Abstract
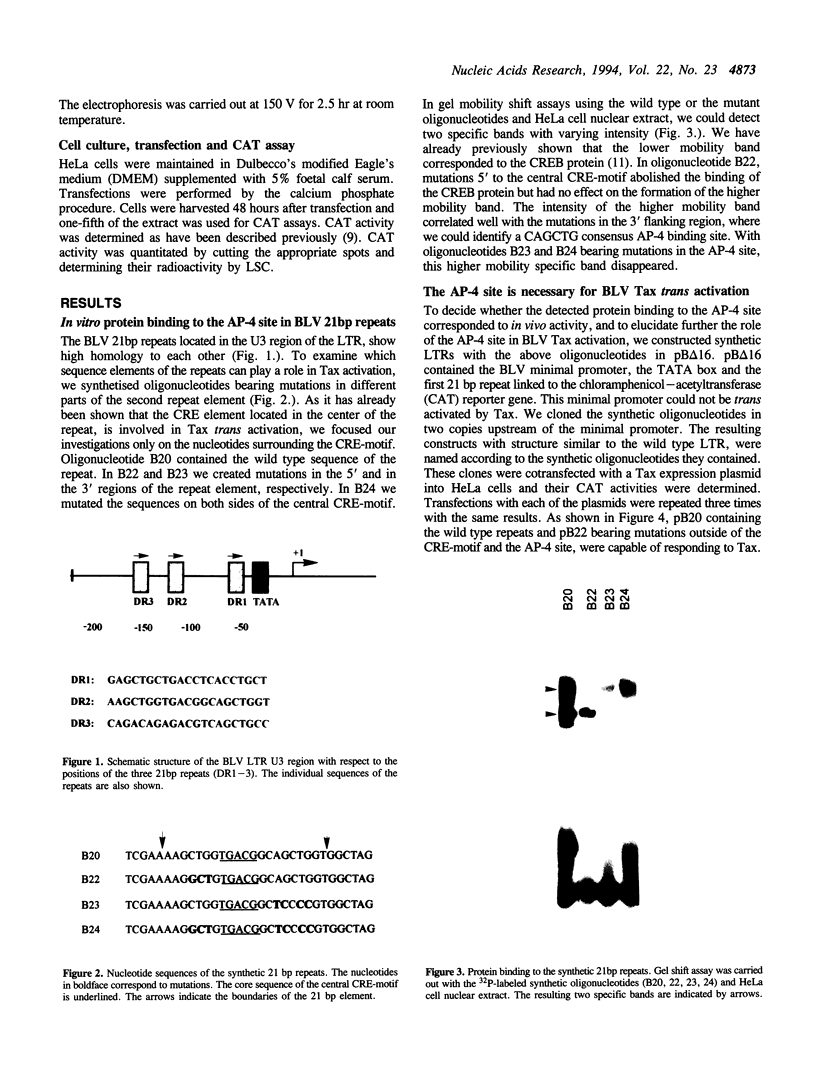
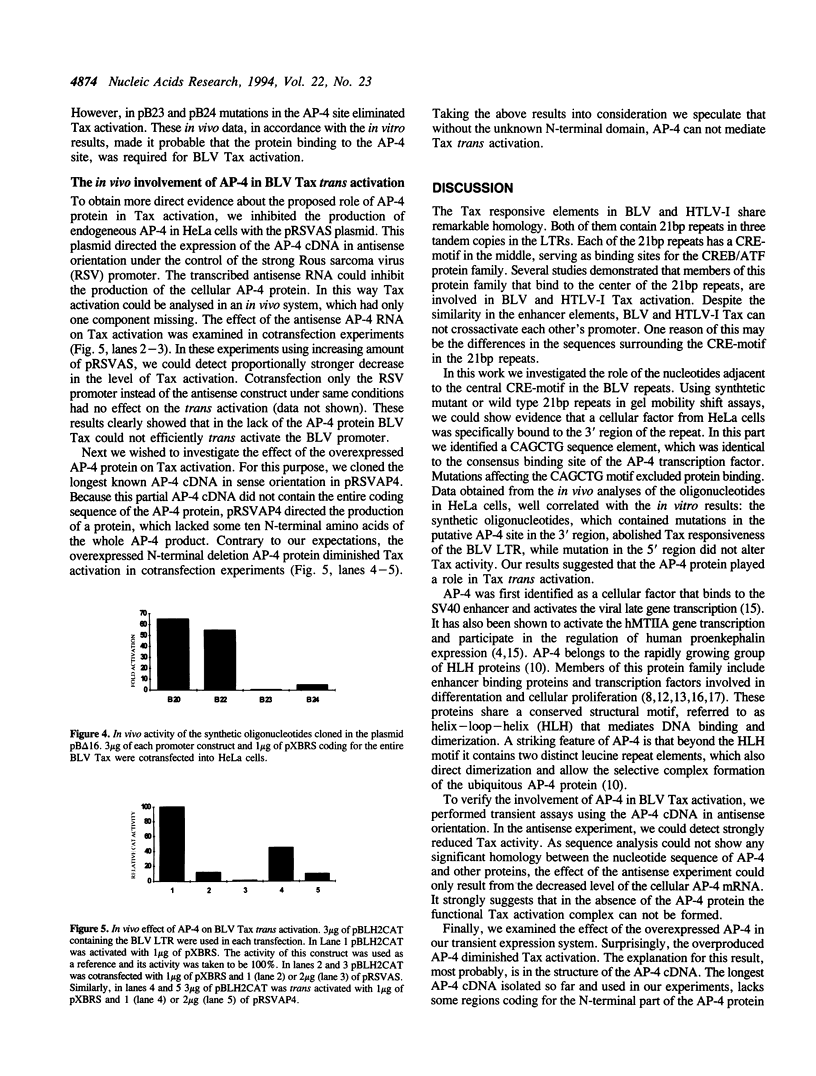
Three 21bp repeats can be found in the bovine leukemia virus long terminal repeat, which are crucial for the LTR directed gene expression by the trans activator protein Tax. Previous studies demonstrated that the major target of the Tax directed activation are the CRE-like elements in the center of these repeats. In this work we report that another motif of the 21bp repeats is also required for the Tax activation. Gel retardation--with the wild type or mutant 21bp repeats--revealed that cellular factors from HeLa cells were specifically bound to the center (CRE-like element) and the 3' region of the repeats, which contains a CAGCTG consensus AP-4 binding site. In vivo analysis using the synthetic 21bp repeats indicated that beyond the consensus CRE-like motif, the AP-4 site is also essential for Tax activation. To determine the role of AP-4 in BLV Tax trans activation, we used the AP-4 cDNA in antisense transient assays. In the in vivo experiments the antisense AP-4 RNA resulted in strongly decreased Tax activation. On the basis of these results we conclude that AP-4 is a good candidate of cellular factors involved in BLV Tax trans activation.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brady J., Jeang K. T., Duvall J., Khoury G. Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type I long terminal repeat. J Virol. 1987 Jul;61(7):2175–2181. doi: 10.1128/jvi.61.7.2175-2181.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burny A., Cleuter Y., Kettmann R., Mammerickx M., Marbaix G., Portetelle D., Van den Broeke A., Willems L., Thomas R. Bovine leukaemia: facts and hypotheses derived from the study of an infectious cancer. Cancer Surv. 1987;6(1):139–159. [PubMed] [Google Scholar]
- Béraud C., Lombard-Platet G., Michal Y., Jalinot P. Binding of the HTLV-I Tax1 transactivator to the inducible 21 bp enhancer is mediated by the cellular factor HEB1. EMBO J. 1991 Dec;10(12):3795–3803. doi: 10.1002/j.1460-2075.1991.tb04949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comb M., Mermod N., Hyman S. E., Pearlberg J., Ross M. E., Goodman H. M. Proteins bound at adjacent DNA elements act synergistically to regulate human proenkephalin cAMP inducible transcription. EMBO J. 1988 Dec 1;7(12):3793–3805. doi: 10.1002/j.1460-2075.1988.tb03264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J Virol. 1987 Aug;61(8):2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Caradonna S. J., Casey J. W. Bovine leukemia virus long terminal repeat: a cell type-specific promoter. Science. 1985 Jan 18;227(4684):317–320. doi: 10.1126/science.2981431. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson D. G., Olson E. N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989 May;3(5):628–640. doi: 10.1101/gad.3.5.628. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. F., Lüscher B., Admon A., Mermod N., Tjian R. Transcription factor AP-4 contains multiple dimerization domains that regulate dimer specificity. Genes Dev. 1990 Oct;4(10):1741–1752. doi: 10.1101/gad.4.10.1741. [DOI] [PubMed] [Google Scholar]
- Kiss-Toth E., Paca-uccaralertkun S., Unk I., Boros I. Member of the CREB/ATF protein family, but not CREB alpha plays an active role in BLV tax trans activation in vivo. Nucleic Acids Res. 1993 Aug 11;21(16):3677–3682. doi: 10.1093/nar/21.16.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989 Sep 8;58(5):823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- Ludwig S. R., Habera L. F., Dellaporta S. L., Wessler S. R. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott S. J., Lindholm P. F., Brown K. M., Gitlin S. D., Duvall J. F., Radonovich M. F., Brady J. N. A 36-kilodalton cellular transcription factor mediates an indirect interaction of human T-cell leukemia/lymphoma virus type I TAX1 with a responsive element in the viral long terminal repeat. Mol Cell Biol. 1990 Aug;10(8):4192–4201. doi: 10.1128/mcb.10.8.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N., Williams T. J., Tjian R. Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vitro. Nature. 1988 Apr 7;332(6164):557–561. doi: 10.1038/332557a0. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Sagata N., Yasunaga T., Tsuzuku-Kawamura J., Ohishi K., Ogawa Y., Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci U S A. 1985 Feb;82(3):677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., LeBowitz J. H., Baldwin A. S., Jr, Sharp P. A. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell. 1988 Feb 12;52(3):415–423. doi: 10.1016/s0092-8674(88)80034-5. [DOI] [PubMed] [Google Scholar]
- Slamon D. J., Shimotohno K., Cline M. J., Golde D. W., Chen I. S. Identification of the putative transforming protein of the human T-cell leukemia viruses HTLV-I and HTLV-II. Science. 1984 Oct 5;226(4670):61–65. doi: 10.1126/science.6089351. [DOI] [PubMed] [Google Scholar]
- Willems L., Gegonne A., Chen G., Burny A., Kettmann R., Ghysdael J. The bovine leukemia virus p34 is a transactivator protein. EMBO J. 1987 Nov;6(11):3385–3389. doi: 10.1002/j.1460-2075.1987.tb02661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L., Kettmann R., Chen G., Portetelle D., Burny A., Derse D. A cyclic AMP-responsive DNA-binding protein (CREB2) is a cellular transactivator of the bovine leukemia virus long terminal repeat. J Virol. 1992 Feb;66(2):766–772. doi: 10.1128/jvi.66.2.766-772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]



