Abstract
The aims of the present study were to estimate the affinity between 3,5-(E)-bis(3-methoxy-4-hydroxybenzal)-4-piperidinone hydrochloride (C0818) and heat shock protein 90 (Hsp90) and to investigate the inhibitory effects of this compound on Hsp90 ATPase activity. Fluorescence spectroscopy was used to examine the affinity between varying concentrations of C0818 and Hsp90, N-Hsp90, M-Hsp90 and C-Hsp90. Fluorescence intensities were recorded in the range of 290–510 nm at 293, 303 and 310 K, respectively. A colorimetric assay for inorganic phosphate (based on the formation of a phosphomolybdate complex and the subsequent reaction with malachite green) were used to examine the inhibitory effects of C0818 on Hsp90 ATPase activity. The equilibrium dissociation constant KD value of C0818 was found to be 23.412±0.943 μmol/L. The interaction between C0818 and Hsp90 was driven mainly by electrostatic interactions. C0818 showed the strongest affinity with C-Hsp90. These results conclusively demonstrate the inhibitory activity of C0818 on the activity of Hsp90 ATPase.
KEY WORDS: Curcumin derivative, Hsp90, ATPase activity, Fluorescence spectrometry, Interaction
Graphical abstract
The molecular chaperone heat shock protein 90 (Hsp90) plays a key role in the cell by stabilizing a number of client proteins. The ATPase activity of Hsp90 is essential for its chaperone function. In this study, C0818 was found the strongest affinity with C-Hsp90, rather than N-Hsp90 or M-Hsp90, and inhibitory effect on the activity of Hsp90 ATPase.
1. Introduction
Heat shock protein 90 (Hsp90) is an ATP-dependent cellular chaperone responsible for maintaining the stability and activity of a number of client proteins involved in cell signaling, proliferation and survival1, 2. Recent studies have shown that Hsp90 is a key protein for oncogenesis and malignancy, and an emerging target for cancer therapeutics3, 4, 5, 6. Hsp90 inhibitors can simultaneously inhibit multiple signaling pathways that cancer cells depend on for their growth and proliferation. Since Hsp90 inhibitors have significant potential as antitumor agents, the discovery of Hsp90 inhibitors with new chemical scaffolds is relevant to advances in cancer chemotherapy.
The chaperoning of most Hsp90 client proteins is regulated by a dynamic cycle driven by ATP binding to Hsp90 and subsequent hydrolysis of the protein7. The intrinsic ATPase activity of Hsp90 is essential to this activity. Inhibition of the ATPase activity of Hsp90 leads to antitumor activity in vitro and in vivo8, 9.
Curcumin is an active ingredient of the plant turmeric (Curcuma longa). In our previous work10, we found that curcumin is a lead compound in the search for new kinds of Hsp90 inhibitors. We thus synthesized a series of derivatives, some of which showed lead-like properties and were found to be more active than curcumin in Hsp90 inhibition and antitumor action11, 12. In the present paper, we have investigated a novel curcumin derivative, 3,5-(E)-bis(3-methoxy-4-hydroxybenzal)-4-piperidinone hydrochloride (C0818), for its interaction with Hsp90 and its inhibitory effect on the activity of Hsp90 ATPase.
2. Materials and methods
2.1. Materials and reagents
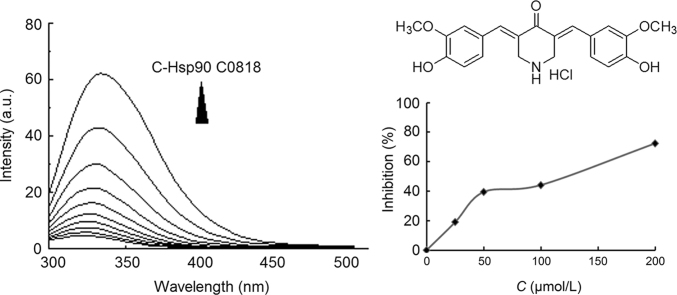
Bacterial strains and plasmids were provided by Prof. Lianru Zhang from School of Life Science, Xiamen University, China. C0818 was designed and synthesized by our laboratory (Fig. 1). Ni2+–nitrilotriacetic acid (NTA) agarose was purchased from General Electric (Little Chalfont, Buckinghamshire, England). ATP was purchased from Sigma–Aldrich (St. Louis, MO, USA). Geldanamycin (GA) was purchased from Shanghai Sangon Biological Engineering (Lot. No. XP0806132012J, Shanghai, China). A stock solution of Hsp90, which was expressed and purified by our laboratory, was prepared in a 10 mmol/L PBS buffer with a pH 7.4, and the applied concentration was fixed at 5.0 μmol/L. C0818 was dissolved in 5% DMSO for fluorescence measurements. The water used in the experiments was thrice-distilled using a Milli-Q Biocel system (Millipore, Bedford, MA, USA). All other reagents were of analytical reagent grade.
Figure 1.
Chemical structure of C0818.
2.2. Cloning, expression, and purification Hsp90
Yeast Hsp90 recombinant plasmid (Hsp90-PET-28a) was used to transform the target constructs of Hsp90 to express in the strain Escherichia coli BL21(DE3). The monoclonal cell was picked and cultured in lysogeny broth (LB) at 37 °C. Various constructs of Hsp90, including histidine (his)-tagged yeast full-length Hsp90 (1–732, 90 kDa), the N-terminal domain of Hsp90 (N-Hsp90, 1–236, 25 kDa), the middle domain of Hsp90 (M-Hsp90, 272–617, 40 kDa) and the C-terminal domain of Hsp90 (C-Hsp90, 629–732, 15 kDa), were induced by IPTG (isopropyl β-d-1-thiogalactopyranoside) and expressed in bacterial strains and purified by Ni2+-NTA (nitrilotriacetic acid) and gel filtration13.
2.3. Fluorescent measurements
Samples were excited at 280 nm and fluorescence intensities were recorded in the range of 290–500 nm at 293, 303 and 310 K, respectively, using a Cary Eclipse (Varian, Palo Alto, CA, USA). Fluorometric titration experiments were performed in 2.0 mL of 5.0 μmol/L Hsp90 solution (10 mmol/L PBS buffer, pH 7.6) with successive additions of C0818 solution (in 0.2% DMSO) from 5.0 to 50 μmol/L. All tests were performed in triplicate14.
2.4. Hsp90 ATPase activity assay
The reaction was performed in 100 μL of assay buffer (6 mmol/L MgCl2, 20 mmol/L KCl and 100 mmol/L Tris–HCl, pH 7.4) containing 0.4 μmol/L Hsp90, 1 mmol/L ATP and different concentrations of C0818 or GA or vehicle (DMSO) and incubated at 310 K for 3 h. At the end of the incubation, the ATPase activity of Hsp90 were assessed by malachite green reagent (0.0812% w/v malachite green, 2.32% w/v polyvinyl alcohol and 5.72% w/v ammonium molybdate in 6 mol/L HCl, and argon water mixed in a ratio of 2:1:1:2, v/v/v/v). Cultures were analyzed in triplicate at an absorbance of 620 nm13. The kinetic analysis of the Hsp90 ATPase activity was carried out using a nonlinear regression fit of the experimental points to the Michaelis–Menten equation. To obtain Km and V values, the Eadie–Hofstee linear transformation (V against V/[s]) was used, with the slope=−Km and the intercept on the x axis=V/Km13.
2.5. Statistical analysis
All data were analyzed with two-sided unpaired t-test in the GraphPad software package for Windows (Prism version 5.0) and Origin8.5 software. Values are expressed as means of triplicate or duplicate experiments.
3. Results
3.1. The interaction between C0818 and Hsp90
3.1.1. C0818 quenched the intrinsic fluorescence of Hsp90
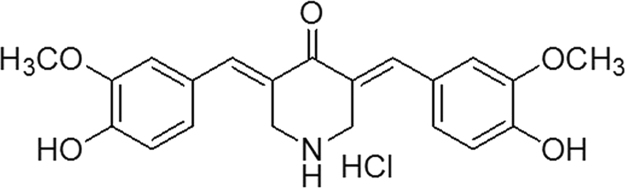
The binding of C0818 to Hsp90 was characterized by fluorescence quenching. At excitation wavelength of 280 nm, the interaction was examined with fluorescence spectrum from 290 to 510 nm. His-tagged Hsp90 displayed maximal fluorescence at 334 nm. When Hsp90 was incubated with increasing concentrations of C0818, the fluorescence intensity gradually decreased with a slight blue shift of λem (Fig. 2A).
Figure 2.
C0818 physically binds to Hsp90. (A) Quenching effect of C0818 (0–50 μmol/L) on Hsp90 endogenous fluorescent in a concentration-dependent manner. The maximal fluorescence displayed at 334 nm. (B) The change trend of Hsp90 fluorescence intensity with C0818 concentration. (C) Stern–Volmer plot for the binding of C0818 with Hsp90. (D) Lineweaver–Burk plot for the binding of C0818 with Hsp90.
The dissociation constants (KD) is calculated by the equation:
| (1) |
where F0 is the steady-state fluorescence intensity of Hsp90, F is the fluorescence intensity of Hsp90 in presence of C0818 at different concentrations, and [Q] is the concentration of C0818. The titration curves for Hsp90 yields estimated dissociation constants (KD) of C0818 for Hsp90, which is of 23.41±0.94 μmol/L (Fig. 2B), indicating that C0818 quenched the intrinsic fluorescence of Hsp90 and that there was a binding interaction between them15 (Table 1).
Table 1.
Dissociation constant KD values of C0818 and geldanamycin (GA).
| Compound | Fmax | KD (μmol/L) | R2 |
|---|---|---|---|
| GA | 546.16±7.92 | 24.39±0.79 | 0.999 |
| C0818 | 329.93±5.79 | 23.41±0.94 | 0.999 |
3.1.2. Quenching constant and quenching rate constant
The quenching process can be analyzed by Stern–Volmer equation:
| (2) |
where F0, F and [Q] means the same as in Eq. (1), KSV is the Stern–Volmer quenching constant in static quenching, Kq is the quenching rate constant, and τ0 is endogenous fluorescence lifetime value of biological macromolecules16, which generally is 10−8 s. Ksv and Kq were obtained from the slope of F0/F plotted against [Q] (Fig. 2C) and the results are listed in Table 2.
Table 2.
The Stern–Volmer quenching constant Ksv and bimolecular quenching constant Kq of C0818 and GA.
| Compound | Kq (L/mol/s) | Ksv (L/mol) | R2 |
|---|---|---|---|
| GA | (7.67±0.44)×1012 | (7.67±0.44)×104 | 0.968 |
| C0818 | (1.49±0.12)×1013 | (1.49±0.12)×105 | 0.943 |
The maximum dynamic collisional quenching constant of all kinds of quenching agent with biopolymers is 2.0×1010 L/mol/s, the quenching rate constant Kq of C0818 was greater than 2.0×1010 L/mol/s, indicating that the quenching effect of C0818 on intrinsic fluorescence of Hsp90 is not due to molecules collision in dynamic quenching, but due to the formation of complex in static quenching17.
3.1.3. Number of binding sites and constant
Static quenching complies with the Lineweaver–Burk equation18:
| (3) |
where F0, F and [Q] means the same as in Eq. (1), KA is the apparent binding constant, and n is the binding site. KA and n were obtained from the intercept and slope of lgF0 –F/F plotted against lg[Q] (Fig. 2D), respectively, and the results are listed in Table 3.
Table 3.
The binding constant KA and the binding number n of C0818 and GA.
| Compound | KA (L/mol) | n | R2 |
|---|---|---|---|
| GA | 2.88×105 | 1.14±0.04 | 0.986 |
| C0818 | 1.04×107 | 1.44±0.05 | 0.989 |
3.1.4. Thermodynamic parameters
Over the temperature ranges examined and when the thermodynamic enthalpy change (ΔHθ) did not vary significantly, van׳t Hoff׳s equation was used to obtain the values of ΔHθ and entropy change (ΔSθ):
| (4) |
| (5) |
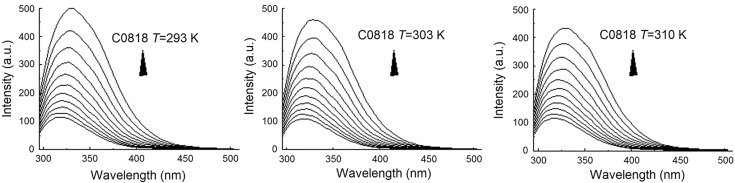
where R is the thermodynamic gas constant and the value is 8.314 J/mol/K, T is thermodynamic temperature, KD is corresponding dissociation constants at different temperatures, ΔGθ is Gibbs free energy change. Fluorescence intensities were recorded in the range of 290–510 nm at 293, 303 and 310 K, respectively (Fig. 3).
Figure 3.
Quenching effect of C0818 (0–50 μmol/L) on Hsp90 endogenous fluorescent at different temperatures (293, 303 and 310 K).
The thermodynamic enthalpy change (ΔHθ), entropy change(ΔSθ) and free energy change (ΔGθ) of C0818–Hsp90 binding were also calculated from the fluorescent spectrum, the results are listed in Table 4. The negative enthalpy change and positive entropy change suggest that electrostatic interaction predominated in stabilizing the C0818–Hsp90 complex19.
Table 4.
Dissociation constant KD and thermodynamic parameters of C0818 and GA at different temperatures.
| Compound | T (K) | KD (μmol/L) | R2 | ΔGθ (kJ/mol) | ΔHθ (kJ/mol) | ΔSθ (J/mol/K) |
|---|---|---|---|---|---|---|
| GA | 293 | 18.63±0.79 | 0.999 | −26.53±0.11 | −11.20±2.42 | 52.32±8.00 |
| 303 | 22.42±0.65 | 0.999 | −26.97±0.07 | |||
| 310 | 24.39±0.80 | 0.999 | −27.37±0.08 | |||
| C0818 | 293 | 39.13±0.91 | 0.999 | −24.72±0.06 | −12.96±0.74 | 40.27±2.45 |
| 303 | 46.44±1.72 | 0.999 | −25.13±0.09 | |||
| 310 | 53.44±2.28 | 0.998 | −25.35±0.11 |
In order to further determine the interaction type between C0818 and Hsp90, a synchronous fluorescence method was used to study the impact of C0818 on Hsp90 conformation.
3.1.5. Impact of C0818 on Hsp90 conformation
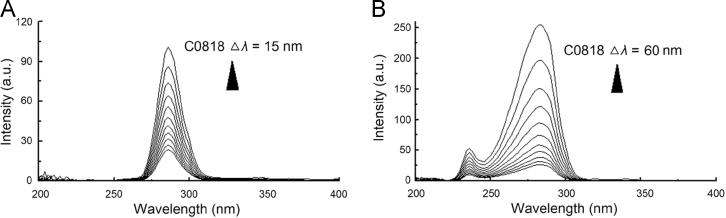
Tyrosine (Tyr) and tryptophan (Trp) are the main source of protein fluorescence. When the D-value (Δλ) between excitation and emission wavelengths was stabilized at 15 or 60 nm, the synchronous fluorescence provided the characteristic information of Tyr or Trp residues.
When Δλ was 15 or 60 nm, increasing concentrations of C0818 led to a dramatic decrease in fluorescence intensity, but the fluorescence peak position of Tyr or Trp residues remained the same (Fig. 4), indicating that the microenvironment of Tyr or Trp residues did not change after binding, and that the hydrophobic microenvironment did not obviously decrease. These results imply that the conformation of Tyr or Trp of Hsp90 did not change.
Figure 4.
Synchronous fluorescence spectra of C0818 (0–50 μmol/L) with Hsp90. (A) ∆λ = 15 nm; (B) ∆λ = 60 nm.
3.1.6. The domain of Hsp90 binding C0818
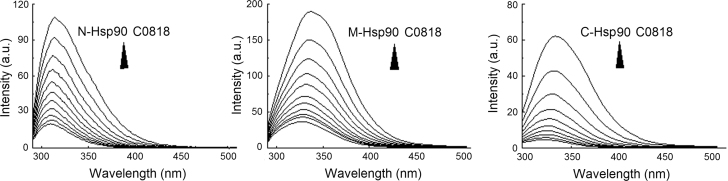
The domain of Hsp90 involved in binding C0818 was determined by constructing three truncation mutants: N-Hsp90, M-Hsp90 and C-Hsp90, corresponding to its ATP-binding domain, its co-chaperone binding domain and its dimerization domain. The fluorescence intensity of C-Hsp90, but not N-Hsp90 or M-Hsp90, was the most obviously quenched with increasing concentrations of C0818, indicating that C0818 may interact with the C-terminal domain (Fig. 5). The dissociation constant KD values are listed in Table 5.
Figure 5.
Quenching effect of C0818 (0–50 μmol/L) on endogenous fluorescent of N-Hsp90, M-Hsp90 and C-Hsp90.
Table 5.
Dissociation constant KD of C0818 and GA.
| Interaction | Fmax | KD (μmol/L) | R2 |
|---|---|---|---|
| N-Hsp90–GA | 221.22±4.91 | 5.84±0.61 | 0.991 |
| M-Hsp90–GA | 158.20±1.68 | 42.13±0.80 | 0.999 |
| C-Hsp90–GA | 67.91±0.63 | 36.72±0.64 | 0.999 |
| N-Hsp90–C0818 | 159.48±3.15 | 38.15±1.40 | 0.999 |
| M-Hsp90–C0818 | 232.46±3.46 | 24.52±0.82 | 0.999 |
| C-Hsp90–C0818 | 76.51±1.35 | 13.51±0.68 | 0.998 |
3.2. C0818 inhibits the ATPase activity of Hsp90
To characterize the inhibition of Hsp90 by C0818 binding, a colorimetric assay for inorganic phosphate was used to measure the inhibitory effect of C0818 on the ATPase activity of Hsp90. This assay is based on the formation of a phosphomolybdate complex and a subsequent reaction with malachite green. When the concentration of ATP was 1 mmol/L, the IC50 values of C0818 and GA were 120.74 and 0.41 μmol/L, respectively. Fig. 6 demonstrates the inhibition of Hsp90 ATPase activity by C0818. GA is a potent antiproliferative agent but is not an ideal clinical agent due to its marked hepatotoxicity and narrow therapeutic window9. C0818, a small-molecule inhibitor, may have potential as a therapeutic agent with improved pharmacological properties compared to GA.
Figure 6.
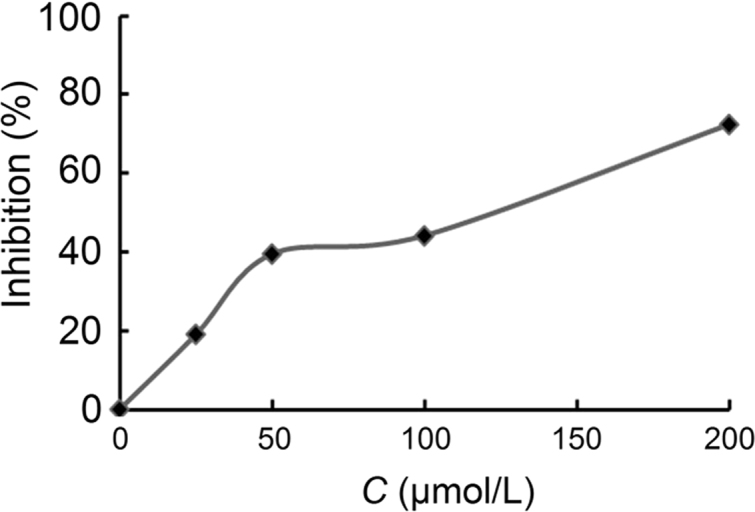
The inhibition rate of Hsp90 ATPase activity under different concentrations of C0818 (0–200 μmol/L).
4. Discussion
Quenching of the fluorescence from a macromolecule can be classified as static quenching or dynamic quenching. In static quenching, the stability of new complexes of quenching agent and fluorescent material results in the decrease of the quenching constant of fluorescence. In dynamic quenching, the excited state molecules of quenching agent and fluorescent material collide with each other, resulting in energy transfer20.
Our data suggest that C0818 is an Hsp90 inhibitor. Compared with the known inhibitor GA, C0818 has a novel scaffold unrelated to that of any other known Hsp90 inhibitor. Spectroscopic methods were used to study the interaction between C0818 and full-length Hsp90, N-Hsp90, M-Hsp90 and C-Hsp90. From the binding energy and dissociation constants, we demonstrated that C0818 is an inhibitor of Hsp90 and that it binds to the C-terminal dimerization domain of Hsp90. The quenching effect of C0818 on Hsp90 intrinsic fluorescence can thus be classified as static quenching. The thermodynamic parameters and the synchronous fluorescence suggest that electrostatic interactions predominate in stabilizing the C0818–Hsp90 complex.
C0818 inhibits the ATPase activity of Hsp90, so the binding of C0818 inhibits the catalysis of ATP hydrolysis. The binding of Hsp90 pockets arrests the catalytic cycle of Hsp90 in the ADP-bound conformation. The affinity-based screen showed that C0818 binds to Hsp90, therefore confirming the interaction between Hsp90 and C0818. In the assay measuring ATPase activity of Hsp90, C0818 demonstrated dose-dependent inhibition of the ATPase activity of Hsp90. As expected, the positive control GA also showed dose-dependent inhibition in this system.
Acknowledgments
The authors gratefully acknowledge the National Science and Technology Foundation of China for Key Projects of “Major New Drugs Innovation and Development” (2012ZX09103-101-028), Fujian Provincial Health and Family Planning Commission of China (2015-1-72), the Projects of Industry-Academy Cooperation for Science and Technology of Fujian Province, Chian (2016Y4005) for this project.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Wegele H., Müller L., Buchner J. Hsp70 and Hsp90-a relay team for protein folding. In: Adrian R.H., Helmreich E., Holzer H., Jung R., Kramer K., Krayer O., editors. Reviews of Physiology, Biochemistry and Pharmacology. Springer; Berlin Heidelberg: 2004. pp. 1–44. [DOI] [PubMed] [Google Scholar]
- 2.Richter K., Hendershot L.M., Freeman B.C. The cellular world according to Hsp90. Nat Struct Mol Biol. 2007;14:90–94. doi: 10.1038/nsmb0207-90. [DOI] [PubMed] [Google Scholar]
- 3.Noguchi M., Yu D., Hirayama R., Ninomiya Y., Sekine E., Kubota N. Inhibition of homologous recombination repair in irradiated tumor cells pretreated with Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Biochem Biophys Res Commun. 2006;351:658–663. doi: 10.1016/j.bbrc.2006.10.094. [DOI] [PubMed] [Google Scholar]
- 4.Soga S., Neckers L.M., Schulte T.W., Shiotsu Y., Akasaka K., Narumi H. KF25706, a novel oxime derivative of radicicol, exhibits in vivo antitumor activity via selective depletion of Hsp90 binding signaling molecules. Cancer Res. 1999;59:2931–2938. [PubMed] [Google Scholar]
- 5.Harashima K., Akimoto T., Nonaka T., Tsuzuki K., Mitsuhashi N., Nakano T. Heat shock protein 90 (Hsp90) chaperone complex inhibitor, radicicol potentiated radiation-induced cell killing in a hormone-sensitive prostate cancer cell line through degradation of the androgen receptor. Int J Radiat Biol. 2005;81:63–76. doi: 10.1080/09553000400029460. [DOI] [PubMed] [Google Scholar]
- 6.Neckers L. Heat shock protein 90: the cancer chaperone. J Biosci. 2007;32:517–530. doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
- 7.Taipale M., Jarosz D.F., Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 8.Schulte T.W., Neckers L.M. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother Pharmacol. 1998;42:273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 9.Supko J.G., Hickman R.L., Grever M.R., Malspeis L. Preclinical pharmacologic evaluation of geldanamycin as an antitumor agent. Cancer Chemother Pharmacol. 1995;36:305–315. doi: 10.1007/BF00689048. [DOI] [PubMed] [Google Scholar]
- 10.Wu L.X., Xu J.H., Huang X.W., Zhang K.Z., Wen C.X., Chen Y.Z. Down-regulation of P210bcr/abl by curcumin involves disrupting the molecular chaperone functions of Hsp90. Acta Pharmacol Sin. 2006;27:694–699. doi: 10.1111/j.1745-7254.2006.00326.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen C., Lui Y., Chen Y.Z., Xu J.H. C086, a novel analog of curcumin, induces growth inhibition and down-regulation of NF-κB in colon cancer cells and xenograft tumors. Cancer Biol Ther. 2011;12:797–807. doi: 10.4161/cbt.12.9.17671. [DOI] [PubMed] [Google Scholar]
- 12.Wu L.X., Yu J., Chen R.J., Liu Y., Lou L.G., Wu Y. Dual inhibition of Bcr-Abl and Hsp90 by C086 potently inhibits the proliferation of imatinib-resistant CML Cells. Clin Cancer Res. 2015;21:833–843. doi: 10.1158/1078-0432.CCR-13-3317. [DOI] [PubMed] [Google Scholar]
- 13.Chen J.J., Guo Q.J., He X.M., Yang S.X., Chen C., Zhang L.R. Study on a screening model for inhibitor of Hsp90 ATPase activity. J Xiamen Univ Nat Sci. 2010;49:711–716. [Google Scholar]
- 14.Gao P.Z., Wu H., Guo J., Xu Y.M. Study on the interaction between breviscapinum and bovine serum albumin by fluorescence spectrometry. Chin J Mod Appl Pharm. 2012;29:106–109. [Google Scholar]
- 15.Zhang G.J., Keita B., Brochon J.C., de Oliveira P., Nadjo L., Craescu C.T. Molecular interaction and energy transfer between human serum albumin and polyoxometalates. J Phys Chem B. 2007;111:1809–1814. doi: 10.1021/jp063758z. [DOI] [PubMed] [Google Scholar]
- 16.Chen G.Z., Huang X.Z., Xu J.G., Wang Z.B., Zheng Z.X. Science Press; Beijing: 1990. Principles of fluorescent spectroscopy. [Google Scholar]
- 17.Pastukhov A.V., Levchenko L.A., Sadkov A.P. Spectroscopic study on binding of rutin to human serum albumin. J Mol Struct. 2006;842:60–66. [Google Scholar]
- 18.Yan C.N., Tong J.Q., Xiong D., Liu Y., Pan Z.T. Studies on the binding reaction features between pefloxacin and bovine serum albumin by fluorescence spectrophtometry. Chin J Analyt Chem. 2006;34:796–800. [Google Scholar]
- 19.Ross P.D., Subramanians S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 20.Fleming K. Introduction to fluorescence theory and methods. Techniq Biophys. 2005;8:1–11. [Google Scholar]