstract
Improved methodologies for modeling cardiac disease phenotypes and accurately screening the efficacy and toxicity of potential therapeutic compounds are actively being sought to advance drug development and improve disease modeling capabilities. To that end, much recent effort has been devoted to the development of novel engineered biomimetic cardiac tissue platforms that accurately recapitulate the structure and function of the human myocardium. Within the field of cardiac engineering, induced pluripotent stem cells (iPSCs) are an exciting tool that offer the potential to advance the current state of the art, as they are derived from somatic cells, enabling the development of personalized medical strategies and patient specific disease models. Here we review different aspects of iPSC-based cardiac engineering technologies. We highlight methods for producing iPSC-derived cardiomyocytes (iPSC-CMs) and discuss their application to compound efficacy/toxicity screening and in vitro modeling of prevalent cardiac diseases. Special attention is paid to the application of micro- and nano-engineering techniques for the development of novel iPSC-CM based platforms and their potential to advance current preclinical screening modalities.
Keywords: induced pluripotent stem cells, cardiac differentiation, tissue engineering, disease modeling, drug screening
1. Introduction: Induced pluripotent stem cells
Advances in bioengineering and in vitro culture technologies have led to a rapid expansion of myocardial model development for use in drug efficacy/toxicity testing (Navarrete et al., 2013), disease modeling (Moretti et al., 2010, Wang et al., 2014), and mechanistic studies of cardiac development (Paige et al., 2012). However, the widespread adoption of such techniques for generating engineered human cardiac constructs that accurately model the in vivo tissue is predicated on the establishment of reliable sources of human cardiomyocytes. To that end, a number of recent studies have been performed assessing the suitability of a variety of different cell sources, including bone marrow-derived stem cells (Valarmathi et al., 2011), embryonic stem cells (ESCs) (Clements and Thomas, 2014), and induced pluripotent stem cells (iPSCs) (Mathur et al., 2015) for use in producing cardiac cells that accurately recapitulate the phenotype of their native counterparts. This review article will focus on iPSCs for potential cardiac engineering strategies, due to the significant advantages they offer over alternative cell sources. Specifically, induced pluripotent stem cells are capable of differentiating down multiple disparate lineages, easy to expand, readily available, and do not require the destruction of embryos, reducing ethical concerns and criticisms associated with their use in research. Furthermore, the isolation of cells from patients opens the door to the potential development of patient specific disease models and individualized medicine applications, which will be discussed in more detail later.
The production of iPSCs from somatic cells began with the ground-breaking work of Dr. Shinya Yamanaka’s research group, who used a gammaretrovirus to randomly express four transcription factors responsible for pluripotency (OCT4, SOX2, KLF4, and c-MYC (OSKC)) in mouse and human fibroblasts (Takahashi et al., 2007, Takahashi and Yamanaka, 2006). Since the publication of these landmark papers, multiple methods have been developed for producing iPSCs more efficiently. The reprogramming process to convert somatic cells to iPSCs can be performed using cells from multiple different tissue sources, including skin fibroblasts (Takahashi, Tanabe, 2007), extra-embryonic tissues from umbilical cord and placenta (Cai et al., 2010), mononuclear cells from peripheral blood (Loh et al., 2009), and even urine-derived cells (Xue et al., 2013, Zhou et al., 2012). Following the establishment of iPSCs as a viable cell source, a number of methods have since been developed to improve the efficiency of iPSC generation, including viral and lentiviral integration, non-integrating viral vectors, and protein-and small molecule-based reprogramming (Table 1). An in-depth discussion of the different methods for deriving iPSCs is beyond the scope of this review, but has been discussed in detail elsewhere (Malik and Rao, 2013, Raab et al., 2014, Sommer and Mostoslavsky, 2013).
Table 1.
Examples of methods to reprogram somatic cells into induced pluripotent stem cells.
| Method | Notes on Method | Advantages and Disadvantages |
Reference |
|---|---|---|---|
| Viral Integration |
|
|
(Romli et al., 2013, Sommer et al., 2009, Takahashi, Tanabe, 2007, Takahashi and Yamanaka, 2006) |
| Non-integrating viral vector |
|
|
(Stadtfeld et al., 2008) |
| Recombinant reprogramming protein |
|
|
(Kim et al., 2009, Zhou, Wu, 2009) |
|
|
(Cho et al., 2010) | |
| Small Molecules |
|
|
(Feng et al., 2009, Huangfu et al., 2008, Ma et al., 2013b, Mikkelsen et al., 2008) |
Despite the potential for integrating transgene sequences to negatively impact translational studies, integrating retro- and lentiviral reprogramming methods remain commonplace in cardiac modeling studies (Davis et al., 2012, Itzhaki et al., 2011, Lee et al., 2015, Malik and Rao, 2013, Sun et al., 2012, Wang, McCain, 2014). This is in part due to the fact that viral reprogramming technologies has been available for several decades, which means the expertise is relatively widespread and the methods are well characterized and reliable (Rao and Malik, 2012). Other methods used for cardiac modeling so far include the use of Sendai virus vectors (Churko et al., 2013), which obviate translational issues associated with transgene integration into the host cell’s genome. Additionally, the use of episomal plasmids (Burridge et al., 2011), co-MIP (Diecke et al., 2015), microRNAs (Li et al., 2011), and direct protein delivery (Zhou et al., 2009) have all been shown to be capable of producing iPSCs that can be differentiated into beating cardiomyocytes.
Enthusiasm for the use of iPSC lines in advancing clinical and basic science research, concerns have been raised regarding whether such cells are identical to ESCs. On the one hand, it has been argued that iPSCs and ESCs are nearly identical, since both cell types exhibit similar phenotypes, dependencies, and behavior in vitro (Kong et al., 2010). Additionally, analysis performed over a significant number of clones highlights a considerable overlap in terms of cellular properties between iPSC and ESC sources, making it difficult to distinguish them without in-depth testing (Yamanaka, 2012). On the other hand, microarray research has demonstrated that hundreds of genes, as well as DNA methylation patterns, are differentially expressed between iPSCs and ESCs (Chin et al., 2009, Newman and Cooper, 2010). Overall, measurement of a range of properties of iPSC and ESC lines, including gene expression, DNA methylation, microRNA expression, differentiation propensity, and complementation activity in embryos, suggest that their properties do vary (Chin, Mason, 2009, Wilson et al., 2009).
Although specific differences have been reported between iPSC and ESC lines, there is little conclusive evidence that cardiomyocytes produced from these cell sources differ in any meaningful way, once differentiated. Therefore, despite distinctive dissimilarities in undifferentiated stem cell sources, the high degree of overall comparability between iPSC- and ESC-derived cardiomyocytes and the reproducibility of the cardiac differentiation methods routinely employed, coupled with the advantages of iPSCs in terms of disease modeling and personalized medicine applications, make iPSCs exciting candidates for application in both clinical and basic cardiac research applications.
2. Differentiation of iPSCs into Human Cardiomyocytes
Based on methods developed using embryonic stem cells, human iPSCs have been found to be capable of differentiating into beating cardiomyocytes through exposure to a variety of stimuli (Shiba et al., 2009, Yoshida and Yamanaka, 2011) (Table 2). Differentiation via embryoid body formation and exposure to serum-containing medium was the first method devised for producing beating cardiomyocytes from stem cell sources (Xu, 2012, Xu et al., 2002). Originally devised using ESCs, methods for producing embryoid bodies using iPSCs have since been described, and such cells have shown a propensity to differentiate successfully into cardiomyocytes (Zhang et al., 2009). Such cells stain positive for sarcomeric myosin light and heavy chains, cardiac troponin, and α-actinin following 50+ days in culture. In addition, electrophysiological recordings from embryoid bodies demonstrated that cells differentiated in this manner were capable of generating action potentials characteristic of atrial, ventricular, and nodal cells. Moreover, these cells undergo rate adaption in response to broad field electrical stimulation and were capable of generating visible contractions in response to stimulation. While embryoid body methodologies are well established, the purity of cardiomyocytes differentiated in this manner is often low (≤1%), limiting their utility for downstream applications.
Table 2.
Methods for differentiating iPSCs into cardiomyocytes (Burridge et al., 2012).
| Method | Pluripotent Culture | Pre-differentiation Culture |
Differentiation Format |
Mesoderm Induction Factors |
Cardiac Specification Factors |
Cardiac Differentiation Factors |
References |
|---|---|---|---|---|---|---|---|
| Suspension EB in StemPro34 |
Knock-out Serum Replacement (KSR)/ FGF2 |
KSR/FGF2 | StemPro34 | Activin A, BMP4, FGF2 |
VEGFA, DKK1 | VEGFA, FGF2 | (Yang, Soonpaa, 2008) |
| VEGFA, DKK1, SB431542, dorsomorphin |
VEGFA, FGF2 | (Kattman et al., 2011) | |||||
| IWR1 | Tri-iodothyronine | (Willems et al., 2011) | |||||
| Forced Aggregation EB |
Colonies on MEFs | Colonies on MEFs | IMDM or F12 and PVA |
Activin A, FGF2 | 20% FBS and DMEM | 20% FBS and DMEM |
(Burridge et al., 2007) |
| Monolayer on Geltrex | Passaged one day prior | RPMI and PVA | BMP4, FGF2 | RPMI and FBS or RPMI-INS |
RPMI-INS | (Burridge, Thompson, 2011) | |
| KSR/ FGF2 on MEF | KSR/ FGF2 on low density MEFs |
LIAPEL | Activin A, BMP4, FGF2, VEGFA, SCF |
LI-BEL | LI-BEL | (Elliott et al., 2011) | |
| Monolayer Differentiation |
Monolayer on Matrigel with MEFs |
Monolayer on Matrigel with MEFs |
RPMI plus B27 | Activin A, BMP4 | RPMI plus B27 | RPMI plus B27 | (Laflamme et al., 2007) |
| BMP4, FGF2, Activin A |
NOGGIN, RAi, DKK1 | DKK1 | (Zhang, Jiang, 2011) | ||||
| Monolayer on Matrigel with MEFs |
Matrigel overlay on confluent monolayer |
RPMI plus B27 (− insulin) |
Activin A, BMP4, FGF2 |
VEGFA, DKK1 | VEGFA, FGF2 | (Uosaki et al., 2011) | |
| KSR/ FGF2 on MEFs | KSR/ FGF2 on low density MEFs |
LI-APEL | Activin A, BMP4, FGF2, VEGFA, SCF |
LI-BEL | LI-BEL | (Elliott, Braam, 2011) | |
| mTeSR1 | mTeSR1 | RPMI plus B27 | Activin A, BMP4 | IWR1 or IWP4 | RPMI plus B27 | (Hudson et al., 2012) | |
| mTeSR1 plus Y27632 | mTeSR1 | RPMI plus B27 (− insulin) |
CHIR99021 | IWR2 or IWP4 | RPMI plus B27 | (Lian, Hsiao, 2012) | |
| Chemically defined E8 medium on a synthetic vitronectin peptide matrix |
Chemically defined E8 medium on a synthetic vitronectin peptide matrix |
CDM3 | CHIR99021, Wnt- C59 |
CDM3 | CDM3 | (Burridge, Matsa, 2014) |
Given the complex diffusional barriers present in embryoid bodies, the use of monolayer culture methods have been developed as a means to offer more readily controllable and reproducible environments in which to apply growth factors and other interventions for iPSC-derived cardiomyocyte production (Mummery et al., 2012). Monolayer stem cell cultures can be differentiated to yield cell populations with high cardiomyocyte purity using Matrigel-coated plates in conjunction with mouse-embryonic-fibroblast (MEF)-conditioned medium. In such systems, the serial application of activin A and BMP-4 growth factors in RPMI-B27 medium has proven efficacious in yielding greater than 50% contracting cardiomyocytes (Yang et al., 2008, Zhu et al., 2011). Furthermore, this monolayer-based method can be combined with a matrix sandwich technique, where iPSCs are cultured as monolayers on Matrigel and overlaid with Matrigel-containing medium combined with the growth factors activin A, BMP-4, and basic fibroblast growth factor (bFGF). This combined protocol has been shown to yield extremely high cardiomyocyte purities of up to 98%, and the resulting cell population is able to progressively mature over 30 days in culture (Zhang et al., 2012). However, it has been demonstrated that this matrix sandwich method only works well for certain cell lines, and new lots of growth factors require optimization to maintain high purity yields.
Temporal regulation of Wnt/ β-catenin signaling, either using shRNAs or small molecules (Lian et al., 2012, Lian et al., 2013), has also been shown to be a reliable method for promoting robust cardiomyocyte differentiation from iPSCs maintained as monolayer cultures. Cardiomyocytes generated in this manner exhibit a high degree of purity (~85%), and have been shown to possess mature electrophysiological properties following 30 days in culture.
Finally, while the described methods for differentiating cardiomyocytes from iPSCs are efficient, they typically rely on complex medium compositions. Culture conditions like these hinder detailed analysis and elucidation of the molecular mechanisms underlying cardiomyogenesis. To address this, Burridge and colleagues recently used human iPSCs (hiPSCs), derived under chemically defined conditions on synthetic matrices, to develop an optimized cardiac differentiation strategy using a chemically defined medium consisting of just three components (CDM3) (Burridge et al., 2014). Use of CDM3 in conjunction with lactate selection (Tohyama et al., 2013) methods led to the generation of 80 to 95% TNNT2+ cells; as high as any achieved with more complex culture systems. This achievement is of particular significance since many differentiation strategies use the cocktail B27 to maintain iPSC-derived cardiomyocytes (iPSC-CMs) during differentiation and afterwards. This study highlighted that many of the factors present in B27 are unnecessary and may in fact skew subtype specification during differentiation. The development of a fully-defined culture environment enables analysis of the specific pathways mediating cardiac differentiation. For example, the authors of this study conducted experiments that identified FGF, activin-Nodal, BMP and Wnt signaling as essential for mesoderm induction, as inhibition of these pathways hindered efficient cardiac differentiation.
Although not reliant on iPSCs, methods for producing cardiomyocytes directly from somatic cell sources, termed “transdifferentiation,” have also been developed (Efe et al., 2011). In such studies, the provision of appropriate developmental cues after an initial epigenetic ‘activation phase’ enabled researchers to hijack conventional reprogramming pathways and specifically shift the outcome towards cardiogenesis. Epigenetically activated cells were successfully generated from mouse fibroblasts via temporal expression of the 4 OSKC iPSC transcription factors. At this stage, cells can be reprogrammed to iPSCs or other differentiated cells through exposure to the corresponding medium conditions. To yield a large number of contracting cardiomyocytes, the authors took epigenetically activated cells and treated them with bone morphogenetic protein-4 (BMP-4) in a chemically defined medium for 5 days. Using this method, the authors saw the incidence of beating, in terms of total colony number, reach as high as 90%. It was demonstrated that a greater number of cardiomyocytes can be induced if cells do not have to go through an iPSC intermediate state. In fact, cardiogenic transdifferentiation was actually impaired with the prolonged overexpression of the OSKC transcription factors. Such methods hold promise for shortening the lead times associated with production of pure cardiomyocyte populations from patient samples. Methods such as this may therefore have a significant impact on personalized medicine strategies in future.
All of the described methods have been demonstrated as successful means to generate iPSC-CMs. However, the phenotype of the contaminating non-cardiac cells remains poorly defined and the resulting cardiac population is typically a mix of atrial, ventricular and nodal cells. This ambiguity regarding the exact make-up of the cardiac population can have significant implications for the use of such cells in the downstream modeling of disease states or drug testing. How cardiac cells interact with non-cardiac cells, and the interplay between different cardiac subtypes, can have a substantial impact on the functionality of engineered tissues at baseline, as well as in response to chemical or pathological challenge. An inability to account for this variability may limit the level of insight that can be gleaned from study of such constructs, in terms of predicting how native myocardial tissue will behave in specific contexts. To address this shortcoming, recent work has focused on improving our ability to derive specific cardiac subtypes from pluripotent sources. Based on the fact that iPSCs typically retain the epigenetic hallmarks of their parental somatic line (Rohani et al., 2014), there is evidence that iPSCs derived from ventricular cardiac tissue bear a significantly improved ability to adopt a cardiac progenitor phenotype. Moreover, the complete differentiation of these cells down the cardiac lineage leads to far higher purity of ventricular cells in the resulting population (Xu et al., 2012). This in turn suggests that manipulation of epigenetic factors may enable more controlled differentiation to form pure populations of a given myocardial sub-population. In addition, recent reports suggest that both embryonic and induced pluripotent stem cells can be directed to become either atrial-like or ventricular-like cardiomyocytes by modulating the retinoic acid and Wnt signaling pathways (Devalla et al., 2015, Karakikes et al., 2014, Zhang et al., 2011). However, a greater understanding of the molecular mechanisms that underpin cardiomyocyte subtype specification is necessary to refine current differentiation protocols to the point where lineage-specific development is possible.
3. Applications of iPSC-CMs in Preclinical Screening
The potential applications for cardiac tissues engineered from iPSCs are vast. Although many see the development of such models as a potential means to generate autologous transplant material, such applications remain some way off. Engineered cardiac tissues have more immediate relevance to investigators seeking to better understand cardiac development (Paige, Thomas, 2012), since such cells possess an innate immature phenotype. Integration of iPSC-derived cells with platforms and culture strategies that promote cardiomyocyte maturation will enable more comprehensive and detailed studies of the signaling pathways that underpin myocardial development. In addition, such experiments will provide greater understanding of the molecular, structural, and functional changes that these cells undergo during different stages of development.
A second application for iPSC-CMs is in preclinical drug screening. As mentioned above, such cells have already been used to demonstrate improved electrophysiological analyses in terms of identifying false positive and negative data (Navarrete, Liang, 2013). The advancement of more biomimetic screening platforms will likely lead to the development of screening assays with significantly better predictive power, in terms of informing on the selection of compounds progressing to clinical trials (Chen et al., 2014). The increased availability of robust methods for generating iPSC lines also opens the door to the development of patient-specific disease modeling platforms with which to study inheritable diseases such as long QT syndrome, amyotrophic lateral sclerosis, Down’s syndrome, spinal muscular atrophy, Duchenne and Becker muscular dystrophies, and Huntington’s disease (Dimos et al., 2008, Ebert et al., 2009, Itzhaki, Maizels, 2011, Park et al., 2008). In the following sections, the applications of iPSCs in preclinical drug screening and modeling cardiac pathologies are discussed in more detail.
3.1. Drug Screening
Perhaps the most attainable short-term goal for iPSC-CM technology is the use of cardiac tissue analogues as in vitro screening assays (Figure 1A–D). Induced PSC-CMs are ideal candidates for applications in drug screening and development. The Comprehensive In vitro Proarrhythmia Assay (CIPA) initiative was recently established with the intention to advance safety pharmacology from more traditional pharmacodynamic methodologies towards a combination of in silico and in vitro compound toxicity assessment (Blinova et al., 2016, Cavero and Holzgrefe, 2014, Fermini et al., 2016, Sager et al., 2014). Since its inception, the program has garnered support from both government agencies and pharmaceutical companies alike, and the inclusion of in vitro screening in the proposed strategies serves to highlight the importance the drug development industry is now placing on the development of accurate biomimetic cardiac tissue models.
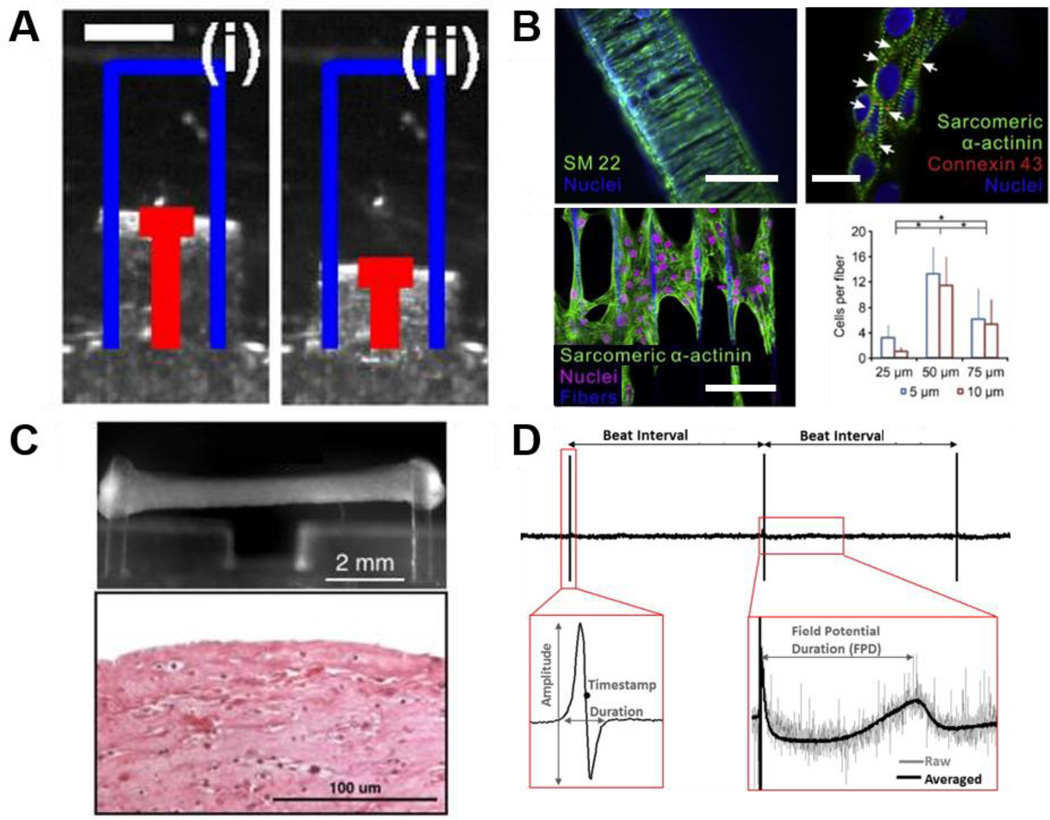
Figure 1. 2D and 3D technologies for high throughput cardiotoxicity screening.
Drug dose response studies can be performed based on cardiac contractility platforms. A. Images of a muscular thin film (MTF) platform incorporating flexible cantilevers supporting cultured cardiomyocytes. Measurement of changes in cantilever position between diastole (i) and during peak systole (ii) enables quantification of force generated in cultured cardiomyocytes. Scale bar = 1 mm. Reprinted with permission (McCain, Sheehy, 2013). B. A three-dimensional filamentous model of human cardiac tissue (top left) used for analysis of contractile function through measurement of filament displacement. In this image, both cardiomyocytes and myofibroblasts were stained for expression of SM22 to show cell distribution throughout the construct. Confocal images of cardiomyocytes aligned on a single fiber show advanced sarcomere structure stained by sarcomeric α-actinin and intercellular gap junctions stained by connexin 43 (white arrows, top right). Confocal images for iPSC-CMs growing on the middle layer of a filamentous matrix and aligned along the fiber direction (bottom left). In such constructs, the formation of 3D cardiac tissue was quantified by cell number on the middle layer relative to the fiber number, and matrices with 50 μm fiber spacing were found to result in the highest value of cell per fiber (bottom right). Reprinted with permission (Ma, Koo, 2014). C. Human engineered cardiac tissues (hECTs) have been shown to mimic key aspects of the newborn human heart and thus allow functional testing for drug screening purposes. Such model systems usually incorporate a culture mold with some form of integrated endposts (top). The illustrated example shows an attached hECT on a mold from a side view. Analysis of post deflection, taking into account post length and stiffness, facilitates measurement of contractile force produced by the engineered tissue. An image is also provided showing a longitudinal section of an hECT stained with hematoxylin and eosin after 12 days in culture (bottom). Reprinted with permission (Cashman et al., 2016). D. Representative field potential waveforms collected from iPSC-CM monolayers using microelectrode arrays (MEAs) and details of the parameters extracted from the raw signal and the time averaged signal. Such analysis can be used to study drug-induced changes in cardiac electrophysiological function for prediction of compound efficacy and toxicity. Reprinted with permission (Gilchrist et al., 2015).
Since primary cardiomyocytes are hard to isolate and have a short lifespan in culture before they begin to dedifferentiate, genetically transformed and/or immortalized cell lines are often utilized for drug screening applications. However, these cells lack important cardiac characteristics and ion channels, and do not fully recapitulate the cellular context of cardiomyocytes (Davis et al., 2011). In contrast, undifferentiated iPSCs can proliferate for extended periods in culture, while iPSC-CMs express the correct electrical and physiological properties of the developing heart, mimicking primary cardiac cell functional performance. Compared with native heart tissue, cardiac cells derived from iPSCs typically exhibit slower conduction velocities and shorter action potential durations, and this is attributed to a lack of maturity in these cultured cells. Efforts to improve cardiac electrophysiological function through application of suitable maturation stimuli is a heavily investigated field and discussed in more detail later. Regardless, the long-term functionality of iPSC-CMs, and their capacity to recapitulate the arrhythmogenic behavior of native cardiac muscle in response to treatment with a wide range of proarrhythmic compounds (Blinova, Stohlman, 2016), highlights their suitability for drug screening applications.
In pharmaceutical development, a major concern and regulatory hurdle is drug-induced cardiac toxicity. Historically, drug-induced arrhythmias have contributed heavily to reported cases of adverse drug reactions (Classen et al., 1997). Consequentially, the FDA and the pharmaceutical industry in general have recently established preclinical drug screening guidelines to test for cardiac toxicity in all new chemical entities prior to market release (Liang et al., 2013). The current in vitro standard for preclinical assessment is the analysis of the hERG channel response in heterologous cell types. Although this method is reliable in terms of isolating compounds with potential arrhythmogenic properties, the high safety margins associated with the hERG assay actually reduce confidence in its ability to predict arrhythmogenic potential accurately (Gintant, 2011). As such, issues with false positives are a significant potential problem and may have led to a number of harmless compounds being kept in development abeyance, despite their therapeutic potential (Kramer et al., 2013). The need for more predictive preclinical cardiac assays is strong, and human iPSC-CMs offer an excellent model for screening new compounds for potentially toxic effects. For example, drugs can be tested for adverse cardiac effects by measuring the disruption of iPSC-CM electrophysiological properties, as discussed previously (Maury et al., 2012). There are data reporting the electrophysiological capacity and responsiveness of iPSC-CMs in response to several cardiac and non-cardiac drugs for reference in research (Inoue and Yamanaka, 2011, Tanaka et al., 2009, Yokoo et al., 2009). It has also been shown that individuals with pre-existing heart conditions are more susceptible to cardiotoxic drugs, which is reflected in disease-specific iPSC-CMs from measurements of action potential duration and drug-induced arrhythmia (Liang, Lan, 2013). Similarly, recent studies have shown that patients with certain genetic backgrounds exhibit increased cardiotoxic sensitivity to treatment with doxorubicin, and this susceptibility is recapitulated in iPSC-derived cardiomyocyte cultures (Burridge et al., 2016, Maillet et al., 2016). Such data highlight the potential for iPSC-CMs to be utilized in patient specific drug screens, potentially allowing doctors to personalize a patient’s treatment options based on their cells response to exposure to different compounds.
As an electromechanical organ, the heart has a number of different outputs that are important for proper function. Therefore, a plethora of screening assays exist that can be utilized to investigate the functional performance of iPSC-CMs for drug-induced cardiotoxicity (Kijlstra et al., 2015). Cellular electrophysiology can be measured directly via patch clamp or indirectly through field potentials as already discussed. Patch clamp methods suffer from low-throughput and therefore are not ideal candidates for pharmaceutical development and drug screening. Microelectrode arrays (MEAs), however, are widely applied in industry and offer the advantage of being noninvasive and can therefore be used to measure cellular responses longitudinally. The extracellular field potential of a cardiomyocyte (or cardiac tissue) correlates directly with the intracellular action potential. Thus, the field potential duration (FPD) can be used as an analogue for the cardiac action potential duration. Navarette et al. have successfully utilized MEAs to record drug-specific cardiotoxic effects in iPSC-CMs by looking at the changes in FPD (Navarrete, Liang, 2013). Importantly, MEAs also offer the added advantage of being able to investigate tissue level electrophysiological parameters, such as conduction velocity, that single cell assays inherently miss. Liang et al. recently performed a drug evaluation study using MEAs and demonstrated that the compound cisapride had no discernable effect on wild-type cardiomyocytes, but did have a pronounced effect on QT prolongation in cells from patients who suffer from long-QT syndrome (Liang, Lan, 2013). Cisapride is known to act on hERG channels (Margulis and Sorota, 2008), and so fails to pass conventional preclinical screens. Therefore, the results obtained by Liang and colleagues demonstrate the importance of screening on cardiomyocytes directly, since the effect of the drug on hERG channels may be counteracted by its effect on other ion channels. Moreover, the presented data demonstrate the value in screening across both wild type and diseased cells since compound action may vary across distinct genetic groups. Such data could prove invaluable in targeting new chemical entities to the correct subpopulation of people for whom they are safe and hold the potential to alleviate specific symptoms.
As the basic function of the heart, cell contraction and beating can also be measured to screen for cardiotoxic effects. Impedance assays (Scott et al., 2014), muscle thin films (Agarwal et al., 2013), and video motion tracking (Huebsch et al., 2014) have all been validated as means to show changes in cardiomyocyte contractile function. Mathur and colleagues recently developed a microfluidic platform for analyzing the beat characteristics of 3D engineered cardiac tissues (Mathur, Loskill, 2015). Using a custom-made motion-tracking software, they were able to demonstrate physiologically relevant changes in cardiac contraction kinetics in response to increasing doses of a number of cardiotoxic compounds, including verapamil and metoprolol. The collected data showed better recapitulation of myocardial tissue responses than is achieved from analysis of 2D monolayers, indicating the importance of mature tissue structures when seeking to model drug responses effectively in vitro.
Cardiac excitation-contraction coupling (ECC) is another important facet of cardiac function. Calcium (Ca2+) acts as a hub in ECC and can be measured in vitro using a number of different intracellular Ca2+ indicator dyes with high signal-to-noise ratios. Although Ca2+ measurements are typically used as a convenient surrogate for assessing changes in voltage-related properties of cells, a number of different reports have shown the ability to discover drug-induced cardiotoxicity in vitro through analysis of Ca2+ imaging data. Advances in optical mapping and image processing are allowing more intricate systems and measurements to be made in a high-throughput manner. Using 2D monolayers of stem cell derived cardiomyocytes and a custom analysis method termed kinetic image cytometry (KIC), Cerignoli et al. were able to track the Ca2+ transients of all the cells in a monolayer virtually simultaneously (Cerignoli et al., 2012). This allowed for detailed investigation of Ca2+ synchronicity between cells. When the bioactive form of triiodothyronine was added to the cells, distinct disturbances in the Ca2+ transients were identified. Additionally, the predicted effects of verapamil and cisapride (L-type Ca2+ channel and K+ channel blockers, respectively) could be detected using the KIC method. Furthermore, Lee et al. have demonstrated that it is possible to simultaneously record intracellular Ca2+ and membrane voltage using optical mapping techniques on iPSC-CMs (Lee et al., 2012).
3.2. Disease modeling
Induced PSCs can be derived from patients with specific diseases of interest and reprogrammed into cells that retain those disease-specific traits. These cells can then be used for disease modeling and/or screening drugs in vitro to assess their ability to correct the observed disease phenotype (Ebert and Svendsen, 2010) (Figure 2). Below we discuss the various iPSC models of cardiac diseases that have been published to date.
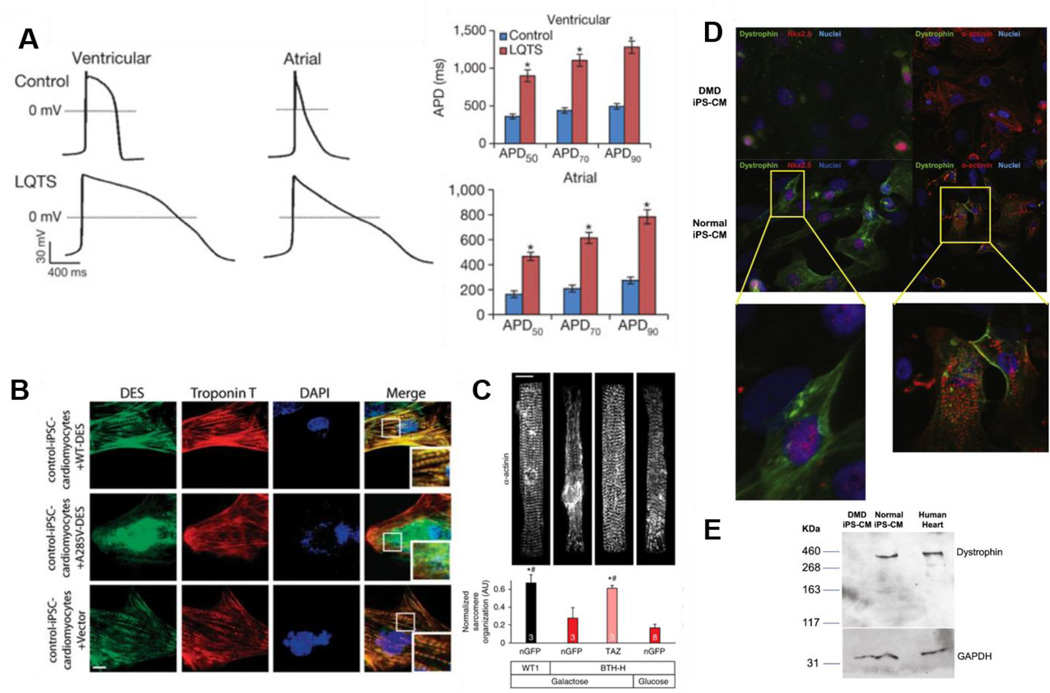
Figure 2. Characterization of iPSCs derived from patients with genetic cardiomyopathies for use in disease modeling applications.
A. Long Q-T syndrome (LQTS) cardiomyopathy modeling. Action potentials were recorded from control and LQTS human iPSC-CMs with ventricular-like and atrial-like morphologies. The graphs indicated prolonged action potential duration (APD) – to reach 50%, 70%, and 90% of repolarization – in both ventricular-like and atrial-like LQTS iPSC-CMs when compared to control. Reprinted with permission (Itzhaki, Maizels, 2011). B. Immunofluorescence staining for desmin (DES: green) and cardiac Troponin-T (red) performed in control iPSC-CMs transduced with wild-type desmin (WT-DES: top), mutant desmin (A285V-DES: middle) and vector alone (bottom) using lentivirus. Forced expression of the mutant A285V-DES in control iPSC-CMs produced a phenotype that included diffuse isolated aggregations of desmin-positive protein (green, middle right) as observed in iPSC-CMs from dilated cardiomyopathy patients, suggesting that A285V-DES exerts a dominant-negative effect. Reprinted with permission (Tse, Ho, 2013). C. Sarcomere organization of patient-derived Barth syndrome cardiomyopathy and control iPSC-CMs. Sarcomere organization was tested in the indicated culture medium and after transfection with the indicated modified RNA. P < 0.05 vs: *, Barth syndrome cells + nGFP modified RNA, in galactose supplemented medium; #, Barth syndrome cells + nGFP modified RNA, in glucose supplemented medium. The presented data suggest treatment with TAZ modified RNA restores sarcomere regularity. Reprinted with permission (Wang, McCain, 2014). D. Cardiomyocytes derived from normal and DMD iPSCs and probed with antibodies against dystrophin (green), together with cardiac specific protein Nkx2-5 or sarcomeric α-actinin (red). Dystrophin staining was demonstrated in normal cardiomyocytes, but not in cardiomyocytes derived from DMD patients. E. Western blot for full length dystrophin detected in the lysates from normal iPSC-CMs (lane 2) and human heart tissue (lane 3) but absent in the lysate from DMD patient iPSC-CMs (lane 1). Molecular weight markers (in kDa) are shown on the left. D and E reprinted with permission (Guan, Mack, 2014).
3.2.1. Ion Channelopathies
Ion channelopathies are perhaps the form of cardiac disease with the most well-established iPSC-based disease models. This is due, in part, to their known impact on measurable single cell electrophysiological endpoints, such as action potential duration (APD). Long QT Syndrome (LQTS) is the most common ion channelopathy, with a prevalence of 1:2000 (Schwartz et al., 2009). Characterized by a delayed repolarization of ventricular cardiomyocytes, and thus a prolonged QT interval on the electrocardiogram (ECG), LQTS is associated with a high risk of ventricular tachyarrhythmia and sudden cardiac death (Figure 2A). LQTS is divided into at least 10 different subtypes according to the underlying genetic or channel mutation. LQTS type 1 (LQT1) is caused by a mutation in the KCNQ1 gene, which encodes for a subunit of the ion channel responsible for the adrenergic-sensitive, slow outward potassium current, IKs. Moretti et al became pioneers of cardiac disease modeling using iPSCs with their 2010 publication in The New England Journal of Medicine in which they modeled the LQT1 disease phenotype using hiPSC-CMs. They demonstrated that ventricular and atrial-type hiPSC-CMs had significantly longer APDs than their healthy control counterparts did, while nodal-type hiPSC-CMs exhibited no such change in APD (Moretti, Bellin, 2010). The authors were also able to demonstrate that beta-blockade helped to protect against catecholamine-induced tachyarrhythmia, highlighting the potential utility of such disease models in helping to test therapies in an in vitro setting prior to their advancement to clinical trials.
Various groups have since published other LQTS subtype disease models, including another LQT1 model by Egashira et al. in which they used MEAs to measure iPSC-CM FPD in response to small molecule inhibitors, thereby demonstrating the role of IKs deficiency in the establishment of the LQT1 phenotype (Egashira et al., 2012). Three groups have published models of LQT2, which is due to a missense mutation in the KCNH2 (hERG) gene (Itzhaki, Maizels, 2011, Lahti et al., 2012, Matsa et al., 2011), causing a reduction in the cardiac potassium current, IKr. Of these, Itzhaki et al. and Matsa et al. screened therapeutic agents for their ability to abolish or reduce the arrhythmogenic phenotype of the disease. Additionally, two groups have investigated a LQT3 disease model, caused by a gain-of-function mutation in the SCN5A gene responsible for the INaL current (Ma et al., 2013a, Terrenoire et al., 2013). Both groups utilized patch clamp methods to characterize abnormal Na+ current inactivation in diseased hiPSC-CMs compared to control, and showed that Na+ channel blockers helped to alleviate the disease phenotype. Furthermore, an iPSC-derived cardiomyocyte model using cells from a patient carrying an SCN5A-1795insD mutation have been published (Davis, Casini, 2012). This mutation gives rise to both LQT3 and Brugada Syndrome phenotypes, as well as conduction defects caused by both gain- and loss-of-function effects on the cardiac Na+ channel, respectively. Finally, Yazawa et al. established a model of LQT8 from Timothy syndrome patients, caused by a mutation in the CACNA1C gene responsible for the Ca2+ influx through L-type Ca2+ channels. Similar to the other LQTS models described, Yazawa et al were able to demonstrate a prolongation of the APD and an alleviation of the arrhythmogenic phenotype with the addition of a therapeutic compound, in this case roscovitine.
Catecholaminergic polymorphic ventricular tachycardia (CPVT) is another inherited channelopathy that is characterized by adrenergically mediated polymorphic ventricular tachycardia in patients without a structural cardiac pathology. Overall, CPVT is caused by improper Ca2+ handling, either through spontaneous Ca2+ release through the ryanodine receptor (RyR2), or through insufficient Ca2+ sequestration by calsequestrin (CASQ2) (Liu et al., 2008). To date, there have been six reports of CPVT disease models using iPSCs. Itzhaki et al., Fatima et al., Jung et al., Pasquale et al., and Paavola et al each developed models of the more prevalent CPVT1 (Di Pasquale et al., 2013, Fatima et al., 2011, Itzhaki et al., 2012, Jung et al., 2012, Paavola et al., 2015), resulting from a dominant mutation in RyR2, while Novak et al developed a model CPVT2 (Novak et al., 2012), the autosomal recessive form of the disease caused by a mutation in CASQ2. For CPVT1, all groups demonstrated aberrant Ca2+ handling and a propensity for delayed after-depolarizations, but each treated the hiPSC-CMs with different compounds to reduce the disease phenotype. Itzhaki et al., utilized beta-blockers to reduce Ca2+ overload, Fatima et al used forskolin to increase cAMP levels, Jung et al found that dantrolene reduced the level of Ca2+ spark activity, and Pasquale et al applied a CaMKII inhibitor to reduce the disease phenotype. Meanwhile, Paavola et al used β-agonists to slow the rate of depolarization in RyR2-mutant cells, suggesting that such treatment could constitute a marker for arrhythmogenicity. The diversity of these studies and findings highlight the fact that in order for disease modeling to be translationally relevant, a concerted effort to find the most promising therapy with consistent results across multiple cell lines is needed before attempting to utilize the treatment in patients.
Taken together, these reports demonstrate that iPSC technologies can be used to reliably recapitulate the pathophysiological hallmarks of arrhythmogenic disease states in vitro, and that these systems can be used as screening systems for new drugs or therapeutic agents. While this is an exciting prospect for the near future, to date investigators have largely restricted their studies to demonstrating that iPSCs isolated from well-characterized monogeneic diseases exhibit perturbed functional phenotypes when differentiated into cardiomyocytes in vitro. For the field of iPSC cardiac disease modeling to grow, such cell types need to be employed in novel mechanistic studies to help delineate the underlying causes of the observed phenotype, or as testbeds with which to evaluate novel therapeutic regimens. Without progression to the use of these cells in these sorts of studies, iPSC-based cardiac disease modeling will fail to have any meaningful impact on translational health sciences and the improvement of patient care.
3.2.2. Structural Cardiomyopathies
Hypertrophic cardiomyopathy (HCM) is the most common inheritable heart disease, yet pharmacologic targets for treatment of the disease remain elusive. Sudden cardiac death associated with HCM is almost exclusively a result of ventricular arrhythmias degenerating into ventricular fibrillation. Like many of the channelopathies described above, a number of genetic mutations behind HCM have been identified, but the remodeling mechanisms behind the pathophysiology are complex and unknown. Therefore, HCM represents a very attractive target for in vitro disease modeling. When iPSC-CMs were generated from two patients with one of the most common mutations present in hypertrophic cardiomyopathy patients (LEOPARD syndrome mutation; Thr468Met in the PTPN11 gene), the resulting cells showed increased sarcomere organization and were larger than those of wild-type controls (Carvajal-Vergara et al., 2010). Lan et al. have produced an iPSC model of HCM and again demonstrated aberrant electromechanical function (Lan et al., 2013). Using patch clamp methods, Lan et al. showed an increased propensity for delayed after depolarizations in HCM cardiomyocytes compared to controls. Using confocal line scanning techniques with fluo-4 to look at Ca2+ transients, the group also showed aberrant Ca2+ handling that was alleviated with the addition of the L-type Ca2+ channel blocker, verapamil.
Familial dilated cardiomyopathy (DCM) is characterized by an increase in chamber size and a thinning of the chamber walls, resulting in volume overload and systolic dysfunction. Sun et al. were the first group to develop an iPSC-CM model of DCM, resulting from a mutation in the TNNT2 gene (Sun, Yazawa, 2012). Since then, two other models of DCM have been published. Siu et al. created hiPSC-CMs from patients with a mutation in LMNA (lamin A/C) (Siu et al., 2012) while Tse et al. used whole exome sequencing to find a mutation in the desmin gene in patients with DCM (Tse et al., 2013) (Figure 2B). In these reports, DCM iPSC-CMs exhibit a predictable increase in cell size and abnormal sarcomere structure and organization. Similar to the iPSC models of LQTS, successful derivation of iPSC-CMs from patients carrying structural gene mutations serves to highlight the power of such technologies to recreate the pathologic phenotype of the disease. Such work has now opened the door for further studies looking at possible mechanisms and, subsequently, the identification of more effective therapeutic targets.
Duchenne muscular dystrophy (DMD) is an X-linked genetic disorder affecting approximately 1 in 3500 males. There are a vast array of mutations associated with DMD, but all result in a knockout of the dystrophin protein. This renders muscle cells particularly prone to mechanical stress and rupture. Over time, this results in muscle scarring and degeneration, eventually leading to death. With improved palliative treatment, DMD patients are living longer, resulting in increased incidence rates of cardiomyopathy and heart failure. Guan et al. were able to develop a model of DMD from urine-derived iPSCs, and found that cardiomyocytes derived from these cells exhibited abnormal Ca2+ handling compared to controls, an increased sensitivity to hypotonic stress, and altered contractile mechanics (Guan et al., 2014) (Figure 2D,E). Similarly, Lin and colleagues demonstrated that cardiomyocytes derived from DMD patient iPSCs exhibited characteristic dystrophin deficiency, as well as elevated levels of resting Ca2+, mitochondrial damage and cell apoptosis. Additionally, they demonstrated that treatment with the membrane sealant Poloxamer 188 significantly decreased resting cytosolic Ca2+ levels, repressed caspase-3 (CASP3) activation and consequently suppressed apoptosis in these cells (Lin et al., 2015).
3.2.3. Other Cardiomyopathies
A number of reports have presented the development of iPSC-based models for inherited diseases that do not traditionally fit as channelopathies or structural cardiomyopathies. Pompe disease is a result of a mutation in the GAA gene encoding for the lysosomal glycogen-degrading enzyme, acid alpha-glucosidase. As a result, myocytes have an accumulation of membrane-bound glycogen, increased cytoplasmic glycogen particles, mitochondrial aberrance, and progressive autophagic buildup (Huang et al., 2011). Huang et al. were able to establish an iPSC model of Pompe disease and produced “cardiomyocyte-like cells” (CMLCs). These Pompe-CMLCs exhibited increased glycogen content, and could be rescued by treatment with recombinant GAA or L-carnitine (Huang, Chen, 2011).
Freidreich’s ataxia (FRDA) is a recessive neurodegenerative disease, that is also associated with HCM, resulting from a mutation in the gene encoding frataxin (involved in mitochondrial function). Hick et al. used an iPSC model of FRDA to investigate both the neural and cardiac pathologies associated with Freidreich’s ataxia (Hick et al., 2013). Both the iPSC-derived neurons and cardiomyocytes demonstrated signs of mitochondrial dysfunction, as measured via mitochondrial membrane potential and structural abnormalities. Barth syndrome is another inherited mitochondrial disorder, resulting from a mutation in the TAZ gene encoding for the protein tafazzin. Wang et al. were able to show that hiPSC-CMs with the TAZ mutation exhibit abnormal sarcomere structure and mitochondrial function, and were able to apply various cardiac tissue engineering approaches to characterize the cardiac dysfunction (Wang, McCain, 2014) (Figure 2C).
Arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C) has also been modeled using iPSC-CMs. ARVD/C is characterized by fatty infiltration (especially within the right ventricle) and is usually caused by mutations in desmosomal proteins (Calkins, 2015). As with many cardiomyopathies, ARVD/C is also associated with an elevated risk of arrhythmia. Induced PSC models of ARVD/C exhibit abnormal nuclear translocation of junction plakoglobin proteins (Pkg), as well as very low β-catenin activity compared with wild-type controls (Caspi et al., 2013, Kim et al., 2013a, Malik and Rao, 2013). Mutant Pkp2 proteins in these cells fail to anchor Pkg to the sarcolemmal membrane, resulting in Pkg nuclear translocation and downregulation of β-catenin activity. Despite this clear in vitro phenotype, no exaggerated lipogenesis or apoptosis in H9 hESC- or mutant iPSC-CMs was found following 3 months in culture (Malik and Rao, 2013). The authors suggest that this observation is consistent with the delayed, adult-onset clinical course of ARVD/C. If accurate, this result serves to highlight the inadequacy of immature iPSC-derived cells for modeling late-onset disease states, and the need for suitable maturation strategies in vitro.
Finally, combination studies using iPSC-CMs for simultaneous disease modeling and drug screening have been performed. Sharma and colleagues infected cardiomyocytes with Coxsackievirus B3 in order to induce a myocarditis phenotype for use as an antiviral drug screening platform (Sharma et al., 2014). Through a combination of bioluminescence imaging, calcium imaging, cell metabolism and vitality assays, and gene expression studies, the authors were able to assess the effect of a number of compounds, including interferon-β1, ribavirin, pyrrolidine dithiocarbamate, and fluoxetine, for their capacity to abrogate CVB3-Luc proliferation and influence cell function and survival in cultured hiPSC-CMs. Studies such as these demonstrate how drug screening and disease modeling applications for iPSC-CMs can be combined to further our understanding of cardiac pathologies and help identify potential drug candidates for improving patient well-being.
3.2.4. Current Limitations of iPSC-CMs for Modeling Cardiomyopathy
While cells differentiated from pluripotent sources using the methods described so far possess a definite cardiac or cardiomyopathic phenotypes, they typically exhibit immature structural and functional properties. This creates problems with regard to the use of these cells in clinically relevant settings (testing novel therapies or evaluating the efficacy and/or toxicity of new drugs), since cell therapy, cardiotoxicity screening, and human heart disease models depend on the cells’ ability to recapitulate the properties of their adult in vivo counterparts accurately (Poon et al., 2011). As such, various methods have been investigated as means to improve cardiomyocyte maturation in vitro. For example, a recent study showed that treating human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) with the growth hormone tri-iodo-l-thyronine (T3) increased maturation in terms of contractile output and structural development (Yang et al., 2014). However, it has been suggested that this approach is somewhat limited in practice, since the common cardiac medium supplement, B27, contains T3 anyway (Wirth et al., 2009). Promoting complete cardiomyocyte maturation from stem cell sources has yet to be achieved, but is likely to require combinatorial approaches mimicking various aspects of the native myocardial niche.
Despite recent advances in the derivation of iPSC-CMs, the lack of cell and tissue maturity, as well as low induction efficiencies, remain significant hurdles to the more widespread application of these cells in preclinical and potential clinical applications. To that end, methods to engineer more accurate and biomimetic cardiac niches in vitro to improve iPSC-CM development and maturation have become a focal point of many bioengineering research programs worldwide. Currently, the most advanced systems rely on the application of complex bioengineering strategies (Kamakura et al., 2013), which are discussed in more detail in the next section. Examples of improved drug screening outcomes or more accurate modeling of disease phenotypes in response to specific bioengineering solutions are provided.
4. Biomimetic Strategies for Human Cardiac Tissue Engineering
The central tenet of tissue engineering is that by generating an environment that more closely recapitulates the in vivo niche, it is possible to develop in vitro culture systems that promote the physiological and functional maturation of cells in a more biomimetic manner. Previous work has shown that long-term culture of cardiomyocytes leads to the development of phenotypically mature cells with more adult-like structural properties, gene expression profiles, and functional characteristics (Lundy et al., 2013). This is not surprising given the time scale of cardiac development in vivo. However, these timescales are impractical for high throughput screening technologies or evaluation of patient responses to specific drugs/therapies, where the shortest time frame possible is desired. In terms of promoting cardiac maturation in vitro, it is likely that no perfect surrogate for a six month to 1-year culture period will be found. Nevertheless, bioengineering techniques can be applied to manipulate cultured cells and enhance development as much as possible. Integration of such advanced culture systems with methods to evaluate functional performance enables the development of platforms capable of providing predictive data on human tissue responses to chemical, mechanical, or pathological challenges. Such technology holds huge potential for advancing preclinical drug screening and disease modeling applications (Ebrahimkhani et al., 2014). The validation of biomimetic human tissue models for predictive mechanistic studies will enable movement away from flat tissue culture plastic-based models (which may offer inadequate representations of the native biological environment) and animal models (which can produce data that often does not translate adequately to human studies). Below, various methods for improving the maturation or physiological accuracy of engineered cardiac tissues are presented and examples of the use of these systems for enhancing preclinical screens are provided.
4.1. Substrate Topography and Stiffness
Engineering cardiac tissues for in vitro modeling purposes requires overcoming a plethora of hurdles, such as cell selection, cell localization, phenotypic maturation, regulation of endogenous regeneration, development of electromechanical function, and optimization of mechanical properties (Caspi et al., 2007, Radisic et al., 2006, Vunjak-Novakovic et al., 2011, Zimmermann et al., 2006). Native cardiac tissue consists of arrays of cells aligned in parallel to facilitate directionally controlled macroscopic contraction (Sosnovik et al., 2009, Streeter and Hanna, 1973, Streeter et al., 1969), whereas conventional in vitro culture platforms lack any form of anisotropic signal, leading to random alignment of the developing cells (Kim et al., 2012a). Therefore, in order to promote correct physiological and functional development, cardiac engineering strategies must promote the uniaxial alignment of the cultured cells to mimic native tissue structure (Sosnovik, Wang, 2009, Streeter and Hanna, 1973, Streeter, Spotnitz, 1969), and enable the subsequent maturation of cardiomyocytes toward an adult phenotype (Kim, Kshitiz, 2012a).
To improve the performance of cultured iPSC-CMs, investigators make use of stem cells’ ability to sense and respond to exogenous microenvironmental stimuli. These signals can influence cell fate, proliferation, and differentiation. It has been suggested that cell adhesion proteins (including integrins, cadherins, and non-muscle myosin II) play a pivotal role in sensing other cells as well as properties of the extracellular matrix (ECM) such as elasticity, porosity, and topography (Wen et al., 2014). In response, cells conform to the microenvironment by modulating intracellular mechanics (Li et al., 2010), reorganizing cytoskeletal architecture, and inducing changes in transcriptional regulation (Engler et al., 2006, McBeath et al., 2004). Knowing that stem cells sense and respond to their environment, incorporation of biomimetic substrates and exposure to correct physiological stimuli has been shown by many to be able to improve the differentiation efficiency of iPSCs down the cardiac lineage, and the subsequent maturation of the developed cardiomyocytes toward an adult phenotype.
One example of an attempt to mimic the native myocardial niche in vitro is the utilization of biomimetic nano-structured surfaces to improve cell adhesion, proliferation, and migration, as well as cardiomyogenic differentiation and structural development (Carson et al., 2016, Kim, Kshitiz, 2012a). Parallel nano-grooves and nano-ridges help maintain the cell polarity and alignment important for organ development by providing contact guidance which influences the organization of microtubules, focal contacts, and actin filaments in parallel with the underlying topography (Kim et al., 2012b). Investigations with cardiomyocytes on dynamically shifting topographies demonstrate the capacity for such cells to reorient themselves in real-time, according to changes in substrate cues, mirroring cells’ capacity to remodel in vivo in response to damage or alterations in mechanical strain (Mengsteab et al., 2016). Similarly, cells cultured on ECM fibers and microgrooves aligned with electrical stimulation patterns have also shown increased elongation, alignment, and electrical functionality (Kim et al., 2010, Kim et al., 2013b) (Figure 3A–C). In addition to improving cellular alignment, nanotopographic cues that match the dimensions of native ECM fibers have been shown to promote substantial improvements in cardiomyocyte maturation, leading to more adult-like cellular morphologies and functional parameters (Kim et al., 2006, Kim, Lipke, 2010, Kim, Jiao, 2013b). This observation lends credence to the theory that accurate recapitulation of the native cellular niche, in this case through recreation of ECM alignment and fiber dimensions, promotes the development of seeded cells toward a more mature phenotype (Kim, Lipke, 2010). Nanotopographic substrates have been used to stratify cardiomyopathic disease phenotypes, where iPSC-CMs from DMD patients were shown to exhibit a reduced capacity to align in parallel with the underlying topography (Macadangdang et al., 2015). The structural deficiencies characteristic of DMD cardiomyocytes prevents these cells from responding to physical cues in a manner comparable to that of wild type controls. The authors highlight altered actin dynamics as a possible mechanism behind the lack of responsiveness of DMD cells to topographic substrates and suggest topographic well plates as a potential preclinical platform with which to test the capacity for novel therapeutics to alleviate or reverse the DMD phenotype.
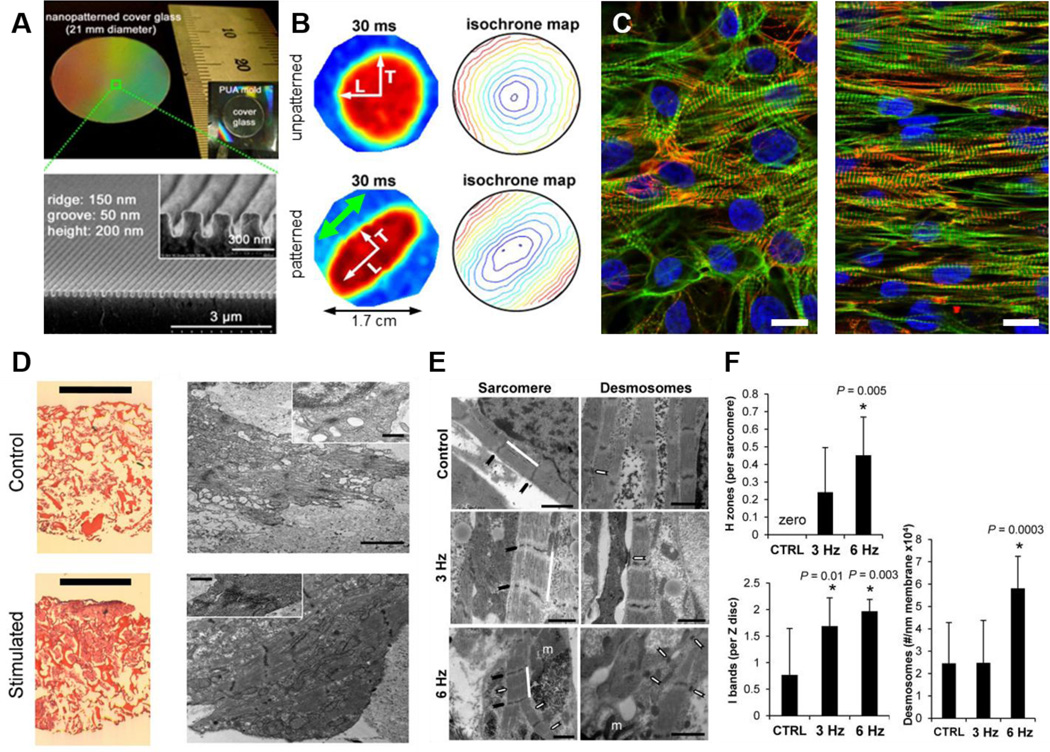
Figure 3. Bioengineering of the myocardial niche.
A. Low magnification image of a glass coverslip patterned with nanogrooves (top). Cross-sectional scanning electron microscopy (SEM) of the same surface illustrating the nanopatterned substratum (bottom). Reprinted with permission (Kim, Lipke, 2010). B. Optical mapping data show anisotropic propagation of action potentials in monolayers cultured on nanofabricated substrata, and isotropic propagation in cells cultured on flat surfaces. Point stimulation at 3 Hz was applied to the center of the cardiac monolayers as indicated by white arrows at 0 ms. The green arrow indicates the alignment of cardiomyocytes on the patterned substrates. Reprinted with permission (Kim, Lipke, 2010). C. Human stem cell-derived cardiomyocytes on flat (left), and nanopatterned (right) substrates highlighting the improvement in structural alignment in cells maintained on suitable topographies. Cells were stained for actin (green), actinin (red), and nuclei (blue). Scale bars: 50 μm. Reprinted with permission (Macadangdang et al., 2014). D. Cardiac constructs maintained without (control), and with (stimulated) electrical stimulation. (Left) Hematoxylin and Eosin (H&E) staining of unstimulated engineered tissue constructs and constructs stimulated with monophasic square wave pulses of 3 V amplitude, 3 Hz frequency and 2 ms duration. Scale bar = 1 mm. (Right) Transmission electron microscopy images of the tissues presented in the H&E images, with insets of sarcomeres. Scale bar = 2 µm in main image, 500 nm in inset. Reprinted with permission (Tandon, Marsano, 2011). E. Ultrastructural analysis of engineered cardiac biowires highlights that electrical stimulation at 6 Hz induces cardiomyocyte self-organization. Representative images of non-stimulated (control), electrically stimulated biowires illustrate sarcomere structure (Sarcomere panel: white bar; Z disks, black arrow; H zones, white arrows; m, mitochondria), and presence of desmosomes (Desmosomes panel, white arrows). Scale bar = 1 μm. Reprinted with permission. (Nunes, Miklas, 2013). F. Morphometric analysis (average ± s.d.) showing ratio of H zones to sarcomeres (CTRL vs. 6 Hz, P = 0.005), ratio of I bands to Z disks (CTRL vs. 3 Hz, P = 0.01; CTRL vs. 6 Hz, P = 0.003), and number of desmosomes per membrane length (CTRL vs. 6 Hz, P = 0.0003). In normal adult cells, the ratio of H zones to sarcomeres is 1 and the ratio of I bands to Z disks is 2. Reprinted with permission (Nunes, Miklas, 2013).
While a culture substrate’s anisotropic properties can have a profound impact on cellular alignment and maturation, its rigidity can also influence cell shape, which is crucial for modulating cell survival and differentiation (Park et al., 2012). It has been shown that substrate rigidities matching the estimated physiological range of the native heart tissue ECM can control the proliferation, differentiation, and morphogenesis of endothelial cells through endogenous regulation of the activating protein p190RhoGAP (Kshitiz et al., 2012). Similarly, studies have demonstrated that isolated embryonic cardiomyocytes cultured on matrices that mimic the elasticity of the developing myocardial microenvironment are optimal for transmitting contractile work to the matrix and for promoting striation development and contractile function (Engler et al., 2008). The cited study also highlighted that matrices with mechanical properties representative of a post-infarct fibrotic scar tissue causes cells to overstrain themselves, leading to a lack of striated myofibrils and the cessation of contraction over time. Data such as these demonstrate how modulation of substrate stiffness can be used to recreate pathological phenotypes in cardiac cells for downstream applications. Furthermore, dynamic hydrogels with time-dependent stiffening properties support greater cardiac progenitor cell differentiation and maturation down the cardiac lineage compared to cells maintained in static hydrogels lacking specific biomechanical cues (Young and Engler, 2011).
4.2. 3D Tissue Culture
An important factor to consider when attempting to faithfully re-create the myocardial niche, is the inclusion of a 3D culture environment to more closely mimic cell-cell and cell matrix mechanics and structural organization. The majority of novel platforms designed to mimic the three-dimensional structure of the mammalian myocardium rely on manipulation of mechanical signals to facilitate the organization and anisotropic alignment of cultured cardiomyocytes within an exogenous matrix. Typically, cells are suspended in a scaffold matrix cultured within a platform that infers directional and structural parameters upon the developing tissue (Liau et al., 2011, Turnbull et al., 2014, van Spreeuwel et al., 2014, Zhang et al., 2013). Cardiomyocytes have repeatedly demonstrated the capacity to respond to these physical stimuli, and orient themselves along the lines of principal strain within such cultures.
Scaffolds can be engineered from polymeric materials of synthetic origin, which include polylactide (PLA), polyglycolide (PGA), lactide and glycolide copolymer (PLGA), poly(ε-caprolactone) (PCL), and poly(N-isopropylacrylamide) (PNIPAAm), as well as combinations of these polymers (Chun et al., 2015). Alternatively, they can be engineered from natural polymers and polymer combinations, such as collagen, gelatin, Matrigel, cellulose, chitosan, hyaluronic acid, or silk fibroin. Examples of synthetic, three-dimensional cardiac tissues include disk-shaped PLGA constructs, honeycomb scaffolds, and electrospun matrices, while examples of natural constructs include hydrogel scaffolds (circular hydrogel molds mechanically and electrically stimulated), and decellularized native tissue (Buikema et al., 2013, Vunjak-Novakovic, Lui, 2011) (Table 3). The use of both biological and synthetic scaffolds have been extensively employed in various cardiac engineering strategies, and each possesses inherent advantages and disadvantages (Chen, 2008, El-Sherbiny and Yacoub, 2013, Eschenhagen and Zimmermann, 2005, Park et al., 2005a, Radhakrishnan et al., 2014). Synthetic scaffolds are more controllable and reproducible, and often exhibit stronger mechanical properties, which can be advantageous in vivo for repairing damaged tissue. Biological scaffolds offer a more accurate representation of the native cardiac ECM. They therefore potentially enable activation of cell signaling pathways to aid cellular development through presentation of antigens recognized by specific cell surface receptors. More in depth discussions of the benefits and weaknesses of different scaffold techniques are discussed elsewhere in detail (El-Sherbiny and Yacoub, 2013, Radhakrishnan, Krishnan, 2014). Due to the wide array of scaffolding approaches investigated so far (each with inherent advantages and disadvantages), a single “optimal” scaffold has yet to be established, highlighting that there currently remains substantial room for further development. Nevertheless, 3D cardiac drug screens with moderate throughput (Bielawski et al., 2016, Hansen et al., 2010) are currently in development and represent the means to reliably quantify contractile force outputs in a manner not possible in 2D culture. Such platforms constitute a viable option for further refinement towards effective, next generation preclinical analysis platforms for evaluating new chemical entities.
Table 3.
Examples of different scaffold types for cardiac regeneration.
| Categories | Examples of Scaffolds | References |
|---|---|---|
| 2-dimensional scaffolds |
|
(Bursac et al., 2002, McDevitt et al., 2002) |
| 3-dimensional scaffolds |
|
(Shimizu et al., 2002, Vunjak-Novakovic, Lui, 2011) (Lim and Mao, 2009) (Buikema, van der Meer, 2013, Radisic et al., 2003) (Engelmayr et al., 2008) |
| Hydrogel- based scaffolds |
|
(Zimmermann et al., 2002) (Radisic et al., 2004, Tandon et al., 2009) |
| Decellularized native tissue |
|
(Ott et al., 2008, Pati et al., 2014) |
Although strategies for generating aligned cardiac tissues in vitro typically rely on the application of an exogenous scaffold material, the development of detachable cell monolayers through manipulation of tunable stimuli-responsive polymers has recently led to the possibility of generating three-dimensional scaffold-free tissue constructs with physiologically relevant cell densities (Isenberg et al., 2008, Jiao et al., 2014, Yang et al., 2007). Detachment of these cell populations can be achieved through a controlled shift in an exogenous stimulus, such as temperature, and the subsequent stacking of cell monolayers represents a reliable method for generating cell dense 3D constructs for various in vivo and in vitro applications. Such tissue constructs do not require integration with scaffold materials, leading to greater cell-cell contact and improved cellular communication. Furthermore, the integration of nanotopographic substrate cues and stimuli-responsive substrates has been shown to facilitate the production of cell dense, scaffold-free constructs with controllable cellular orientation and anisotropic contraction patterns (Jiao, Trosper, 2014). Oxygen diffusion gradients within such cell dense constructs limit the thicknesses that can be achieved using such methods. However, studies have shown that co-culture with endothelial cells can create vascular beds within 3D, cell sheet-based constructs, thereby enabling the synthesis of thicker cardiac tissues for downstream applications (Sekine et al., 2013).
Cardiac spheroids represent an alternative method for generating scaffold-free 3D tissues, albeit without the structural alignment present in cell sheet engineering techniques. Recent work from Beauchamp and colleagues details methods for generating 3D spheroid cultures of cardiomyocytes derived from pluripotent stem cell sources (Beauchamp et al., 2015). These constructs are free of necrotic cores and exhibit spontaneous contractile activity, calcium transients, and sarcomeric development . Similar 3D spheroid microtissues have been used for studying cardiac disease development in Chagas patients (Fares et al., 2013). Specifically, investigators used spheroid tissues of cardiac cells to investigate the matrix altering effects of cardiac exposure to 5% sera from patients with Chagas disease. Although not using iPSCs, this study demonstrated that exposure to trypanosome- infected blood caused an increase in cardiac ECM component expression in 3D cardiac cultures. Moreover, the data suggest that MMP-2 and MMP-9 are correlated with the cardiac spheroid remodeling induced by sera of patients with Chagas disease. Given the importance of ECM production for studying matrix remodeling in disease states, these results highlight the value of 3D culture systems for investigating pathological mechanisms associated with ECM production and regulation.
4.3. Electromechanical Conditioning
Given the electrically and mechanically active nature of cardiac tissue, perhaps the most important factors to consider when seeking to develop biomimetic functional profiles in engineered myocardial tissue are physiological representations of mechanical strain and electrical activation (Wang et al., 2013). Studies have demonstrated that prolonged exposure of engineered cardiac tissue to continuous pacing promotes improvements in contractile force production, sarcomere structural development (Figure 3D), and Ca2+ responsiveness, as well as a reduction in spontaneous beating activity (Ahadian et al., 2013, Barash et al., 2010, Hirt et al., 2014, Lasher et al., 2012, Maidhof et al., 2012, Tandon et al., 2011). Furthermore, treatment of constructs with a combination of electrical pacing and insulin-like growth factor 1 (IGF-1) supplementation has been shown to further improve the contractile properties and connexin-43 expression in engineered heart constructs over and above levels observed in response to either factor applied independently (Park et al., 2014). The integration of three-dimensional cardiac biowires (derived from pluripotent stem cell sources) with long-term stimulation protocols for up to 4 weeks in vitro promotes improvements in sarcomeric banding (Figure 3E,F), excitation thresholds, and conduction velocities (Nunes et al., 2013, Xiao et al., 2014). Cells from these preparations also exhibit larger hERG currents, more negative resting membrane potentials, and larger capacitances (suggesting larger cells) than unstimulated controls, further indicating the improved functionality of cardiomyocytes exposed to long-term electrical stimulation (Nunes, Miklas, 2013).
Similarly, studies of engineered cardiac construct responses to mechanical conditioning have indicated that application of uniaxial cyclic stain regimes promote physiological hypertrophy and cardiomyocyte proliferation over above levels achievable in unstrained constructs (Tulloch et al., 2011). Additionally, work with primary cells has also indicated that cyclic stretch promotes cardiomyocyte proliferation through activation of p38-mitogen-activated protein kinase (Clause et al., 2009).
Given the apparent importance of mechanical strain in promoting cardiomyocyte development, it is not surprising that modulation of this stimuli can have a negative impact on cardiac development in vitro. McCain et al. designed and built a system to mimic mechanical overload in cultured cardiomyocytes by applying cyclic stretch to engineered laminar ventricular tissue on a stretchable chip (McCain et al., 2013). The authors showed that the gene expression profiles activated in response to cyclic stretch were characteristic of pathological remodeling, and included a decrease in α- to β-myosin heavy chain ratios, as well as maladaptive changes to myocyte shape and sarcomere alignment. Examined calcium transients were found to resemble those reported in failing myocytes and peak systolic stress was significantly reduced. As such, the presented data demonstrate how application of pathologically relevant stretch regimens can create tissues with disease relevant phenotypes.
While electrical pacing and mechanical stretch have been shown to have profound impacts on cardiac development in isolation, exposure of engineered cardiac tissues to combined electrical and mechanical stimuli can be used to further improve structural and functional maturation (Feng et al., 2005, Morgan and Black, 2014). In such protocols, application of the electrical stimulus was timed to occur after the beginning of mechanical stimulation in order to mimic the biophysical environment present during isovolumetric contraction. Specifically, application of combined electromechanical stimuli was found to improve construct function by promoting the expression of proteins responsible for cell-cell communication and contractility, namely SERCA2a, troponin T, and Akt (Morgan and Black, 2014). These data serve to highlight the significant effect electrical and mechanical stimulation can have on developing engineered cardiac constructs. Furthermore, they demonstrate that combinatorial stimulation programs can have further additive effects on cardiomyocyte maturation, highlighting the need for a multi-stimulus approach when seeking to model adult myocardial tissue effectively.
4.4. Biochemical Modulation of Cardiac Phenotype
In addition to recreation of the physical myocardial niche, biomolecular and biophysical studies have looked at small molecule treatment (Huang et al., 2008, Yang, Rodriguez, 2014) to further improve the phenotype of cultured cardiomyocytes. Using such methods, researchers have shown that treatment of engineered cardiac tissues with either insulin-like growth factor 1 or the β2 agonist clenbuterol led to significant increases in contractile force production in a time dependent manner (Huang, Khait, 2008). In addition, multiple adrenergic receptor agonists have long been known to enhance hypertrophy of cultured cardiac cells (Deng et al., 2000, Foldes et al., 2011). Although more typically associated with models of pathological hypertrophy, the possibility of optimizing treatment with such factors to bring about the formation of a non-pathological hypertrophied phenotype is an exciting potential avenue of research.
Treatment of stem cell-derived cardiomyocytes with the thyroid hormone T3 has been shown to upregulate markers of the cardiac contractile apparatus, correlating with improved force production and calcium handling in these cells (Yang, Rodriguez, 2014). Such treatment mimics the spike in thyroid hormone release after birth, which has been linked to correct cardiac development in humans. As such, this treatment offers an excellent example of mimicking cardiac developmental cues in vitro to bring about the establishment of a more mature cardiac state.
Lastly, microRNA modulation has been shown to modulate cardiomyocyte maturation. Specifically, miRNA-499 overexpression in embryonic stem cell-derived cardiovascular progenitors promotes a significant increase in the yield of ventricular cardiomyocytes, and an increase in contractile protein expression (Fu et al., 2011). By contrast, miRNA-1 transduction does not alter cardiomyocyte yield but shortens action potential duration and promotes the development of a hyperpolarized resting membrane potential as signs of functional maturation (Fu, Rushing, 2011). Also, miRNA-1 but not −499 has been shown to augment immature Ca2+ transient amplitudes and kinetics (Fu, Rushing, 2011). Similarly, miR208 has been highlighted as a potential candidate that also may promote cardiac maturation, due to its involvement in thyroid hormone responsiveness and the myosin isoform switch (Callis et al., 2009, van Rooij et al., 2007).
More recently, the Let-7 family of miRNAs was found to be the most highly upregulated miRNA during cardiac maturation in vitro. Based on this observation, overexpression of Let-7 family members in human ESC-derived cardiomyocytes leads to enhanced cell size, sarcomere length, force of contraction, and respiratory capacity (Kuppusamy et al., 2015). The authors of this study used large-scale expression data, target analysis, and metabolic flux assays to suggest that Let-7-driven cardiomyocyte maturation could be a result of down-regulation of the phosphoinositide 3 kinase (PI3K)/AKT protein kinase/insulin pathway and an up-regulation of fatty acid metabolism.
Although miRNAs can be used to promote cardiac development, they also hold the potential to recreate cardiomyopathic phenotypes. While forced overexpression of stress-inducible miRNAs is sufficient to induce hypertrophy in cultured cardiomyocytes, overexpression of miR-195, which is up-regulated during cardiac hypertrophy, results in pathological cardiac growth and heart failure in transgenic mice (van Rooij et al., 2006). Although yet to be applied in iPSC cultures, such methods constitute a viable option for future studies seeking to use iPSC-CMs to model pathological cardiac development.
4.5. Multiple Cell Type Culture
Although the majority of engineered cardiac tissues focus on the form and function of cardiomyocytes, accurate recapitulation of the myocardial niche requires integration of multiple cell types, including cardiac fibroblasts and endothelial cells. Additionally, the integration of supporting vascular cell types in 3D engineered cardiac tissues would enable to researchers to model cardiac vascularization in order to study coronary disease and infarction in vitro (Amano et al., 2016). Given the prevalence of heart disease in the world today, such models would likely yield data that would be of tremendous value to modern healthcare professionals and would offer a useful in vitro model for the high throughput evaluation of novel heart disease therapeutics.
Co-culture of engineered cardiac tissues with endothelial cells has been shown to improve production of vessel forming cytokines and can lead to the generation of capillary networks capable of linking engineered tissues to the host vasculature during in vivo studies (Sekine et al., 2008, Sekine, Shimizu, 2013). In addition, data has been published suggesting that maintaining co-cultures of endothelial cells and cardiomyocytes can induce improvements in cardiomyocyte viability and function (Vuorenpaa et al., 2014). Electrophysiological studies of cardiomyocyte and cardiac fibroblast co-cultures suggest that the development of mechanical interactions between these cell types during cardiac injury may act to impair cardiac conduction through increased activation of mechanosensitive channels in response to tension applied to the myocyte by the myofibroblast (Thompson et al., 2011). Such studies demonstrate how interplay between different myocardial cell types can lead to significant changes in overall function in engineered tissues. Moreover, the presented data illustrate the importance of such interactions when seeking to understand changes in performance in disease states and ischemic tissues.
In addition to co-culture with supporting myocardial cell types, co-culture of iPSCs with primary cardiac cells has also been shown to increase yields of iPSC-CMs (Ou et al., 2016). Such data suggests cardiac cells may help modify their in vitro niche to promote the development of an environment better to suited to promoting cardiac differentiation.
4.6. Instrumented Cardiac Functional Assays
Once recapitulation of adult tissue structure and function is achieved, measurement of functional performance is a further important consideration if the system is to be effectively utilized in a broad spectrum of studies. Functional performance should be quantifiable in order to enable comparison to the output of the native tissue, and to validate the improvement of the engineered construct over conventional cell culture methods. In the case of cardiac muscle, this means the developed system must possess the means to measure contractile and/or electrophysiological performance. Desire to accurately investigate functional outputs in engineered cardiac constructs has led to the development of novel cardiac biological microelectromechanical systems (BioMEMS) capable of monitoring these parameters in real-time.
The potentially fatal nature of channelopathies in human cardiac disease (Chockalingam et al., 2015), coupled with the importance of assessing changes in ion channel performance as a means of predicting drug-induced arrhythmogenesis (Sanguinetti and Mitcheson, 2005, Singh, 1993), has led to extensive characterization of iPSC-CM electrophysiology in vitro. Patch clamp recordings offer the means to gain high-resolution data pertaining to the functional maturity of single cells (Scheel et al., 2014), as well as a means to characterize cardiomyocyte subtype, response to stimuli, and pathology. However, such methods are invasive, extremely low throughput, and incapable of providing measurement of whole tissue function. In their place, cardiac screening technologies are increasingly focusing on the use of either microelectrode arrays (MEAs) or optical mapping techniques to assess the electrophysiological output of cultured cardiomyocytes. MEAs permit recording of electrical field potentials from cardiac monolayers similar to whole heart analysis by electrocardiography (Clements and Thomas, 2014, Navarrete, Liang, 2013, Riedel et al., 2014), whereas the use of either genetically encoded or exogenously applied voltage-sensitive dyes enable real-time monitoring of action potential propagation through cultured cells (Kijlstra, Hu, 2015). Ca2+ sensitive dyes have also been employed for analysis of cardiac function (Burridge, Thompson, 2011, Joshi-Mukherjee et al., 2013). However, it should be noted that Ca2+ dyes do not directly report electrophysiological endpoints, and are actually used as a surrogate for direct voltage measurements in most cases.
Using MEAs and optical mapping, estimations of cardiac conduction velocity, action potential duration, and beat frequency/regularity can be made, providing in depth analysis of the health and functional stability of engineered cardiac tissues. Through assessment of parameters such as these, investigators have demonstrated the capacity for in vitro models of human cardiac tissue to recapitulate physiological responses to cardiotoxic compounds (Navarrete, Liang, 2013, Riedel, Jou, 2014). Specifically, the use of MEAs has been shown to be advantageous in correctly selecting out false positive and negative compounds as identified using more conventional ion channel studies. The ability for such screening systems to provide more accurate appraisals of compound action in vitro holds promise for the use of these technologies for improving the efficacy of current preclinical screening protocols.
In addition to measurement of electrophysiological performance, assessment of contraction within in vitro platforms represents a vital secondary metric for accurately predicting whole heart function. The majority of engineered myocardial models measure contractile function either through direct measurement of force using a transducer (Sondergaard et al., 2012), or through analysis of fixed post deflection (Hansen, Eder, 2010, Ribeiro et al., 2014, Rodriguez et al., 2011, Rodriguez et al., 2014) or substrate deformation in response to cardiomyocyte contraction (Kijlstra, Hu, 2015, Ma et al., 2014). Use of microscale cantilevers is becoming increasingly common as a means to monitor cardiomyocyte contractile properties reliably and reproducibly in a multiplexed, and therefore higher throughput, manner (Agarwal, Goss, 2013, McCain, Sheehy, 2013, Park et al., 2005b, Shim et al., 2012, Stancescu et al., 2015) (Figure 1A). Deflection recordings can be used to extrapolate measurements of contractile performance in such systems and so provide feedback on the functional development of the engineered tissue.
More recently, Pioner et al. presented the means to isolate individual myofibrils from hiPSC-CMs in order to assess their mechanical and kinetic properties (Pioner et al., 2016). Moreover, the authors demonstrated the feasibility of using this system in conjunction with hiPSC-CMs to study the development of contractile abnormalities for disease-associated mutations in sarcomere proteins. Among other metrics, analysis revealed a marked impairment of maximal isometric tension generated by myofibrils from cells derived from patients carrying a MYH7 mutation. The collected data highlight the capacity for this novel functional assay to stratify disease phenotypes and potentially screen for novel therapeutics to alleviate symptoms associated with myofibrillar mutations.
Although many of these in vitro cardiac assays focus on a single cell type, integration of heart-on-a-chip models with microfluidics (Giridharan, 2014, Giridharan et al., 2010) has opened the possibility of linking multiple in vitro tissue models to create body-on-a-chip devices capable of modeling tissue-tissue cross talk in response to drug treatment or specific pathologies (Esch et al., 2014, Oleaga et al., 2016, Smith et al., 2015, Sung et al., 2013). The importance of cardiac tissue for accurate predictions of system toxicity makes integration of heart-on-a-chip models into such complex systems a vital component for effective modeling of whole-body responses to compound exposure (Sung, Esch, 2013). However, the ability for such complex systems to accurately model system responses to drug treatment has yet to be demonstrated effectively.
5. Unmet Needs and Future Perspectives
Although iPSCs are good candidates for drug screening and disease modeling applications, they are not without limitations. As detailed in this review, iPSC-CMs are developmentally immature, often making it difficult to relate data collected from such sources to that of their fully mature counterparts in the adult human heart. The late onset nature of many of the diseases discussed means that phenotypically immature iPSC-CMs likely fail to recapitulate disease development accurately, since they are incapable of reaching the stage of development where the disease typically manifests in vivo. Moreover, complex cellular interactions in human metabolic processes often cannot be recapitulated in these culture systems, not to mention the confounding factor of human genetic variation between cells of different donors. A large panel of patient-derived iPSCs needs to be assessed in order to take into account the range of responses that a given drug may elicit when applied to a heterogeneous human population. Such a library could also be used to refine clinical trial designs by indicating whether a stratified subpopulation will respond uniformly to a specific drug (Grskovic et al., 2011).
For these reasons, and despite their substantial promise, patient-specific iPSC models have not yet replaced animal models and traditional cell line assays in the development of new drug compounds. The persistent issue with maturity and accurate recapitulation of adult-onset disease phenotypes has led to an explosion of bioengineering strategies to improve engineered cardiac tissue development in vitro. Such techniques seek to mimic multiple aspects of the cardiac microenvironmental niche, using manipulation of ECM alignment and topography, small molecule doping, and electromechanical stimulation to improve the differentiation, development, and maturation of cultured cardiomyocytes.
Looking to the future, integration of cardiomyocyte culture systems with other cell types to model cellular interactions within the heart as well as between organs will enable more precise comparisons with the in vivo condition to be made. Such research may lead to the establishment of new in vitro disease models for conditions that do not directly target cardiomyocytes. For example, co-culture of cardiomyocytes and patient fibroblasts within a suitable tissue engineered platform may allow researchers to model cardiac defects brought about by connective tissue disorders such as Marfan syndrome (Quarto et al., 2012) and scleroderma (Wang et al., 2016). Although not a cardiac-specific condition, Marfan syndrome can cause defective aortic media, defective valve cusps, interatrial communication, and sunken chest (pectus excavatum) (McKusick, 1955). The establishment of a suitable Marfan syndrome iPSC-based model of the myocardium would enable in-depth analysis of how connective tissue physiology contributes to the formation of physiologically-correct cardiac structures during development. Regardless of the disease, improvements in our ability to model the intricate cell-cell and cell-matrix interactions that occur within the mature and developing myocardium will likely lead to more accurate and complex models of the human heart for numerous downstream applications.
Despite the wide array of iPSC-based cardiac disease models produced to date, recent improvements in gene editing technologies also hold promise for furthering our capacity to model cardiac disease effectively. The clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas9 system uses short guide RNAs to drive CRISPR-associated endonucleases (Cas9) to a target locus for precision editing of genetic sequences (Mali et al., 2013). This revolutionary technology is more efficient, easier to use, and simpler to generate than earlier gene editing methods that require the synthesis of custom proteins for each DNA target. As such, CRISPR/Cas9 technology is now widely used to edit the genome of a wide variety of cell types, including iPSCs (Byrne and Church, 2015). Recent work has demonstrated the feasibility of creating inducible gene knock out lines through a combination of CRISPR/Cas9 gene editing and Cre/LoxP or Flp/FRT recombination (Chen et al., 2015). In doing so, the authors were able to create iPSC lines in which precise temporal control of SOX2, PAX6, OTX2, and AGO2 could be exerted. The potential for such technology to advance cardiac disease modeling is clear. Pluripotent cells could be driven down the cardiac lineage and matured before knocking out the gene of interest and observing the changing phenotype. Alternatively, knock out of the gene at an early stage would enable study of how expression of this gene influences cardiac development. Work with iPSC-CMs and skeletal muscle cells from DMD patients has shown that CRISPR also offers a powerful tool for correcting deleterious mutations. Cardiomyocytes derived from DMD iPSCs were edited with CRISPR/Cas9 to restore the reading frame of dystrophin, leading to improved membrane integrity and restoration of the dystrophin glycoprotein complex (Young et al., 2016). Combination of iPSC, gene editing, and combinatorial tissue engineering technologies to create more accurate models of cardiac disease is a daunting prospect. However, the successful combination of such strategies will have a dramatic effect on our capacity to generate meaning preclinical assessment of novel therapies. Furthermore, such model systems will simultaneously expand our capacity to study the mechanisms underlying disease onset and development accurately.
6. Concluding Remarks
The widespread adoption of human induced pluripotent stem cells in preclinical screening has the potential to make a significant contribution to the understanding of cardiac disease onset and progression, and to the development of new drugs for the betterment of human health. The capacity to model individual genetic backgrounds allows for analysis of patient specific reactions to different treatment modalities, as well as identification of consistent pathophysiological traits as suitable drug targets for novel therapeutic interventions. In addition, the use of iPSC-CMs in combination with novel biomaterials and tissue engineering technologies to enhance structural and functional development have enabled more comprehensive analysis of cardiac disease phenotypes and drug-induced functional changes in vitro. However, several concerns need to be addressed before such iPSC-based technologies can be effectively applied to more widespread commercial settings. The adoption of iPSC-based tissue engineering strategies for improved cardiac modeling will require the development of standardized protocols capable of addressing issues with iPSC interline variability, cardiomyocyte alignment and maturation, high-throughput assessment of functional phenotypes, and effective recapitulation of disease states. Work to date serves to highlight the vast array of technologies available to improve iPSC-cardiac development, and focus must now be shifted toward the integration of these technologies in order to produce physiologically and functionally accurate representations of adult myocardial tissue.
Acknowledgments
This work was supported by a Wallace H. Coulter Foundation Translational Research Partnership Award, a Washington State Life Science Discovery Fund grant, an American Heart Association (AHA 13SDG14560076) Scientist Development Grant, an NIH R21 grant (1R21EB020132-01A1), and an International Collaborative R&D Program Grant with KIAT funded by the MOTIE (N0000894) awarded to D-H. Kim. M.A. Laflamme acknowledges NIH support for his research (RO1 HL117991), as well as support from the McEwen Centre for Regenerative Medicine, the Peter Munk Cardiac Centre, and the Ontario Institute for Regenerative Medicine. A.S.T. Smith is supported by an NIH T32 post-doctoral fellowship (2T32HL007312-36A1), and J. Macadangdang is the recipient of an NIH bioengineering cardiovascular training grant fellowship (5T32EB001650-12).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal A, Goss JA, Cho A, McCain ML, Parker KK. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab on a chip. 2013;13:3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahadian S, Ostrovidov S, Hosseini V, Kaji H, Ramalingam M, Bae H, et al. Electrical stimulation as a biomimicry tool for regulating muscle cell behavior. Organogenesis. 2013;9:87–92. doi: 10.4161/org.25121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y, Nishiguchi A, Matsusaki M, Iseoka H, Miyagawa S, Sawa Y, et al. Development of vascularized iPSC derived 3D–cardiomyocyte tissues by filtration Layer-by-Layer technique and their application for pharmaceutical assays. Acta biomaterialia. 2016;33:110–121. doi: 10.1016/j.actbio.2016.01.033. [DOI] [PubMed] [Google Scholar]
- Barash Y, Dvir T, Tandeitnik P, Ruvinov E, Guterman H, Cohen S. Electric field stimulation integrated into perfusion bioreactor for cardiac tissue engineering. Tissue engineering Part C, Methods. 2010;16:1417–1426. doi: 10.1089/ten.TEC.2010.0068. [DOI] [PubMed] [Google Scholar]
- Beauchamp P, Moritz W, Kelm JM, Ullrich ND, Agarkova I, Anson BD, et al. Development and Characterization of a Scaffold-Free 3D Spheroid Model of Induced Pluripotent Stem Cell-Derived Human Cardiomyocytes. Tissue engineering Part C, Methods. 2015;21:852–861. doi: 10.1089/ten.TEC.2014.0376. [DOI] [PubMed] [Google Scholar]
- Bielawski KS, Leonard A, Bhandari S, Murry CE, Sniadecki NJ. Real-Time Force and Frequency Analysis of Engineered Human Heart Tissue Derived from Induced Pluripotent Stem Cells Using Magnetic Sensing. Tissue engineering Part C, Methods. 2016 doi: 10.1089/ten.tec.2016.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinova K, Stohlman J, Vicente J, Chan D, Johannesen L, Hortigon-Vinagre MP, et al. Comprehensive Translational Assessment of Human Induced Pluripotent Stem Cell Derived Cardiomyocytes for Evaluating Drug-Induced Arrhythmias. Toxicological sciences : an official journal of the Society of Toxicology. 2016 doi: 10.1093/toxsci/kfw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema J, van der Meer P, Sluijter JP, Domian IJ. Engineering Myocardial Tissue: The Convergence of Stem Cells Biology and Tissue Engineering Technology. Stem Cells. 2013 doi: 10.1002/stem.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Anderson D, Priddle H, Barbadillo Munoz MD, Chamberlain S, Allegrucci C, et al. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25:929–938. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Li YF, Matsa E, Wu H, Ong S-G, Sharma A, et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med. 2016;22:547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, et al. Chemically defined generation of human cardiomyocytes. Nat Meth. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, Peters A, et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursac N, Parker KK, Iravanian S, Tung L. Cardiomyocyte cultures with controlled macroscopic anisotropy: a model for functional electrophysiological studies of cardiac muscle. Circ Res. 2002;91:e45–e54. doi: 10.1161/01.res.0000047530.88338.eb. [DOI] [PubMed] [Google Scholar]
- Byrne SM, Church GM. Crispr-mediated Gene Targeting of Human Induced Pluripotent Stem Cells. Current protocols in stem cell biology. 2015;35:5A. doi: 10.1002/9780470151808.sc05a08s35. 8 1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Li W, Su H, Qin D, Yang J, Zhu F, et al. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J Biol Chem. 2010;285:11227–11234. doi: 10.1074/jbc.M109.086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins H. Arrhythmogenic right ventricular dysplasia/cardiomyopathy-three decades of progress. Circulation journal : official journal of the Japanese Circulation Society. 2015;79:901–913. doi: 10.1253/circj.CJ-15-0288. [DOI] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. The Journal of clinical investigation. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson D, Hnilova M, Yang X, Nemeth CL, Tsui JH, Smith AS, et al. Nanotopography-Induced Structural Anisotropy and Sarcomere Development in Human Cardiomyocytes Derived from Induced Pluripotent Stem Cells. ACS applied materials & interfaces. 2016 doi: 10.1021/acsami.5b11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashman TJ, Josowitz R, Johnson BV, Gelb BD, Costa KD. Human Engineered Cardiac Tissues Created Using Induced Pluripotent Stem Cells Reveal Functional Characteristics of BRAF-Mediated Hypertrophic Cardiomyopathy. PLoS One. 2016;11:e0146697. doi: 10.1371/journal.pone.0146697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M, et al. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circulation Cardiovascular genetics. 2013;6:557–568. doi: 10.1161/CIRCGENETICS.113.000188. [DOI] [PubMed] [Google Scholar]
- Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, Habib IH, et al. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res. 2007;100:263–272. doi: 10.1161/01.RES.0000257776.05673.ff. [DOI] [PubMed] [Google Scholar]
- Cavero I, Holzgrefe H. Comprehensive in vitro Proarrhythmia Assay, a novel in vitro/in silico paradigm to detect ventricular proarrhythmic liability: a visionary 21st century initiative. Expert opinion on drug safety. 2014;13:745–758. doi: 10.1517/14740338.2014.915311. [DOI] [PubMed] [Google Scholar]
- Cerignoli F, Charlot D, Whittaker R, Ingermanson R, Gehalot P, Savchenko A, et al. High throughput measurement of Ca2+ dynamics for drug risk assessment in human stem cell-derived cardiomyocytes by kinetic image cytometry. Journal of Pharmacological and Toxicological Methods. 2012;66:246–256. doi: 10.1016/j.vascn.2012.08.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Lee E, Tu R, Santiago K, Grosberg A, Fowlkes C, et al. Integrated platform for functional monitoring of biomimetic heart sheets derived from human pluripotent stem cells. Biomaterials. 2014;35:675–683. doi: 10.1016/j.biomaterials.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QZ. Biomaterials in cardiac tissue engineering: ten years of research survey. In: Harding SC, Ali NN, Lyon AR, Boccaccini AR, editors. Materials Science and Engineering: R: Reports. 2008. pp. 1–37. [Google Scholar]
- Chen Y, Cao J, Xiong M, Petersen AJ, Dong Y, Tao Y, et al. Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17:233–244. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Lee CS, Kwon YW, Paek JS, Lee SH, Hur J, et al. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood. 2010;116:386–395. doi: 10.1182/blood-2010-02-269589. [DOI] [PubMed] [Google Scholar]
- Chockalingam P, Mizusawa Y, Wilde AA. Channelopathies - emerging trends in the management of inherited arrhythmias. Indian pacing and electrophysiology journal. 2015;15:43–54. doi: 10.1016/S0972-6292(16)30841-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun YW, Balikov DA, Feaster TK, Williams CH, Sheng CC, Lee JB, et al. Combinatorial polymer matrices enhance in vitro maturation of human induced pluripotent stem cell-derived cardiomyocytes. Biomaterials. 2015;67:52–64. doi: 10.1016/j.biomaterials.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churko JM, Burridge PW, Wu JC. Generation of human iPSCs from human peripheral blood mononuclear cells using non-integrative Sendai virus in chemically defined conditions. Methods in molecular biology (Clifton, NJ) 2013;1036:81–88. doi: 10.1007/978-1-62703-511-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 1997;277:301–306. [PubMed] [Google Scholar]
- Clause KC, Tinney JP, Liu LJ, Keller BB, Tobita K. Engineered early embryonic cardiac tissue increases cardiomyocyte proliferation by cyclic mechanical stretch via p38-MAP kinase phosphorylation. Tissue engineering Part A. 2009;15:1373–1380. doi: 10.1089/ten.tea.2008.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements M, Thomas N. High-throughput multi-parameter profiling of electrophysiological drug effects in human embryonic stem cell derived cardiomyocytes using multi-electrode arrays. Toxicological sciences : an official journal of the Society of Toxicology. 2014;140:445–461. doi: 10.1093/toxsci/kfu084. [DOI] [PubMed] [Google Scholar]
- Davis RP, Casini S, van den Berg CW, Hoekstra M, Remme CA, Dambrot C, et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125:3079–3091. doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- Davis RP, van den Berg CW, Casini S, Braam SR, Mummery CL. Pluripotent stem cell models of cardiac disease and their implication for drug discovery and development. Trends Mol Med. 2011;17:475–484. doi: 10.1016/j.molmed.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Deng XF, Rokosh DG, Simpson PC. Autonomous and growth factor-induced hypertrophy in cultured neonatal mouse cardiac myocytes. Comparison with rat. Circ Res. 2000;87:781–788. doi: 10.1161/01.res.87.9.781. [DOI] [PubMed] [Google Scholar]
- Devalla HD, Schwach V, Ford JW, Milnes JT, El-Haou S, Jackson C, et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pasquale E, Lodola F, Miragoli M, Denegri M, Avelino-Cruz JE, Buonocore M, et al. CaMKII inhibition rectifies arrhythmic phenotype in a patient-specific model of catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis. 2013;4:e843. doi: 10.1038/cddis.2013.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diecke S, Lu J, Lee J, Termglinchan V, Kooreman NG, Burridge PW, et al. Novel codon-optimized mini-intronic plasmid for efficient, inexpensive, and xeno-free induction of pluripotency. Scientific reports. 2015;5:8081. doi: 10.1038/srep08081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science (New York, NY) 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Svendsen CN. Human stem cells and drug screening: opportunities and challenges. Nat Rev Drug Discov. 2010;9:367–372. doi: 10.1038/nrd3000. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, Thomson JA, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimkhani MR, Young CL, Lauffenburger DA, Griffith LG, Borenstein JT. Approaches to in vitro tissue regeneration with application for human disease modeling and drug development. Drug Discov Today. 2014;19:754–762. doi: 10.1016/j.drudis.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nature cell biology. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- Egashira T, Yuasa S, Suzuki T, Aizawa Y, Yamakawa H, Matsuhashi T, et al. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc Res. 2012;95:419–429. doi: 10.1093/cvr/cvs206. [DOI] [PubMed] [Google Scholar]
- El-Sherbiny IM, Yacoub MH. Hydrogel scaffolds for tissue engineering: Progress and challenges. Glob Cardiol Sci Pract. 2013;2013:316–342. doi: 10.5339/gcsp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nature methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Engelmayr GC, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nat Mater. 2008;7:1003–1010. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang H-Y, Speicher DW, et al. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. Journal of cell science. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Esch MB, Smith AS, Prot JM, Oleaga C, Hickman JJ, Shuler ML. How multi-organ microdevices can help foster drug development. Adv Drug Deliv Rev. 2014;69–70:158–169. doi: 10.1016/j.addr.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenhagen T, Zimmermann WH. Engineering myocardial tissue. Circ Res. 2005;97:1220–1231. doi: 10.1161/01.RES.0000196562.73231.7d. [DOI] [PubMed] [Google Scholar]
- Fares RC, Gomes Jde A, Garzoni LR, Waghabi MC, Saraiva RM, Medeiros NI, et al. Matrix metalloproteinases 2 and 9 are differentially expressed in patients with indeterminate and cardiac clinical forms of Chagas disease. Infection and immunity. 2013;81:3600–3608. doi: 10.1128/IAI.00153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnáiz-Cot JJ, et al. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell Physiol Biochem. 2011;28:579–592. doi: 10.1159/000335753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Feng Z, Matsumoto T, Nomura Y, Nakamura T. An electro-tensile bioreactor for 3-D culturing of cardiomyocytes. A bioreactor system that simulates the myocardium’s electrical and mechanical response in vivo. IEEE engineering in medicine and biology magazine : the quarterly magazine of the Engineering in Medicine & Biology Society. 2005;24:73–79. doi: 10.1109/memb.2005.1463399. [DOI] [PubMed] [Google Scholar]
- Fermini B, Hancox JC, Abi-Gerges N, Bridgland-Taylor M, Chaudhary KW, Colatsky T, et al. A New Perspective in the Field of Cardiac Safety Testing through the Comprehensive In Vitro Proarrhythmia Assay Paradigm. Journal of biomolecular screening. 2016;21:1–11. doi: 10.1177/1087057115594589. [DOI] [PubMed] [Google Scholar]
- Foldes G, Mioulane M, Wright JS, Liu AQ, Novak P, Merkely B, et al. Modulation of human embryonic stem cell-derived cardiomyocyte growth: a testbed for studying human cardiac hypertrophy? J Mol Cell Cardiol. 2011;50:367–376. doi: 10.1016/j.yjmcc.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu JD, Rushing SN, Lieu DK, Chan CW, Kong CW, Geng L, et al. Distinct roles of microRNA-1 and −499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PLoS One. 2011;6:e27417. doi: 10.1371/journal.pone.0027417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist KH, Lewis GF, Gay EA, Sellgren KL, Grego S. High-throughput cardiac safety evaluation and multi-parameter arrhythmia profiling of cardiomyocytes using microelectrode arrays. Toxicology and Applied Pharmacology. 2015;288:249–257. doi: 10.1016/j.taap.2015.07.024. [DOI] [PubMed] [Google Scholar]
- Gintant G. An evaluation of hERG current assay performance: Translating preclinical safety studies to clinical QT prolongation. Pharmacology & Therapeutics. 2011;129:109–119. doi: 10.1016/j.pharmthera.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Giridharan GA. Effects of physiologic mechanical stimulation on embryonic chick cardiomyocytes using a microfluidic cardiac cell culture model. Analytical chemistry. 2014 doi: 10.1021/ac503716z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giridharan GA, Nguyen MD, Estrada R, Parichehreh V, Hamid T, Ismahil MA, et al. Microfluidic cardiac cell culture model (muCCCM) Analytical chemistry. 2010;82:7581–7587. doi: 10.1021/ac1012893. [DOI] [PubMed] [Google Scholar]
- Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nat Rev Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- Guan X, Mack DL, Moreno CM, Strande JL, Mathieu J, Shi Y, et al. Dystrophin-deficient cardiomyocytes derived from human urine: new biologic reagents for drug discovery. Stem cell research. 2014;12:467–480. doi: 10.1016/j.scr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Eder A, Bonstrup M, Flato M, Mewe M, Schaaf S, et al. Development of a drug screening platform based on engineered heart tissue. Circ Res. 2010;107:35–44. doi: 10.1161/CIRCRESAHA.109.211458. [DOI] [PubMed] [Google Scholar]
- Hick A, Wattenhofer-Donzé M, Chintawar S, Tropel P, Simard JP, Vaucamps N, et al. Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich’s ataxia. Dis Model Mech. 2013;6:608–621. doi: 10.1242/dmm.010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt MN, Boeddinghaus J, Mitchell A, Schaaf S, Bornchen C, Muller C, et al. Functional improvement and maturation of rat and human engineered heart tissue by chronic electrical stimulation. J Mol Cell Cardiol. 2014;74:151–161. doi: 10.1016/j.yjmcc.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Huang HP, Chen PH, Hwu WL, Chuang CY, Chien YH, Stone L, et al. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum Mol Genet. 2011;20:4851–4864. doi: 10.1093/hmg/ddr424. [DOI] [PubMed] [Google Scholar]
- Huang YC, Khait L, Birla RK. Modulating the functional performance of bioengineered heart muscle using growth factor stimulation. Ann Biomed Eng. 2008;36:1372–1382. doi: 10.1007/s10439-008-9517-9. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson J, Titmarsh D, Hidalgo A, Wolvetang E, Cooper-White J. Primitive cardiac cells from human embryonic stem cells. Stem Cells Dev. 2012;21:1513–1523. doi: 10.1089/scd.2011.0254. [DOI] [PubMed] [Google Scholar]
- Huebsch N, Loskill P, Mandegar MA, Marks NC, Sheehan AS, Ma Z, et al. Automated video-based analysis of contractility and calcium flux in human iPS-derived cardiomyocytes cultured over different spatial scales. Tissue engineering Part C, Methods. 2014 doi: 10.1089/ten.tec.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Yamanaka S. The use of induced pluripotent stem cells in drug development. Clin Pharmacol Ther. 2011;89:655–661. doi: 10.1038/clpt.2011.38. [DOI] [PubMed] [Google Scholar]
- Isenberg BC, Tsuda Y, Williams C, Shimizu T, Yamato M, Okano T, et al. A thermoresponsive, microtextured substrate for cell sheet engineering with defined structural organization. Biomaterials. 2008;29:2565–2572. doi: 10.1016/j.biomaterials.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, et al. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol. 2012;60:990–1000. doi: 10.1016/j.jacc.2012.02.066. [DOI] [PubMed] [Google Scholar]
- Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A. Modelling the long QT syndrome with induced pluripotent stem cells. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- Jiao A, Trosper NE, Yang HS, Kim J, Tsui JH, Frankel SD, et al. Thermoresponsive nanofabricated substratum for the engineering of three-dimensional tissues with layer-by-layer architectural control. ACS Nano. 2014;8:4430–4439. doi: 10.1021/nn4063962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Mukherjee R, Dick IE, Liu T, O’Rourke B, Yue DT, Tung L. Structural and functional plasticity in long-term cultures of adult ventricular myocytes. J Mol Cell Cardiol. 2013;65:76–87. doi: 10.1016/j.yjmcc.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CB, Moretti A, Mederos y, Schnitzler M, Iop L, Storch U, Bellin M, et al. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med. 2012;4:180–191. doi: 10.1002/emmm.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura T, Makiyama T, Sasaki K, Yoshida Y, Wuriyanghai Y, Chen J, et al. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circulation journal : official journal of the Japanese Circulation Society. 2013;77:1307–1314. doi: 10.1253/circj.cj-12-0987. [DOI] [PubMed] [Google Scholar]
- Karakikes I, Senyei GD, Hansen J, Kong CW, Azeloglu EU, Stillitano F, et al. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem cells translational medicine. 2014;3:18–31. doi: 10.5966/sctm.2013-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kijlstra JD, Hu D, Mittal N, Kausel E, van der Meer P, Garakani A, et al. Integrated Analysis of Contractile Kinetics, Force Generation, and Electrical Activity in Single Human Stem Cell-Derived Cardiomyocytes. Stem Cell Reports. 2015;5:1226–1238. doi: 10.1016/j.stemcr.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013a;494:105–110. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Kim P, Song I, Cha JM, Lee SH, Kim B, et al. Guided three-dimensional growth of functional cardiomyocytes on polyethylene glycol nanostructures. Langmuir. 2006;22:5419–5426. doi: 10.1021/la060283u. [DOI] [PubMed] [Google Scholar]
- Kim DH, Kshitiz, Smith RR, Kim P, Ahn EH, Kim HN, et al. Nanopatterned cardiac cell patches promote stem cell niche formation and myocardial regeneration. Integr Biol (Camb) 2012a;4:1019–1033. doi: 10.1039/c2ib20067h. [DOI] [PubMed] [Google Scholar]
- Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, et al. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc Natl Acad Sci U S A. 2010;107:565–570. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Provenzano PP, Smith CL, Levchenko A. Matrix nanotopography as a regulator of cell function. J Cell Biol. 2012b;197:351–360. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HN, Jiao A, Hwang NS, Kim MS, Kang dH, Kim DH, et al. Nanotopography-guided tissue engineering and regenerative medicine. Adv Drug Deliv Rev. 2013b;65:536–558. doi: 10.1016/j.addr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong CW, Akar FG, Li RA. Translational potential of human embryonic and induced pluripotent stem cells for myocardial repair: insights from experimental models. Thromb Haemost. 2010;104:30–38. doi: 10.1160/TH10-03-0189. [DOI] [PubMed] [Google Scholar]
- Kramer J, Obejero-Paz CA, Myatt G, Kuryshev YA, Bruening-Wright A, Verducci JS, et al. MICE Models: Superior to the HERG Model in Predicting Torsade de Pointes. Scientific reports. 2013;3:2100. doi: 10.1038/srep02100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshitiz, Hubbi ME, Ahn EH, Downey J, Afzal J, Kim DH, et al. Matrix rigidity controls endothelial differentiation and morphogenesis of cardiac precursors. Sci Signal. 2012;5:ra41. doi: 10.1126/scisignal.2003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, et al. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci U S A. 2015;112:E2785–E2794. doi: 10.1073/pnas.1424042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotech. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkelä E, et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5:220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasher RA, Pahnke AQ, Johnson JM, Sachse FB, Hitchcock RW. Electrical stimulation directs engineered cardiac tissue to an age-matched native phenotype. Journal of tissue engineering. 2012;3:2041731412455354. doi: 10.1177/2041731412455354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Klos M, Bollensdorff C, Hou L, Ewart P, Kamp TJ, et al. Simultaneous voltage and calcium mapping of genetically purified human induced pluripotent stem cell-derived cardiac myocyte monolayers. Circ Res. 2012;110:1556–1563. doi: 10.1161/CIRCRESAHA.111.262535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ran X, Lai KWH, Lau VYM, Siu DCW, Tse HF. Generation and Characterization of Patient-Specific iPSC Model for Cardiovascular Disease. Humana Press; 2015. pp. 1–23. [DOI] [PubMed] [Google Scholar]
- Li D, Zhou J, Wang L, Shin ME, Su P, Lei X, et al. Integrated biochemical and mechanical signals regulate multifaceted human embryonic stem cell functions. J Cell Biol. 2010;191:631–644. doi: 10.1083/jcb.201006094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. 2011 doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, et al. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. 2013;127:1677–1691. doi: 10.1161/CIRCULATIONAHA.113.001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liau B, Christoforou N, Leong KW, Bursac N. Pluripotent stem cell-derived cardiac tissue patch with advanced structure and function. Biomaterials. 2011;32:9180–9187. doi: 10.1016/j.biomaterials.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SH, Mao HQ. Electrospun scaffolds for stem cell engineering. Adv Drug Deliv Rev. 2009;61:1084–1096. doi: 10.1016/j.addr.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Lin B, Li Y, Han L, Kaplan AD, Ao Y, Kalra S, et al. Modeling and study of the mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from individuals with Duchenne muscular dystrophy. Disease Models & Mechanisms. 2015;8:457–466. doi: 10.1242/dmm.019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Ruan Y, Priori SG. Catecholaminergic polymorphic ventricular tachycardia. Progress in cardiovascular diseases. 2008;51:23–30. doi: 10.1016/j.pcad.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Wei H, Zhao Y, Lu J, Li G, Sahib NB, et al. Modeling type 3 long QT syndrome with cardiomyocytes derived from patient-specific induced pluripotent stem cells. Int J Cardiol. 2013a;168:5277–5286. doi: 10.1016/j.ijcard.2013.08.015. [DOI] [PubMed] [Google Scholar]
- Ma T, Xie M, Laurent T, Ding S. Progress in the reprogramming of somatic cells. Circ Res. 2013b;112:562–574. doi: 10.1161/CIRCRESAHA.111.249235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Koo S, Finnegan MA, Loskill P, Huebsch N, Marks NC, et al. Three-dimensional filamentous human diseased cardiac tissue model. Biomaterials. 2014;35:1367–1377. doi: 10.1016/j.biomaterials.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macadangdang J, Guan X, Smith AS, Lucero R, Czerniecki S, Childers MK, et al. Nanopatterned Human iPSC-based Model of a Dystrophin-Null Cardiomyopathic Phenotype. Cell Mol Bioeng. 2015;8:320–322. doi: 10.1007/s12195-015-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macadangdang J, Lee HJ, Carson D, Jiao A, Fugate J, Pabon L, et al. Capillary force lithography for cardiac tissue engineering. Journal of visualized experiments : JoVE. 2014 doi: 10.3791/50039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidhof R, Tandon N, Lee EJ, Luo J, Duan Y, Yeager K, et al. Biomimetic perfusion and electrical stimulation applied in concert improved the assembly of engineered cardiac tissue. Journal of tissue engineering and regenerative medicine. 2012;6:e12–e23. doi: 10.1002/term.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet A, Tan K, Chai X, Sadananda SN, Mehta A, Ooi J, et al. Modeling Doxorubicin-Induced Cardiotoxicity in Human Pluripotent Stem Cell Derived-Cardiomyocytes. Scientific reports. 2016;6:25333. doi: 10.1038/srep25333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-Guided Human Genome Engineering via Cas9. Science (New York, NY) 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik N, Rao MS. A review of the methods for human iPSC derivation. Methods in molecular biology (Clifton, NJ) 2013;997:23–33. doi: 10.1007/978-1-62703-348-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis M, Sorota S. Additive effects of combined application of multiple hERG blockers. Journal of cardiovascular pharmacology. 2008;51:549–552. doi: 10.1097/FJC.0b013e31817532ee. [DOI] [PubMed] [Google Scholar]
- Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Scientific reports. 2015;5:8883. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, et al. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J. 2011;32:952–962. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury Y, Gauthier M, Peschanski M, Martinat C. Human pluripotent stem cells for disease modelling and drug screening. Bioessays. 2012;34:61–71. doi: 10.1002/bies.201100071. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- McCain ML, Sheehy SP, Grosberg A, Goss JA, Parker KK. Recapitulating maladaptive, multiscale remodeling of failing myocardium on a chip. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9770–9775. doi: 10.1073/pnas.1304913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt TC, Angello JC, Whitney ML, Reinecke H, Hauschka SD, Murry CE, et al. In vitro generation of differentiated cardiac myofibers on micropatterned laminin surfaces. Journal of biomedical materials research. 2002;60:472–479. doi: 10.1002/jbm.1292. [DOI] [PubMed] [Google Scholar]
- McKusick VA. The Cardiovascular Aspects of Marfan’s Syndrome: A Heritable Disorder of Connective Tissue. Circulation. 1955;11:321–342. doi: 10.1161/01.cir.11.3.321. [DOI] [PubMed] [Google Scholar]
- Mengsteab PY, Uto K, Smith AS, Frankel S, Fisher E, Nawas Z, et al. Spatiotemporal control of cardiac anisotropy using dynamic nanotopographic cues. Biomaterials. 2016;86:1–10. doi: 10.1016/j.biomaterials.2016.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- Morgan KY, Black LD., 3rd Mimicking isovolumic contraction with combined electromechanical stimulation improves the development of engineered cardiac constructs. Tissue engineering Part A. 2014;20:1654–1667. doi: 10.1089/ten.tea.2013.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery CL, Zhang J, Ng ES, Elliott DA, Elefanty AG, Kamp TJ. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete EG, Liang P, Lan F, Sanchez-Freire V, Simmons C, Gong T, et al. Screening drug-induced arrhythmia [corrected] using human induced pluripotent stem cell-derived cardiomyocytes and low-impedance microelectrode arrays. Circulation. 2013;128:S3–S13. doi: 10.1161/CIRCULATIONAHA.112.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Cooper JB. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7:258–262. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Novak A, Lorber A, Itskovitz-Eldor J, Binah O. Modeling Catecholaminergic Polymorphic Ventricular Tachycardia using Induced Pluripotent Stem Cell-derived Cardiomyocytes. Rambam Maimonides Med J. 2012;3:e0015. doi: 10.5041/RMMJ.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, et al. Biowire: a new platform for maturation of human pluripotent stem cell derived cardiomyocytes. Nature methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleaga C, Bernabini C, Smith AS, Srinivasan B, Jackson M, McLamb W, et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Scientific reports. 2016;6:20030. doi: 10.1038/srep20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- Ou D, Wang Q, Huang Y, Zeng D, Wei T, Ding L, et al. Co-culture with neonatal cardiomyocytes enhances the proliferation of iPSC-derived cardiomyocytes via FAK/JNK signaling. BMC Developmental Biology. 2016;16:11. doi: 10.1186/s12861-016-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola J, Vaananen H, Larsson K, Penttinen K, Toivonen L, Kontula K, et al. Slowed depolarization and irregular repolarization in catecholaminergic polymorphic ventricular tachycardia: a study from cellular Ca2+ transients and action potentials to clinical monophasic action potentials and electrocardiography. Europace : European pacing, arrhythmias, and cardiac electrophysiology : journal of the working groups on cardiac pacing, arrhythmias, and cardiac cellular electrophysiology of the European Society of Cardiology. 2015 doi: 10.1093/europace/euv380. [DOI] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Larson BL, Kolewe ME, Vunjak-Novakovic G, Freed LE. Biomimetic scaffold combined with electrical stimulation and growth factor promotes tissue engineered cardiac development. Experimental cell research. 2014;321:297–306. doi: 10.1016/j.yexcr.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Radisic M, Lim JO, Chang BH, Vunjak-Novakovic G. A novel composite scaffold for cardiac tissue engineering. In Vitro Cell Dev Biol Anim. 2005a;41:188–196. doi: 10.1290/0411071.1. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim HN, Kim DH, Levchenko A, Suh KY. Quantitative analysis of the combined effect of substrate rigidity and topographic guidance on cell morphology. IEEE Trans Nanobioscience. 2012;11:28–36. doi: 10.1109/TNB.2011.2165728. [DOI] [PubMed] [Google Scholar]
- Park J, Ryu J, Choi SK, Seo E, Cha JM, Ryu S, et al. Real-time measurement of the contractile forces of self-organized cardiomyocytes on hybrid biopolymer microcantilevers. Analytical chemistry. 2005b;77:6571–6580. doi: 10.1021/ac0507800. [DOI] [PubMed] [Google Scholar]
- Pati F, Jang J, Ha DH, Won Kim S, Rhie JW, Shim JH, et al. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nature communications. 2014;5:3935. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioner JM, Racca AW, Klaiman JM, Yang KC, Guan X, Pabon L, et al. Isolation and Mechanical Measurements of Myofibrils from Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cell Reports. 2016;6:885–896. doi: 10.1016/j.stemcr.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon E, Kong CW, Li RA. Human pluripotent stem cell-based approaches for myocardial repair: from the electrophysiological perspective. Mol Pharm. 2011;8:1495–1504. doi: 10.1021/mp2002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarto N, Leonard B, Li S, Marchand M, Anderson E, Behr B, et al. Skeletogenic phenotype of human Marfan embryonic stem cells faithfully phenocopied by patient-specific induced-pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:215–220. doi: 10.1073/pnas.1113442109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab S, Klingenstein M, Liebau S, Linta L. A Comparative View on Human Somatic Cell Sources for iPSC Generation. Stem cells international. 2014;2014:768391. doi: 10.1155/2014/768391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan J, Krishnan UM, Sethuraman S. Hydrogel based injectable scaffolds for cardiac tissue regeneration. Biotechnol Adv. 2014;32:449–461. doi: 10.1016/j.biotechadv.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Radisic M, Euloth M, Yang L, Langer R, Freed LE, Vunjak-Novakovic G. High-density seeding of myocyte cells for cardiac tissue engineering. Biotechnol Bioeng. 2003;82:403–414. doi: 10.1002/bit.10594. [DOI] [PubMed] [Google Scholar]
- Radisic M, Park H, Chen F, Salazar-Lazzaro JE, Wang Y, Dennis R, et al. Biomimetic approach to cardiac tissue engineering: oxygen carriers and channeled scaffolds. Tissue engineering. 2006;12:2077–2091. doi: 10.1089/ten.2006.12.2077. [DOI] [PubMed] [Google Scholar]
- Radisic M, Park H, Shing H, Consi T, Schoen FJ, Langer R, et al. Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. Proc Natl Acad Sci U S A. 2004;101:18129–18134. doi: 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MS, Malik N. Assessing iPSC Reprogramming Methods for Their Suitability in Translational Medicine. Journal of cellular biochemistry. 2012;113:3061–3068. doi: 10.1002/jcb.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AJ, Zaleta-Rivera K, Ashley EA, Pruitt BL. Stable, covalent attachment of laminin to microposts improves the contractility of mouse neonatal cardiomyocytes. ACS applied materials & interfaces. 2014;6:15516–15526. doi: 10.1021/am5042324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel M, Jou CJ, Lai S, Lux RL, Moreno AP, Spitzer KW, et al. Functional and pharmacological analysis of cardiomyocytes differentiated from human peripheral blood mononuclear-derived pluripotent stem cells. Stem Cell Reports. 2014;3:131–141. doi: 10.1016/j.stemcr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AG, Han SJ, Regnier M, Sniadecki NJ. Substrate stiffness increases twitch power of neonatal cardiomyocytes in correlation with changes in myofibril structure and intracellular calcium. Biophysical journal. 2011;101:2455–2464. doi: 10.1016/j.bpj.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez ML, Graham BT, Pabon LM, Han SJ, Murry CE, Sniadecki NJ. Measuring the Contractile Forces of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes With Arrays of Microposts. Journal of Biomechanical Engineering. 2014;136:051005. doi: 10.1115/1.4027145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani L, Johnson AA, Arnold A, Stolzing A. The aging signature: a hallmark of induced pluripotent stem cells? Aging Cell. 2014;13:2–7. doi: 10.1111/acel.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romli F, Alitheen NB, Hamid M, Ismail R, Nik Abd Rahman NM. Current techniques in reprogramming cell potency. J Cell Biochem. 2013;114:1230–1237. doi: 10.1002/jcb.24477. [DOI] [PubMed] [Google Scholar]
- Sager PT, Gintant G, Turner JR, Pettit S, Stockbridge N. Rechanneling the cardiac proarrhythmia safety paradigm: A meeting report from the Cardiac Safety Research Consortium. American Heart Journal. 2014;167:292–300. doi: 10.1016/j.ahj.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Mitcheson JS. Predicting drug-hERG channel interactions that cause acquired long QT syndrome. Trends in pharmacological sciences. 2005;26:119–124. doi: 10.1016/j.tips.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Scheel O, Frech S, Amuzescu B, Eisfeld J, Lin KH, Knott T. Action potential characterization of human induced pluripotent stem cell-derived cardiomyocytes using automated patch-clamp technology. Assay and drug development technologies. 2014;12:457–469. doi: 10.1089/adt.2014.601. [DOI] [PubMed] [Google Scholar]
- Schwartz PJ, Stramba-Badiale M, Crotti L, Pedrazzini M, Besana A, Bosi G, et al. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120:1761–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CW, Zhang X, Abi-Gerges N, Lamore SD, Abassi YA, Peters MF. An Impedance-Based Cellular Assay Using Human iPSC-Derived Cardiomyocytes to Quantify Modulators of Cardiac Contractility. Toxicological sciences : an official journal of the Society of Toxicology. 2014;142:331–338. doi: 10.1093/toxsci/kfu186. [DOI] [PubMed] [Google Scholar]
- Sekine H, Shimizu T, Hobo K, Sekiya S, Yang J, Yamato M, et al. Endothelial Cell Coculture Within Tissue-Engineered Cardiomyocyte Sheets Enhances Neovascularization and Improves Cardiac Function of Ischemic Hearts. Circulation. 2008;118:S145–S152. doi: 10.1161/CIRCULATIONAHA.107.757286. [DOI] [PubMed] [Google Scholar]
- Sekine H, Shimizu T, Sakaguchi K, Dobashi I, Wada M, Yamato M, et al. In vitro fabrication of functional three-dimensional tissues with perfusable blood vessels. Nature communications. 2013;4:1399. doi: 10.1038/ncomms2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, et al. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ Res. 2014;115:556–566. doi: 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y, Hauch KD, Laflamme MA. Cardiac applications for human pluripotent stem cells. Curr Pharm Des. 2009;15:2791–2806. doi: 10.2174/138161209788923804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J, Grosberg A, Nawroth JC, Parker KK, Bertoldi K. Modeling of cardiac muscle thin films: pre-stretch, passive and active behavior. Journal of biomechanics. 2012;45:832–841. doi: 10.1016/j.jbiomech.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Yamato M, Isoi Y, Akutsu T, Setomaru T, Abe K, et al. Fabrication of pulsatile cardiac tissue grafts using a novel 3-dimensional cell sheet manipulation technique and temperature-responsive cell culture surfaces. Circ Res. 2002;90:e40. doi: 10.1161/hh0302.105722. [DOI] [PubMed] [Google Scholar]
- Singh BN. Choice and chance in drug therapy of cardiac arrhythmias: technique versus drug-specific responses in evaluation of efficacy. The American journal of cardiology. 1993;72:114F–124F. doi: 10.1016/0002-9149(93)90974-h. [DOI] [PubMed] [Google Scholar]
- Siu CW, Lee YK, Ho JC, Lai WH, Chan YC, Ng KM, et al. Modeling of lamin A/C mutation premature cardiac aging using patient-specific induced pluripotent stem cells. Aging (Albany NY) 2012;4:803–822. doi: 10.18632/aging.100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AST, Long CJ, McAleer C, Guo X, Esch M, Prot JM, et al. Chapter 7 ‘Body-on-a-Chip’ Technology and Supporting Microfluidics. Human-based Systems for Translational Research: The Royal Society of Chemistry. 2015:132–161. [Google Scholar]
- Sommer CA, Mostoslavsky G. The evolving field of induced pluripotency: recent progress and future challenges. Journal of cellular physiology. 2013;228:267–275. doi: 10.1002/jcp.24155. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondergaard CS, Mathews G, Wang L, Jeffreys A, Sahota A, Wood M, et al. Contractile and electrophysiologic characterization of optimized self-organizing engineered heart tissue. Ann Thorac Surg. 2012;94:1241–1248. doi: 10.1016/j.athoracsur.2012.04.098. discussion 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovik DE, Wang R, Dai G, Wang T, Aikawa E, Novikov M, et al. Diffusion spectrum MRI tractography reveals the presence of a complex network of residual myofibers in infarcted myocardium. Circ Cardiovasc Imaging. 2009;2:206–212. doi: 10.1161/CIRCIMAGING.108.815050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science (New York, NY) 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancescu M, Molnar P, McAleer CW, McLamb W, Long CJ, Oleaga C, et al. A phenotypic in vitro model for the main determinants of human whole heart function. Biomaterials. 2015;60:20–30. doi: 10.1016/j.biomaterials.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter DD, Hanna WT. Engineering mechanics for successive states in canine left ventricular myocardium. II. Fiber angle and sarcomere length. Circ Res. 1973;33:656–664. doi: 10.1161/01.res.33.6.656. [DOI] [PubMed] [Google Scholar]
- Streeter DD, Spotnitz HM, Patel DP, Ross J, Sonnenblick EH. Fiber orientation in the canine left ventricle during diastole and systole. Circ Res. 1969;24:339–347. doi: 10.1161/01.res.24.3.339. [DOI] [PubMed] [Google Scholar]
- Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JH, Esch MB, Prot JM, Long CJ, Smith A, Hickman JJ, et al. Microfabricated mammalian organ systems and their integration into models of whole animals and humans. Lab on a chip. 2013;13:1201–1212. doi: 10.1039/c3lc41017j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Tohyama S, Murata M, Nomura F, Kaneko T, Chen H, et al. In vitro pharmacologic testing using human induced pluripotent stem cell-derived cardiomyocytes. Biochem Biophys Res Commun. 2009;385:497–502. doi: 10.1016/j.bbrc.2009.05.073. [DOI] [PubMed] [Google Scholar]
- Tandon N, Cannizzaro C, Chao PH, Maidhof R, Marsano A, Au HT, et al. Electrical stimulation systems for cardiac tissue engineering. Nat Protoc. 2009;4:155–173. doi: 10.1038/nprot.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon N, Marsano A, Maidhof R, Wan L, Park H, Vunjak-Novakovic G. Optimization of electrical stimulation parameters for cardiac tissue engineering. Journal of tissue engineering and regenerative medicine. 2011;5:e115–e125. doi: 10.1002/term.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrenoire C, Wang K, Tung KW, Chung WK, Pass RH, Lu JT, et al. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J Gen Physiol. 2013;141:61–72. doi: 10.1085/jgp.201210899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SA, Copeland CR, Reich DH, Tung L. Mechanical Coupling Between Myofibroblasts and Cardiomyocytes Slows Electrical Conduction in Fibrotic Cell Monolayers. Circulation. 2011;123:2083–2093. doi: 10.1161/CIRCULATIONAHA.110.015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Tse HF, Ho JC, Choi SW, Lee YK, Butler AW, Ng KM, et al. Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing. Hum Mol Genet. 2013;22:1395–1403. doi: 10.1093/hmg/dds556. [DOI] [PubMed] [Google Scholar]
- Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, et al. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull IC, Karakikes I, Serrao GW, Backeris P, Lee JJ, Xie C, et al. Advancing functional engineered cardiac tissues toward a preclinical model of human myocardium. FASEB J. 2014;28:644–654. doi: 10.1096/fj.13-228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uosaki H, Fukushima H, Takeuchi A, Matsuoka S, Nakatsuji N, Yamanaka S, et al. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS One. 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valarmathi MT, Fuseler JW, Goodwin RL, Davis JM, Potts JD. The mechanical coupling of adult marrow stromal stem cells during cardiac regeneration assessed in a 2-D co-culture model. Biomaterials. 2011;32:2834–2850. doi: 10.1016/j.biomaterials.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proceedings of the National Academy of Sciences. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science (New York, NY) 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- van Spreeuwel AC, Bax NA, Bastiaens AJ, Foolen J, Loerakker S, Borochin M, et al. The influence of matrix (an)isotropy on cardiomyocyte contraction in engineered cardiac microtissues. Integr Biol (Camb) 2014;6:422–429. doi: 10.1039/c3ib40219c. [DOI] [PubMed] [Google Scholar]
- Vunjak-Novakovic G, Lui KO, Tandon N, Chien KR. Bioengineering heart muscle: a paradigm for regenerative medicine. Annu Rev Biomed Eng. 2011;13:245–267. doi: 10.1146/annurev-bioeng-071910-124701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorenpaa H, Ikonen L, Kujala K, Huttala O, Sarkanen JR, Ylikomi T, et al. Novel in vitro cardiovascular constructs composed of vascular-like networks and cardiomyocytes. In Vitro Cell Dev Biol Anim. 2014;50:275–286. doi: 10.1007/s11626-013-9703-4. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang G, To F, Butler JR, Claude A, McLaughlin RM, et al. Myocardial scaffold-based cardiac tissue engineering: application of coordinated mechanical and electrical stimulations. Langmuir. 2013;29:11109–11117. doi: 10.1021/la401702w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Nakamura K, Jinnin M, Kudo H, Goto M, Era T, et al. Establishment and gene expression analysis of disease-derived induced pluripotent stem cells of scleroderma. Journal of dermatological science. 2016 doi: 10.1016/j.jdermsci.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, et al. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth EK, Roth S, Blechschmidt C, Hölter SM, Becker L, Racz I, et al. Neuronal 3′,3,5-Triiodothyronine (T3) Uptake and Behavioral Phenotype of Mice Deficient in Mct8, the Neuronal T3 Transporter Mutated in Allan-Herndon-Dudley Syndrome. The Journal of Neuroscience. 2009;29:9439–9449. doi: 10.1523/JNEUROSCI.6055-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Zhang B, Liu H, Miklas JW, Gagliardi M, Pahnke A, et al. Microfabricated perfusable cardiac biowire: a platform that mimics native cardiac bundle. Lab on a chip. 2014;14:869–882. doi: 10.1039/c3lc51123e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. Differentiation and enrichment of cardiomyocytes from human pluripotent stem cells. J Mol Cell Cardiol. 2012;52:1203–1212. doi: 10.1016/j.yjmcc.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- Xu H, Yi BA, Wu H, Bock C, Gu H, Lui KO, et al. Highly efficient derivation of ventricular cardiomyocytes from induced pluripotent stem cells with a distinct epigenetic signature. Cell research. 2012;22:142–154. doi: 10.1038/cr.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Cai X, Wang L, Liao B, Zhang H, Shan Y, et al. Generating a non-integrating human induced pluripotent stem cell bank from urine-derived cells. PLoS One. 2013;8:e70573. doi: 10.1371/journal.pone.0070573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Yang J, Yamato M, Shimizu T, Sekine H, Ohashi K, Kanzaki M, et al. Reconstruction of functional tissues with cell sheet engineering. Biomaterials. 2007;28:5033–5043. doi: 10.1016/j.biomaterials.2007.07.052. [DOI] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yang X, Rodriguez M, Pabon L, Fischer KA, Reinecke H, Regnier M, et al. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J Mol Cell Cardiol. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo N, Baba S, Kaichi S, Niwa A, Mima T, Doi H, et al. The effects of cardioactive drugs on cardiomyocytes derived from human induced pluripotent stem cells. Biochem Biophys Res Commun. 2009;387:482–488. doi: 10.1016/j.bbrc.2009.07.052. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Yamanaka S. iPS cells: a source of cardiac regeneration. J Mol Cell Cardiol. 2011;50:327–332. doi: 10.1016/j.yjmcc.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Young Courtney S, Hicks Michael R, Ermolova Natalia V, Nakano H, Jan M, Younesi S, et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JL, Engler AJ. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials. 2011;32:1002–1009. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shadrin IY, Lam J, Xian HQ, Snodgrass HR, Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials. 2013;34:5813–5820. doi: 10.1016/j.biomaterials.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X, et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell research. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Benda C, Dunzinger S, Huang Y, Ho JC, Yang J, et al. Generation of human induced pluripotent stem cells from urine samples. Nat Protoc. 2012;7:2080–2089. doi: 10.1038/nprot.2012.115. [DOI] [PubMed] [Google Scholar]
- Zhu WZ, Van Biber B, Laflamme MA. Methods for the derivation and use of cardiomyocytes from human pluripotent stem cells. Methods in molecular biology (Clifton, NJ) 2011;767:419–431. doi: 10.1007/978-1-61779-201-4_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann WH, Melnychenko I, Wasmeier G, Didié M, Naito H, Nixdorff U, et al. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med. 2006;12:452–458. doi: 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]
- Zimmermann WH, Schneiderbanger K, Schubert P, Didié M, Münzel F, Heubach JF, et al. Tissue engineering of a differentiated cardiac muscle construct. Circ Res. 2002;90:223–230. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]